

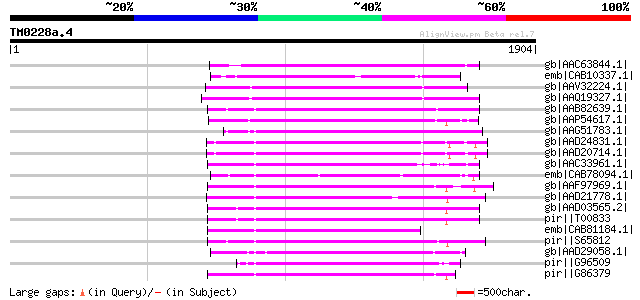
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0228a.4
(1904 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC63844.1| putative non-LTR retroelement reverse transcripta... 649 0.0
emb|CAB10337.1| reverse transcriptase like protein [Arabidopsis ... 620 e-175
gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cult... 525 e-147
gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultiva... 500 e-139
gb|AAB82639.1| putative non-LTR retroelement reverse transcripta... 497 e-138
gb|AAP54617.1| putative non-LTR retroelement reverse transcripta... 495 e-138
gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Ara... 483 e-134
gb|AAD24831.1| putative non-LTR retroelement reverse transcripta... 481 e-133
gb|AAD20714.1| putative non-LTR retroelement reverse transcripta... 480 e-133
gb|AAC33961.1| contains similarity to reverse trancriptase (Pfam... 468 e-130
emb|CAB78094.1| RNA-directed DNA polymerase-like protein [Arabid... 467 e-129
gb|AAF97969.1| F21J9.30 [Arabidopsis thaliana] 458 e-127
gb|AAD21778.1| putative non-LTR retroelement reverse transcripta... 454 e-125
gb|AAD03565.2| putative non-LTR retroelement reverse transcripta... 449 e-124
pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabi... 449 e-124
emb|CAB81184.1| putative protein [Arabidopsis thaliana] gi|45393... 442 e-122
pir||S65812 RNA-directed DNA polymerase (EC 2.7.7.49) (clone DW1... 440 e-121
gb|AAD29058.1| putative non-LTR retroelement reverse transcripta... 437 e-120
pir||G96509 protein F27F5.21 [imported] - Arabidopsis thaliana g... 436 e-120
pir||G86379 protein F5A9.24 [imported] - Arabidopsis thaliana gi... 429 e-118
>gb|AAC63844.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25408124|pir||C84716 hypothetical protein
At2g31080 [imported] - Arabidopsis thaliana
Length = 1231
Score = 649 bits (1675), Expect = 0.0
Identities = 388/998 (38%), Positives = 537/998 (52%), Gaps = 66/998 (6%)
Query: 724 NFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYSDHSPLL 783
+ IDLGF G+KFTW R VAKRLDR RL + EA V HL + SDH+PL
Sbjct: 70 SLIDLGFKGNKFTWRRGRQESTVVAKRLDRVFVCAHARLKWQEAVVSHLPFMASDHAPLY 129
Query: 784 LRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGNLQDKLQRVQEAATEFNKTV 843
++ P R R ++N+ V
Sbjct: 130 VQLE-PLQQRKLR-----------------------------------------KWNREV 147
Query: 844 FGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEMLWFQKSRENW 903
FG+IH RK ++ ++ +Q S+ L E L KE +L QEE LWFQKSRE +
Sbjct: 148 FGDIHVRKEKLVADIKEVQDLLGVVLSDDLLAKEEVLLKEMDLVLEQEETLWFQKSREKY 207
Query: 904 VRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTNLFSL-DVQV 962
+ GDRNT FFH T++RR+RN+I L +D W TD L+ A +Y L+SL DV
Sbjct: 208 IELGDRNTTFFHTSTIIRRRRNRIESLKGDDDRWVTDKVELEAMALTYYKRLYSLEDVSE 267
Query: 963 RRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHIL 1022
RN + +I E+ L+ + EV SA+ SM FKAPGPDG+ FY+Q W +
Sbjct: 268 VRNMLPTGGFASISEAEKAALLQAFTKAEVVSAVKSMGRFKAPGPDGYQPVFYQQCWETV 327
Query: 1023 GEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVN 1082
G + V F+ G LLVLI KV P +++ RP+SLCNV +K+ITK++V
Sbjct: 328 GPSVTRFVLEFFETGVLPASTNDALLVLIAKVAKPERIQQFRPVSLCNVLFKIITKMMVT 387
Query: 1083 RFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRV 1142
R + ++ KL+ P Q SFI GR + DNI+L QE +H++ +K G + LK+DLEKAYDRV
Sbjct: 388 RLKNVISKLIGPAQASFIPGRLSIDNIVLVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRV 447
Query: 1143 NWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLF 1202
WDFL+ TL G IM GV P++SVLWNG + SF RGLRQGDPLSPYLF
Sbjct: 448 RWDFLQETLEAAGLSEGWTSRIMAGVTDPSMSVLWNGERTDSFVPARGLRQGDPLSPYLF 507
Query: 1203 VLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLA 1262
VLC+ERL I+ V + +W+P+ VS G +SH+ FADD++LF +AS Q+R+I+ L
Sbjct: 508 VLCLERLCHLIEASVGKREWKPIAVSCGGSKLSHVCFADDLILFAEASVAQIRIIRRVLE 567
Query: 1263 DFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQGRVKRDH 1322
FCEASG KV+LEKS++ FS V + ++ +S GI LGKYLG+P+LQ R+ ++
Sbjct: 568 RFCEASGQKVSLEKSKIFFSHNVSREMEQLISEESGIGCTKELGKYLGMPILQKRMNKET 627
Query: 1323 FATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVCNHVDKMVRN 1382
F ++E+V+ RLA WK L+ AGR+ L K++LSS+PVH M ++ LP S + +D+ R
Sbjct: 628 FGEVLERVSARLAGWKGRSLSLAGRITLTKAVLSSIPVHVMSAILLPVSTLDTLDRYSRT 687
Query: 1383 CIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFLNEKGKLWVQA 1442
+WG + HL++W+ + + KA+G +GLRSAR N+AL+ K+ L +K LW +
Sbjct: 688 FLWGSTMEKKKQHLLSWRKICKPKAEGGIGLRSARDMNKALVAKVGWRLLQDKESLWARV 747
Query: 1443 LSQKYIKSGSILTEELR--PGGSYVWRGI-LKARDSVSDGFGPRLGAGSS-SLWYSNWLG 1498
+ +KY G T L+ P S WR + + R+ V G G G G + W WL
Sbjct: 748 VRKKYKVGGVQDTSWLKPQPRWSSTWRSVAVGLREVVVKGVGWVPGDGCTIRFWLDRWLL 807
Query: 1499 TGKIAS-RIPFVHITDTTLTVADIWRQGA-WNFDRLYTLLPQEIREEISSVQIPSIQTGE 1556
+ + + AD W G+ WN + L LP+ ++ + SV +
Sbjct: 808 QEPLVELGTDMIPEGERIKVAADYWLPGSGWNLEILGLYLPETVKRRLLSVVVQVFLGNG 867
Query: 1557 DRILWREASDDIYSASSAYQFLNED--GLPETG-FWKLIWKASAPEKVRFFLWLVGRDAL 1613
D I W+ D ++ SAY L D P G F+ IWK PE+VR F+WLV ++ +
Sbjct: 868 DEISWKGTQDGAFTVRSAYSLLQGDVGDRPNMGSFFNRIWKLITPERVRVFIWLVSQNVI 927
Query: 1614 PTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCPG--SVWVRGQLGRLLPWISD----S 1667
TN +R R HL+ A C C E HVLR CP +W RLLP S
Sbjct: 928 MTNVERVRRHLSENAICSVCNGAEETILHVLRDCPAMEPIW-----RRLLPLRRHHEFFS 982
Query: 1668 ATFRHWLFGHLQNRDN---TLFCAMAWNLWKWRNSFVF 1702
+ WLF ++ TLF W WKWR VF
Sbjct: 983 QSLLEWLFTNMDPVKGIWPTLFGMGIWWAWKWRCCDVF 1020
>emb|CAB10337.1| reverse transcriptase like protein [Arabidopsis thaliana]
gi|7268307|emb|CAB78601.1| reverse transcriptase like
protein [Arabidopsis thaliana] gi|7485171|pir||G71420
hypothetical protein - Arabidopsis thaliana
Length = 929
Score = 620 bits (1599), Expect = e-175
Identities = 358/915 (39%), Positives = 510/915 (55%), Gaps = 61/915 (6%)
Query: 728 LGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYSDHSPLLLRCH 787
+GF G++FTW R + VAKRLDR L RL + EA L C
Sbjct: 1 MGFKGNRFTWRRGLVESTFVAKRLDRVLFCAHARLKWQEA----------------LLCP 44
Query: 788 APTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGNLQDK--LQRVQEAATEFNKTVFG 845
A D RPFRF+AAW+SH F+ ++ SW D L L R++ ++NK VFG
Sbjct: 45 AQNVDARRRPFRFEAAWLSHEGFKELLTASW---DTGLSTPVALNRLRWQLKKWNKEVFG 101
Query: 846 NIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEMLWFQKSRENWVR 905
NIH RK +V L+ +Q + ++ L E L KE+ +L+QEE LWFQKSRE +
Sbjct: 102 NIHVRKEKVVSDLKAVQDLLEVVQTDDLLMKEDTLLKEFDVLLHQEETLWFQKSREKLLA 161
Query: 906 FGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTNLFSL-DVQVRR 964
GDRNT FFH TV+RR+RN+I L + W T+ L++ A +Y L+SL DV V R
Sbjct: 162 LGDRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVTEKEALEKLAMDYYRKLYSLEDVSVVR 221
Query: 965 NRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGE 1024
+ P + EE++ L P + +EV A+ SM FKAPGPDG+ FY+Q W +GE
Sbjct: 222 GTLPTEGFPRLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVGE 281
Query: 1025 DLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRF 1084
+ V F++G LLVL+ KV P ++ RP+SLCNV +K+ITK++V R
Sbjct: 282 SVSKFVMEFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIRL 341
Query: 1085 RPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTGGGLVALKIDLEKAYDRVNW 1144
+ ++ KL+ P Q SFI GR + DNI++ QE +H++ +K G + LK+DLEKAYDR+ W
Sbjct: 342 KNVISKLIGPAQASFIPGRLSFDNIVVVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRIRW 401
Query: 1145 DFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVL 1204
DFL TL G + IM V P +S+LWNG K SF +RGLRQGDP+SPYLFVL
Sbjct: 402 DFLAETLEAAGLSEGWIKRIMECVAGPEMSLLWNGEKTDSFTPERGLRQGDPISPYLFVL 461
Query: 1205 CMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADF 1264
C+ERL QI+ V G W+ + +S+ G +SH+ FADD++LF +AS Q
Sbjct: 462 CIERLCHQIETAVGRGDWKSISISQGGPKVSHVCFADDLILFAEASVAQ----------- 510
Query: 1265 CEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQGRVKRDHFA 1324
KV+LEKS++ FS V + + ++ GI LGKYLG+P+LQ R+ +D F
Sbjct: 511 ------KVSLEKSKIFFSNNVSRDLEGLITAETGIGSTRELGKYLGMPVLQKRINKDTFG 564
Query: 1325 TIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVCNHVDKMVRNCI 1384
++E+V++RL+ WK+ L+ AGR+ L K++L S+P+H M S+ LP S+ +DK+ RN +
Sbjct: 565 EVLERVSSRLSGWKSRSLSLAGRITLTKAVLMSIPIHTMSSILLPASLLEQLDKVSRNFL 624
Query: 1385 WGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFLNEKGKLWVQALS 1444
WG R HL++WK V + KA G LGLR+++ N ALL K+ LN+K LW + L
Sbjct: 625 WGSTVEKRKQHLLSWKKVCRPKAAGGLGLRASKDMNRALLAKVGWRLLNDKVSLWARVLR 684
Query: 1445 QKYIKSGSILTEELRPGGSY--VWRGILKARDSVSDGFGPRLGAGSSSLWYSNWLGTGKI 1502
+KY + + L P ++ WR I G G R G W+ +
Sbjct: 685 RKYKVTDVHDSSWLVPKATWSSTWRSI---------GVGLREGVA------KGWILHEPL 729
Query: 1503 ASRIP-FVHITDTTLTVADIWRQG-AWNFDRLYTLLPQEIREEISSVQIPSIQTGEDRIL 1560
+R + + V + W +G W+ +L LP+ + + + +V I + DRI
Sbjct: 730 CTRATCLLSPEELNARVEEFWTEGVGWDMVKLGQCLPRSVTDRLHAVVIKGVLGLRDRIS 789
Query: 1561 WREASDDIYSASSAYQFL--NEDGLP-ETGFWKLIWKASAPEKVRFFLWLVGRDALPTNA 1617
W+ SD ++ SAY L E+ P F+K IW APE+VR FLWLVG+ + TN
Sbjct: 790 WQGTSDGDFTVGSAYVLLTQEEESKPCMESFFKRIWGVIAPERVRVFLWLVGQQVIMTNV 849
Query: 1618 KRHRHHLATTASCIR 1632
+R R H+ C R
Sbjct: 850 ERVRRHIGDIEVCQR 864
>gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|45267888|gb|AAS55787.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 1936
Score = 525 bits (1352), Expect = e-147
Identities = 327/972 (33%), Positives = 483/972 (49%), Gaps = 31/972 (3%)
Query: 710 QQRASNMLNMLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYV 769
+ + N + L C+ DLGF G T++ R +G R V RLDRA+ D WR FPEA V
Sbjct: 791 ETQMQNFRDALYDCDLQDLGFKGVPHTYDNRRDGWRNVKVRLDRAVADDKWRDLFPEAQV 850
Query: 770 EHLARVYSDHSPLLLRCHAPTGDRAARP-FRFQAAWVSHPSFESVVNTSW-----RAQDG 823
HL SDHSP+LL R + ++ W P V+ +W + G
Sbjct: 851 SHLVSPCSDHSPILLEFIVKDTTRPRQKCLHYEIVWEREPESVQVIEEAWINAGVKTDLG 910
Query: 824 NLQDKLQRVQEAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKE 883
++ L RV A ++KT N+ + + ++L+ + + R S R
Sbjct: 911 DINIALGRVMSALRSWSKTKVKNVGKELEKARKKLEDLIASNAARSSI------RQATDH 964
Query: 884 YGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPST 943
++LY+EEMLW Q+SR NW++ GDRNT+FFH++ V R K+NKI L E+G + S
Sbjct: 965 MNEMLYREEMLWLQRSRVNWLKEGDRNTRFFHSRAVWRAKKNKISKLRDENGAIHSTTSV 1024
Query: 944 LQEEAKRFYTNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFK 1003
L+ A ++ ++ D + + + +KL EE+ AI + K
Sbjct: 1025 LETMATEYFQGVYKADPSLNPESVTRLFQEKVTDAMNEKLCQEFKEEEIAQAIFQIGPLK 1084
Query: 1004 APGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDL 1063
+P PDGF A FY++ W L D+ V + FQ+G + T +VLIPK D P LKD
Sbjct: 1085 SPRPDGFPARFYQRNWGTLKSDIILAVRNFFQSGLMPKGVNDTAIVLIPKKDQPIDLKDY 1144
Query: 1064 RPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIH-TR 1122
RPISLCNV YK+++K LVNR RP+L LVS Q +FI GR DN +LA E H+I +
Sbjct: 1145 RPISLCNVVYKVVSKCLVNRLRPILDDLVSKEQSAFIQGRMITDNALLAFECFHSIQKNK 1204
Query: 1123 KTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKL 1182
K A K+DL KAYDRV+W FL L GF R V IM V T SV +NG+ L
Sbjct: 1205 KANSAACAYKLDLSKAYDRVDWRFLELALNKLGFAHRWVSWIMLCVTTVRYSVKFNGTLL 1264
Query: 1183 PSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADD 1242
SF RGLRQG+PLSP+LF+ + L++ ++ V + P+++ + GIS+L FADD
Sbjct: 1265 RSFAPTRGLRQGEPLSPFLFLFVADGLSLLLKEKVAQNSLTPLKICRQAPGISYLLFADD 1324
Query: 1243 VLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRA 1302
LLF +A K + ++K L ++ + +G +N K +LF + ++ + + R
Sbjct: 1325 TLLFFKAEKKEAEVVKEVLTNYAQGTGQLINPAKCSILFGEASPSSVSEDIRNTLQVERD 1384
Query: 1303 SNLGKYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHA 1362
+ +YLG P +GR+ + F ++ K+ R+ W N L+ G+ L K+++ ++PV+
Sbjct: 1385 NFEDRYLGFPTPEGRMHKGRFQSLQAKIAKRVIQWGENFLSSGGKEILIKAVIQAIPVYV 1444
Query: 1363 MQSLWLPDSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEA 1422
M PDSV + + KM RN WG R H AW +T++K G LG R + N+A
Sbjct: 1445 MGLFKFPDSVYDELTKMTRNFWWGADNGRRRTHWRAWDSLTKAKINGGLGFRDYKLFNQA 1504
Query: 1423 LLGKLVHSFLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGP 1482
LL + + L Q L KY GS+ S W GI D + G
Sbjct: 1505 LLTRQAWRLIEFPNSLCAQVLKAKYFPHGSLTDTTFSANASPTWHGIEYGLDLLKKGIIW 1564
Query: 1483 RLGAGSS-SLWYSNWLGTGKIASRIPFVHITDTTLT-VAD-IWRQGAWNFDRLYTLLPQE 1539
R+G G+S +W W+ + SR P + L V+D I G W+ ++ +
Sbjct: 1565 RIGNGNSVRIWRDPWI--PRDLSRRPVSSKANCRLKWVSDLIAEDGTWDSAKINQYFLKI 1622
Query: 1540 IREEISSVQIPSIQTGEDRILWREASDDIYSASSAYQF------LNEDGLPETG----FW 1589
+ I + I S + ED I W +S SAY+ +N + W
Sbjct: 1623 DADIIQKICI-SARLEEDFIAWHPDKTGRFSVRSAYKLALQLADMNNCSSSSSSRLNKSW 1681
Query: 1590 KLIWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRC-- 1647
+LIWK + P+KVR F W V ++L T + + +L C C EDA H L RC
Sbjct: 1682 ELIWKCNVPQKVRIFAWRVASNSLATMENKKKRNLERFDVCGICDREKEDAGHALCRCVH 1741
Query: 1648 PGSVWVRGQLGR 1659
S+WV + G+
Sbjct: 1742 ANSLWVNLEKGK 1753
>gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultivar-group)]
Length = 2367
Score = 500 bits (1288), Expect = e-139
Identities = 324/1034 (31%), Positives = 500/1034 (48%), Gaps = 42/1034 (4%)
Query: 696 RYVHCQRLLAGTSQQQRASNMLNMLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRAL 755
++ H R G Q Q + ++L+ C DLGF G T++ + G R V RLDR +
Sbjct: 874 QFEHFSRTPRGEPQMQ---DFRDVLQDCELHDLGFKGVPHTYDNKREGWRNVKVRLDRVV 930
Query: 756 GDVAWRLCFPEAYVEHLARVYSDHSPLLLRCHAPTGDRAARP-FRFQAAWVSHPSFESVV 814
D WR + A V HL SDH P+LL + + ++ W P V+
Sbjct: 931 ADDKWRDIYSTAQVVHLVSPCSDHCPILLNLVVKDPHQLRQKCLHYEIVWEREPEATQVI 990
Query: 815 NTSW-----RAQDGNLQDKLQRVQEAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRD 869
+W +A G++ L +V A +++ N+ R + ++L + + + R
Sbjct: 991 EEAWVVAGEKADLGDINKALAKVMTALRSWSRAKVKNVGRELEKARKKLAELIESNADRT 1050
Query: 870 SESLARLERDLQKEYGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHG 929
+ R+ ++LY+EEMLW Q+SR NW++ DRNTKFFH++ V R K+NKI
Sbjct: 1051 ------VIRNATDHMNELLYREEMLWLQRSRVNWLKDEDRNTKFFHSRAVWRAKKNKISK 1104
Query: 930 LFLEDGNWCTDPSTLQEE--AKRFYTNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPV 987
L D N ST++ E A ++ ++++ D + + + +KL
Sbjct: 1105 L--RDANETVHSSTMKLESMATEYFQDVYTADPNLNPETVTRLIQEKVTDIMNEKLCEDF 1162
Query: 988 SLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATL 1047
+ +E+ AI + K+PGPDGF A FY++ W + D+ V FQ G + T
Sbjct: 1163 TEDEISQAIFQIGPLKSPGPDGFPARFYQRNWGTIKADIIGAVRRFFQTGLMPEGVNDTA 1222
Query: 1048 LVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRD 1107
+VLIPK + P L+D RPISLCNV YK+++K LVNR RP+L LVS Q +F+ GR D
Sbjct: 1223 IVLIPKKEQPVDLRDFRPISLCNVVYKVVSKCLVNRLRPILDDLVSVEQSAFVQGRMITD 1282
Query: 1108 NIILAQEVMHTIH-TRKTGGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMW 1166
N +LA E H + +K A K+DL KAYDRV+W FL + GF R V+ IM
Sbjct: 1283 NALLAFECFHAMQKNKKANHAACAYKLDLSKAYDRVDWRFLEMAMNKLGFARRWVNWIMK 1342
Query: 1167 GVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVR 1226
V + V +NG+ L SF RGLRQGDPL P+LF+ + L++ ++ V + P +
Sbjct: 1343 CVTSVRYMVKFNGTLLQSFAPTRGLRQGDPLLPFLFLFVADGLSLLLKEKVAQNSLTPFK 1402
Query: 1227 VSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVG 1286
V + GISHL FADD LLF +A + + ++K L+ + +G +N K +L
Sbjct: 1403 VCRAAPGISHLLFADDTLLFFKAHQREAEVVKEVLSSYAMGTGQLINPAKCSILMGGAST 1462
Query: 1287 QRRQRELSGLVGISRASNLGKYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAG 1346
+S ++ + R +YLG P +GR+ + F ++ K+ R+ W N L+ G
Sbjct: 1463 PAVSEAISEILQVERDRFEDRYLGFPTPEGRMHKGRFQSLQAKIWKRVIQWGENHLSTGG 1522
Query: 1347 RVCLAKSILSSLPVHAMQSLWLPDSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSK 1406
+ L K+++ ++PV+ M LP+SV + + K+ +N W R H AW +T+ K
Sbjct: 1523 KEVLIKAVIQAIPVYVMGIFKLPESVIDDLTKLTKNFWWDSMNGQRKTHWKAWDSLTKPK 1582
Query: 1407 AKGRLGLRSARKNNEALLGKLVHSFLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVW 1466
+ G LG R R N+ALL + + L + L KY GS++ S W
Sbjct: 1583 SLGGLGFRDYRLFNQALLARQAWRLITYPDSLCARVLKAKYFPHGSLIDTSFGSNSSPAW 1642
Query: 1467 RGILKARDSVSDGFGPRLGAGSS-SLWYSNWLGTGKIASRIPFVHITDTTLT-VAD-IWR 1523
R I D + G R+G G+S +W +WL + SR P + L V+D I
Sbjct: 1643 RSIEYGLDLLKKGIIWRVGNGNSIRIWRDSWL--PRDHSRRPITGKANCRLKWVSDLITE 1700
Query: 1524 QGAWNFDRLYTLLPQEIREEISSVQIPSIQTGEDRILWREASDDIYSASSAY----QFLN 1579
G+W+ +++ E I ++ I S ++ ED I W + ++S SAY Q +N
Sbjct: 1701 DGSWDVPKIHQYFHNLDAEVILNICISS-RSEEDFIAWHPDKNGMFSVRSAYRLAAQLVN 1759
Query: 1580 EDGLPETG------FWKLIWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRC 1633
+ +G W++IWK P+KV+ F W V + L T + + L + C C
Sbjct: 1760 IEESSSSGTNNINKAWEMIWKCKVPQKVKIFAWRVASNCLATMVNKKKRKLEQSDMCQIC 1819
Query: 1634 GAVVEDAEHVLRRC--PGSVW-VRGQLGRLLPWISDSATFRHWLFGHLQ---NRDNTLFC 1687
ED H L RC +W + G + I S R WLF L+ + +F
Sbjct: 1820 DRENEDDAHALCRCIQASQLWSCMHKSGSVSVDIKASVLGRFWLFDCLEKIPEYEQAMFL 1879
Query: 1688 AMAWNLWKWRNSFV 1701
W W RN +
Sbjct: 1880 MTLWRNWYVRNELI 1893
>gb|AAB82639.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25408936|pir||A84888 hypothetical protein
At2g45230 [imported] - Arabidopsis thaliana
Length = 1374
Score = 497 bits (1280), Expect = e-138
Identities = 309/1022 (30%), Positives = 502/1022 (48%), Gaps = 47/1022 (4%)
Query: 719 MLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYSD 778
ML +C ++ G +F+W N V RLDR + + AW FP+A +L ++ SD
Sbjct: 163 MLNSCGLWEVNHSGYQFSWYGNRNDE-LVQCRLDRTVANQAWMELFPQAKATYLQKICSD 221
Query: 779 HSPLLLRCHAPTGD--RAARPFRFQAAWVSHPSFESVVNTSWRAQDGN----LQDKLQRV 832
HSPL+ GD R F++ WV F+ ++ W Q + +K+
Sbjct: 222 HSPLINNL---VGDNWRKWAGFKYDKRWVQREGFKDLLCNFWSQQSTKTNALMMEKIASC 278
Query: 833 QEAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEE 892
+ +++ + + R + ++ +L ++ + D LARL+++L +EY + EE
Sbjct: 279 RREISKWKRVSKPSSAVRIQELQFKLDAATKQIPF-DRRELARLKKELSQEYNN----EE 333
Query: 893 MLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFY 952
W +KSR W+R GDRNTK+FHA T RR +N+I L E+G T L A+ ++
Sbjct: 334 QFWQEKSRIMWMRNGDRNTKYFHAATKNRRAQNRIQKLIDEEGREWTSDEDLGRVAEAYF 393
Query: 953 TNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHA 1012
LF+ + +EN TP + + + L+AP++ EEV+ A S+ K PGPDG +
Sbjct: 394 KKLFASEDVGYTVEELENLTPLVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNG 453
Query: 1013 YFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVS 1072
+ Y+Q+W +G+ + MV + F++G + T + LIPK+ K+ D RPISLCNV
Sbjct: 454 FLYQQFWETMGDQITEMVQAFFRSGSIEEGMNKTNICLIPKILKAEKMTDFRPISLCNVI 513
Query: 1073 YKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTR-KTGGGLVAL 1131
YK+I K++ NR + +L L+S Q +F+ GR DNI++A E++H + + K +A+
Sbjct: 514 YKVIGKLMANRLKKILPSLISETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAI 573
Query: 1132 KIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGL 1191
K D+ KAYDRV W FL + GF + LIM V++ VL NG+ RGL
Sbjct: 574 KTDISKAYDRVEWPFLEKAMRGLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGL 633
Query: 1192 RQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASK 1251
RQGDPLSPYLFV+C E L +Q+ ++ + ++V++ ISHL FADD + +C+ +
Sbjct: 634 RQGDPLSPYLFVICTEMLVKMLQSAEQKNQITGLKVARGAPPISHLLFADDSMFYCKVND 693
Query: 1252 DQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGL 1311
+ L I + ++ ASG +VN KS + F K + + R+ + +GI R G YLGL
Sbjct: 694 EALGQIIRIIEEYSLASGQRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGL 753
Query: 1312 PLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDS 1371
P K + + +++ ++ W++N L+ G+ L K++ +LP + M +P +
Sbjct: 754 PESFQGSKVATLSYLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKT 813
Query: 1372 VCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSF 1431
+C ++ ++ W R H AW +++ KA G LG + N ALLGK +
Sbjct: 814 ICQQIESVMAEFWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRM 873
Query: 1432 LNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSS-S 1490
+ EK L + +Y L L S+ W+ I +A+ + G +G G + +
Sbjct: 874 ITEKDSLMAKVFKSRYFSKSDPLNAPLGSRPSFAWKSIYEAQVLIKQGIRAVIGNGETIN 933
Query: 1491 LWYSNWLGT--GKIASRIPFVHI-----TDTTLTVADIWRQGA--WNFDRLYTLLPQEIR 1541
+W W+G K A + H+ ++ V D+ WN++ + L P +
Sbjct: 934 VWTDPWIGAKPAKAAQAVKRSHLVSQYAANSIHVVKDLLLPDGRDWNWNLVSLLFPDNTQ 993
Query: 1542 EEISSVQIPSIQTGEDRILWREASDDIYSASSAY----QFLNEDGLPE-------TGFWK 1590
E I +++ P + DR W + YS S Y + +N+ P+ ++
Sbjct: 994 ENILALR-PGGKETRDRFTWEYSRSGHYSVKSGYWVMTEIINQRNNPQEVLQPSLDPIFQ 1052
Query: 1591 LIWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCPGS 1650
IWK P K+ FLW + L + HLA SC+RC + E H+L +CP +
Sbjct: 1053 QIWKLDVPPKIHHFLWRCVNNCLSVASNLAYRHLAREKSCVRCPSHGETVNHLLFKCPFA 1112
Query: 1651 --VWVRGQLGRLLPWISDSATFR---HWLFGHL----QNRDNTLFCAMAWNLWKWRNSFV 1701
W L + FR H L H ++ + L + W LWK RN V
Sbjct: 1113 RLTWAISPLPAPPGGEWAESLFRNMHHVLSVHKSQPEESDHHALIPWILWRLWKNRNDLV 1172
Query: 1702 FE 1703
F+
Sbjct: 1173 FK 1174
>gb|AAP54617.1| putative non-LTR retroelement reverse transcriptase [Oryza sativa
(japonica cultivar-group)] gi|37536056|ref|NP_922330.1|
putative non-LTR retroelement reverse transcriptase
[Oryza sativa (japonica cultivar-group)]
gi|10140689|gb|AAG13524.1| putative non-LTR retroelement
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1382
Score = 495 bits (1275), Expect = e-138
Identities = 313/1023 (30%), Positives = 510/1023 (49%), Gaps = 57/1023 (5%)
Query: 720 LEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYSDH 779
L+ C IDLGF+G KFTW + + RLDRA+ + + F + VE++ SDH
Sbjct: 170 LDDCGLIDLGFVGPKFTWSNKQDANSNSKVRLDRAVANGEFSRYFEDCLVENVITTSSDH 229
Query: 780 SPLLL----RCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGN------LQDKL 829
+ + R H + FRF+AAW+ + VV SWR + L
Sbjct: 230 YAISIDLSRRNHGQRRIPIQQGFRFEAAWLRAEDYREVVENSWRISSAGCVGLRGVWSVL 289
Query: 830 QRVQEAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILY 889
Q+V + +++K FG++ R+ ++ER+L+ ++Q ++ + + E+ ++++ ++
Sbjct: 290 QQVAVSLKDWSKASFGSVRRKILKMERKLKSLRQSPV---NDVVIQEEKLIEQQLCELFE 346
Query: 890 QEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAK 949
+EE++ Q+SR +W+R GDRNT FFHA+ RR+ N+I L +DG+ C ++ A+
Sbjct: 347 KEEIMARQRSRVDWLREGDRNTAFFHARASARRRTNRIKELVRDDGSRCISQEGIKRMAE 406
Query: 950 RFYTNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDG 1009
FY NLFS + +++ + +L + EE+++A+ M S KAPGPDG
Sbjct: 407 VFYENLFSSEPCDSMEEVLDAIPNKVGDFINGELGKQYTNEEIKTALFQMGSTKAPGPDG 466
Query: 1010 FHAYFYKQYWHILGEDLHHMVASAFQNGGANPE-LLATLLVLIPKVDNPTKLKDLRPISL 1068
F A FY+ +W IL E + + V F G PE L +++VLIPKV+N + L RPISL
Sbjct: 467 FPALFYQTHWGILEEHICNAVRG-FLLGEEIPEGLCDSVVVLIPKVNNASHLSKFRPISL 525
Query: 1069 CNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTGGGL 1128
CNV YK+ +KVL NR +P L +VS Q +F+ GR D+ ++A E +HTI +
Sbjct: 526 CNVLYKIASKVLANRLKPFLPDIVSEFQSAFVPGRLITDSALVAYECLHTIRKQHNKNPF 585
Query: 1129 VALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQ 1188
ALKID+ KAYDRV W +L L GF ++ +M V + +V NG
Sbjct: 586 FALKIDMMKAYDRVEWAYLSGCLSKLGFSQDWINTVMRCVSSVRYAVKINGELTKPVVPS 645
Query: 1189 RGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQ 1248
RG+RQGDP+SPYLF+LC E L+ + G+ + ++ + G ISHL FADD + F +
Sbjct: 646 RGIRQGDPISPYLFLLCTEGLSCLLHKKEVAGELQGIKNGRHGPPISHLLFADDSIFFAK 705
Query: 1249 ASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKY 1308
A ++ +K+TL +C ASG K+NL KS + F K + + + + Y
Sbjct: 706 ADSRNVQALKNTLRSYCSASGQKINLHKSSIFFGKRCPDAVKISVKSCLQVDNEVLQDSY 765
Query: 1309 LGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWL 1368
LG+P G + F + E++ R+ W + L++AG + K++ ++P + M +
Sbjct: 766 LGMPTEIGLATTNFFKFLPERIWKRVNGWTDRPLSRAGMETMLKAVAQAIPNYVMSCFRI 825
Query: 1369 PDSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLV 1428
P S+C + + + WG + H +W ++ K G +G R N+A+LG+
Sbjct: 826 PVSICEKMKTCIADHWWGFEDGKKKMHWKSWSWLSTPKFLGGMGFREFTTFNQAMLGRQC 885
Query: 1429 HSFLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGS 1488
L + L + L +Y + S S+ WR +L R+ ++ G +G G
Sbjct: 886 WRLLTDPDSLCSRVLKGRYFPNSSFWEAAQPKSPSFTWRSLLFGRELLAKGVRWGVGDGK 945
Query: 1489 S-SLWYSNWLGTGK---IASRIPFVHITDTTLTVADIWRQGAWNFDRLYTLLPQEIREEI 1544
+ ++ NW+ + + + PF TD T++ W+ D + +L P +I +EI
Sbjct: 946 TIKIFSDNWIPGFRPQLVTTLSPFP--TDATVSCLMNEDARCWDGDLIRSLFPVDIAKEI 1003
Query: 1545 SSVQIPSIQTGE-DRILWREASDDIYSASSAYQFLNEDG----------------LPETG 1587
+QIP + G+ D W +YS SAY + L
Sbjct: 1004 --LQIPISRHGDADFASWPHDKLGLYSVRSAYNLARSEAFFADQSNSGRGMASRLLESQK 1061
Query: 1588 FWKLIWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRC 1647
WK +WK +AP K++ LW + L T + R H+ +T C+ C + EHV C
Sbjct: 1062 DWKGLWKINAPGKMKITLWRAAHECLATGFQLRRRHIPSTDGCVFCNR-DDTVEHVFLFC 1120
Query: 1648 P--GSVW--VRG----QLGRLLPWISDSATFRHWLFGHLQ---NRDNTLFCAMAWNLWKW 1696
P +W ++G +LGR + +T R W+F L+ + NTL W++W+
Sbjct: 1121 PFAAQIWEEIKGKCAVKLGR-----NGFSTMRQWIFDFLKRGSSHANTLLAVTFWHIWEA 1175
Query: 1697 RNS 1699
RN+
Sbjct: 1176 RNN 1178
>gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
gi|25405244|pir||E96519 probable reverse transcriptase,
16838-20266 [imported] - Arabidopsis thaliana
Length = 1142
Score = 483 bits (1244), Expect = e-134
Identities = 307/959 (32%), Positives = 466/959 (48%), Gaps = 30/959 (3%)
Query: 777 SDHSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRA----QDGNLQDKLQRV 832
SDHSP++ A R + FRF W+ ++ W ++G +KL
Sbjct: 4 SDHSPVIATI-ADKIPRGKQNFRFDKRWIGKDGLLEAISQGWNLDSGFREGQFVEKLTNC 62
Query: 833 QEAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEE 892
+ A +++ K++ + ++ L Q++ D R E + L L++ Y D EE
Sbjct: 63 RRAISKWRKSLIPFGRQTIEDLKAELDVAQRDDD-RSREEITELTLRLKEAYRD----EE 117
Query: 893 MLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFY 952
W+QKSR W++ GD N+KFFHA T RR RN+I GL E+G W + +Q A ++
Sbjct: 118 QYWYQKSRSLWMKLGDNNSKFFHALTKQRRARNRITGLHDENGIWSIEDDDIQNIAVSYF 177
Query: 953 TNLFSL-DVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFH 1011
NLF+ + QV + E Q I D L A + EVR+A+ + KAPGPDG
Sbjct: 178 QNLFTTANPQVFDEALGEVQV-LITDRINDLLTADATECEVRAALFMIHPEKAPGPDGMT 236
Query: 1012 AYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNV 1071
A F+++ W I+ DL +V S Q G + L T + LIPK + PT++ +LRPISLCNV
Sbjct: 237 ALFFQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPISLCNV 296
Query: 1072 SYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKT-GGGLVA 1130
YK+I+K+L R + +L L+S Q +F+ GR DNI++AQE+ H + T + +A
Sbjct: 297 GYKVISKILCQRLKTVLPNLISETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKDKFMA 356
Query: 1131 LKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRG 1190
+K D+ KAYD+V W+F+ A L GF + + IMW + T VL NG +RG
Sbjct: 357 IKTDMSKAYDQVEWNFIEALLRKMGFCEKWISWIMWCITTVQYKVLINGQPKGLIIPERG 416
Query: 1191 LRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQAS 1250
LRQGDPLSPYLF+LC E L I+ + ++V+ +SHL FADD L FC+A+
Sbjct: 417 LRQGDPLSPYLFILCTEVLIANIRKAERQNLITGIKVATPSPAVSHLLFADDSLFFCKAN 476
Query: 1251 KDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLG 1310
K+Q +I L + SG ++N KS + F V + ++ ++GI +G YLG
Sbjct: 477 KEQCGIILEILKQYESVSGQQINFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMGSYLG 536
Query: 1311 LPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPD 1370
LP G K F+ + +++ +R+ W L+K G+ + KS+ ++LP + M LP
Sbjct: 537 LPESLGGSKTKVFSFVRDRLQSRINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCFRLPK 596
Query: 1371 SVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHS 1430
++ + + V W G++R H +AW + SK+ G LG R+ N ALL K +
Sbjct: 597 AITSKLTSAVAKFWWSSNGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWR 656
Query: 1431 FLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSS- 1489
+ L+ + +Y + + L SY WR ++ AR V G R+G+G+S
Sbjct: 657 LITAPDSLFAKVFKGRYFRKSNPLDSIKSYSPSYGWRSMISARSLVYKGLIKRVGSGASI 716
Query: 1490 SLWYSNWLGTGKIASRIPFVHITDTTLTVADI--WRQGAWNFDRLYTLLPQEIREEISSV 1547
S+W W+ I D +L V + R WN D L L E IS++
Sbjct: 717 SVWNDPWIPAQFPRPAKYGGSIVDPSLKVKSLIDSRSNFWNIDLLKELFDPEDVPLISAL 776
Query: 1548 QIPSIQTGEDRILWREASDDIYSASSAYQF----LNED----GLPETGFWKLIWKASAPE 1599
I + ED + W Y+ S Y LNE G T IWK P
Sbjct: 777 PIGN-PNMEDTLGWHFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKAYIWKVQCPP 835
Query: 1600 KVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRC--PGSVWVRGQL 1657
K+R FLW + +P + + + C+ CGA E H L +C +W Q+
Sbjct: 836 KLRHFLWQILSGCVPVSENLRKRGILCDKGCVSCGASEESINHTLFQCHPARQIWALSQI 895
Query: 1658 GR---LLPWISDSATFRHWLFGHLQNRDNTLFCAMAWNLWKWRNSFVFEQVPWQPGDVM 1713
+ P S H + D+ + + W +WK RN VFE V P +++
Sbjct: 896 PTAPGIFPSNSIFTNLDHLFWRIPSGVDSAPYPWIIWYIWKARNEKVFENVDKDPMEIL 954
>gb|AAD24831.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25408166|pir||G84721 hypothetical protein
At2g31520 [imported] - Arabidopsis thaliana
Length = 1524
Score = 481 bits (1237), Expect = e-133
Identities = 327/1064 (30%), Positives = 503/1064 (46%), Gaps = 70/1064 (6%)
Query: 715 NMLNMLEACNFIDLGFMGSKFTW--ERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHL 772
N NM+ C+ D+ G +F+W ER + V LDR + AW FP A E L
Sbjct: 314 NFRNMVSHCDIEDMRSKGDRFSWVGERHTH---TVKCCLDRVFINSAWTATFPYAETEFL 370
Query: 773 ARVYSDHSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGN----LQDK 828
SDH P+L+ + + R ++ FRF + P+F+ +V TSWR + + ++
Sbjct: 371 DFTGSDHKPVLVHFNE-SFPRRSKLFRFDNRLIDIPTFKRIVQTSWRTNRNSRSTPITER 429
Query: 829 LQRVQEAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDIL 888
+ ++A N +R ++++ L + D + + +L+ L K + D
Sbjct: 430 ISSCRQAMARLKHASNLNSEQRIKKLQSSLNRAMESTRRVDRQLIPQLQESLAKAFSD-- 487
Query: 889 YQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEA 948
EE+ W QKSR W++ GD+NT +FHA T R +N+++ + + G T + A
Sbjct: 488 --EEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHA 545
Query: 949 KRFYTNLFSLD-VQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGP 1007
+ F+TN+FS + ++V + ++ D L S E+ AI + KAPGP
Sbjct: 546 QDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLD-LTKEFSDTEIYDAICQIGDDKAPGP 604
Query: 1008 DGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPIS 1067
DG A FYK W I+G D+ V F+ P + T + +IPK+ NPT L D RPI+
Sbjct: 605 DGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIA 664
Query: 1068 LCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRK-TGG 1126
LCNV YK+I+K LVNR + L+ +VS Q +FI GR DN+++A EVMH++ RK
Sbjct: 665 LCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSK 724
Query: 1127 GLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFP 1186
+A+K D+ KAYDRV WDFL T+ FGF + + IM V++ SVL NGS
Sbjct: 725 TYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYIT 784
Query: 1187 LQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLF 1246
RG+RQGDPLSPYLF+LC + L+ I G R VR+ I+HL FADD L F
Sbjct: 785 PTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAITHLQFADDSLFF 844
Query: 1247 CQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLG 1306
CQA+ + +K + SG K+N++KS + F V Q +L ++ I G
Sbjct: 845 CQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSKLKQILEIPNQGGGG 904
Query: 1307 KYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSL 1366
KYLGLP GR K++ F II++V R ++W L+ AG+ + KS+ ++PV+AM
Sbjct: 905 KYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCF 964
Query: 1367 WLPDSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGK 1426
LP + + ++ ++ N W K N R VAWK + SK +G LG R K N+ALL K
Sbjct: 965 KLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAK 1024
Query: 1427 LVHSFLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGA 1486
+ L+ + + +Y K SIL ++R SY W +L + G +G
Sbjct: 1025 QAWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLLDGIALLKKGTRHLIGD 1084
Query: 1487 GSSSLWYSNWLGTGKIASRIP----FVHITDTTLTVADIW-RQGA---WNFDRLYTLLPQ 1538
G + +G I P T +T+ +++ R+G+ W+ ++ + Q
Sbjct: 1085 GQNIR-----IGLDNIVDSHPPRPLNTEETYKEMTINNLFERKGSYYFWDDSKISQFVDQ 1139
Query: 1539 EIREEISSVQIPSIQTGEDRILWREASDDIYSASSAYQFLNEDGLPETGFWKL------- 1591
I + + + D+I+W + Y+ S Y L D P T +
Sbjct: 1140 SDHGFIHRIYLAKSKK-PDKIIWNYNTTGEYTVRSGYWLLTHD--PSTNIPAINPPHGSI 1196
Query: 1592 -----IWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRR 1646
IW K++ FLW AL T + + C RC E H L
Sbjct: 1197 DLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPICPRCHRENESINHALFT 1256
Query: 1647 CPGSVWVRGQLGRLLPWISDSATFRHWLFGH--LQNRDNTLFCA---------------M 1689
CP + W+SDS+ R+ L + +N N L +
Sbjct: 1257 CP--------FATMAWWLSDSSLIRNQLMSNDFEENISNILNFVQDTTMSDFHKLLPVWL 1308
Query: 1690 AWNLWKWRNSFVFEQVPWQPGDVMRRIHLDTVEFHRLAQGTQRS 1733
W +WK RN+ VF + P + +T ++ Q +++
Sbjct: 1309 IWRIWKARNNVVFNKFRESPSKTVLSAKAETHDWLNATQSHKKT 1352
>gb|AAD20714.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25412331|pir||G84649 hypothetical protein
At2g25550 [imported] - Arabidopsis thaliana
Length = 1750
Score = 480 bits (1235), Expect = e-133
Identities = 328/1065 (30%), Positives = 505/1065 (46%), Gaps = 72/1065 (6%)
Query: 715 NMLNMLEACNFIDLGFMGSKFTW--ERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHL 772
N NM+ C+ D+ G +F+W ER + V LDR + AW FP A +E L
Sbjct: 540 NFRNMVSHCDIEDMRSKGDRFSWVGERHTH---TVKCCLDRVFINSAWTATFPYAEIEFL 596
Query: 773 ARVYSDHSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGN----LQDK 828
SDH P+L+ + + R ++ FRF + P+F+ +V TSWR + + ++
Sbjct: 597 DFTGSDHKPVLVHFNE-SFPRRSKLFRFDNRLIDIPTFKRIVQTSWRTNRNSRSTPITER 655
Query: 829 LQRVQEAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDIL 888
+ ++A N +R ++++ L + D + + +L+ L K + D
Sbjct: 656 ISSCRQAMARLKHASNLNSEQRIKKLQSSLNRAMESTRRVDRQLIPQLQESLAKAFSD-- 713
Query: 889 YQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEA 948
EE+ W QKSR W++ GD+NT +FHA T R +N+++ + + G T + A
Sbjct: 714 --EEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHA 771
Query: 949 KRFYTNLFSLD-VQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGP 1007
+ F+TN+FS + ++V + ++ D L S E+ AI + KAPGP
Sbjct: 772 QDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLD-LTKEFSDTEIYDAICQIGDDKAPGP 830
Query: 1008 DGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPIS 1067
DG A FYK W I+G D+ V F+ P + T + +IPK+ NPT L D RPI+
Sbjct: 831 DGLTARFYKNCWDIVGYDVILEVKKFFETSFMKPSINHTNICMIPKITNPTTLSDYRPIA 890
Query: 1068 LCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRK-TGG 1126
LCNV YK+I+K LVNR + L+ +VS Q +FI GR DN+++A EVMH++ RK
Sbjct: 891 LCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSK 950
Query: 1127 GLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFP 1186
+A+K D+ KAYDRV WDFL T+ FGF + + IM V++ SVL NGS
Sbjct: 951 TYMAVKTDVSKAYDRVEWDFLETTMRLFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYIT 1010
Query: 1187 LQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLF 1246
RG+RQGDPLSPYLF+LC + L+ I G R VR+ I+HL FADD L F
Sbjct: 1011 PTRGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAITHLQFADDSLFF 1070
Query: 1247 CQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLG 1306
CQA+ + +K + SG K+N++KS + F V Q L ++ I G
Sbjct: 1071 CQANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSRLKQILEIPNQGGGG 1130
Query: 1307 KYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSL 1366
KYLGLP GR K++ F II++V R ++W L+ AG+ + KS+ ++PV+AM
Sbjct: 1131 KYLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCF 1190
Query: 1367 WLPDSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGK 1426
LP + + ++ ++ N W K N R VAWK + SK +G LG R K N+ALL K
Sbjct: 1191 KLPKGIVSEIESLLMNFWWEKASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAK 1250
Query: 1427 LVHSFLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGA 1486
+ L+ + + +Y K SIL ++R SY W +L + G +G
Sbjct: 1251 QAWRLIQYPNSLFARVMKARYFKDVSILDAKVRKQQSYGWASLLDGIALLKKGTRHLIGD 1310
Query: 1487 GSSSLWYSNWLGTGKIASRIP----FVHITDTTLTVADIW-RQGA---WNFDRLYTLLPQ 1538
G + +G I P T +T+ +++ R+G+ W+ ++ + Q
Sbjct: 1311 GQNIR-----IGLDNIVDSHPPRPLNTEETYKEMTINNLFERKGSYYFWDDSKISQFVDQ 1365
Query: 1539 EIREEISSVQIPSIQTGEDRILWREASDDIYSASSAYQFLNEDGLPETGFWKL------- 1591
I + + + D+I+W + Y+ S Y L D P T +
Sbjct: 1366 SDHGFIHRIYLAKSKK-PDKIIWNYNTTGEYTVRSGYWLLTHD--PSTNIPAINPPHGSI 1422
Query: 1592 -----IWKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRR 1646
IW K++ FLW AL T + + SC RC E H L
Sbjct: 1423 DLKTRIWNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPSCPRCHRENESINHALFT 1482
Query: 1647 CPGSVWVRGQLGRLLPW-ISDSATFRHWLFGH--LQNRDNTLFCA--------------- 1688
CP + + W +SDS+ R+ L + +N N L
Sbjct: 1483 CPFAT---------MAWRLSDSSLIRNQLMSNDFEENISNILNFVQDTTMSDFHKLLPVW 1533
Query: 1689 MAWNLWKWRNSFVFEQVPWQPGDVMRRIHLDTVEFHRLAQGTQRS 1733
+ W +WK RN+ VF + P + +T ++ Q +++
Sbjct: 1534 LIWRIWKARNNVVFNKFRESPSKTVLSAKAETHDWLNATQSHKKT 1578
>gb|AAC33961.1| contains similarity to reverse trancriptase (Pfam: rvt.hmm, score:
42.57) [Arabidopsis thaliana] gi|7486711|pir||T01893
hypothetical protein F8M12.22 - Arabidopsis thaliana
Length = 1662
Score = 468 bits (1204), Expect = e-130
Identities = 320/1008 (31%), Positives = 472/1008 (46%), Gaps = 88/1008 (8%)
Query: 718 NMLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYS 777
NM+ C+ D+ +G +F+W + V LDRA + FP A +E L S
Sbjct: 543 NMVSTCDLKDIRSIGDRFSWVGERHSH-TVKCCLDRAFINSEGAFLFPFAELEFLEFTGS 601
Query: 778 DHSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWR----AQDGNLQDKLQRVQ 833
DH PL L T R RPFRF + P F++ V W Q +L D+++ +
Sbjct: 602 DHKPLFLSLEK-TETRKMRPFRFDKRLLEVPHFKTYVKAGWNKAINGQRKHLPDQVRTCR 660
Query: 834 EAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEM 893
+A + N R +++ L + + +++ ++R+L Y D EE
Sbjct: 661 QAMAKLKHKSNLNSRIRINQLQAALDKAMSSVNRTERRTISHIQRELTVAYRD----EER 716
Query: 894 LWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYT 953
W QKSR W++ GDRNT+FFHA T R N++ + E+G + A+ F+T
Sbjct: 717 YWQQKSRNQWMKEGDRNTEFFHACTKTRFSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFT 776
Query: 954 NLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAY 1013
++ + + P + + D L +S E+ +AI + KAPGPDG A
Sbjct: 777 KVYESNGRPVSIIDFAGFKPIVTEQINDDLTKDLSDLEIYNAICHIGDDKAPGPDGLTAR 836
Query: 1014 FYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSY 1073
FYK W I+G D+ V F+ + T + +IPK+ NP L D RPI+LCNV Y
Sbjct: 837 FYKSCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSDYRPIALCNVLY 896
Query: 1074 KLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRK-TGGGLVALK 1132
K+I+K LV R + L +VS Q +FI GR DN+++A E+MH++ TRK +A+K
Sbjct: 897 KIISKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVK 956
Query: 1133 IDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLR 1192
D+ KAYDRV W+FL T+ FGF + IM V++ SVL NG + QRG+R
Sbjct: 957 TDVSKAYDRVEWNFLETTMRLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIR 1016
Query: 1193 QGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKD 1252
QGDPLSPYLF+LC + L I+N V EG R +R+ G++HL FADD L FCQ++
Sbjct: 1017 QGDPLSPYLFILCADILNHLIKNRVAEGDIRGIRIGNGVPGVTHLQFADDSLFFCQSNVR 1076
Query: 1253 QLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLP 1312
+ +K + SG K+N+ KS + F V Q L ++GI GKYLGLP
Sbjct: 1077 NCQALKDVFDVYEYYSGQKINMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLP 1136
Query: 1313 LLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSV 1372
GR KRD F IIE+V R +SW L+ AG+ + KS+ S+PV+AM LP ++
Sbjct: 1137 EQFGRKKRDMFNYIIERVKKRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNI 1196
Query: 1373 CNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFL 1432
+ ++ ++ N W K R +AWK + SK +G LG R K N+ALL K V +
Sbjct: 1197 VSEIEALLMNFWWEKNAKKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMI 1256
Query: 1433 NEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSSSLW 1492
N L+ + + +Y + SIL + + SY W +L D + G
Sbjct: 1257 NNPNSLFARIMKARYFREDSILDAKRQRYQSYGWTSMLAGLDVIKKG------------- 1303
Query: 1493 YSNWL-GTGKIASRIPFVHITDTTLTVADIWRQGAWNFDRLYTLLPQEIREEISSVQIPS 1551
S ++ G GK S WN + L+ + + + +
Sbjct: 1304 -SRFIVGDGKTGS-------------------YRYWNAHLISQLVSPDDHRFVMNHHLSR 1343
Query: 1552 IQTGEDRILWREASDDIYSASSAYQFLNEDGLPETGFWKLIWKASAPEKVRFFLWLVGRD 1611
I +D+++W YS+S Y +WK K+++ LW
Sbjct: 1344 I-VHQDKLVWN------YSSSGDY---------------TLWKLPIIPKIKYMLWRTISK 1381
Query: 1612 ALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCP--GSVWVRGQLGRLLPWI----- 1664
ALPT ++ + C RC E HVL CP S+W PW+
Sbjct: 1382 ALPTRSRLLTRGMDIDPHCPRCPTEEETINHVLFTCPYAASIWGLSN----FPWLPGHTF 1437
Query: 1665 -SDSATFRHWLFGHLQNRDNTLFCA-------MAWNLWKWRNSFVFEQ 1704
D+ +L N NTL + W LWK RN+ VF +
Sbjct: 1438 SQDTEENISFLINSFSN--NTLNTEQRLAPFWLIWRLWKARNNLVFNK 1483
>emb|CAB78094.1| RNA-directed DNA polymerase-like protein [Arabidopsis thaliana]
gi|4538901|emb|CAB39638.1| RNA-directed DNA
polymerase-like protein [Arabidopsis thaliana]
gi|7485606|pir||T04018 hypothetical protein F17A8.60 -
Arabidopsis thaliana
Length = 1274
Score = 467 bits (1201), Expect = e-129
Identities = 308/1004 (30%), Positives = 483/1004 (47%), Gaps = 52/1004 (5%)
Query: 727 DLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYSDHSPLLLRC 786
D+ G+ +W R + RLDRALG+ +W FP + E+L SDH PL+
Sbjct: 115 DINHTGNSLSW-RGTRYSHFIKSRLDRALGNCSWSELFPMSKCEYLRFEGSDHRPLVTYF 173
Query: 787 HAPTGDRAARPFRFQAAWVSHPSFESVVNTSWR-AQDGNLQDKLQRVQEAATEFNKTVFG 845
AP R+ +PFRF ++V W A+ ++ K+ R +++ ++ K
Sbjct: 174 GAPPLKRS-KPFRFDRRLREKEEIRALVKEVWELARQDSVLYKISRCRQSIIKWTKEQNS 232
Query: 846 NIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEMLWFQKSRENWVR 905
N + ++ ++ L+ D D + + ++L+ Y QEE+ W Q SR W+
Sbjct: 233 NSAKAIKKAQQALESALSA-DIPDPSLIGSITQELEAAYR----QEELFWKQWSRVQWLN 287
Query: 906 FGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTNLFSLDVQVRRN 965
GDRN +FHA T RR N + + G + + ++ N+F+
Sbjct: 288 SGDRNKGYFHATTRTRRMLNNLSVIEDGSGQEFHEEEQIASTISSYFQNIFTTSNNSDLQ 347
Query: 966 RIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYWHILGED 1025
+ E +P I ++L+ SL E++ A+ S+ + KAPGPDGF A F+ YW I+ D
Sbjct: 348 VVQEALSPIISSHCNEELIKISSLLEIKEALFSISADKAPGPDGFSASFFHAYWDIIEAD 407
Query: 1026 LHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKVLVNRFR 1085
+ + S F + +P L T + LIPK+ P K+ D RPI+LCNV YK++ K+L R +
Sbjct: 408 VSRDIRSFFVDSCLSPRLNETHVTLIPKISAPRKVSDYRPIALCNVQYKIVAKILTRRLQ 467
Query: 1086 PLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTGGGL---VALKIDLEKAYDRV 1142
P L +L+S Q +F+ GR DN+++ E++H + R +G +A+K D+ KAYDR+
Sbjct: 468 PWLSELISLHQSAFVPGRAIADNVLITHEILHFL--RVSGAKKYCSMAIKTDMSKAYDRI 525
Query: 1143 NWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLSPYLF 1202
W+FL+ L GF + + +M V T + S L NGS S RGLRQGDPLSPYLF
Sbjct: 526 KWNFLQEVLMRLGFHDKWIRWVMQCVCTVSYSFLINGSPQGSVVPSRGLRQGDPLSPYLF 585
Query: 1203 VLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIKHTLA 1262
+LC E L+ + E+G +RV++ ++HL FADD + FC+ + + + L
Sbjct: 586 ILCTEVLSGLCRKAQEKGVMVGIRVARGSPQVNHLLFADDTMFFCKTNPTCCGALSNILK 645
Query: 1263 DFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQGRVKRDH 1322
+ ASG +NL KS + FS Q +R + + I +GKYLGLP GR KRD
Sbjct: 646 KYELASGQSINLAKSAITFSSKTPQDIKRRVKLSLRIDNEGGIGKYLGLPEHFGRRKRDI 705
Query: 1323 FATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVCNHVDKMVRN 1382
F++I++++ R SW L+ AG+ L K++LSS+P +AM LP S+C + ++
Sbjct: 706 FSSIVDRIRQRSHSWSIRFLSSAGKQILLKAVLSSMPSYAMMCFKLPASLCKQIQSVLTR 765
Query: 1383 CIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFLNEKGKLWVQA 1442
W + R V+W +T +G LG R + KL L E L +
Sbjct: 766 FWWDSKPDKRKMAWVSWDKLTLPINEGGLGFRE-------IEAKLSWRILKEPHSLLSRV 818
Query: 1443 LSQKYIKSGSILTEELRPG-GSYVWRGILKARDSVSDGFGPRLGAGSS-SLWYSNWLGTG 1500
L KY + S + P S+ WRGIL RD + G G +G G S ++W WL
Sbjct: 819 LLGKYCNTSSFMDCSASPSFASHGWRGILAGRDLLRKGLGWSIGQGDSINVWTEAWLSPS 878
Query: 1501 KIASRIPFVHITDTTLTVADIWRQG--AWNFDRLYTLLPQEIREEISSVQIPSIQTGEDR 1558
+ I T+ L+V D+ +WN + + LPQ ++I + I ++ +D
Sbjct: 879 SPQTPIGPPTETNKDLSVHDLICHDVKSWNVEAIRKHLPQ-YEDQIRKITINALPL-QDS 936
Query: 1559 ILWREASDDIYSASSAYQFLNEDGLP----ETGFWKLIWKASAPEKVRFFLWLVGRDALP 1614
++W Y+ + Y + P + + K IWK KV+ FLW + ALP
Sbjct: 937 LVWLPVKSGEYTTKTGYALAKLNSFPASQLDFNWQKNIWKIHTSPKVKHFLWKAMKGALP 996
Query: 1615 TNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCP--GSVWVRGQLGRLLPWISDSATFRH 1672
R ++ +C RCG E + H++ CP VW L P + + + H
Sbjct: 997 VGEALSRRNIEAEVTCKRCGQ-TESSLHLMLLCPYAKKVW------ELAPVLFNPSEATH 1049
Query: 1673 WLFGHL-------------QNRDNTLFCAMAWNLWKWRNSFVFE 1703
L L+ + W+LWK RN +F+
Sbjct: 1050 SSVALLLVDAKRMVALPPTGLGSAPLYPWLLWHLWKARNRLIFD 1093
>gb|AAF97969.1| F21J9.30 [Arabidopsis thaliana]
Length = 1270
Score = 458 bits (1178), Expect = e-127
Identities = 321/1068 (30%), Positives = 510/1068 (47%), Gaps = 70/1068 (6%)
Query: 719 MLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYSD 778
M++ C+ +++ G+ FTW R G + RLDRA G+ W FP + L SD
Sbjct: 126 MIKGCDLVEMPAHGNGFTWAGR-RGDHWIQCRLDRAFGNKEWFCFFPVSNQTFLDFRGSD 184
Query: 779 HSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSW-RAQDG---NLQDKLQRVQE 834
H P+L++ + + D FRF ++ + + +W R + G ++ D+L+ ++
Sbjct: 185 HRPVLIKLMS-SQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGKHGTNISVADRLRACRK 243
Query: 835 AATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEML 894
+ + + K N + ++E L+ +Q W + ++ L++DL K Y + EE
Sbjct: 244 SLSSWKKQNNLNSLDKINQLEAALEK-EQSLVWPIFQRVSVLKKDLAKAYRE----EEAY 298
Query: 895 WFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTN 954
W QKSR+ W+R G+RN+K+FHA R+R +I L +GN T + E A ++ N
Sbjct: 299 WKQKSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGN 358
Query: 955 LFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYF 1014
LF P + + LV VS +E++ A+ S+K APGPDG A F
Sbjct: 359 LFKSSNPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALF 418
Query: 1015 YKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYK 1074
++ YW +G + V F +G E T L LIPK +PT++ DLRPISLC+V YK
Sbjct: 419 FQHYWSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYK 478
Query: 1075 LITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTR-KTGGGLVALKI 1133
+I+K++ R +P L ++VS Q +F+ R DNI++A E++H++ + +A+K
Sbjct: 479 IISKIMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKS 538
Query: 1134 DLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQ 1193
D+ KAYDRV W +LR+ L GF + V+ IM V + SVL N LQRGLRQ
Sbjct: 539 DMSKAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQ 598
Query: 1194 GDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQ 1253
GDPLSP+LFVLC E L + EG ++ S++G + HL FADD L C+AS++Q
Sbjct: 599 GDPLSPFLFVLCTEGLTHLLNKAQWEGALEGIQFSENGPMVHHLLFADDSLFLCKASREQ 658
Query: 1254 LRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPL 1313
+++ L + A+G +NL KS + F + V ++ + + +GI G YLGLP
Sbjct: 659 SLVLQKILKVYGNATGQTINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPE 718
Query: 1314 LQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVC 1373
K D + +++ +L W L++ G+ L KS+ ++PV AM LP + C
Sbjct: 719 CFSGSKVDMLHYLKDRLKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTC 778
Query: 1374 NHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFLN 1433
+++ + + W ++R H +W+ + K G LG R + N+ALL K L+
Sbjct: 779 ENLESAMASFWWDSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLH 838
Query: 1434 EKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSS-SLW 1492
L + L +Y + L L S+ WR IL R+ +S G R+G G+S +W
Sbjct: 839 FPDCLLSRLLKSRYFDATDFLDAALSQRPSFGWRSILFGRELLSKGLQKRVGDGASLFVW 898
Query: 1493 YSNWLGTGKIASRIPFVHITDTTLTVADIW--RQGAWNFDRLYTL-LPQEIREEISSVQI 1549
W+ + I D TL V + R G W+ + L+ L LP++I + I
Sbjct: 899 IDPWIDDNGFRAPWRKNLIYDVTLKVKALLNPRTGFWDEEVLHDLFLPEDI---LRIKAI 955
Query: 1550 PSIQTGEDRILWREASDDIYSASSAYQFLNE----------DGLPET-GFWKLIWKASAP 1598
+ + D +W+ +S SAY + P T G +W
Sbjct: 956 KPVISQADFFVWKLNKSGDFSVKSAYWLAYQTKSQNLRSEVSMQPSTLGLKTQVWNLQTD 1015
Query: 1599 EKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCPGS--VWVRGQ 1656
K++ FLW V CG + E H L CP S +W
Sbjct: 1016 PKIKIFLWKV------------------------CGELGESTNHTLFLCPLSRQIWALSD 1051
Query: 1657 LGRLLPWISDSATFRHWLFGH-LQNRDN--------TLFCAMAWNLWKWRNSFVFEQVPW 1707
S+ + + + H L+N+DN +F + W +WK RNSF+FE + +
Sbjct: 1052 YPFPPDGFSNGSIYSN--INHLLENKDNKEWPINLRKIFPWILWRIWKNRNSFIFEGISY 1109
Query: 1708 QPGDVMRRIHLDTVEFHR---LAQGTQRSNPIASDGINSVMLYTDGSW 1752
D + +I D VE+ L NP S+G++ V ++ W
Sbjct: 1110 PATDTVIKIRDDVVEWFEAQCLDGEGSALNPPLSNGVHFVGSVSENLW 1157
>gb|AAD21778.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25410938|pir||G84429 hypothetical protein
At2g01840 [imported] - Arabidopsis thaliana
Length = 1715
Score = 454 bits (1169), Expect = e-125
Identities = 299/1037 (28%), Positives = 484/1037 (45%), Gaps = 57/1037 (5%)
Query: 715 NMLNMLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLAR 774
N NM+ CN DL G+ ++W + + LDR + W+ FP E L
Sbjct: 521 NFTNMINCCNMKDLKSKGNPYSWVGK-RQNETIESCLDRVFINSDWQASFPAFETEFLPI 579
Query: 775 VYSDHSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRA----QDGNLQDKLQ 830
SDH+P+++ + + FR+ F V W G +KL
Sbjct: 580 AGSDHAPVIIDIAEEVCTKRGQ-FRYDRRHFQFEDFVDSVQRGWNRGRSDSHGGYYEKLH 638
Query: 831 RVQEAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQ 890
++ ++ + N + ++ R+ +++H +++ RL +DL + Y D
Sbjct: 639 CCRQELAKWKRRTKTNTAEKIETLKYRVDAAERDHTL-PHQTILRLRQDLNQAYRD---- 693
Query: 891 EEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKR 950
EE+ W KSR W+ GDRNT FF+A T +R+ RN+I + G T+ + A+
Sbjct: 694 EELYWHLKSRNRWMLLGDRNTMFFYASTKLRKSRNRIKAITDAQGIENFRDDTIGKVAEN 753
Query: 951 FYTNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGF 1010
++ +LF+ I+ P + + +L+ V+ +EVR A+ ++ + +APG DGF
Sbjct: 754 YFADLFTTTQTSDWEEIISGIAPKVTEQMNHELLQSVTDQEVRDAVFAIGADRAPGFDGF 813
Query: 1011 HAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCN 1070
A FY W ++G D+ MV F++ + ++ T + LIPK+ +P + D RPISLC
Sbjct: 814 TAAFYHHLWDLIGNDVCLMVRHFFESDVMDNQINQTQICLIPKIIDPKHMSDYRPISLCT 873
Query: 1071 VSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKT-GGGLV 1129
SYK+I+K+L+ R + L ++S Q +F+ G+ DN+++A E++H++ +R+ G V
Sbjct: 874 ASYKIISKILIKRLKQCLGDVISDSQAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYV 933
Query: 1130 ALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQR 1189
A+K D+ KAYDRV W+FL + GF PR V IM V + + VL NGS R
Sbjct: 934 AVKTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSR 993
Query: 1190 GLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQA 1249
G+RQGDPLSPYLF+ C E L+ ++ + ++++KD + ISHL FADD L FC+A
Sbjct: 994 GIRQGDPLSPYLFLFCAEVLSNMLRKAEVNKQIHGMKITKDCLAISHLLFADDSLFFCRA 1053
Query: 1250 SKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYL 1309
S + + + EASG K+N KS ++F + + R++ L L+GI GKYL
Sbjct: 1054 SNQNIEQLALIFKKYEEASGQKINYAKSSIIFGQKIPTMRRQRLHRLLGIDNVRGGGKYL 1113
Query: 1310 GLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLP 1369
GLP GR K + F I+ KV R W N L+ AG+ + K+I +LPV++M LP
Sbjct: 1114 GLPEQLGRRKVELFEYIVTKVKERTEGWAYNYLSPAGKEIVIKAIAMALPVYSMNCFLLP 1173
Query: 1370 DSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVH 1429
+CN ++ ++ WGK + +G LG + + N ALL K
Sbjct: 1174 TLICNEINSLITAFWWGK------------------ENEGDLGFKDLHQFNRALLAKQAW 1215
Query: 1430 SFLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAG-S 1488
L L + Y + + L SY W I + + + G RLG G +
Sbjct: 1216 RILTNPQSLLARLYKGLYYPNTTYLRANKGGHASYGWNSIQEGKLLLQQGLRVRLGDGQT 1275
Query: 1489 SSLWYSNWLGTGKIASRIPFVHITDTTLTVADIWRQGAWNFDRLY---TLLPQEIREEIS 1545
+ +W WL T + R I D + VAD+WR+ +D + L P++ ++++
Sbjct: 1276 TKIWEDPWLPT--LPPRPARGPILDEDMKVADLWRENKREWDPVIFEGVLNPED--QQLA 1331
Query: 1546 SVQIPSIQTGEDRILWREASDDIYSASSAYQFLNEDGLPE----------TGFWKLIWKA 1595
S D W + Y+ S Y L E + IW+
Sbjct: 1332 KSLYLSNYAARDSYKWAYTRNTQYTVRSGYWVATHVNLTEEEIINPLEGDVPLKQEIWRL 1391
Query: 1596 SAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCPGS--VWV 1653
K++ F+W AL T + ++ +C RC E H++ C + VW
Sbjct: 1392 KITPKIKHFIWRCLSGALSTTTQLRNRNIPADPTCQRCCNADETINHIIFTCSYAQVVWR 1451
Query: 1654 RGQL---GRLLPWISDSATFRHWLFG----HLQNRDNTLFCAMAWNLWKWRNSFVFEQVP 1706
RL + R L G +L + + + W LWK RN ++F+Q+
Sbjct: 1452 SANFSGSNRLCFTDNLEENIRLILQGKKNQNLPILNGLMPFWIMWRLWKSRNEYLFQQLD 1511
Query: 1707 WQPGDVMRRIHLDTVEF 1723
P V ++ + E+
Sbjct: 1512 RFPWKVAQKAEQEATEW 1528
>gb|AAD03565.2| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411819|pir||H84557 hypothetical protein
At2g17910 [imported] - Arabidopsis thaliana
Length = 1344
Score = 449 bits (1156), Expect = e-124
Identities = 308/1016 (30%), Positives = 474/1016 (46%), Gaps = 42/1016 (4%)
Query: 718 NMLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYS 777
+ML C+ +LG G+ FTW N + K LDR G+ AW FP A+ L + S
Sbjct: 140 HMLLNCSMHELGSTGNSFTWGGNRNDQWVQCK-LDRCFGNPAWFSIFPNAHQWFLEKFGS 198
Query: 778 DHSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGNLQDKLQRVQEAAT 837
DH P+L++ + FR+ P V++ SW + + +
Sbjct: 199 DHRPVLVK-FTNDNELFRGQFRYDKRLDDDPYCIEVIHRSWNSA---MSQGTHSSFFSLI 254
Query: 838 EFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDS------ESLARLERDLQKEYGDILYQE 891
E + + H + R++ ++++ D S + ++ L YGD E
Sbjct: 255 ECRRAISVWKHSSDTNAQSRIKRLRKDLDAEKSIQIPCWPRIEYIKDQLSLAYGD----E 310
Query: 892 EMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRF 951
E+ W QKSR+ W+ GD+NT FFHA R +N++ L E+ T S + A F
Sbjct: 311 ELFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTRNSDKGKIASSF 370
Query: 952 YTNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFH 1011
+ NLF+ + N +E + E L+ V+ EV +A+ S+ APGPDGF
Sbjct: 371 FENLFTSTYILTHNNHLEGLQAKVTSEMNHNLIQEVTELEVYNAVFSINKESAPGPDGFT 430
Query: 1012 AYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNV 1071
A F++Q+W ++ + + F+ G + T + LIPK+ +P ++ DLRPISLC+V
Sbjct: 431 ALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSV 490
Query: 1072 SYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTR-KTGGGLVA 1130
YK+I+K+L R + L +VS Q +F+ R DNI++A E++H++ T + +A
Sbjct: 491 LYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMA 550
Query: 1131 LKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRG 1190
K D+ KAYDRV W FL + GF + + IM V + + SVL NG RG
Sbjct: 551 FKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRG 610
Query: 1191 LRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQAS 1250
+RQGDPLSP LFVLC E L + + GK ++ V ++HL FADD LL C+A+
Sbjct: 611 IRQGDPLSPALFVLCTEALIHILNKAEQAGKITGIQFQDKKVSVNHLLFADDTLLMCKAT 670
Query: 1251 KDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLG 1310
K + + L+ + + SG +NL KS + F K V + + + GIS GKYLG
Sbjct: 671 KQECEELMQCLSQYGQLSGQMINLNKSAITFGKNVDIQIKDWIKSRSGISLEGGTGKYLG 730
Query: 1311 LPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPD 1370
LP KRD F I EK+ +RL W L++ G+ L KSI +LPV+ M LP
Sbjct: 731 LPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPK 790
Query: 1371 SVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHS 1430
++C + ++ + W R H ++W+ +T K +G G + + N+ALL K
Sbjct: 791 NLCQKLTTVMMDFWWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWR 850
Query: 1431 FLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSSS 1490
L EKG L+ + +Y + L+ SY WR IL R+ + G +G G +
Sbjct: 851 VLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSILFGRELLMQGLRTVIGNGQKT 910
Query: 1491 -LWYSNWLGTG---KIASRIPFVHITDTTLTVADIWRQGAWNFDRLYTLLPQEIREEISS 1546
+W WL G + +R F+++ + D + WN + L L P + EI
Sbjct: 911 FVWTDKWLHDGSNRRPLNRRRFINVDLKVSQLIDPTSRN-WNLNMLRDLFPWK-DVEIIL 968
Query: 1547 VQIPSIQTGEDRILWREASDDIYSASSAYQFLNED-----------GLPETGFWKLIWKA 1595
Q P + ED W + + +YS + Y+FL++ + IW
Sbjct: 969 KQRP-LFFKEDSFCWLHSHNGLYSVKTGYEFLSKQVHHRLYQEAKVKPSVNSLFDKIWNL 1027
Query: 1596 SAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCP--GSVWV 1653
K+R FLW A+P + + + C+ C E H+L CP VW
Sbjct: 1028 HTAPKIRIFLWKALHGAIPVEDRLRTRGIRSDDGCLMCDTENETINHILFECPLARQVWA 1087
Query: 1654 RGQLGRLLPWISDSA-TFRHWLFGHLQNRD---NTLFCA--MAWNLWKWRNSFVFE 1703
L S+S T L Q D + F + + W LWK RN+ +FE
Sbjct: 1088 ITHLSSAGSEFSNSVYTNMSRLIDLTQQNDLPHHLRFVSPWILWFLWKNRNALLFE 1143
>pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabidopsis thaliana
(fragment)
Length = 1365
Score = 449 bits (1156), Expect = e-124
Identities = 308/1016 (30%), Positives = 474/1016 (46%), Gaps = 42/1016 (4%)
Query: 718 NMLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYS 777
+ML C+ +LG G+ FTW N + K LDR G+ AW FP A+ L + S
Sbjct: 161 HMLLNCSMHELGSTGNSFTWGGNRNDQWVQCK-LDRCFGNPAWFSIFPNAHQWFLEKFGS 219
Query: 778 DHSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGNLQDKLQRVQEAAT 837
DH P+L++ + FR+ P V++ SW + + +
Sbjct: 220 DHRPVLVK-FTNDNELFRGQFRYDKRLDDDPYCIEVIHRSWNSA---MSQGTHSSFFSLI 275
Query: 838 EFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDS------ESLARLERDLQKEYGDILYQE 891
E + + H + R++ ++++ D S + ++ L YGD E
Sbjct: 276 ECRRAISVWKHSSDTNAQSRIKRLRKDLDAEKSIQIPCWPRIEYIKDQLSLAYGD----E 331
Query: 892 EMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRF 951
E+ W QKSR+ W+ GD+NT FFHA R +N++ L E+ T S + A F
Sbjct: 332 ELFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTRNSDKGKIASSF 391
Query: 952 YTNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFH 1011
+ NLF+ + N +E + E L+ V+ EV +A+ S+ APGPDGF
Sbjct: 392 FENLFTSTYILTHNNHLEGLQAKVTSEMNHNLIQEVTELEVYNAVFSINKESAPGPDGFT 451
Query: 1012 AYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNV 1071
A F++Q+W ++ + + F+ G + T + LIPK+ +P ++ DLRPISLC+V
Sbjct: 452 ALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSV 511
Query: 1072 SYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTR-KTGGGLVA 1130
YK+I+K+L R + L +VS Q +F+ R DNI++A E++H++ T + +A
Sbjct: 512 LYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMA 571
Query: 1131 LKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRG 1190
K D+ KAYDRV W FL + GF + + IM V + + SVL NG RG
Sbjct: 572 FKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRG 631
Query: 1191 LRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQAS 1250
+RQGDPLSP LFVLC E L + + GK ++ V ++HL FADD LL C+A+
Sbjct: 632 IRQGDPLSPALFVLCTEALIHILNKAEQAGKITGIQFQDKKVSVNHLLFADDTLLMCKAT 691
Query: 1251 KDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLG 1310
K + + L+ + + SG +NL KS + F K V + + + GIS GKYLG
Sbjct: 692 KQECEELMQCLSQYGQLSGQMINLNKSAITFGKNVDIQIKDWIKSRSGISLEGGTGKYLG 751
Query: 1311 LPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPD 1370
LP KRD F I EK+ +RL W L++ G+ L KSI +LPV+ M LP
Sbjct: 752 LPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPK 811
Query: 1371 SVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHS 1430
++C + ++ + W R H ++W+ +T K +G G + + N+ALL K
Sbjct: 812 NLCQKLTTVMMDFWWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWR 871
Query: 1431 FLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSSS 1490
L EKG L+ + +Y + L+ SY WR IL R+ + G +G G +
Sbjct: 872 VLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSILFGRELLMQGLRTVIGNGQKT 931
Query: 1491 -LWYSNWLGTG---KIASRIPFVHITDTTLTVADIWRQGAWNFDRLYTLLPQEIREEISS 1546
+W WL G + +R F+++ + D + WN + L L P + EI
Sbjct: 932 FVWTDKWLHDGSNRRPLNRRRFINVDLKVSQLIDPTSRN-WNLNMLRDLFPWK-DVEIIL 989
Query: 1547 VQIPSIQTGEDRILWREASDDIYSASSAYQFLNED-----------GLPETGFWKLIWKA 1595
Q P + ED W + + +YS + Y+FL++ + IW
Sbjct: 990 KQRP-LFFKEDSFCWLHSHNGLYSVKTGYEFLSKQVHHRLYQEAKVKPSVNSLFDKIWNL 1048
Query: 1596 SAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRCP--GSVWV 1653
K+R FLW A+P + + + C+ C E H+L CP VW
Sbjct: 1049 HTAPKIRIFLWKALHGAIPVEDRLRTRGIRSDDGCLMCDTENETINHILFECPLARQVWA 1108
Query: 1654 RGQLGRLLPWISDSA-TFRHWLFGHLQNRD---NTLFCA--MAWNLWKWRNSFVFE 1703
L S+S T L Q D + F + + W LWK RN+ +FE
Sbjct: 1109 ITHLSSAGSEFSNSVYTNMSRLIDLTQQNDLPHHLRFVSPWILWFLWKNRNALLFE 1164
>emb|CAB81184.1| putative protein [Arabidopsis thaliana] gi|4539357|emb|CAB40051.1|
putative protein [Arabidopsis thaliana]
gi|7486147|pir||T04278 hypothetical protein F25I24.40 -
Arabidopsis thaliana
Length = 1294
Score = 442 bits (1136), Expect = e-122
Identities = 271/777 (34%), Positives = 398/777 (50%), Gaps = 11/777 (1%)
Query: 718 NMLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYS 777
NM+ C+ D+ +G +F+W + V LDRA + FP A +E L S
Sbjct: 523 NMVSTCDLKDIRSIGDRFSWVGERHSH-TVKCCLDRAFINSEGAFLFPFAELEFLEFTGS 581
Query: 778 DHSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWR----AQDGNLQDKLQRVQ 833
DH PL L T R RPFRF + P F++ V W Q +L D+++ +
Sbjct: 582 DHKPLFLSLEK-TETRKMRPFRFDKRLLEVPHFKTYVKAGWNKAINGQRKHLPDQVRTCR 640
Query: 834 EAATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEM 893
+A + N R +++ L + + +++ ++R+L Y D EE
Sbjct: 641 QAMAKLKHKSNLNSRIRINQLQAALDKAMSSVNRTERRTISHIQRELTVAYRD----EER 696
Query: 894 LWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYT 953
W QKSR W++ GDRNT+FFHA T R N++ + E+G + A+ F+T
Sbjct: 697 YWQQKSRNQWMKEGDRNTEFFHACTKTRFSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFT 756
Query: 954 NLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAY 1013
++ + + P + + D L +S E+ +AI + KAPGPDG A
Sbjct: 757 KVYESNGRPVSIIDFAGFKPIVTEQINDDLTKDLSDLEIYNAICHIGDDKAPGPDGLTAR 816
Query: 1014 FYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSY 1073
FYK W I+G D+ V F+ + T + +IPK+ NP L D RPI+LCNV Y
Sbjct: 817 FYKSCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSDYRPIALCNVLY 876
Query: 1074 KLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRK-TGGGLVALK 1132
K+I+K LV R + L +VS Q +FI GR DN+++A E+MH++ TRK +A+K
Sbjct: 877 KIISKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVK 936
Query: 1133 IDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLR 1192
D+ KAYDRV W+FL T+ FGF + IM V++ SVL NG + QRG+R
Sbjct: 937 TDVSKAYDRVEWNFLETTMRLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIR 996
Query: 1193 QGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKD 1252
QGDPLSPYLF+LC + L I+N V EG R +R+ G++HL FADD L FCQ++
Sbjct: 997 QGDPLSPYLFILCADILNHLIKNRVAEGDIRGIRIGNGVPGVTHLQFADDSLFFCQSNVR 1056
Query: 1253 QLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLP 1312
+ +K + SG K+N+ KS + F V Q L ++GI GKYLGLP
Sbjct: 1057 NCQALKDVFDVYEYYSGQKINMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLP 1116
Query: 1313 LLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSV 1372
GR KRD F IIE+V R +SW L+ AG+ + KS+ S+PV+AM LP ++
Sbjct: 1117 EQFGRKKRDMFNYIIERVKKRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNI 1176
Query: 1373 CNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFL 1432
+ ++ ++ N W K R +AWK + SK +G LG R K N+ALL K V +
Sbjct: 1177 VSEIEALLMNFWWEKNAKKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMI 1236
Query: 1433 NEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSS 1489
N L+ + + +Y + SIL + + SY W +L D + G +G G +
Sbjct: 1237 NNPNSLFARIMKARYFREDSILDAKRQRYQSYGWTSMLAGLDVIKKGSRFIVGDGKT 1293
>pir||S65812 RNA-directed DNA polymerase (EC 2.7.7.49) (clone DW15) - Arabidopsis
thaliana retrotransposon Ta11-1 gi|976278|gb|AAA75254.1|
reverse transcriptase
Length = 1333
Score = 440 bits (1132), Expect = e-121
Identities = 307/1036 (29%), Positives = 483/1036 (45%), Gaps = 41/1036 (3%)
Query: 718 NMLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYS 777
+ML++C+ ++L +G+ FTW + N + RLDR G+ W FP + E L + S
Sbjct: 164 DMLDSCDMLELPSIGNPFTWGGKTN-EMWIQSRLDRCFGNKNWFRFFPISNQEFLDKRGS 222
Query: 778 DHSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSWRAQDGN----LQDKLQRVQ 833
DH P+L+R T + FRF + P+ + + +W N + DKL+ +
Sbjct: 223 DHRPVLVRL-TKTKEEYRGNFRFDKRLFNQPNVKETIVQAWNGSQRNENLLVLDKLKHCR 281
Query: 834 EAATEFNKTVFGNIHRRKRRVERRLQ-GIQQEHDWRDSESLARLERDLQKEYGDILYQEE 892
A + + K NI+ R + R ++Q + ++ + L+ DL K D EE
Sbjct: 282 SALSRWKKE--NNINSSTRITQARAALELEQSSGFPRADLVFSLKNDLCKANHD----EE 335
Query: 893 MLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFY 952
+ W QKSR W+ GD+NT FFHA R + I L +G + D A+ ++
Sbjct: 336 VFWSQKSRAKWMHSGDKNTSFFHASVKDNRGKQHIDQLCDVNGLFHKDEMNKGAIAEAYF 395
Query: 953 TNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHA 1012
++LF + E+ P + + L+A VS E+R A+ +++S APG DGF
Sbjct: 396 SDLFKSTDPSSFVDLFEDYQPRVTESMNNTLIAAVSKNEIREAVFAIRSSSAPGVDGFTG 455
Query: 1013 YFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVS 1072
+F+++YW I+ + + + F G T L L+PK P K+ DLRPISLC+V
Sbjct: 456 FFFQKYWSIICLQVTKEIQNFFLLGYFPKSWNFTHLCLLPKKKKPDKMTDLRPISLCSVL 515
Query: 1073 YKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKT-GGGLVAL 1131
YK+I+K++V R +P L LVSP Q +F+ R DNI++A EV+H + T K+ G +A+
Sbjct: 516 YKIISKIMVRRLQPFLPDLVSPNQSAFVAERLIFDNILIAHEVVHGLRTHKSVSKGFIAI 575
Query: 1132 KIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGL 1191
K ++ KA+DRV W+++RA L GF + V IM+ + + + SVL N + RGL
Sbjct: 576 KSNMSKAFDRVEWNYVRALLDALGFHQKWVGWIMFMISSVSYSVLINDKAFGNIVPSRGL 635
Query: 1192 RQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASK 1251
RQGDPLSP+LFVLC E L + +G +R S++G I HL FADD L C+A K
Sbjct: 636 RQGDPLSPFLFVLCSEGLTHLMNRAERQGLLSGIRFSENGPAIHHLLFADDSLFMCKAVK 695
Query: 1252 DQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGL 1311
+++ +IK + + +G ++N +KS + +V + + + +GI+ YLGL
Sbjct: 696 EEVTVIKSIFKVYGDVTGQRINYDKSSITLGALVDEDCKVWIQAELGITNEGGASTYLGL 755
Query: 1312 PLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDS 1371
P K I +++ TRL+ W L+ G+ L K+ +L +AM L +
Sbjct: 756 PECFSGSKVQLLDYIKDRLKTRLSGWFARTLSMGGKETLLKAFALALLFYAMSCFKLTKT 815
Query: 1372 VCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSF 1431
C ++ + + W + R H V+ + + SK G LG R N+ALL K
Sbjct: 816 TCVNMTSAMSDFWWNALEHKRKTHWVSCEKMCLSKENGGLGFRDIESFNQALLAKQAWRL 875
Query: 1432 LNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSS-S 1490
L L+ + +Y L EL+ SY WR IL RD + GF ++G GSS S
Sbjct: 876 LQFPNSLFARFFKSRYYDEEDFLDAELKATPSYAWRSILHGRDLLIKGFRKKVGNGSSTS 935
Query: 1491 LWYSNWLGTGKIASRIPFVHITDTTLTVADI--WRQGAWNFDRLYTLL-PQEIREEISSV 1547
+W W+ + + L V D+ DRL L P +I +
Sbjct: 936 VWMDPWIYDNDPRLPLQKHFSVNLDLRVHDLINVEDRCRRRDRLEELFYPADIEIIVKRN 995
Query: 1548 QIPSIQTGEDRILWREASDDIYSASSAYQFLNEDGLPE-----------TGFWKLIWKAS 1596
+ S+ +D +W + YS S Y + PE G + IW
Sbjct: 996 PVVSM---DDFWVWLHSKSGEYSVKSGYWLAFQTNKPELIREARVQPSTNGLKEKIWSTL 1052
Query: 1597 APEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRC--PGSVWVR 1654
K++ FLW + ALP + R + C CG E HVL C VW
Sbjct: 1053 TSPKIKLFLWRILSSALPVAYQIIRRGMPIDPRCQVCGEEGESINHVLFTCSLARQVWAL 1112
Query: 1655 GQLGRLLPWISDSATFRHWLF-------GHLQNRDNTLFCAMAWNLWKWRNSFVFEQVPW 1707
+ +S+ F + + G + + + + W LWK R+ FE +
Sbjct: 1113 SGVPTSQFGFQNSSIFANIQYLLELKGKGLIPEQIKKSWPWVLWRLWKNRDKLFFEGTIF 1172
Query: 1708 QPGDVMRRIHLDTVEF 1723
P + +I D E+
Sbjct: 1173 SPLKSIEKIRDDVQEW 1188
>gb|AAD29058.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411151|pir||H84465 hypothetical protein
At2g05200 [imported] - Arabidopsis thaliana
Length = 1229
Score = 437 bits (1124), Expect = e-120
Identities = 288/943 (30%), Positives = 463/943 (48%), Gaps = 33/943 (3%)
Query: 727 DLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYSDHSPLLLRC 786
DL + G+ F+W R + V +RLDRA+ + +W FP E+L SDH PL++
Sbjct: 69 DLKYSGNPFSW-RGMRYDWFVRQRLDRAMSNNSWLESFPSGRSEYLRFEGSDHRPLVVFV 127
Query: 787 HAPTGDRAARPFRFQAAWVSHPSFESVVNTSW-RAQDGNLQDKLQRVQEAATEFNKTVFG 845
R + FRF + +++ +W A D ++ K+ + + + +
Sbjct: 128 DEARVKRRGQ-FRFDNRLRDNDVVNALIQETWTNAGDASVLTKMNQCRREIINWTRLQNL 186
Query: 846 N----IHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEMLWFQKSRE 901
N I + ++ +E L D + ++ L L+ Y EE W Q+SR
Sbjct: 187 NSAELIEKTQKALEEALTA-----DPPNPTTIGALTATLEHAYK----LEEQFWKQRSRV 237
Query: 902 NWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCT--DPSTLQEEAKRFYTNLFSLD 959
W+ GDRNT +FHA T RR +N++ +ED N + + + ++ +F+ +
Sbjct: 238 LWLHSGDRNTGYFHAVTRNRRTQNRL--TVMEDINGVAQHEEHQISQIISGYFQQIFTSE 295
Query: 960 VQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYFYKQYW 1019
+ + E P + + D L + EEV+ A+ S+ + KAPGPDGF A FY YW
Sbjct: 296 SDGDFSVVDEAIEPMVSQGDNDFLTRIPNDEEVKDAVFSINASKAPGPDGFTAGFYHSYW 355
Query: 1020 HILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYKLITKV 1079
HI+ D+ + F + + T + LIPK P K+ D RPI+LCN+ YK++ K+
Sbjct: 356 HIISTDVGREIRLFFTSKNFPRRMNETHIRLIPKDLGPRKVADYRPIALCNIFYKIVAKI 415
Query: 1080 LVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTRKTGGGL-VALKIDLEKA 1138
+ R + +L KL+S Q +F+ GR DN+++ EV+H + T +A+K D+ KA
Sbjct: 416 MTKRMQLILPKLISENQSAFVPGRVISDNVLITHEVLHFLRTSSAKKHCSMAVKTDMSKA 475
Query: 1139 YDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQGDPLS 1198
YDRV WDFL+ L FGF +D ++ V + + S L NG+ RGLRQGDPLS
Sbjct: 476 YDRVEWDFLKKVLQRFGFHSIWIDWVLECVTSVSYSFLINGTPQGKVVPTRGLRQGDPLS 535
Query: 1199 PYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQLRLIK 1258
P LF+LC E L+ + VRVS +G ++HL FADD + F ++ + +
Sbjct: 536 PCLFILCTEVLSGLCTRAQRLRQLPGVRVSINGPRVNHLLFADDTMFFSKSDPESCNKLS 595
Query: 1259 HTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPLLQGRV 1318
L+ + +ASG +N KS + FS + + ++ ++ I + GKYLGLP GR
Sbjct: 596 EILSRYGKASGQSINFHKSSVTFSSKTPRSVKGQVKRILKIRKEGGTGKYLGLPEHFGRR 655
Query: 1319 KRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVCNHVDK 1378
KRD F II+K+ + SW + L++AG+ + K++L+S+P+++M LP ++C +
Sbjct: 656 KRDIFGAIIDKIRQKSHSWASRFLSQAGKQVMLKAVLASMPLYSMSCFKLPSALCRKIQS 715
Query: 1379 MVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFLNEKGKL 1438
++ W + R VAW +T K G LG R + N++LL KL LN L
Sbjct: 716 LLTRFWWDTKPDVRKTSWVAWSKLTNPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPESL 775
Query: 1439 WVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSS-SLWYSNWL 1497
+ L KY S S + +L S+ WR I+ R+ + +G G + G S+W WL
Sbjct: 776 LSRILLGKYCHSSSFMECKLPSQPSHGWRSIIAGREILKEGLGWLITNGEKVSIWNDPWL 835
Query: 1498 GTGKIASRIPFVHITDTTLTVADIWRQGA--WNFDRLYTLLPQEIREEISSVQIPSIQTG 1555
K I L V+ + Q W+++++ +LP I + PS G
Sbjct: 836 SISKPLVPIGPALREHQDLRVSALINQNTLQWDWNKIAVILP-NYENLIKQLPAPS-SRG 893
Query: 1556 EDRILWREASDDIYSASSAYQFLNEDG--LPETGF-WKL-IWKASAPEKVRFFLWLVGRD 1611
D++ W Y++ S Y + +P+T F W+ +WK K++ +W +
Sbjct: 894 VDKLAWLPVKSGQYTSRSGYGIASVASIPIPQTQFNWQSNLWKLQTLPKIKHLMWKAAME 953
Query: 1612 ALPTNAKRHRHHLATTASCIRCGAVVEDAEHVLRRC--PGSVW 1652
ALP + R H++ +A+C RCGA E H+ C VW
Sbjct: 954 ALPVGIQLVRRHISPSAACHRCGA-PESTTHLFFHCEFAAQVW 995
>pir||G96509 protein F27F5.21 [imported] - Arabidopsis thaliana
gi|7767672|gb|AAF69169.1| F27F5.21 [Arabidopsis thaliana]
Length = 1023
Score = 436 bits (1122), Expect = e-120
Identities = 270/821 (32%), Positives = 408/821 (48%), Gaps = 44/821 (5%)
Query: 821 QDGNLQDKLQRVQEAATEFNKTVFGN----IHRRKRRVERRLQGIQQEHDWRDSESLARL 876
++GN+ D++ V A E N+ + + +H + ++ Q D R SE + L
Sbjct: 119 KEGNIIDRV--VGAAKDEINEKLMKHDEFIVHIKNEFIDAA-----QRDDSRTSEEITDL 171
Query: 877 ERDLQKEYGDILYQEEMLWFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGN 936
L++ Y D EE W+QKSR W++ GD N+K+FHA T RR RN+I GL E+G
Sbjct: 172 TIRLKEAYID----EEQYWYQKSRSLWMKVGDNNSKYFHALTKQRRARNRIIGLHDENGK 227
Query: 937 WCTDPSTLQEEAKRFYTNLFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAI 996
W + +Q A ++ LF+ + I + D L P + +EVR+A+
Sbjct: 228 WFAEDKDVQHIAVSYFETLFNKSNPQNIAESLSEVKTLITDQINDFLTTPATEDEVRAAL 287
Query: 997 MSMKSFKAPGPDGFHAYFYKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDN 1056
M KAPGPDG A F+++ W + D+ +V S Q+G + +L T + LIPK +
Sbjct: 288 FLMHPEKAPGPDGMTALFFQKPWATIKMDMLFLVNSFLQDGVFDKQLNTTNICLIPKTER 347
Query: 1057 PTKLKDLRPISLCNVSYKLITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVM 1116
PT++ +LRPISLCNV YK+I+K+L R + +L L+S +F+ GR DNI++AQE+
Sbjct: 348 PTRMTELRPISLCNVGYKIISKILCQRLKKILPSLISETHSAFVEGRLISDNILIAQEMF 407
Query: 1117 HTIHTRKT-GGGLVALKIDLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSV 1175
H + T + G +A+K D+ KAYDRV W+F+ L GF + + IMW + T V
Sbjct: 408 HGLRTNPSCKGKFMAIKTDMSKAYDRVEWNFIEELLKKMGFCEKWISWIMWCITTVQYRV 467
Query: 1176 LWNGSKLPSFPLQRGLRQGDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGIS 1235
L NG +RGLRQGDPLSPYLF+LC E L I+ +E ++V+ +S
Sbjct: 468 LLNGQPRGLIVPKRGLRQGDPLSPYLFILCTEVLIANIRKAEQEKLITGIKVATASPSVS 527
Query: 1236 HLFFADDVLLFCQASKDQLRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSG 1295
HL FA+D L FC+A+K+Q +I L + SG +N KS + F VG + E+
Sbjct: 528 HLLFANDSLFFCKANKEQCGVILGILKQYEAVSGQMINFSKSSIQFGHKVGDDIKAEIKS 587
Query: 1296 LVGISRASNLGKYLGLPLLQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSIL 1355
+GI +G YLGLP G K F+ + +++ TR+ W L+K G+ + KS++
Sbjct: 588 ALGIHSIGGMGSYLGLPESLGGSKTRIFSFVRDRLQTRINGWSAKFLSKGGKEVMIKSVV 647
Query: 1356 SSLPVHAMQSLWLPDSVCNHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRS 1415
+ LP + M LP ++ + + V W G +R H +AW + SKA G LG RS
Sbjct: 648 AVLPTYVMSCFRLPKTITSKLTSAVAKFWWSSDGESRGMHWMAWNKLCSSKADGGLGFRS 707
Query: 1416 ARKNNEALLGKLVHSFLNEKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDS 1475
N ALL K + + L+ + +Y + + L + SY WR I+ AR
Sbjct: 708 VNDFNSALLAKQLWRLITVPDSLFAKVFKGRYFRKSNPLDKLKSYSPSYGWRSIISARSL 767
Query: 1476 VSDGFGPRLGAGSS-SLWYSNWLGTGKIASRIPFVHITDTTLTVADI--WRQGAWNFDRL 1532
V G R+G+G+S S+W W+ I + D +L V ++ R WN D L
Sbjct: 768 VKKGLINRVGSGASISVWEDPWIPAQFPRPAISNGSVIDPSLKVNNLIDSRSNFWNIDFL 827
Query: 1533 YTLLPQEIREEISSVQIPSIQTGEDRILWREASDDIYSASSAYQFLNEDGLPETGFWKLI 1592
+ E IS++ + S T ED + DI + + I
Sbjct: 828 NEIFVPEDVLLISAMHLGS-STKEDSL-------DIKTLKAH-----------------I 862
Query: 1593 WKASAPEKVRFFLWLVGRDALPTNAKRHRHHLATTASCIRC 1633
WK P K+R FLW V +P + + + C RC
Sbjct: 863 WKVQCPPKLRHFLWQVLSGCVPVTENLRKRGINCDSGCARC 903
>pir||G86379 protein F5A9.24 [imported] - Arabidopsis thaliana
gi|9945082|gb|AAG03119.1| F5A9.24 [Arabidopsis thaliana]
Length = 1254
Score = 429 bits (1104), Expect = e-118
Identities = 286/916 (31%), Positives = 453/916 (49%), Gaps = 30/916 (3%)
Query: 719 MLEACNFIDLGFMGSKFTWERRVNGRRAVAKRLDRALGDVAWRLCFPEAYVEHLARVYSD 778
M++ C+ +++ G+ FTW R G + RLDRA G+ W FP + L SD
Sbjct: 129 MIKGCDLVEMPAHGNGFTWAGR-RGDHWIQCRLDRAFGNKEWFCFFPVSNQTFLDFRGSD 187
Query: 779 HSPLLLRCHAPTGDRAARPFRFQAAWVSHPSFESVVNTSW-RAQDG---NLQDKLQRVQE 834
H P+L++ + + D FRF ++ + + +W R + G ++ D+L+ ++
Sbjct: 188 HRPVLIKLMS-SQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGKHGTNISVADRLRACRK 246
Query: 835 AATEFNKTVFGNIHRRKRRVERRLQGIQQEHDWRDSESLARLERDLQKEYGDILYQEEML 894
+ + + K N + ++E L+ +Q W + ++ L++DL K Y + EE
Sbjct: 247 SLSSWKKQNNLNSLDKINQLEAALEK-EQSLVWPIFQRVSVLKKDLAKAYRE----EEAY 301
Query: 895 WFQKSRENWVRFGDRNTKFFHAQTVVRRKRNKIHGLFLEDGNWCTDPSTLQEEAKRFYTN 954
W QKSR+ W+R G+RN+K+FHA R+R +I L +GN T + E A ++ N
Sbjct: 302 WKQKSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGN 361
Query: 955 LFSLDVQVRRNRIMENQTPTIPMEERDKLVAPVSLEEVRSAIMSMKSFKAPGPDGFHAYF 1014
LF P + + LV VS +E++ A+ S+K APGPDG A F
Sbjct: 362 LFKSSNPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALF 421
Query: 1015 YKQYWHILGEDLHHMVASAFQNGGANPELLATLLVLIPKVDNPTKLKDLRPISLCNVSYK 1074
++ YW +G + V F +G E T L LIPK +PT++ DLRPISLC+V YK
Sbjct: 422 FQHYWSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYK 481
Query: 1075 LITKVLVNRFRPLLHKLVSPLQGSFILGRGTRDNIILAQEVMHTIHTR-KTGGGLVALKI 1133
+I+K++ R +P L ++VS Q +F+ R DNI++A E++H++ + +A+K
Sbjct: 482 IISKIMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKS 541
Query: 1134 DLEKAYDRVNWDFLRATLYDFGFPPRTVDLIMWGVRTPAVSVLWNGSKLPSFPLQRGLRQ 1193
D+ KAYDRV W +LR+ L GF + V+ IM V + SVL N LQRGLRQ
Sbjct: 542 DMSKAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQ 601
Query: 1194 GDPLSPYLFVLCMERLAIQIQNLVEEGKWRPVRVSKDGVGISHLFFADDVLLFCQASKDQ 1253
GDPLSP+LFVLC E L + EG ++ S++G + HL FADD L C+AS++Q
Sbjct: 602 GDPLSPFLFVLCTEGLTHLLNKAQWEGALEGIQFSENGPMVHHLLFADDSLFLCKASREQ 661
Query: 1254 LRLIKHTLADFCEASGMKVNLEKSRMLFSKVVGQRRQRELSGLVGISRASNLGKYLGLPL 1313
+++ L + A+G +NL KS + F + V ++ + + +GI G YLGLP
Sbjct: 662 SLVLQKILKVYGNATGQTINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPE 721
Query: 1314 LQGRVKRDHFATIIEKVNTRLASWKNNLLNKAGRVCLAKSILSSLPVHAMQSLWLPDSVC 1373
K D + +++ +L W L++ G+ L KS+ ++PV AM LP + C
Sbjct: 722 CFSGSKVDMLHYLKDRLKEKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTC 781
Query: 1374 NHVDKMVRNCIWGKGGNARSWHLVAWKDVTQSKAKGRLGLRSARKNNEALLGKLVHSFLN 1433
+++ + + W ++R H +W+ + K G LG R + N+ALL K L+
Sbjct: 782 ENLESAMASFWWDSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLH 841
Query: 1434 EKGKLWVQALSQKYIKSGSILTEELRPGGSYVWRGILKARDSVSDGFGPRLGAGSS-SLW 1492
L + L +Y + L L S+ WR IL R+ +S G R+G G+S +W
Sbjct: 842 FPDCLLSRLLKSRYFDATDFLDAALSQRPSFGWRSILFGRELLSKGLQKRVGDGASLFVW 901
Query: 1493 YSNWLGTGKIASRIPFVHITDTTLTVADIW--RQGAWNFDRLYTL-LPQEIREEISSVQI 1549
W+ + I D TL V + R G W+ + L+ L LP++I + I
Sbjct: 902 IDPWIDDNGFRAPWRKNLIYDVTLKVKALLNPRTGFWDEEVLHDLFLPEDI---LRIKAI 958
Query: 1550 PSIQTGEDRILWREASDDIYSASSAYQFLNE----------DGLPET-GFWKLIWKASAP 1598
+ + D +W+ +S SAY + P T G +W
Sbjct: 959 KPVISQADFFVWKLNKSGDFSVKSAYWLAYQTKSQNLRSEVSMQPSTLGLKTQVWNLQTD 1018
Query: 1599 EKVRFFLWLVGRDALP 1614
K++ FLW V LP
Sbjct: 1019 PKIKIFLWKVLSGILP 1034
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.336 0.145 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,087,568,701
Number of Sequences: 2540612
Number of extensions: 127175035
Number of successful extensions: 399877
Number of sequences better than 10.0: 1369
Number of HSP's better than 10.0 without gapping: 792
Number of HSP's successfully gapped in prelim test: 577
Number of HSP's that attempted gapping in prelim test: 395394
Number of HSP's gapped (non-prelim): 2148
length of query: 1904
length of database: 863,360,394
effective HSP length: 143
effective length of query: 1761
effective length of database: 500,052,878
effective search space: 880593118158
effective search space used: 880593118158
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 83 (36.6 bits)
Lotus: description of TM0228a.4


