

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0230.5
(406 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
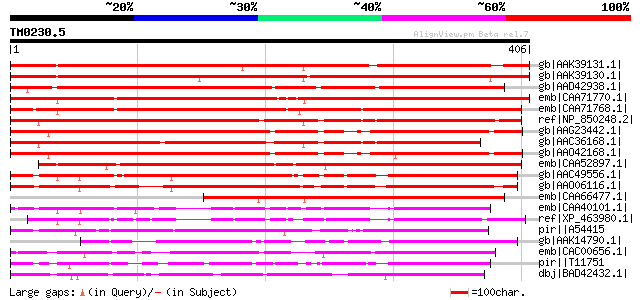
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK39131.1| bZIP transcription factor 3 [Phaseolus vulgaris] 675 0.0
gb|AAK39130.1| bZIP transcription factor 2 [Phaseolus vulgaris] 648 0.0
gb|AAD42938.1| G-Box binding protein 2 [Catharanthus roseus] 573 e-162
emb|CAA71770.1| bZIP DNA-binding protein [Petroselinum crispum] ... 552 e-156
emb|CAA71768.1| bZIP DNA-binding protein [Petroselinum crispum] ... 532 e-150
ref|NP_850248.2| bZIP transcription factor family protein [Arabi... 490 e-137
gb|AAG23442.1| G-Box binding protein, putative [Arabidopsis thal... 464 e-129
gb|AAC36168.1| putative G-box binding bZIP transcription factor ... 461 e-128
gb|AAO42168.1| putative G-Box binding protein [Arabidopsis thali... 459 e-128
emb|CAA52897.1| G-box binding protein [Lycopersicon esculentum] ... 441 e-122
gb|AAC49556.1| DNA-binding factor of bZIP class gi|7489475|pir||... 416 e-115
gb|AAO06116.1| bZIP transcription factor ZIP1 [Hordeum vulgare s... 358 1e-97
emb|CAA66477.1| transcription factor [Vicia faba] gi|7488851|pir... 340 6e-92
emb|CAA40101.1| HBP-1a [Triticum aestivum] gi|122771|sp|P23922|H... 313 6e-84
ref|XP_463980.1| putative transcription factor HBP-1a [Oryza sat... 308 3e-82
pir||A54415 transcription factor HBP-1a(c14) - wheat gi|497895|d... 266 9e-70
gb|AAK14790.1| G-box binding factor bZIP transcription factor [C... 230 7e-59
emb|CAC00656.1| common plant regulatory factor 5 [Petroselinum c... 221 4e-56
pir||T11751 transcription repressor ROM1 - kidney bean gi|135485... 206 8e-52
dbj|BAD42432.1| bZip transcription factor [Psophocarpus tetragon... 197 4e-49
>gb|AAK39131.1| bZIP transcription factor 3 [Phaseolus vulgaris]
Length = 397
Score = 675 bits (1742), Expect = 0.0
Identities = 338/412 (82%), Positives = 361/412 (87%), Gaps = 21/412 (5%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQA 60
MGSSEMDKTPKEKESKTPP T QEQ TT+TGT+NP DW FQAYSP+PPHGFLAS+PQA
Sbjct: 1 MGSSEMDKTPKEKESKTPPPTSQEQSSTTATGTINP-DWPGFQAYSPIPPHGFLASSPQA 59
Query: 61 HPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGN 120
HPYMWGVQ MPPYGTPPHPYVAMYP GGIYAHPS+PPGSYPFSPFAMPSPNGI EASGN
Sbjct: 60 HPYMWGVQQFMPPYGTPPHPYVAMYPPGGIYAHPSMPPGSYPFSPFAMPSPNGIAEASGN 119
Query: 121 TPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSKSGESGS 180
TPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMIT KNNEHGKT GTSANG HSKSG+S S
Sbjct: 120 TPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITGKNNEHGKTRGTSANGIHSKSGDSAS 179
Query: 181 --EGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTP----HQTMSM 234
EGTSEGSDANSQNDS++KSGGR+DSFEDEPSQNGT+ +T+QNGG++TP +Q + +
Sbjct: 180 EGEGTSEGSDANSQNDSQMKSGGRQDSFEDEPSQNGTSAYTSQNGGISTPATVVNQNVPI 239
Query: 235 VPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQ 294
+PISAGGAPGAVPGP TNLNIGMDYWGTP SNIPAL VPSTAVA GSRD VQ
Sbjct: 240 IPISAGGAPGAVPGPTTNLNIGMDYWGTPAPSNIPALGRKVPSTAVA------GSRDSVQ 293
Query: 295 SQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIR 354
SQ WLQDERE+KRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIR
Sbjct: 294 SQLWLQDEREIKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIR 353
Query: 355 SDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSGQTEAVQGGH 406
SDYEQLL+EN ALKERLGELP N DQH ++AQQ+GQTE VQGGH
Sbjct: 354 SDYEQLLSENTALKERLGELPAN--------DQHVGNEAQQNGQTEGVQGGH 397
>gb|AAK39130.1| bZIP transcription factor 2 [Phaseolus vulgaris]
Length = 417
Score = 648 bits (1672), Expect = 0.0
Identities = 327/419 (78%), Positives = 357/419 (85%), Gaps = 15/419 (3%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQ-PPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQ 59
MGSS+MDKTPKEKESKTPP T QEQ PPTT T+NP DWS+FQAYSPMPPHGFLAS+PQ
Sbjct: 1 MGSSDMDKTPKEKESKTPPATSQEQSPPTTGMATINP-DWSNFQAYSPMPPHGFLASSPQ 59
Query: 60 AHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASG 119
AHPYMWGVQHIMPPYGTP HPYVAMYPHGGIYAHPSIPPGSYPFSPFAM SPNGI + SG
Sbjct: 60 AHPYMWGVQHIMPPYGTPAHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMASPNGIADTSG 119
Query: 120 NTPGSMEADGKPPEVKEKLPIKRSKGSL--GSLNM-ITRKNNEHGKTPGTSANGAHSKSG 176
N PGS+E KPPEVKEKLP+KRSKGS GSLNM IT KNN+ GKT G SANG HSKSG
Sbjct: 120 NNPGSIEVGAKPPEVKEKLPVKRSKGSASGGSLNMWITGKNNDLGKTTGESANGIHSKSG 179
Query: 177 ESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTP----HQTM 232
+S S+GTSEGSD NSQNDS+LKSG R+DSFEDEPSQNG++ H QNG + P +QTM
Sbjct: 180 DSASDGTSEGSDENSQNDSQLKSGERQDSFEDEPSQNGSSAHAPQNGVHSRPQTVVNQTM 239
Query: 233 SMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDG 292
+PIS APGAVPGP TNLNIGMDYWGTPTSS IPALHG V STAVAGGM T GSRDG
Sbjct: 240 P-IPISTASAPGAVPGPTTNLNIGMDYWGTPTSSAIPALHGKVSSTAVAGGMITAGSRDG 298
Query: 293 VQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSR 352
VQSQ WLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRA+ LKEENA+LR+EVSR
Sbjct: 299 VQSQIWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRADVLKEENATLRAEVSR 358
Query: 353 IRSDYEQLLTENAALKERLGEL-----PGNDDLRSGKSDQHTHDDAQQSGQTEAVQGGH 406
IRS++EQL +ENA+LKERLGE+ PGN+D+RSG++DQH +D QQS Q EAVQG H
Sbjct: 359 IRSEFEQLRSENASLKERLGEIPGVATPGNEDVRSGQNDQHVSNDTQQSRQKEAVQGVH 417
>gb|AAD42938.1| G-Box binding protein 2 [Catharanthus roseus]
Length = 394
Score = 573 bits (1478), Expect = e-162
Identities = 289/391 (73%), Positives = 321/391 (81%), Gaps = 14/391 (3%)
Query: 1 MGSSEMDKTPKE----KESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLAS 56
MGSSE+DK+ KE KE+K P + QEQP TS GT PDW+ FQAYSP+PPHGFLAS
Sbjct: 1 MGSSEIDKSSKEAKEAKEAKETPPSSQEQPAATSAGT---PDWTGFQAYSPIPPHGFLAS 57
Query: 57 NPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVE 116
+PQAHPYMWGVQH+MPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGI E
Sbjct: 58 SPQAHPYMWGVQHLMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIAE 117
Query: 117 ASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSKSG 176
S NTPG+ME DGK E KEKLPIKRS+GSLGSLNMIT KNN+ GKT G SANGA SKS
Sbjct: 118 PSVNTPGNMEVDGKASEGKEKLPIKRSRGSLGSLNMITGKNNDAGKTSGASANGACSKSA 177
Query: 177 ESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMSMVP 236
ES SEG+SEGSDANSQN+S++KSG R+DS E SQNG+ H +QNGG NTPH ++MVP
Sbjct: 178 ESASEGSSEGSDANSQNESQMKSGNRQDS--GETSQNGSGAHGSQNGGTNTPHSMVAMVP 235
Query: 237 ISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQ 296
+SA G V GPATNLNIGMDYWGT S +P + G VPST V GGM +RD VQ+Q
Sbjct: 236 LSAS---GGVTGPATNLNIGMDYWGTAASPTVPVVRGKVPSTPVGGGMVP--ARDPVQAQ 290
Query: 297 HWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSD 356
W+QDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEEN SLR+EVS IRS+
Sbjct: 291 LWIQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENNSLRAEVSLIRSE 350
Query: 357 YEQLLTENAALKERLGELPGNDDLRSGKSDQ 387
YEQLL +NAALKERLGE G DD RS +++Q
Sbjct: 351 YEQLLAQNAALKERLGEASGQDDPRSSRNEQ 381
>emb|CAA71770.1| bZIP DNA-binding protein [Petroselinum crispum]
gi|7488943|pir||T14911 bZIP DNA-binding protein -
parsley
Length = 420
Score = 552 bits (1423), Expect = e-156
Identities = 285/424 (67%), Positives = 330/424 (77%), Gaps = 22/424 (5%)
Query: 1 MGSSEMDKTPKE-KESKTPPL-TPQEQPPTTSTGTVNP----------PDWSSFQAYSPM 48
MGSS MDK+PK+ KE+K P + T QEQP + PDWS FQAYSPM
Sbjct: 1 MGSSGMDKSPKDIKEAKEPKIPTSQEQPSPAAAAAAAAAAAAAAGPVTPDWSGFQAYSPM 60
Query: 49 PPHGFLASNPQA-HPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFA 107
PPHG++AS+PQA HPYMWGVQH+MPPYGTPPHPYV MYPHGGIYAHPS+PPGSYPFSPFA
Sbjct: 61 PPHGYMASSPQAPHPYMWGVQHMMPPYGTPPHPYV-MYPHGGIYAHPSMPPGSYPFSPFA 119
Query: 108 MPSPNGIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTS 167
+PSPNG+ EA GNTPGS EADGK E KEKLPIKRSKGSLGSLNMIT KNNE KT G +
Sbjct: 120 IPSPNGVAEAFGNTPGSTEADGKVSEGKEKLPIKRSKGSLGSLNMITGKNNEASKTLGAA 179
Query: 168 ANGAHSKSGESGSEGTSE-GSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLN 226
ANG +SKSG+S S+G+SE GSDANSQNDS++KSG R+DS E E SQNG A H QNG N
Sbjct: 180 ANGGYSKSGDSASDGSSEEGSDANSQNDSQIKSGSRQDSLEGE-SQNGNA-HGLQNGQ-N 236
Query: 227 TPH----QTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAG 282
H Q +S+VPI+A G GA+PGP TNLNIGMDYWG TSS +PA+ G V S + G
Sbjct: 237 ANHSMVNQQISIVPITAAGTAGAIPGPMTNLNIGMDYWGGVTSSAVPAMRGKVTSPPITG 296
Query: 283 GMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEE 342
G+ T G+RD VQSQ WLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAE L+EE
Sbjct: 297 GIVTAGARDNVQSQLWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLQEE 356
Query: 343 NASLRSEVSRIRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSGQTEAV 402
NASLR+E+ R RS+YE+ L +NA LKE++G++ G +D G++DQHT D+ Q++G E
Sbjct: 357 NASLRAELGRARSEYEKALAQNAILKEKVGDVAGQEDQWPGRNDQHTGDEGQETGHIEPG 416
Query: 403 QGGH 406
Q GH
Sbjct: 417 QSGH 420
>emb|CAA71768.1| bZIP DNA-binding protein [Petroselinum crispum]
gi|7488942|pir||T14909 bZIP DNA-binding protein -
parsley
Length = 407
Score = 532 bits (1371), Expect = e-150
Identities = 278/410 (67%), Positives = 322/410 (77%), Gaps = 15/410 (3%)
Query: 1 MGSSEMDKTPKE-KESKTPPLTPQEQPPTTSTGTVNP--PDWSSFQAYSPMPPHGFLASN 57
MGSSEM+K+ KE KE KTP T QEQ G P PDWS FQAYSPMPPHG++AS+
Sbjct: 1 MGSSEMEKSSKETKEPKTP--TSQEQVSPVVAGPAGPVTPDWSGFQAYSPMPPHGYMASS 58
Query: 58 PQA-HPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIV- 115
PQA HPYMWGVQH+MPPYGTPPHPYV MYPHGGIYAHPS+PPGSYPFSPFAMPSPNG+
Sbjct: 59 PQAPHPYMWGVQHMMPPYGTPPHPYV-MYPHGGIYAHPSMPPGSYPFSPFAMPSPNGVAA 117
Query: 116 EASGNTPGSMEADG-KPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSK 174
EASGNTPGSMEADG K E KEKLPIKRSKGSLGSLNMIT K NE K G + NG +SK
Sbjct: 118 EASGNTPGSMEADGGKVSEGKEKLPIKRSKGSLGSLNMITGKTNEASKPSGAATNGGYSK 177
Query: 175 SGESGSEGTSE-GSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGL---NTPHQ 230
SGES SEG+SE GSDANSQNDS++KSG R+DS E S NG A H QNG + +Q
Sbjct: 178 SGESASEGSSEEGSDANSQNDSQIKSGSRQDSLEAGASHNGNA-HGLQNGQYANNSMVNQ 236
Query: 231 TMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSR 290
+S+VP+S G +PGPATNLNIGMDYWG TSS IPA+ G V S + GG + G+R
Sbjct: 237 PISVVPLSTAGPTAVLPGPATNLNIGMDYWGGATSSAIPAMRGQV-SPPITGGTVSAGAR 295
Query: 291 DGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEV 350
D VQSQ WLQDERELKRQ+RKQSNRESARRSRLRKQAECDELAQRAEALKEENASLR+E+
Sbjct: 296 DNVQSQLWLQDERELKRQKRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRAEL 355
Query: 351 SRIRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSGQTE 400
SR R++YE+++ +N LKE++ E+PG +D G++DQH + ++++G TE
Sbjct: 356 SRFRTEYEKIVAQNEVLKEKIREVPGQEDQWPGRNDQHNGNGSRETGHTE 405
>ref|NP_850248.2| bZIP transcription factor family protein [Arabidopsis thaliana]
gi|63003876|gb|AAY25467.1| At2g35530 [Arabidopsis
thaliana]
Length = 409
Score = 490 bits (1261), Expect = e-137
Identities = 252/410 (61%), Positives = 302/410 (73%), Gaps = 21/410 (5%)
Query: 1 MGSSEMDKTPKEKESKTPPLT----PQEQPPTTSTGT-VNPPDWSSFQAYSPMPP-HGFL 54
M S+EM+K+ KEKE KTPP + P Q P+++ + PDWS FQAYSPMPP HG++
Sbjct: 1 MASNEMEKSSKEKEPKTPPPSSTAPPSSQEPSSAVSAGMATPDWSGFQAYSPMPPPHGYV 60
Query: 55 ASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGI 114
AS+PQ HPYMWGVQH+MPPYGTPPHPYVAMYP GG+YAHPS+PPGSYP+SP+AMPSPNG+
Sbjct: 61 ASSPQPHPYMWGVQHMMPPYGTPPHPYVAMYPPGGMYAHPSMPPGSYPYSPYAMPSPNGM 120
Query: 115 VEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSK 174
E SGNT G + D K EVKEKLPIKRS+GSLGSLNMIT KNNE GK G SANGA+SK
Sbjct: 121 TEVSGNTTGGTDGDAKQSEVKEKLPIKRSRGSLGSLNMITGKNNEPGKNSGASANGAYSK 180
Query: 175 SGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTP----HQ 230
SGES S+G+SEGSD NSQNDS G + S+NG + + QNG TP Q
Sbjct: 181 SGESASDGSSEGSDGNSQNDS---GSGLDGKDAEAASENGGSANGPQNGSAGTPILPVSQ 237
Query: 231 TMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSR 290
T+ ++P++A G VPGP TNLNIGMDYWG PTS+ IP +HG V ST V G +A GSR
Sbjct: 238 TVPIMPMTAAG----VPGPPTNLNIGMDYWGAPTSAGIPGMHGKV-STPVPGVVAP-GSR 291
Query: 291 DGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEV 350
DG SQ WLQD+RELKRQRRKQSNRESARRSRLRKQAECDELAQRAE L EEN +LR+E+
Sbjct: 292 DGGHSQPWLQDDRELKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLNEENTNLRAEI 351
Query: 351 SRIRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSGQTE 400
++++S E+L TEN +LK++L P + + + H D Q+G E
Sbjct: 352 NKLKSQCEELTTENTSLKDQLSLFPPLEGI--SMDNDHQEPDTNQTGAAE 399
>gb|AAG23442.1| G-Box binding protein, putative [Arabidopsis thaliana]
gi|25344489|pir||H86445 probable G-Box binding protein
[imported] - Arabidopsis thaliana
Length = 387
Score = 464 bits (1195), Expect = e-129
Identities = 244/412 (59%), Positives = 289/412 (69%), Gaps = 36/412 (8%)
Query: 1 MGSSEMDKTPKEKESKT-PPLTPQEQPPTT---------STGTVNPPDWSSFQAYSPMPP 50
MGSSEM+K+ KEKE KT PP T P T S G DWS FQAYSPMPP
Sbjct: 1 MGSSEMEKSGKEKEPKTTPPSTSSSAPATVVSQEPSSAVSAGVAVTQDWSGFQAYSPMPP 60
Query: 51 HGFLASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPS 110
HG++AS+PQ HPYMWGVQH+MPPYGTPPHPYV MYP GG+YAHPS+PPGSYP+SP+AMPS
Sbjct: 61 HGYVASSPQPHPYMWGVQHMMPPYGTPPHPYVTMYPPGGMYAHPSLPPGSYPYSPYAMPS 120
Query: 111 PNGIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANG 170
PNG+ EASGNT +E DGKP + KEKLPIKRSKGSLGSLNMI KNNE GK G SANG
Sbjct: 121 PNGMAEASGNTGSVIEGDGKPSDGKEKLPIKRSKGSLGSLNMIIGKNNEAGKNSGASANG 180
Query: 171 AHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTP-H 229
A SKS ESGS+G+S+GSDANSQNDS + G+ + S++G + H G N P +
Sbjct: 181 ACSKSAESGSDGSSDGSDANSQNDSGSRHNGKDG---ETASESGGSAHGPPRNGSNLPVN 237
Query: 230 QTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGS 289
QT++++P+SA G VPGP TNLNIGMDYW HGNV G G
Sbjct: 238 QTVAIMPVSATG----VPGPPTNLNIGMDYWSG---------HGNV------SGAVPGVV 278
Query: 290 RDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSE 349
DG QSQ WLQDERE+KRQRRKQSNRESARRSRLRKQAECDELAQRAE L EN+SLR+E
Sbjct: 279 VDGSQSQPWLQDEREIKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLNGENSSLRAE 338
Query: 350 VSRIRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSGQTEA 401
+++++S YE+LL EN++LK + P L G D++ + + + Q A
Sbjct: 339 INKLKSQYEELLAENSSLKNKFSSAP---SLEGGDLDKNEQEPQRSTRQDVA 387
>gb|AAC36168.1| putative G-box binding bZIP transcription factor [Arabidopsis
thaliana] gi|25344493|pir||G84769 hypothetical protein
At2g35530 [imported] - Arabidopsis thaliana
Length = 368
Score = 461 bits (1187), Expect = e-128
Identities = 240/378 (63%), Positives = 284/378 (74%), Gaps = 24/378 (6%)
Query: 1 MGSSEMDKTPKEKESKTPPLT----PQEQPPTTSTGT-VNPPDWSSFQAYSPMPP-HGFL 54
M S+EM+K+ KEKE KTPP + P Q P+++ + PDWS FQAYSPMPP HG++
Sbjct: 1 MASNEMEKSSKEKEPKTPPPSSTAPPSSQEPSSAVSAGMATPDWSGFQAYSPMPPPHGYV 60
Query: 55 ASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGI 114
AS+PQ HPYMWGVQH+MPPYGTPPHPYVAMYP GG+YAHPS+PPGSYP+SP+AMPSPNG+
Sbjct: 61 ASSPQPHPYMWGVQHMMPPYGTPPHPYVAMYPPGGMYAHPSMPPGSYPYSPYAMPSPNGM 120
Query: 115 VEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSK 174
E S + D K EVKEKLPIKRS+GSLGSLNMIT KNNE GK G SANGA+SK
Sbjct: 121 TEVS---VSGTDGDAKQSEVKEKLPIKRSRGSLGSLNMITGKNNEPGKNSGASANGAYSK 177
Query: 175 SGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTP----HQ 230
SGES S+G+SEGSD NSQN S L + S+NG + + QNG TP Q
Sbjct: 178 SGESASDGSSEGSDGNSQNSSLLFFHS-----AEAASENGGSANGPQNGSAGTPILPVSQ 232
Query: 231 TMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSR 290
T+ ++P++A G VPGP TNLNIGMDYWG PTS+ IP +HG V ST V G +A GSR
Sbjct: 233 TVPIMPMTAAG----VPGPPTNLNIGMDYWGAPTSAGIPGMHGKV-STPVPGVVAP-GSR 286
Query: 291 DGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEV 350
DG SQ WLQD+RELKRQRRKQSNRESARRSRLRKQAECDELAQRAE L EEN +LR+E+
Sbjct: 287 DGGHSQPWLQDDRELKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLNEENTNLRAEI 346
Query: 351 SRIRSDYEQLLTENAALK 368
++++S E+L TEN +LK
Sbjct: 347 NKLKSQCEELTTENTSLK 364
>gb|AAO42168.1| putative G-Box binding protein [Arabidopsis thaliana]
gi|42562458|ref|NP_174494.2| bZIP transcription factor
family protein [Arabidopsis thaliana]
Length = 389
Score = 459 bits (1182), Expect = e-128
Identities = 244/414 (58%), Positives = 289/414 (68%), Gaps = 38/414 (9%)
Query: 1 MGSSEMDKTPKEKESKT-PPLTPQEQPPTT---------STGTVNPPDWSSFQAYSPMPP 50
MGSSEM+K+ KEKE KT PP T P T S G DWS FQAYSPMPP
Sbjct: 1 MGSSEMEKSGKEKEPKTTPPSTSSSAPATVVSQEPSSAVSAGVAVTQDWSGFQAYSPMPP 60
Query: 51 HGFLASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPS 110
HG++AS+PQ HPYMWGVQH+MPPYGTPPHPYV MYP GG+YAHPS+PPGSYP+SP+AMPS
Sbjct: 61 HGYVASSPQPHPYMWGVQHMMPPYGTPPHPYVTMYPPGGMYAHPSLPPGSYPYSPYAMPS 120
Query: 111 PNGIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANG 170
PNG+ EASGNT +E DGKP + KEKLPIKRSKGSLGSLNMI KNNE GK G SANG
Sbjct: 121 PNGMAEASGNTGSVIEGDGKPSDGKEKLPIKRSKGSLGSLNMIIGKNNEAGKNSGASANG 180
Query: 171 AHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTP-H 229
A SKS ESGS+G+S+GSDANSQNDS + G+ + S++G + H G N P +
Sbjct: 181 ACSKSAESGSDGSSDGSDANSQNDSGSRHNGKDG---ETASESGGSAHGPPRNGSNLPVN 237
Query: 230 QTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGS 289
QT++++P+SA G VPGP TNLNIGMDYW HGNV G G
Sbjct: 238 QTVAIMPVSATG----VPGPPTNLNIGMDYWSG---------HGNV------SGAVPGVV 278
Query: 290 RDGVQSQHWLQ--DERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLR 347
DG QSQ WLQ DERE+KRQRRKQSNRESARRSRLRKQAECDELAQRAE L EN+SLR
Sbjct: 279 VDGSQSQPWLQVSDEREIKRQRRKQSNRESARRSRLRKQAECDELAQRAEVLNGENSSLR 338
Query: 348 SEVSRIRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSGQTEA 401
+E+++++S YE+LL EN++LK + P L G D++ + + + Q A
Sbjct: 339 AEINKLKSQYEELLAENSSLKNKFSSAP---SLEGGDLDKNEQEPQRSTRQDVA 389
>emb|CAA52897.1| G-box binding protein [Lycopersicon esculentum]
gi|629667|pir||S42394 G-box-binding protein - tomato
Length = 406
Score = 441 bits (1135), Expect = e-122
Identities = 236/382 (61%), Positives = 280/382 (72%), Gaps = 10/382 (2%)
Query: 23 QEQPPTTSTGTVNPPDWSSFQAYSPMPPHGFLASNPQAHPYMWGVQHIMPPY--GTPPHP 80
QEQ +T+TGT NP +W FQ YSPMPPHGF+AS+PQAHPYMWGVQH++ + T
Sbjct: 12 QEQASSTATGTANP-EWPGFQGYSPMPPHGFMASSPQAHPYMWGVQHLITLWYSTTSLCH 70
Query: 81 YVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNTPGSMEADGKPPEVKEKLPI 140
YV+ HGGIYAHPS+PPGSYPFSPFAMPSPNG+ E + NT + E DGK EVKEKLPI
Sbjct: 71 YVS---HGGIYAHPSMPPGSYPFSPFAMPSPNGVAEVAVNTSSNTELDGKSSEVKEKLPI 127
Query: 141 KRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSKSGESGSEGTSEGSDANSQNDSELKSG 200
KR KGSLGSLNMIT KN E GKT G SANG +SKS ESGSEG+SEGSDANSQN+S +KS
Sbjct: 128 KRLKGSLGSLNMITGKNTELGKTSGASANGVYSKSAESGSEGSSEGSDANSQNESPMKSA 187
Query: 201 GRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMSMVPISAGGAPGA--VPGPATNLNIGMD 258
GR+DS E E SQN + H++Q + PH + + A + G P L
Sbjct: 188 GRQDSAE-ERSQNRNSAHSSQM-RTSAPHSLSTKLCNHANVSCGCWRYSWPHNQLEYWYG 245
Query: 259 YWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESA 318
+ ++IPA+HG VPS +VAGG+ GSRD VQSQ W+QDERELKRQRRKQSNRESA
Sbjct: 246 LLECRSFADIPAIHGKVPSASVAGGIGNAGSRDIVQSQMWIQDERELKRQRRKQSNRESA 305
Query: 319 RRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGELPGND 378
RRSRLRKQAECDELAQRAE LKEENASLR+E+S +RS+++QL ++NA+LKERLGE+ G D
Sbjct: 306 RRSRLRKQAECDELAQRAEVLKEENASLRAELSCLRSEHDQLASQNASLKERLGEVSGRD 365
Query: 379 DLRSGKSDQHTHDDAQQSGQTE 400
D R + H + D Q S QTE
Sbjct: 366 DPRPSRKYIHLNKDTQTSSQTE 387
>gb|AAC49556.1| DNA-binding factor of bZIP class gi|7489475|pir||T03241 G-box
binding factor 1A - rice
Length = 390
Score = 416 bits (1069), Expect = e-115
Identities = 230/411 (55%), Positives = 283/411 (67%), Gaps = 36/411 (8%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNP---PDWSSFQAYSPMPPHGF---- 53
MGSS D K ++ P QEQ P S+ T P PDW++FQ Y P+PPHGF
Sbjct: 1 MGSSGADAPTKTSKASAP----QEQQPPASSSTATPAVYPDWANFQGYPPIPPHGFFPSP 56
Query: 54 LASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
+AS+PQ HPYMWG Q ++PPYGTPP PYV MYP G +YAHPS+PPG++PF+P+AM SPNG
Sbjct: 57 VASSPQGHPYMWGAQPMIPPYGTPPPPYV-MYPPG-VYAHPSMPPGAHPFTPYAMASPNG 114
Query: 114 IVEASGNTPGSM-----EADGKPPEVKEKLPIKRSKGSLGSLNMITRKNN-EHGKTPGTS 167
+ +G T + E DGK E KEK PIKRSKGSLGSLNMIT KN+ EHGKT G S
Sbjct: 115 NADPTGTTTTAAAAAAGETDGKSSEGKEKSPIKRSKGSLGSLNMITGKNSTEHGKTSGAS 174
Query: 168 ANGAHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNT 227
ANGA S+SGESGSE +SEGS+ANSQNDS K G+ E SQNG + +Q LN
Sbjct: 175 ANGAISQSGESGSESSSEGSEANSQNDSHHKESGQEQDGEVRSSQNGVSRSPSQ-AKLN- 232
Query: 228 PHQTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATG 287
QTM+++P+++ G VP P TNLNIGMDYW T+S+ PA+HG TA G M G
Sbjct: 233 --QTMAIMPMTSSGP---VPAPTTNLNIGMDYWAN-TASSTPAIHGKATPTAAPGSMVPG 286
Query: 288 GSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLR 347
+ W+QDERELKRQRRKQSNRESARRSRLRKQAEC+ELAQRAE LK+EN SLR
Sbjct: 287 --------EQWVQDERELKRQRRKQSNRESARRSRLRKQAECEELAQRAEVLKQENTSLR 338
Query: 348 SEVSRIRSDYEQLLTENAALKERLGELP-GNDDLRSGKSDQHTHDDAQQSG 397
EV+RIR +Y++LL++N++LKE+L + D+ QH+ DD+Q+ G
Sbjct: 339 DEVNRIRKEYDELLSKNSSLKEKLEDKQHKTDEAGVDNKLQHSGDDSQKKG 389
>gb|AAO06116.1| bZIP transcription factor ZIP1 [Hordeum vulgare subsp. vulgare]
Length = 367
Score = 358 bits (920), Expect = 1e-97
Identities = 206/405 (50%), Positives = 260/405 (63%), Gaps = 54/405 (13%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNP-PDWSSFQAYSPMPPHGF----LA 55
M SSE + K ++ TP ++QPPT+ST T PDW++FQ Y P+PPHGF +
Sbjct: 1 MRSSEAETPAKANKASTPQ---EQQPPTSSTTTPTVYPDWTNFQGYPPIPPHGFFPSPVV 57
Query: 56 SNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIV 115
SNPQ HPYMWG Q +MPPYGTPP+ +YP GG+YAHPS+ PG+
Sbjct: 58 SNPQGHPYMWGPQPMMPPYGTPPY---VIYPPGGVYAHPSMRPGTT-------------- 100
Query: 116 EASGNTPGSM--EADGKPPEVKEKLPIKRSKGSLGSLNMITRKNN-EHGKTPGTSANGAH 172
TP + E +GK + EK PIKRSKGSLGSLNMIT KN EHGKT G SANG
Sbjct: 101 ----TTPATAGGETNGKSSDGIEKSPIKRSKGSLGSLNMITGKNCVEHGKTSGASANGTI 156
Query: 173 SKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTM 232
S+SGESGSE +SEGS+ANSQND + K G+ + SQNG + +Q T +
Sbjct: 157 SQSGESGSESSSEGSEANSQNDLQHKESGQEQDGDVRSSQNGVSPSPSQAQLKQT--SAI 214
Query: 233 SMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDG 292
+P S G VPGP TNL IGMDYW T+S+ PALHG V TA+ G +A
Sbjct: 215 MQMPSS-----GPVPGPTTNLKIGMDYWAN-TASSSPALHGKVTPTAIPGDLAP------ 262
Query: 293 VQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSR 352
++ W+QDERELKRQ+RKQSNR+SARRSRLRKQAEC+ELAQRAE LK+ENASL+ EVSR
Sbjct: 263 --TEPWMQDERELKRQKRKQSNRDSARRSRLRKQAECEELAQRAEVLKQENASLKDEVSR 320
Query: 353 IRSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSG 397
IR +Y++LL++N++LK+ +G+ D + H+ Q+SG
Sbjct: 321 IRKEYDELLSKNSSLKDNIGDKQHKTD------EAGLHNKLQRSG 359
>emb|CAA66477.1| transcription factor [Vicia faba] gi|7488851|pir||T12092
G-box-binding protein - fava bean
Length = 257
Score = 340 bits (871), Expect = 6e-92
Identities = 173/243 (71%), Positives = 196/243 (80%), Gaps = 7/243 (2%)
Query: 152 MITRKNNEHGKTPGTSANGAHSKSGESGSEGTSEGSDANSQN---DSELKSGGRRDSFED 208
M+T KNNE KTPG SANG HSKSGES SEG+SEGSD NS N DS+LKSG R+DSFED
Sbjct: 1 MVTGKNNELSKTPGASANGIHSKSGESASEGSSEGSDENSHNVLQDSQLKSGERQDSFED 60
Query: 209 EPSQNGTAVHTTQNGGLNTPH----QTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPT 264
EPSQNG++ QNGGLNTPH QTMS+VP+S G AV GP TNLNIGMDYWGTP
Sbjct: 61 EPSQNGSSAQAPQNGGLNTPHTVVNQTMSVVPMSVAGPIAAVAGPTTNLNIGMDYWGTPA 120
Query: 265 SSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLR 324
SS IPA+HG VPSTAVAGGM G RDGV SQ WLQDERELKRQRRKQSNRESARRSRLR
Sbjct: 121 SSTIPAMHGKVPSTAVAGGMVNAGPRDGVHSQPWLQDERELKRQRRKQSNRESARRSRLR 180
Query: 325 KQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALKERLGELPGNDDLRSGK 384
KQAECDELAQRA+ L EENASLR+E+SRI+S++ + L ENAALK + GE+ N+++ G+
Sbjct: 181 KQAECDELAQRADVLSEENASLRAELSRIKSEHAKALAENAALKVKQGEILRNEEIVPGQ 240
Query: 385 SDQ 387
+DQ
Sbjct: 241 NDQ 243
>emb|CAA40101.1| HBP-1a [Triticum aestivum] gi|122771|sp|P23922|HBP1A_WHEAT
Transcription factor HBP-1a (Histone-specific
transcription factor HBP1) gi|100838|pir||A41349
histone-specific transcription factor HBP1 - wheat
gi|1199790|dbj|BAA07289.1| transcription factor
HBP-1a(17) [Triticum aestivum] gi|170749|gb|AAA34293.1|
DNA-binding protein
Length = 349
Score = 313 bits (802), Expect = 6e-84
Identities = 190/384 (49%), Positives = 229/384 (59%), Gaps = 68/384 (17%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNP--PDWSSFQAYSPMPPHGFL---A 55
MGS+ D + K SK P QEQPP T++GT P P+W FQ Y MPPHGF
Sbjct: 1 MGSN--DPSTPSKASKPPE---QEQPPATTSGTTAPVYPEWPGFQGYPAMPPHGFFPPPV 55
Query: 56 SNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIP---PGSYPFSPFAMPSPN 112
+ QAHPYMWG QH++PPYGTPP PY+ MYP G +YAHP+ P P YP P
Sbjct: 56 AAGQAHPYMWGPQHMVPPYGTPPPPYM-MYPPGTVYAHPTAPGVHPFHYPMQTNGNLEPA 114
Query: 113 GIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAH 172
G A G PG+ E +GK NE GKT G SANG
Sbjct: 115 G---AQGAAPGAAETNGK---------------------------NEPGKTSGPSANGVT 144
Query: 173 SKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTM 232
S S ESGS+ SEGSDANSQNDS K D E+ +QNG + H++ +G N P M
Sbjct: 145 SNS-ESGSDSESEGSDANSQNDSHSKEN---DVNENGSAQNGVS-HSSSHGTFNKP---M 196
Query: 233 SMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDG 292
+VP+ +G G V GPATNLNIGMDYWG SS +PA+ G VPS + G
Sbjct: 197 PLVPVQSGAVIG-VAGPATNLNIGMDYWGATGSSPVPAMRGKVPSGSARG---------- 245
Query: 293 VQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSR 352
+ W DERELK+Q+RK SNRESARRSRLRKQAEC+EL QRAEALK EN+SLR E+ R
Sbjct: 246 ---EQW--DERELKKQKRKLSNRESARRSRLRKQAECEELGQRAEALKSENSSLRIELDR 300
Query: 353 IRSDYEQLLTENAALKERLGELPG 376
I+ +YE+LL++N +LK +LGE G
Sbjct: 301 IKKEYEELLSKNTSLKAKLGESGG 324
>ref|XP_463980.1| putative transcription factor HBP-1a [Oryza sativa (japonica
cultivar-group)] gi|51963838|ref|XP_506702.1| PREDICTED
P0482F12.32 gene product [Oryza sativa (japonica
cultivar-group)] gi|41053045|dbj|BAD07975.1| putative
transcription factor HBP-1a [Oryza sativa (japonica
cultivar-group)] gi|41053088|dbj|BAD08032.1| putative
transcription factor HBP-1a [Oryza sativa (japonica
cultivar-group)]
Length = 347
Score = 308 bits (788), Expect = 3e-82
Identities = 191/395 (48%), Positives = 234/395 (58%), Gaps = 68/395 (17%)
Query: 15 SKTPPLTPQEQPPTTSTGTVNP--PDWSSFQAYSPMPPHGF----LASNPQAHPYMWGVQ 68
SK + EQ P T++GT P P+W FQAYS +PPHGF +A++PQAHPYMWG Q
Sbjct: 10 SKATKASEPEQSPATTSGTTAPVYPEWPGFQAYSAIPPHGFFPPPVAASPQAHPYMWGAQ 69
Query: 69 HIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGIVEASGNTPGSMEAD 128
++PPYGTPP PY+ MYP G +YAHPS P G +PF+ + M + NG VE +G PG+ E +
Sbjct: 70 PMVPPYGTPP-PYM-MYPPGTVYAHPSTP-GVHPFNHYPMLA-NGNVETAGTAPGASEIN 125
Query: 129 GKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSKSGESGSEGTSEGSD 188
GK NE G+T G SANG S S ESGSE SEGSD
Sbjct: 126 GK---------------------------NELGRTSGPSANGITSHS-ESGSESESEGSD 157
Query: 189 ANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMSMVPISAGGAPGAVPG 248
ANSQNDS K D ED SQNG + HT N Q MSM P G G V
Sbjct: 158 ANSQNDSHSKEN---DVKEDGSSQNGIS-HTALN-------QNMSMAPTQTGVVIGGV-A 205
Query: 249 PATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGVQSQHWLQDERELKRQ 308
P TNLNIGMDYWG SS +PA+HG S +V G + W DERELK+Q
Sbjct: 206 PTTNLNIGMDYWGAAGSSPVPAMHGKASSGSVRG-------------EQW--DERELKKQ 250
Query: 309 RRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRIRSDYEQLLTENAALK 368
+RKQSNRESARRSRLRKQAEC+EL+ RA+ L+ EN+SLR+E+ RI+ +YE LL+ NA+LK
Sbjct: 251 KRKQSNRESARRSRLRKQAECEELSVRADNLRAENSSLRAELERIKKEYEALLSHNASLK 310
Query: 369 ERLGELPGNDDLRSGKSDQHTHDDAQQSGQTEAVQ 403
E+ L GN D ++Q+ + Q Q Q
Sbjct: 311 EK---LEGNSDSIPYMNEQNDTNGTHQKQQDSDAQ 342
>pir||A54415 transcription factor HBP-1a(c14) - wheat gi|497895|dbj|BAA02304.1|
transcription factor HBP-1a(c14) [Triticum aestivum]
Length = 381
Score = 266 bits (680), Expect = 9e-70
Identities = 162/389 (41%), Positives = 222/389 (56%), Gaps = 32/389 (8%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYSPMPP---------- 50
MG ++ K ++S T Q + T + PDWS FQAY +P
Sbjct: 1 MGKGDVATRSKSQKSSTA----QNEQSTPTNPPTAYPDWSQFQAYYNVPGTTQMTPPAYF 56
Query: 51 HGFLASNPQAHPYMWGVQHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPS 110
H +A +PQ HPYMWG Q +MPPYGTPP PY MY G Y +PPGS+P+SP+ + +
Sbjct: 57 HSTVAPSPQGHPYMWGPQ-MMPPYGTPP-PYATMYAQGTPYQQAPMPPGSHPYSPYPVQA 114
Query: 111 PNGIVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANG 170
PNG V+ + G E D K + K K P+KRSKGSLGSL+++T K+ P S++
Sbjct: 115 PNGTVQTPTSGAGGSETD-KSNKNKRKTPLKRSKGSLGSLDVVTVKDKTPPAKPLVSSSN 173
Query: 171 AHSKSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQN----GTAVHTTQ-NGGL 225
S ESGS S GS NS++ S K G D ++ GTAV TQ + G
Sbjct: 174 EGSSQSESGSGSYSGGSSTNSKSGSHTKDGSEHGPANDASNKGVTAQGTAVEPTQVSSGP 233
Query: 226 NTPHQTMSMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMA 285
+ M P+ P +PG AT +++G+DYWG PTS +P +HG + A
Sbjct: 234 VVLNPMMPYWPV-----PPPMPGQATGVSMGVDYWGAPTS--VP-MHGK--AVAAPTSAP 283
Query: 286 TGGSRDGVQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENAS 345
+ SRD V S +QDERE+K+Q+RKQSNRESARRSRLRKQAE +E+A RA+ LK+EN+S
Sbjct: 284 SSNSRDIVLSDPVIQDEREVKKQKRKQSNRESARRSRLRKQAEWEEVASRADLLKQENSS 343
Query: 346 LRSEVSRIRSDYEQLLTENAALKERLGEL 374
L+ E+ +++ + L +EN +L E+L L
Sbjct: 344 LKEELKQLQEKCDNLTSENTSLHEKLKAL 372
>gb|AAK14790.1| G-box binding factor bZIP transcription factor [Catharanthus
roseus]
Length = 316
Score = 230 bits (586), Expect = 7e-59
Identities = 146/344 (42%), Positives = 192/344 (55%), Gaps = 43/344 (12%)
Query: 56 SNPQAHPYMWGVQH-IMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGI 114
++P HPYMWG QH +MPPYGTP PY A+YP GG+YAHP++
Sbjct: 9 ASPTPHPYMWGGQHPLMPPYGTPV-PYPALYPPGGVYAHPTM------------------ 49
Query: 115 VEASGNTPGSMEADG-KPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHS 173
G T + E+D K E K++ KRS+G+ G+ ++ K E GK S N +
Sbjct: 50 ATTPGTTQANAESDAVKVSEGKDRPTSKRSRGASGNHGLVAAKVAESGKAASESGNDGAT 109
Query: 174 KSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMS 233
+S ESGSEG+S+GSD N N+ EL SG ++ SFE + TA ++T ++
Sbjct: 110 QSAESGSEGSSDGSDEN--NNHEL-SGTKKGSFEQMLADGATAQNST----------AIA 156
Query: 234 MVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGV 293
P S G P A+P ATNLNIGMD W SS P PS V +A G D
Sbjct: 157 NFPNSVPGNPVAMP--ATNLNIGMDLWNA--SSAAPGAMKMRPSHGVPSAVAPGMVND-- 210
Query: 294 QSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRI 353
W+QDERELKRQ+RKQSNRESARRSRLRKQAEC+EL QR E L EN +LR E+ R+
Sbjct: 211 ---QWIQDERELKRQKRKQSNRESARRSRLRKQAECEELQQRVETLSNENRALRDELQRL 267
Query: 354 RSDYEQLLTENAALKERLGELPGNDDLRSGKSDQHTHDDAQQSG 397
+ E+L +EN ++K+ L + G + + +S T Q G
Sbjct: 268 SEECEKLTSENNSIKDELTRVCGPEAVSKLESSSITKQQLQSRG 311
>emb|CAC00656.1| common plant regulatory factor 5 [Petroselinum crispum]
Length = 352
Score = 221 bits (562), Expect = 4e-56
Identities = 155/391 (39%), Positives = 210/391 (53%), Gaps = 64/391 (16%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSS-FQAY-SPMPPHGFLASN- 57
MG+ E + TP + T P+ + PP+ PDWSS QAY F AS
Sbjct: 1 MGAGE-ENTPSKHSKPTAPVQEVQTPPSY-------PDWSSSMQAYYGAGAAPAFYASTV 52
Query: 58 --PQAHPYMWGVQH-IMPPYGTP-PHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
P HPYMWG QH +MPPYGTP P+P +YP GG+YAHPSI +P +P+
Sbjct: 53 APPTPHPYMWGGQHPLMPPYGTPIPYP---VYPPGGMYAHPSIAT-----NPSIVPTA-- 102
Query: 114 IVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHS 173
E++GK + K++ K+SKG+ G+ + K + GK +S N +
Sbjct: 103 ------------ESEGKAVDGKDRNSTKKSKGASGNGSSGGGKTGDSGKAASSSGNEGGT 150
Query: 174 KSGESGSEGTSEG-SDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTM 232
+S ESGS+G+S+G SD N+ ++ S G++ SF + +A ++
Sbjct: 151 QSAESGSDGSSDGGSDENTNHEF---STGKKGSFHQMLADGASAQNSV------------ 195
Query: 233 SMVPISAGGAPG--AVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSR 290
AG PG V PA NLNIGMD W + N PS AVA G T R
Sbjct: 196 ------AGSVPGNPVVSVPAANLNIGMDLWNASPAGNGSLKVRQNPSAAVAPG--TMIVR 247
Query: 291 DGVQSQHWL-QDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSE 349
DG+ + W+ QDERELKRQ+RKQSNRESARRSRLRKQAEC+EL R E L EN SLR E
Sbjct: 248 DGMMPEQWVNQDERELKRQKRKQSNRESARRSRLRKQAECEELQGRVETLNNENRSLRDE 307
Query: 350 VSRIRSDYEQLLTENAALKERLGELPGNDDL 380
+ R+ + E++ +EN +KE L + G +++
Sbjct: 308 LKRLSEECEKVTSENNTIKEELIRVYGPEEV 338
>pir||T11751 transcription repressor ROM1 - kidney bean
gi|1354857|gb|AAB36514.1| bZIP transcriptional repressor
ROM1
Length = 339
Score = 206 bits (525), Expect = 8e-52
Identities = 149/383 (38%), Positives = 199/383 (51%), Gaps = 59/383 (15%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSS-FQAY-----SPMPPHGFL 54
MG+ E T K S + QE P + PDWSS QAY +P P
Sbjct: 1 MGAGEESTTKSSKSSSSV----QETPTVPAY-----PDWSSSMQAYYAPGAAPPPFFAST 51
Query: 55 ASNPQAHPYMWGVQH-IMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNG 113
++P HPY+WG QH +MPPYGTP PY A+YP G IYAH P +P+
Sbjct: 52 VASPTPHPYLWGSQHPLMPPYGTPV-PYPALYPPGSIYAH----------HPSMAVTPSV 100
Query: 114 IVEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHS 173
+ + S E +GK + K++ K+ KG+ + K E GK S N S
Sbjct: 101 VQQ-------STEIEGKGTDGKDRDSSKKLKGTSANAGS---KAGESGKAGSGSGNDGMS 150
Query: 174 KSGESGSEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPHQTMS 233
+SGESGSEG+S SD N N+ + + ++ SF D +G G ++ S
Sbjct: 151 QSGESGSEGSSNASDEN--NNQQESATNKKGSF-DLMLVDGANAQNNSGGAIS-----QS 202
Query: 234 MVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDGV 293
+P G P V PATNLNIGMD W + A + S A G +A G
Sbjct: 203 SMP----GKP-VVSMPATNLNIGMDLWNASSGGGEAAKMRHNQSGA-PGVVALG------ 250
Query: 294 QSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEVSRI 353
+ W+QDERELKRQ+RKQSNRESARRSRLRKQAEC++L +R E L EN +LR E+ R+
Sbjct: 251 --EQWIQDERELKRQKRKQSNRESARRSRLRKQAECEDLQKRVETLGSENRTLREELQRL 308
Query: 354 RSDYEQLLTENAALKERLGELPG 376
+ E+L +EN+++KE L + G
Sbjct: 309 SEECEKLTSENSSIKEELERMCG 331
>dbj|BAD42432.1| bZip transcription factor [Psophocarpus tetragonolobus]
Length = 424
Score = 197 bits (502), Expect = 4e-49
Identities = 138/381 (36%), Positives = 201/381 (52%), Gaps = 44/381 (11%)
Query: 1 MGSSEMDKTPKEKESKTPPLTPQEQPPTTSTGTVNPPDWSSFQAYSP---MPPH--GFLA 55
MG+SE K+ K +P T Q P+ PDW++ Q Y P +PP+ +A
Sbjct: 1 MGNSEEGKSIKTGSPSSPATTDQTNQPSIHVY----PDWAAMQYYGPRVNIPPYFNSAVA 56
Query: 56 SNPQAHPYMWGV-QHIMPPYGTPPHPYVAMYPHGGIYAHPSIPPGSYPFSPFAMPSPNGI 114
S HPYMWG Q +MPPYG P Y A Y HGG+Y HP++ G +P +PSP
Sbjct: 57 SGHAPHPYMWGSPQAMMPPYGPP---YAAFYSHGGVYTHPAVAIGPHPHGQ-GVPSP--- 109
Query: 115 VEASGNTPGSMEADGKPPEVKEKLPIKRSKGSLGSLNMITRKNNEHGKTPGTSANGAHSK 174
+ TP S+E+ K ++ +K+ KG G I N E ++ GA ++
Sbjct: 110 --PAAGTPSSVESPTKLSGNTDQGLMKKLKGFDGLAMSIGNCNAE------SAERGAENR 161
Query: 175 SGESG-SEGTSEGSDANSQNDSELKSGGRRDSFEDEPSQNGTAVHTTQNGGLNTPH-QTM 232
+S +EG+S+GSD N+ ++++ R+ S E P+ +G TQ G ++ +
Sbjct: 162 LSQSADTEGSSDGSDGNTAGANKMR---RKRSREGTPTTDGEGKTETQEGSVSKETASSR 218
Query: 233 SMVPISAGGAPGAVPGPATNLNIGMDYWGTPTSSNIPALHGNVPSTAVAGGMATGGSRDG 292
++P + G + GP + S AL PST + T +
Sbjct: 219 KIMPATPASVAGNLVGPIVS------------SGMTTALELRNPSTVHSKANNTSAPQPC 266
Query: 293 --VQSQHWLQDERELKRQRRKQSNRESARRSRLRKQAECDELAQRAEALKEENASLRSEV 350
V S+ WLQ+ERELKR+RRKQSNRESARRSRLRKQAE +ELA++ E L EN SL+SE+
Sbjct: 267 AVVPSEAWLQNERELKRERRKQSNRESARRSRLRKQAETEELARKVEMLSTENVSLKSEI 326
Query: 351 SRIRSDYEQLLTENAALKERL 371
+++ EQ+ EN+AL+E+L
Sbjct: 327 TQLTESSEQMRMENSALREKL 347
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.306 0.126 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 853,012,436
Number of Sequences: 2540612
Number of extensions: 43994145
Number of successful extensions: 187245
Number of sequences better than 10.0: 4983
Number of HSP's better than 10.0 without gapping: 1311
Number of HSP's successfully gapped in prelim test: 3867
Number of HSP's that attempted gapping in prelim test: 171586
Number of HSP's gapped (non-prelim): 14176
length of query: 406
length of database: 863,360,394
effective HSP length: 130
effective length of query: 276
effective length of database: 533,080,834
effective search space: 147130310184
effective search space used: 147130310184
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0230.5


