

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0227.9
(273 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
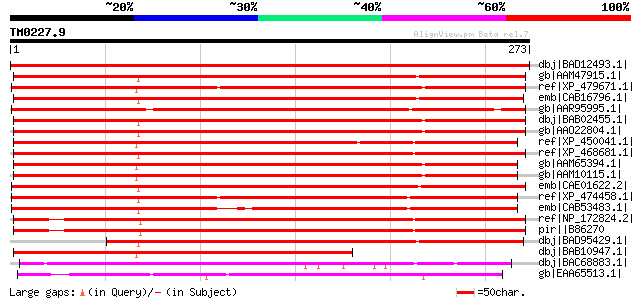
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD12493.1| nodulin of unknown function [Lotus corniculatus ... 572 e-162
gb|AAM47915.1| unknown protein [Arabidopsis thaliana] gi|2026021... 338 1e-91
ref|XP_479671.1| MAP3K-like protein kinase [Oryza sativa (japoni... 329 6e-89
emb|CAB16796.1| MAP3K-like protein kinase [Arabidopsis thaliana]... 325 8e-88
gb|AAR95995.1| hypothetical protein kinase [Musa acuminata] 325 1e-87
dbj|BAB02455.1| MAP3K protein kinase-like protein [Arabidopsis t... 322 9e-87
gb|AAO22804.1| unknown protein [Arabidopsis thaliana] 322 9e-87
ref|XP_450041.1| MAP3K protein kinase-like protein [Oryza sativa... 305 1e-81
ref|XP_468681.1| expressed protein [Oryza sativa (japonica culti... 304 2e-81
gb|AAM65394.1| unknown [Arabidopsis thaliana] 302 6e-81
gb|AAM10115.1| unknown protein [Arabidopsis thaliana] gi|1842515... 302 6e-81
emb|CAE01622.2| OSJNBa0042L16.16 [Oryza sativa (japonica cultiva... 301 2e-80
ref|XP_474458.1| OSJNBa0039K24.18 [Oryza sativa (japonica cultiv... 292 8e-78
emb|CAB53483.1| CAA303710.1 protein [Oryza sativa] 273 4e-72
ref|NP_172824.2| expressed protein [Arabidopsis thaliana] 268 9e-71
pir||B86270 hypothetical protein F21F23.12 - Arabidopsis thalian... 268 9e-71
dbj|BAD95429.1| MAP3K-like protein kinase [Arabidopsis thaliana] 255 8e-67
dbj|BAB10947.1| unnamed protein product [Arabidopsis thaliana] 213 6e-54
dbj|BAC68883.1| hypothetical protein [Streptomyces avermitilis M... 99 1e-19
gb|EAA65513.1| hypothetical protein AN1330.2 [Aspergillus nidula... 70 8e-11
>dbj|BAD12493.1| nodulin of unknown function [Lotus corniculatus var. japonicus]
Length = 337
Score = 572 bits (1475), Expect = e-162
Identities = 273/273 (100%), Positives = 273/273 (100%)
Query: 1 MELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYED 60
MELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYED
Sbjct: 65 MELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYED 124
Query: 61 TIWLCRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKARL 120
TIWLCRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKARL
Sbjct: 125 TIWLCRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKARL 184
Query: 121 RKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGNV 180
RKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGNV
Sbjct: 185 RKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGNV 244
Query: 181 GIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAGN 240
GIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAGN
Sbjct: 245 GIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAGN 304
Query: 241 RWPNFIAVDFYKRGDGGGAPEALDLANRNLIRL 273
RWPNFIAVDFYKRGDGGGAPEALDLANRNLIRL
Sbjct: 305 RWPNFIAVDFYKRGDGGGAPEALDLANRNLIRL 337
>gb|AAM47915.1| unknown protein [Arabidopsis thaliana] gi|20260218|gb|AAM13007.1|
unknown protein [Arabidopsis thaliana]
gi|18402763|ref|NP_564553.1| expressed protein
[Arabidopsis thaliana] gi|25372963|pir||A96534
hypothetical protein F14J22.5 [imported] - Arabidopsis
thaliana gi|10120435|gb|AAG13060.1| Unknown protein
[Arabidopsis thaliana]
Length = 359
Score = 338 bits (867), Expect = 1e-91
Identities = 161/272 (59%), Positives = 209/272 (76%), Gaps = 4/272 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN+YSWLTTHNSFA G TGS +LA TNQ+DSIT QL NGVRG MLDM+D+++ I
Sbjct: 73 LPFNKYSWLTTHNSFARLGEVSRTGSAILAPTNQQDSITSQLNNGVRGFMLDMYDFQNDI 132
Query: 63 WLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC G C +T FQPA+N+L+E +VFL + E++TI I+D+V S G+ KVFD A
Sbjct: 133 WLCHSFDGTCFNFTAFQPAINILREFQVFLEKNKEEVVTIIIEDYVKSPKGLTKVFDAAG 192
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
LRKF FPVS+MPKNG DWP + M+RKN RL+VFTS++ +EA+EGIAY+W Y+VE+Q+GN
Sbjct: 193 LRKFMFPVSRMPKNGGDWPRLDDMVRKNQRLLVFTSDSHKEATEGIAYQWKYMVENQYGN 252
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K G C NR +S PM++ +KSLVL+N+F + + AC+ NS+ L+ + C++AAG
Sbjct: 253 GGLKVGVCPNRAQSAPMSDKSKSLVLVNHFPDAAD-VIVACKQNSASLLESIKTCYQAAG 311
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
RWPNFIAVDFYKR DGGGAP+A+D+AN NLI
Sbjct: 312 QRWPNFIAVDFYKRSDGGGAPQAVDVANGNLI 343
>ref|XP_479671.1| MAP3K-like protein kinase [Oryza sativa (japonica cultivar-group)]
gi|51963696|ref|XP_506619.1| PREDICTED P0015C07.31 gene
product [Oryza sativa (japonica cultivar-group)]
gi|50725707|dbj|BAD33173.1| MAP3K-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 411
Score = 329 bits (843), Expect = 6e-89
Identities = 156/273 (57%), Positives = 208/273 (76%), Gaps = 5/273 (1%)
Query: 2 ELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDT 61
+LPFN YSWLTTHNS+A G + +TGS ++ TNQED+IT QLKNGVRGLMLD +D+ +
Sbjct: 66 DLPFNNYSWLTTHNSYALAGSSSATGSALITQTNQEDTITAQLKNGVRGLMLDTYDFNND 125
Query: 62 IWLC---RGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKA 118
+WLC +G C +T FQPA+NVLKE+R FL +P+E+ITIF++D+ SG+ + KVF+ +
Sbjct: 126 VWLCHSFQGKCFNFTAFQPAINVLKEIRTFLDGNPSEVITIFLEDYTASGS-LPKVFNAS 184
Query: 119 RLRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFG 178
L K+WFPV+KMPK+G DWP +K MI +N RL+VFTS S+EASEGIAYEW+YVVE+Q+G
Sbjct: 185 GLMKYWFPVAKMPKSGGDWPLLKDMISQNERLLVFTSKKSKEASEGIAYEWSYVVENQYG 244
Query: 179 NVGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAA 238
N G+ G C NR ES M++ ++SLVLMN+F + C +NS+PL++M+ C +
Sbjct: 245 NEGMVEGKCPNRAESPAMDSKSQSLVLMNFFTTDPSQTG-VCANNSAPLVSMLKTCHDLS 303
Query: 239 GNRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
GNRWPN+IAVDFY R DGGGAP A D+AN +L+
Sbjct: 304 GNRWPNYIAVDFYMRSDGGGAPLATDIANGHLV 336
>emb|CAB16796.1| MAP3K-like protein kinase [Arabidopsis thaliana]
gi|7270644|emb|CAB80361.1| MAP3K-like protein kinase
[Arabidopsis thaliana] gi|25407778|pir||D85436
MAP3K-like protein kinase [imported] - Arabidopsis
thaliana
Length = 799
Score = 325 bits (833), Expect = 8e-88
Identities = 154/271 (56%), Positives = 202/271 (73%), Gaps = 4/271 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN+YSWLTTHNS+A G N +TGS +++ NQEDSIT+QLKNGVRG+MLD +D+++ I
Sbjct: 383 LPFNKYSWLTTHNSYAITGANSATGSFLVSPKNQEDSITNQLKNGVRGIMLDTYDFQNDI 442
Query: 63 WLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC G C +T FQPA+N LKE+ FL ++ +EI+TI ++D+V S G+ VF+ +
Sbjct: 443 WLCHSTGGTCFNFTAFQPAINALKEINDFLESNLSEIVTIILEDYVKSQMGLTNVFNASG 502
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L KF P+S+MPK+G+DWPTV M+++N RL+VFTS +EASEG+AY+WNY+VE+Q+GN
Sbjct: 503 LSKFLLPISRMPKDGTDWPTVDDMVKQNQRLVVFTSKKDKEASEGLAYQWNYMVENQYGN 562
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K GSC +R ES ++ ++SLV NYF N +AC DNSSPLI MM C AAG
Sbjct: 563 DGMKDGSCSSRSESSSLDTMSRSLVFQNYFETSPN-STQACADNSSPLIEMMRTCHEAAG 621
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNL 270
RWPNFIAVDFY+R D GGA EA+D AN L
Sbjct: 622 KRWPNFIAVDFYQRSDSGGAAEAVDEANGRL 652
>gb|AAR95995.1| hypothetical protein kinase [Musa acuminata]
Length = 376
Score = 325 bits (832), Expect = 1e-87
Identities = 156/270 (57%), Positives = 202/270 (74%), Gaps = 7/270 (2%)
Query: 2 ELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDT 61
+LPFN+YSWLTTHNSFA G + +TG+ ++ FTNQ D+IT QL NGVRGLMLDM+D+ +
Sbjct: 99 DLPFNKYSWLTTHNSFADAGAHSATGATLITFTNQHDNITSQLNNGVRGLMLDMYDFRND 158
Query: 62 IWLCRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKARLR 121
IWLC QPA+NVLKE+ FL +P+E+ITIFI+D+V S +G++KVF+ + L
Sbjct: 159 IWLCHSTAVYQ---QPAINVLKEIETFLAANPSEVITIFIEDYVKSPSGLSKVFNASGLM 215
Query: 122 KFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGNVG 181
K+WFPV +MPKNGSDWP + MI +N+RL+VFTS AS+EASEGIAYEWNYVVE+Q+G+ G
Sbjct: 216 KYWFPVDQMPKNGSDWPLLSKMIDQNHRLLVFTSVASKEASEGIAYEWNYVVENQYGDEG 275
Query: 182 IKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAGNR 241
+ GSC +R ES PM+ KSLVLMNYFR AC +NS+PL+ M+ C + NR
Sbjct: 276 MTPGSCPSRAESSPMSTTLKSLVLMNYFR-TNPSASSACHNNSAPLLDMLKTCHGLSANR 334
Query: 242 WPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
W NFIAVDFY +GD APEA D+AN +++
Sbjct: 335 WANFIAVDFYMKGD---APEAADVANGHMV 361
>dbj|BAB02455.1| MAP3K protein kinase-like protein [Arabidopsis thaliana]
gi|15230348|ref|NP_188562.1| expressed protein
[Arabidopsis thaliana]
Length = 413
Score = 322 bits (824), Expect = 9e-87
Identities = 150/272 (55%), Positives = 204/272 (74%), Gaps = 4/272 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LP+N+YSWLTTHNSFA G TGS +LA +NQ+DSIT QL NGVRG MLD++D+++ I
Sbjct: 73 LPYNKYSWLTTHNSFARMGAKSGTGSMILAPSNQQDSITSQLLNGVRGFMLDLYDFQNDI 132
Query: 63 WLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC G C YT FQPA+N+LKE +VFL + ++T+ ++D+V S NG+ +VFD +
Sbjct: 133 WLCHSYGGNCFNYTAFQPAVNILKEFQVFLDKNKDVVVTLILEDYVKSPNGLTRVFDASG 192
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
LR F FPVS+MPKNG DWPT+ MI +N RL+VFTSN +EASEGIA+ W Y++E+Q+G+
Sbjct: 193 LRNFMFPVSRMPKNGEDWPTLDDMICQNQRLLVFTSNPQKEASEGIAFMWRYMIENQYGD 252
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K G C NRPES+ M + ++SL+L+NYF + + +C+ NS+PL+ + C A+G
Sbjct: 253 GGMKAGVCTNRPESVAMGDRSRSLILVNYFPDTADVIG-SCKQNSAPLLDTVKNCQEASG 311
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
RWPNFIAVDFYKR DGGGAP+A+D+AN + +
Sbjct: 312 KRWPNFIAVDFYKRSDGGGAPKAVDVANGHAV 343
>gb|AAO22804.1| unknown protein [Arabidopsis thaliana]
Length = 397
Score = 322 bits (824), Expect = 9e-87
Identities = 150/272 (55%), Positives = 204/272 (74%), Gaps = 4/272 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LP+N+YSWLTTHNSFA G TGS +LA +NQ+DSIT QL NGVRG MLD++D+++ I
Sbjct: 57 LPYNKYSWLTTHNSFARMGAKSGTGSMILAPSNQQDSITSQLLNGVRGFMLDLYDFQNDI 116
Query: 63 WLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC G C YT FQPA+N+LKE +VFL + ++T+ ++D+V S NG+ +VFD +
Sbjct: 117 WLCHSYGGNCFNYTAFQPAVNILKEFQVFLDKNKDVVVTLILEDYVKSPNGLTRVFDASG 176
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
LR F FPVS+MPKNG DWPT+ MI +N RL+VFTSN +EASEGIA+ W Y++E+Q+G+
Sbjct: 177 LRNFMFPVSRMPKNGEDWPTLDDMICQNQRLLVFTSNPQKEASEGIAFMWRYMIENQYGD 236
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K G C NRPES+ M + ++SL+L+NYF + + +C+ NS+PL+ + C A+G
Sbjct: 237 GGMKAGVCTNRPESVAMGDRSRSLILVNYFPDTADVIG-SCKQNSAPLLDTVKNCQEASG 295
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
RWPNFIAVDFYKR DGGGAP+A+D+AN + +
Sbjct: 296 KRWPNFIAVDFYKRSDGGGAPKAVDVANGHAV 327
>ref|XP_450041.1| MAP3K protein kinase-like protein [Oryza sativa (japonica
cultivar-group)] gi|51963708|ref|XP_506634.1| PREDICTED
OJ1310_F05.17 gene product [Oryza sativa (japonica
cultivar-group)] gi|46806453|dbj|BAD17589.1| MAP3K
protein kinase-like protein [Oryza sativa (japonica
cultivar-group)] gi|46389989|dbj|BAD16231.1| MAP3K
protein kinase-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 412
Score = 305 bits (780), Expect = 1e-81
Identities = 147/268 (54%), Positives = 197/268 (72%), Gaps = 5/268 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN YSWL THNSF+ G TG + F NQED++T+QL+NGVRGLMLDM+D+ D I
Sbjct: 68 LPFNRYSWLVTHNSFSIIGEPSHTGVERVTFYNQEDTVTNQLRNGVRGLMLDMYDFNDDI 127
Query: 63 WLC---RGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC +G C +T FQPA++ LKEV FL +PTEIITIFI+D+V S G++K+F A
Sbjct: 128 WLCHSLQGQCYNFTAFQPAIDTLKEVEAFLSENPTEIITIFIEDYVHSTMGLSKLFTAAD 187
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L K+W+P+S+MP NG DWP+V M+ KN+RL+VFTS++S+EASEGIAY+W+Y++E++ G+
Sbjct: 188 LTKYWYPISEMPTNGKDWPSVTDMVAKNHRLLVFTSDSSKEASEGIAYQWSYLLENESGD 247
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
GI GSC NR ES P+N+ + SL + NYF + ++EAC++NS L M+ C+ AAG
Sbjct: 248 PGIT-GSCPNRKESQPLNSRSASLFMQNYFPTI-PVENEACKENSVGLPQMVQTCYTAAG 305
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLAN 267
NR PNFIAV++Y R DGGG + D N
Sbjct: 306 NRIPNFIAVNYYMRSDGGGVFDVQDRIN 333
>ref|XP_468681.1| expressed protein [Oryza sativa (japonica cultivar-group)]
gi|41469135|gb|AAS07086.1| expressed protein [Oryza
sativa (japonica cultivar-group)]
Length = 360
Score = 304 bits (778), Expect = 2e-81
Identities = 143/272 (52%), Positives = 192/272 (70%), Gaps = 4/272 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN+Y++LTTHNSFA G TG P + F NQED++TDQL NGVR LMLD +D++ +
Sbjct: 71 LPFNKYAYLTTHNSFAIVGEPSHTGVPRITFDNQEDTVTDQLNNGVRALMLDTYDFKGDV 130
Query: 63 WLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC G C +T F+PAL+ KE+ FL +P+EI+T+ ++D+V + NG+ VF +
Sbjct: 131 WLCHSNGGKCNDFTAFEPALDTFKEIEAFLGANPSEIVTLILEDYVHAPNGLTNVFKASG 190
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L K+WFPVSKMP+ G DWP V M+ N RL+VFTS S++A+EGIAY+WNY+VE+ +G+
Sbjct: 191 LMKYWFPVSKMPQKGKDWPLVSDMVASNQRLLVFTSIRSKQATEGIAYQWNYMVENNYGD 250
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+ G C NR ES P+N+ TKSLVL+NYF +V AC +S L M++ C+ AAG
Sbjct: 251 DGMDAGKCSNRAESAPLNDKTKSLVLVNYFPSV-PVKVTACLQHSKSLTDMVNTCYGAAG 309
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
NRW N +AVD+YKR DGGGA +A DL N L+
Sbjct: 310 NRWANLLAVDYYKRSDGGGAFQATDLLNGRLL 341
>gb|AAM65394.1| unknown [Arabidopsis thaliana]
Length = 426
Score = 302 bits (774), Expect = 6e-81
Identities = 142/268 (52%), Positives = 192/268 (70%), Gaps = 4/268 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN+Y+WL THN+F+ G + F NQED+IT+QL+NGVRGLMLDM+D+ + I
Sbjct: 80 LPFNKYTWLMTHNAFSNANAPLLPGVERITFYNQEDTITNQLQNGVRGLMLDMYDFNNDI 139
Query: 63 WLC---RGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC RG C +T FQPA+N+L+EV FL +PTEI+TI I+D+V G++ +F A
Sbjct: 140 WLCHSLRGQCFNFTXFQPAINILREVEAFLSQNPTEIVTIIIEDYVHRPKGLSTLFANAG 199
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L K+WFPVSKMP+ G DWPTV M+++N+RL+VFTS A++E EG+AY+W Y+VE++ G+
Sbjct: 200 LDKYWFPVSKMPRKGEDWPTVTDMVQENHRLLVFTSVAAKEDEEGVAYQWRYMVENESGD 259
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K GSC NR ES P+N+ + SL LMNYF D AC+++S+PL M+ C ++ G
Sbjct: 260 PGVKRGSCPNRKESQPLNSKSSSLFLMNYFPTYPVEKD-ACKEHSAPLAEMVGTCLKSGG 318
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLAN 267
NR PNF+AV+FY R DGGG E LD N
Sbjct: 319 NRMPNFLAVNFYMRSDGGGVFEILDRMN 346
>gb|AAM10115.1| unknown protein [Arabidopsis thaliana] gi|18425155|ref|NP_569045.1|
expressed protein [Arabidopsis thaliana]
gi|15451188|gb|AAK96865.1| Unknown protein [Arabidopsis
thaliana]
Length = 426
Score = 302 bits (774), Expect = 6e-81
Identities = 142/268 (52%), Positives = 192/268 (70%), Gaps = 4/268 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN+Y+WL THN+F+ G + F NQED+IT+QL+NGVRGLMLDM+D+ + I
Sbjct: 80 LPFNKYTWLMTHNAFSNANAPLLPGVERITFYNQEDTITNQLQNGVRGLMLDMYDFNNDI 139
Query: 63 WLC---RGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC RG C +T FQPA+N+L+EV FL +PTEI+TI I+D+V G++ +F A
Sbjct: 140 WLCHSLRGQCFNFTAFQPAINILREVEAFLSQNPTEIVTIIIEDYVHRPKGLSTLFANAG 199
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L K+WFPVSKMP+ G DWPTV M+++N+RL+VFTS A++E EG+AY+W Y+VE++ G+
Sbjct: 200 LDKYWFPVSKMPRKGEDWPTVTDMVQENHRLLVFTSVAAKEDEEGVAYQWRYMVENESGD 259
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K GSC NR ES P+N+ + SL LMNYF D AC+++S+PL M+ C ++ G
Sbjct: 260 PGVKRGSCPNRKESQPLNSKSSSLFLMNYFPTYPVEKD-ACKEHSAPLAEMVGTCLKSGG 318
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLAN 267
NR PNF+AV+FY R DGGG E LD N
Sbjct: 319 NRMPNFLAVNFYMRSDGGGVFEILDRMN 346
>emb|CAE01622.2| OSJNBa0042L16.16 [Oryza sativa (japonica cultivar-group)]
gi|50924276|ref|XP_472498.1| OSJNBa0042L16.16 [Oryza
sativa (japonica cultivar-group)]
Length = 413
Score = 301 bits (770), Expect = 2e-80
Identities = 141/273 (51%), Positives = 194/273 (70%), Gaps = 4/273 (1%)
Query: 2 ELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDT 61
+LPFN Y+WLTTHNSFA G TG+ + NQ+D+ITDQL NGVRGLMLDM+D+ +
Sbjct: 77 DLPFNRYAWLTTHNSFARLGTRSRTGTAIATAWNQQDTITDQLNNGVRGLMLDMYDFRND 136
Query: 62 IWLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKA 118
IWLC G C +T F PA+ VL E+ FL +P+E++T+F++D+V S G+ +V + +
Sbjct: 137 IWLCHSFGGACQNFTAFVPAVEVLGEIERFLARNPSEVVTVFVEDYVESPMGLTRVLNAS 196
Query: 119 RLRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFG 178
L K+ FP +MPK+G DWP + M+R N+RL++FTS +++EA+EGI YEW+YVVE+Q+G
Sbjct: 197 GLTKYVFPAWRMPKSGGDWPRLSDMVRDNHRLLLFTSKSAKEAAEGIPYEWHYVVENQYG 256
Query: 179 NVGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAA 238
G+ G C NR ES MN+ ++SLVL+NYFR++ N AC+DNS+ L+ M+ C +
Sbjct: 257 TKGMIKGRCPNRAESAAMNDLSRSLVLVNYFRDLPNF-PVACKDNSAELLDMLTTCHDLS 315
Query: 239 GNRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
+RW NFIAVDFYKR D GGA EA D AN L+
Sbjct: 316 ADRWANFIAVDFYKRSDRGGAAEATDRANGGLV 348
>ref|XP_474458.1| OSJNBa0039K24.18 [Oryza sativa (japonica cultivar-group)]
gi|38345515|emb|CAE01799.2| OSJNBa0039K24.18 [Oryza
sativa (japonica cultivar-group)]
Length = 468
Score = 292 bits (747), Expect = 8e-78
Identities = 135/269 (50%), Positives = 193/269 (71%), Gaps = 5/269 (1%)
Query: 2 ELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDT 61
+L FN Y+WLTTHNSFA G TG+P++A NQED++T QLKNGVRGLMLD +D+++
Sbjct: 76 DLAFNRYTWLTTHNSFAIVGSPSRTGTPIIAPPNQEDTVTAQLKNGVRGLMLDAYDFQNE 135
Query: 62 IWLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKA 118
+WLC G C + +Q A++VLKE+ FL +P+E+IT+F++D+ G+ + KV +
Sbjct: 136 VWLCHSFGGKCYNFAAYQRAMDVLKEIGAFLDANPSEVITVFVEDYAGPGS-LGKVVGGS 194
Query: 119 RLRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFG 178
L K+ FP +KMPK G DWP +K MI +N+RL++FTS ++ S+G+AYEW+YV+E+Q+G
Sbjct: 195 GLSKYLFPPAKMPKGGGDWPLLKDMIAQNHRLLMFTSKRGKDGSDGLAYEWDYVLETQYG 254
Query: 179 NVGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAA 238
N G+ GGSC R ES+ M++ +SL+LMN+F + AC +NS+PL+A + C+ A+
Sbjct: 255 NDGLVGGSCPKRAESMAMDSTKQSLILMNFF-STNPSQSWACGNNSAPLVAKLKACYDAS 313
Query: 239 GNRWPNFIAVDFYKRGDGGGAPEALDLAN 267
RWPNFIAVD+Y R GGGAP A D+AN
Sbjct: 314 AKRWPNFIAVDYYMRSKGGGAPLATDVAN 342
>emb|CAB53483.1| CAA303710.1 protein [Oryza sativa]
Length = 416
Score = 273 bits (698), Expect = 4e-72
Identities = 130/269 (48%), Positives = 185/269 (68%), Gaps = 17/269 (6%)
Query: 2 ELPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDT 61
+L FN Y+WLTTHNSFA G TG+P++A NQED++T QLKNGVRGLMLD +D+++
Sbjct: 76 DLAFNRYTWLTTHNSFAIVGSPSRTGTPIIAPPNQEDTVTAQLKNGVRGLMLDAYDFQNE 135
Query: 62 IWLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKA 118
+WLC G C + +Q A++VLKE+ FL +P+E+IT+F++D+ G+
Sbjct: 136 VWLCHSFGGKCYNFAAYQRAMDVLKEIGAFLDANPSEVITVFVEDYAGPGS--------- 186
Query: 119 RLRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFG 178
L K +MPK G DWP +K MI +N+RL++FTS ++ S+G+AYEW+YV+E+Q+G
Sbjct: 187 -LGKSG---GRMPKGGGDWPLLKDMIAQNHRLLMFTSKRGKDGSDGLAYEWDYVLETQYG 242
Query: 179 NVGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAA 238
N G+ GGSC R ES+ M++ +SL+LMN+F + AC +NS+PL+A + C+ A+
Sbjct: 243 NDGLVGGSCPKRAESMAMDSTKQSLILMNFF-STNPSQSWACGNNSAPLVAKLKACYDAS 301
Query: 239 GNRWPNFIAVDFYKRGDGGGAPEALDLAN 267
RWPNFIAVD+Y R GGGAP A D+AN
Sbjct: 302 AKRWPNFIAVDYYMRSKGGGAPLATDVAN 330
>ref|NP_172824.2| expressed protein [Arabidopsis thaliana]
Length = 317
Score = 268 bits (686), Expect = 9e-71
Identities = 128/272 (47%), Positives = 181/272 (66%), Gaps = 11/272 (4%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
+PFN+Y++LTTHNS+A +G L QED+I QL +GVR LMLD +DYE +
Sbjct: 45 MPFNKYAFLTTHNSYAIEG-------KALHVATQEDTIVQQLNSGVRALMLDTYDYEGDV 97
Query: 63 WLCRG---PCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
W C C ++T F A++ KE+ FL +P+EI+T+ ++D+V S NG+ KVF +
Sbjct: 98 WFCHSFDEQCFEFTKFNRAIDTFKEIFAFLTANPSEIVTLILEDYVKSQNGLTKVFTDSG 157
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L+KFWFPV MP G DWP VK M+ N+RLIVFTS S++ +EGIAY+WNY+VE+Q+G+
Sbjct: 158 LKKFWFPVQNMPIGGQDWPLVKDMVANNHRLIVFTSAKSKQETEGIAYQWNYMVENQYGD 217
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K C NR +S + + TK+LV +N+F+ V C +NS L+ M+ C+ AAG
Sbjct: 218 DGVKPDECSNRADSALLTDKTKALVSVNHFKTV-PVKILTCEENSEQLLDMIKTCYVAAG 276
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
NRW NF+AV+FYKR +GGG +A+D N L+
Sbjct: 277 NRWANFVAVNFYKRSNGGGTFQAIDKLNGELL 308
>pir||B86270 hypothetical protein F21F23.12 - Arabidopsis thaliana
gi|8920573|gb|AAF81295.1| Contains similarity to
MAP3K-like protein kinase from Arabidopsis thaliana
gb|Z99707
Length = 346
Score = 268 bits (686), Expect = 9e-71
Identities = 128/272 (47%), Positives = 181/272 (66%), Gaps = 11/272 (4%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
+PFN+Y++LTTHNS+A +G L QED+I QL +GVR LMLD +DYE +
Sbjct: 74 MPFNKYAFLTTHNSYAIEG-------KALHVATQEDTIVQQLNSGVRALMLDTYDYEGDV 126
Query: 63 WLCRG---PCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
W C C ++T F A++ KE+ FL +P+EI+T+ ++D+V S NG+ KVF +
Sbjct: 127 WFCHSFDEQCFEFTKFNRAIDTFKEIFAFLTANPSEIVTLILEDYVKSQNGLTKVFTDSG 186
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L+KFWFPV MP G DWP VK M+ N+RLIVFTS S++ +EGIAY+WNY+VE+Q+G+
Sbjct: 187 LKKFWFPVQNMPIGGQDWPLVKDMVANNHRLIVFTSAKSKQETEGIAYQWNYMVENQYGD 246
Query: 180 VGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLIAMMHVCFRAAG 239
G+K C NR +S + + TK+LV +N+F+ V C +NS L+ M+ C+ AAG
Sbjct: 247 DGVKPDECSNRADSALLTDKTKALVSVNHFKTV-PVKILTCEENSEQLLDMIKTCYVAAG 305
Query: 240 NRWPNFIAVDFYKRGDGGGAPEALDLANRNLI 271
NRW NF+AV+FYKR +GGG +A+D N L+
Sbjct: 306 NRWANFVAVNFYKRSNGGGTFQAIDKLNGELL 337
>dbj|BAD95429.1| MAP3K-like protein kinase [Arabidopsis thaliana]
Length = 291
Score = 255 bits (652), Expect = 8e-67
Identities = 120/222 (54%), Positives = 160/222 (72%), Gaps = 4/222 (1%)
Query: 52 MLDMWDYEDTIWLCR---GPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSG 108
MLD +D+++ IWLC G C +T FQPA+N LKE+ FL ++ +EI+TI ++D+V S
Sbjct: 1 MLDTYDFQNDIWLCHSTGGTCFNFTAFQPAINALKEINDFLESNLSEIVTIILEDYVKSQ 60
Query: 109 NGVNKVFDKARLRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYE 168
G+ VF+ + L KF P+S+MPK+G+DWPTV M+++N RL+VFTS +EASEG+AY+
Sbjct: 61 MGLTNVFNASGLSKFLLPISRMPKDGTDWPTVDDMVKQNQRLVVFTSKKDKEASEGLAYQ 120
Query: 169 WNYVVESQFGNVGIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHDDEACRDNSSPLI 228
WNY+VE+Q+GN G+K GSC +R ES ++ ++SLV NYF N +AC DNSSPLI
Sbjct: 121 WNYMVENQYGNDGMKDGSCSSRSESSSLDTMSRSLVFQNYFETSPN-STQACADNSSPLI 179
Query: 229 AMMHVCFRAAGNRWPNFIAVDFYKRGDGGGAPEALDLANRNL 270
MM C AAG RWPNFIAVDFY+R D GGA EA+D AN L
Sbjct: 180 EMMRTCHEAAGKRWPNFIAVDFYQRSDSGGAAEAVDEANGRL 221
>dbj|BAB10947.1| unnamed protein product [Arabidopsis thaliana]
Length = 365
Score = 213 bits (541), Expect = 6e-54
Identities = 97/181 (53%), Positives = 134/181 (73%), Gaps = 3/181 (1%)
Query: 3 LPFNEYSWLTTHNSFAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTI 62
LPFN+Y+WL THN+F+ G + F NQED+IT+QL+NGVRGLMLDM+D+ + I
Sbjct: 60 LPFNKYTWLMTHNAFSNANAPLLPGVERITFYNQEDTITNQLQNGVRGLMLDMYDFNNDI 119
Query: 63 WLC---RGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKAR 119
WLC RG C +T FQPA+N+L+EV FL +PTEI+TI I+D+V G++ +F A
Sbjct: 120 WLCHSLRGQCFNFTAFQPAINILREVEAFLSQNPTEIVTIIIEDYVHRPKGLSTLFANAG 179
Query: 120 LRKFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTSNASREASEGIAYEWNYVVESQFGN 179
L K+WFPVSKMP+ G DWPTV M+++N+RL+VFTS A++E EG+AY+W Y+VE++ N
Sbjct: 180 LDKYWFPVSKMPRKGEDWPTVTDMVQENHRLLVFTSVAAKEDEEGVAYQWRYMVENECIN 239
Query: 180 V 180
+
Sbjct: 240 L 240
>dbj|BAC68883.1| hypothetical protein [Streptomyces avermitilis MA-4680]
gi|29827714|ref|NP_822348.1| hypothetical protein
SAV1173 [Streptomyces avermitilis MA-4680]
Length = 464
Score = 99.0 bits (245), Expect = 1e-19
Identities = 79/278 (28%), Positives = 133/278 (47%), Gaps = 22/278 (7%)
Query: 6 NEYSWLTTHNSFAAKGVNWSTGSPVLAFT-NQEDSITDQLKNGVRGLMLDMWDYEDTIWL 64
++ ++LT HN++A GV+ P + NQ I QL +GVRG M+D+ D L
Sbjct: 185 DQVTFLTAHNAYA-NGVDGGFAPPFVNLVPNQTRGINQQLTDGVRGFMMDIHQTSDGAIL 243
Query: 65 CRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFIDDHVTSGNGVNKVFDKARLRKFW 124
C CT + ++ + FL HP +++T+F++D+V G +++ + L
Sbjct: 244 CHNSCTLVSKPVALWVDIQRMVDFLKQHPDQVVTVFLEDYVDPGVLRSELARVSGLSDVL 303
Query: 125 FPVSKMPKNGSDWPTVKTMIRKNYRLIVFT--SNASREA------SEGIAYEWNYVVE-- 174
+ + S WP + +I N+RL++FT S +S E+ S G+ Y+ + VE
Sbjct: 304 YRPDRTGVRQSGWPRMADLIAANHRLLIFTDHSRSSDESAGLTRDSFGVMYQREWTVENY 363
Query: 175 -SQFGNVGIKGGSCQNR----PESLPM---NNATKSLVLMNYFRNVRNHDDEACRDNSSP 226
S +G SC +R ++P+ +A + L +MN+FR+ A DN+
Sbjct: 364 WSMGSGLGSSDWSCYSRWYGADTNIPLTYTESAFRPLFVMNHFRDAA-IASTATTDNTKL 422
Query: 227 LIAMMHVCFRAAGNRWPNFIAVDFYKRGDGGGAPEALD 264
C R A + PNF+AVD Y G+ A + L+
Sbjct: 423 ADRAQRFC-RPAARKKPNFLAVDRYDLGNPTSAVDTLN 459
>gb|EAA65513.1| hypothetical protein AN1330.2 [Aspergillus nidulans FGSC A4]
gi|67521746|ref|XP_658934.1| hypothetical protein
AN1330_2 [Aspergillus nidulans FGSC A4]
gi|49086920|ref|XP_405467.1| hypothetical protein
AN1330.2 [Aspergillus nidulans FGSC A4]
Length = 470
Score = 69.7 bits (169), Expect = 8e-11
Identities = 68/272 (25%), Positives = 110/272 (40%), Gaps = 28/272 (10%)
Query: 5 FNEYSWLTTHNS-FAAKGVNWSTGSPVLAFTNQEDSITDQLKNGVRGLMLDMWDYEDTIW 63
++ + + HNS F A+G NQ + QL +GVR L T++
Sbjct: 161 YSNITQVAAHNSPFVAQGN---------VAANQALDVHYQLDDGVRMLQFQTHIMNGTMY 211
Query: 64 LCRGPCTKYTTFQPALNVLKEVRVFLVTHPTEIITIFID--DHVTSGNGVNKVFDKARLR 121
LC C P + L + +L HP +++TI I D+V GN + + + L
Sbjct: 212 LCHTSCDLLNV-GPLEDYLSNITEWLRQHPYDVVTILIGNYDYVDPGNFTTPM-ENSGLM 269
Query: 122 KFWFPVSKMPKNGSDWPTVKTMIRKNYRLIVFTS-NASREASEGIAYEWNYVVESQFGNV 180
F F +P DWPT+ ++I R IVF A++ A + E++ + E+ F
Sbjct: 270 DFVFTPPMIPMGLDDWPTLGSIILSGKRAIVFMDYQANQTAYPWLMDEFSQMWETPFSPT 329
Query: 181 GIKGGSCQNRPESLPMNNATKSLVLMNYFRNVRNHD-------------DEACRDNSSPL 227
RP L +A K + + N+ N+ +E D+
Sbjct: 330 DRDFPCTVQRPPDLAAEDARKRMYMANHNLNIDFSIASLNLLIPNTALLNETNADHGYGS 389
Query: 228 IAMMHVCFRAAGNRWPNFIAVDFYKRGDGGGA 259
+ M NR PNF+ VD+Y G+ G+
Sbjct: 390 VGRMAENCTTLWNRPPNFLLVDYYNEGNFNGS 421
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.136 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 498,677,723
Number of Sequences: 2540612
Number of extensions: 20838471
Number of successful extensions: 36853
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 36749
Number of HSP's gapped (non-prelim): 70
length of query: 273
length of database: 863,360,394
effective HSP length: 126
effective length of query: 147
effective length of database: 543,243,282
effective search space: 79856762454
effective search space used: 79856762454
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0227.9


