

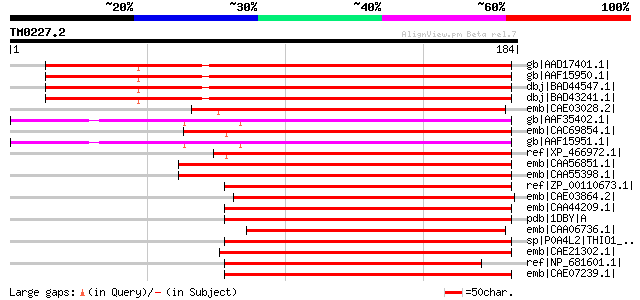
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0227.2
(184 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAD17401.1| putative thioredoxin M [Arabidopsis thaliana] gi|... 174 1e-42
gb|AAF15950.1| thioredoxin m3 [Arabidopsis thaliana] 173 2e-42
dbj|BAD44547.1| putative thioredoxin M [Arabidopsis thaliana] 173 2e-42
dbj|BAD43241.1| putative thioredoxin M [Arabidopsis thaliana] 172 4e-42
emb|CAE03028.2| OSJNBa0084A10.3 [Oryza sativa (japonica cultivar... 132 5e-30
gb|AAF35402.1| thioredoxin m4 [Arabidopsis thaliana] gi|21593734... 121 8e-27
emb|CAC69854.1| putative thioredoxin m2 [Pisum sativum] 121 1e-26
gb|AAF15951.1| thioredoxin m4 [Arabidopsis thaliana] 119 3e-26
ref|XP_466972.1| putative Thioredoxin M-type, chloroplast precur... 118 7e-26
emb|CAA56851.1| thioredoxin m [Chlamydomonas reinhardtii] gi|172... 115 8e-25
emb|CAA55398.1| thioredoxin m [Chlamydomonas reinhardtii] 115 8e-25
ref|ZP_00110673.1| COG0526: Thiol-disulfide isomerase and thiore... 112 4e-24
emb|CAE03864.2| OSJNBa0081C01.10 [Oryza sativa (japonica cultiva... 112 7e-24
emb|CAA44209.1| thioredoxin Ch2 [Chlamydomonas reinhardtii] 111 9e-24
pdb|1DBY|A Chain A, Nmr Structures Of Chloroplast Thioredoxin M ... 111 9e-24
emb|CAA06736.1| thioredoxin M [Oryza sativa] gi|11135471|sp|Q9ZP... 110 2e-23
sp|P0A4L2|THIO1_ANASO Thioredoxin 1 (TRX-1) (Thioredoxin M) gi|6... 109 3e-23
emb|CAE21302.1| Thioredoxin [Prochlorococcus marinus str. MIT 93... 108 6e-23
ref|NP_681601.1| thioredoxin [Thermosynechococcus elongatus BP-1... 108 6e-23
emb|CAE07239.1| Thioredoxin [Synechococcus sp. WH 8102] gi|33865... 108 7e-23
>gb|AAD17401.1| putative thioredoxin M [Arabidopsis thaliana]
gi|18206255|sp|Q9SEU7|TRXM3_ARATH Thioredoxin M-type 3,
chloroplast precursor (TRX-M3)
gi|15226561|ref|NP_179159.1| thioredoxin M-type 3,
chloroplast (TRX-M3) [Arabidopsis thaliana]
gi|51970422|dbj|BAD43903.1| putative thioredoxin M
[Arabidopsis thaliana]
Length = 173
Score = 174 bits (440), Expect = 1e-42
Identities = 89/170 (52%), Positives = 122/170 (71%), Gaps = 3/170 (1%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSF-PFHLRITHKPLHLQNPPHLPRSHKIRS 72
SSSSSS + + H SS S L P SF P LR + + L + R +
Sbjct: 4 SSSSSSICFNPTRFHTARHISSPSRLFPVTSFSPRSLRFSDRRSLLSSSASRLRLSPL-- 61
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+++RA +T+ WE+S+LKS+ PVLVEFY SWCGPCRMVHR+ID+IA +YAG+LNC+L
Sbjct: 62 CVRDSRAAEVTQRSWEDSVLKSETPVLVEFYTSWCGPCRMVHRIIDEIAGDYAGKLNCYL 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+N D D+ +A++YEIKAVPVV+LFK+G+K ++++GTMPKEFY++AIERVL
Sbjct: 122 LNADNDLPVAEEYEIKAVPVVLLFKNGEKRESIMGTMPKEFYISAIERVL 171
>gb|AAF15950.1| thioredoxin m3 [Arabidopsis thaliana]
Length = 173
Score = 173 bits (439), Expect = 2e-42
Identities = 89/170 (52%), Positives = 122/170 (71%), Gaps = 3/170 (1%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSF-PFHLRITHKPLHLQNPPHLPRSHKIRS 72
SSSSSS + + H SS S L P SF P LR + + L + R +
Sbjct: 4 SSSSSSICFNPTRFHTARHISSPSRLFPVTSFSPRSLRFSDRRSLLSSSASRLRLSPL-- 61
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+++RA +T+ WE+S+LKS+ PVLVEFY SWCGPCRMVHR+ID+IA +YAG+LNC+L
Sbjct: 62 CVRDSRAAEVTQRSWEDSVLKSETPVLVEFYTSWCGPCRMVHRIIDEIAGDYAGKLNCYL 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+N D D+ +A++YEIKAVPVV+LFK+G+K ++++GTMPKEFY++AIERVL
Sbjct: 122 LNEDNDLPVAEEYEIKAVPVVLLFKNGEKRESIMGTMPKEFYISAIERVL 171
>dbj|BAD44547.1| putative thioredoxin M [Arabidopsis thaliana]
Length = 173
Score = 173 bits (439), Expect = 2e-42
Identities = 88/170 (51%), Positives = 122/170 (71%), Gaps = 3/170 (1%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSF-PFHLRITHKPLHLQNPPHLPRSHKIRS 72
SSSSSS + + H SS S L P SF P LR + + L + R +
Sbjct: 4 SSSSSSICFNPTRFHTARHISSPSRLFPVTSFSPRSLRFSDRRSLLSSSASRLRLSPL-- 61
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+++RA +T+ WE+S+LKS+ PVLVEFY SWCGPCRMVHR+ID+IA +YAG+LNC+L
Sbjct: 62 CVRDSRAAEVTQRSWEDSVLKSETPVLVEFYTSWCGPCRMVHRIIDEIAGDYAGKLNCYL 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+N D D+ +A++YE+KAVPVV+LFK+G+K ++++GTMPKEFY++AIERVL
Sbjct: 122 LNADNDLPVAEEYEVKAVPVVLLFKNGEKRESIMGTMPKEFYISAIERVL 171
>dbj|BAD43241.1| putative thioredoxin M [Arabidopsis thaliana]
Length = 173
Score = 172 bits (436), Expect = 4e-42
Identities = 88/170 (51%), Positives = 121/170 (70%), Gaps = 3/170 (1%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSF-PFHLRITHKPLHLQNPPHLPRSHKIRS 72
SSSSSS + + H SS S L P SF P LR + + L + R +
Sbjct: 4 SSSSSSICFNPTRFHTARHISSPSRLFPVTSFSPRSLRFSDRRSLLSSSASRLRLSPL-- 61
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+++RA +T+ WE+S+LKS+ PVLVEFY SWCGPCRMVHR+ID+I +YAG+LNC+L
Sbjct: 62 CVRDSRAAEVTQRSWEDSVLKSETPVLVEFYTSWCGPCRMVHRIIDEITGDYAGKLNCYL 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+N D D+ +A++YEIKAVPVV+LFK+G+K ++++GTMPKEFY++AIERVL
Sbjct: 122 LNADNDLPVAEEYEIKAVPVVLLFKNGEKRESIMGTMPKEFYISAIERVL 171
>emb|CAE03028.2| OSJNBa0084A10.3 [Oryza sativa (japonica cultivar-group)]
gi|50924364|ref|XP_472542.1| OSJNBa0084A10.3 [Oryza
sativa (japonica cultivar-group)]
Length = 175
Score = 132 bits (332), Expect = 5e-30
Identities = 59/117 (50%), Positives = 84/117 (71%), Gaps = 3/117 (2%)
Query: 67 SHKIRSLR---QETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKE 123
+H+IR++ A IT W ++ SDIPVL+EF+ASWCGPCRMVHR++D+IA+E
Sbjct: 52 AHRIRTVSCAYSPRGAKTITACSWNEYVICSDIPVLIEFWASWCGPCRMVHRIVDEIAQE 111
Query: 124 YAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
YAGR+ C+ ++TD Q+A Y I+ +P V+LFKDG+K ++ GT+PK YV AIE+
Sbjct: 112 YAGRIKCYKLDTDDYPQVATSYSIERIPTVLLFKDGEKTHSITGTLPKAVYVRAIEK 168
>gb|AAF35402.1| thioredoxin m4 [Arabidopsis thaliana] gi|21593734|gb|AAM65701.1|
thioredoxin m4 [Arabidopsis thaliana]
gi|15795101|dbj|BAB02365.1| thioredoxin m4 [Arabidopsis
thaliana] gi|14030705|gb|AAK53027.1| AT3g15360/MJK13_2
[Arabidopsis thaliana] gi|16974519|gb|AAL31169.1|
AT3g15360/MJK13_2 [Arabidopsis thaliana]
gi|15232567|ref|NP_188155.1| thioredoxin M-type 4,
chloroplast (TRX-M4) [Arabidopsis thaliana]
gi|27735268|sp|Q9SEU6|TRXM4_ARATH Thioredoxin M-type 4,
chloroplast precursor (TRX-M4)
Length = 193
Score = 121 bits (304), Expect = 8e-27
Identities = 68/194 (35%), Positives = 109/194 (56%), Gaps = 15/194 (7%)
Query: 1 MASISTSISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQN 60
MAS+ S+++T S ++S SSS S SR ++P+ F + + L Q+
Sbjct: 1 MASLLDSVTVTRVFSLPIAASVSSSSAAP---SVSRRRISPARFLEFRGLKSSRSLVTQS 57
Query: 61 PP-------HLPRSHKIRSLRQETRATPI-----TKDLWENSILKSDIPVLVEFYASWCG 108
+ R +I Q+T A + + W+ +L+SD+PVLVEF+A WCG
Sbjct: 58 ASLGANRRTRIARGGRIACEAQDTTAAAVEVPNLSDSEWQTKVLESDVPVLVEFWAPWCG 117
Query: 109 PCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGT 168
PCRM+H ++D +AK++AG+ + +NTD A+ Y I++VP V++FK G+K D++IG
Sbjct: 118 PCRMIHPIVDQLAKDFAGKFKFYKINTDESPNTANRYGIRSVPTVIIFKGGEKKDSIIGA 177
Query: 169 MPKEFYVNAIERVL 182
+P+E IER L
Sbjct: 178 VPRETLEKTIERFL 191
>emb|CAC69854.1| putative thioredoxin m2 [Pisum sativum]
Length = 180
Score = 121 bits (303), Expect = 1e-26
Identities = 53/121 (43%), Positives = 81/121 (66%), Gaps = 2/121 (1%)
Query: 64 LPRSHKIRSLRQET--RATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIA 121
+PR ++ ++T IT W++ +++SD PVLVEF+A WCGPCRM+H +ID++A
Sbjct: 60 VPRGGRVLCEARDTAVEVASITDGNWQSLVIESDTPVLVEFWAPWCGPCRMMHPIIDELA 119
Query: 122 KEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERV 181
KEY G+ C+ +NTD A Y I+++P V+ FKDG+K DA+IG++PK + IE+
Sbjct: 120 KEYVGKFKCYKLNTDESPSTATRYGIRSIPTVIFFKDGEKKDAIIGSVPKASLITTIEKF 179
Query: 182 L 182
L
Sbjct: 180 L 180
>gb|AAF15951.1| thioredoxin m4 [Arabidopsis thaliana]
Length = 193
Score = 119 bits (299), Expect = 3e-26
Identities = 67/194 (34%), Positives = 108/194 (55%), Gaps = 15/194 (7%)
Query: 1 MASISTSISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQN 60
MAS+ S+++T S ++S SSS S SR ++P+ F + + L Q+
Sbjct: 1 MASLLDSVTVTRVFSLPIAASVSSSSAAP---SVSRRRISPARFLEFRGLKSSRSLVTQS 57
Query: 61 PP-------HLPRSHKIRSLRQETRATPI-----TKDLWENSILKSDIPVLVEFYASWCG 108
+ R +I Q+T A + + W+ +L+SD+PVLVEF+A WCG
Sbjct: 58 ASLGANRRTRIARGGRIACEAQDTTAAAVEVPNLSDSEWQTKVLESDVPVLVEFWAPWCG 117
Query: 109 PCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGT 168
PCRM+H ++D +AK++AG+ + +NTD + Y I++VP V++FK G+K D++IG
Sbjct: 118 PCRMIHPIVDQLAKDFAGKFKFYKINTDESPNTPNRYGIRSVPTVIIFKGGEKKDSIIGA 177
Query: 169 MPKEFYVNAIERVL 182
+P+E IER L
Sbjct: 178 VPRETLEKTIERFL 191
>ref|XP_466972.1| putative Thioredoxin M-type, chloroplast precursor [Oryza sativa
(japonica cultivar-group)] gi|49388235|dbj|BAD25355.1|
putative Thioredoxin M-type, chloroplast precursor
[Oryza sativa (japonica cultivar-group)]
Length = 173
Score = 118 bits (296), Expect = 7e-26
Identities = 50/110 (45%), Positives = 83/110 (75%), Gaps = 2/110 (1%)
Query: 75 QET--RATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
QET + +TK W++ +++S++PVLVEF+ASWCGPC+M+ VI ++KEY G+LNC+
Sbjct: 62 QETSFQVPDVTKSTWQSLVVESELPVLVEFWASWCGPCKMIDPVIGKLSKEYEGKLNCYK 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+NTD + IA + I+++P +M+FK+G+K DAVIG +P+ V++I++ +
Sbjct: 122 LNTDENPDIATQFGIRSIPTMMIFKNGEKKDAVIGAVPESTLVSSIDKYI 171
>emb|CAA56851.1| thioredoxin m [Chlamydomonas reinhardtii]
gi|1729935|sp|P23400|TRXM_CHLRE Thioredoxin M-type,
chloroplast precursor (TRX-M) (Thioredoxin CH2)
Length = 140
Score = 115 bits (287), Expect = 8e-25
Identities = 49/121 (40%), Positives = 78/121 (63%)
Query: 62 PHLPRSHKIRSLRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIA 121
P R+ RS+ A + D ++N +L+S +PVLV+F+A WCGPCR++ V+D+IA
Sbjct: 19 PAFARAAPRRSVVVRAEAGAVNDDTFKNVVLESSVPVLVDFWAPWCGPCRIIAPVVDEIA 78
Query: 122 KEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERV 181
EY +L C +NTD +A +Y I+++P +M+FK G+KC+ +IG +PK V +E+
Sbjct: 79 GEYKDKLKCVKLNTDESPNVASEYGIRSIPTIMVFKGGKKCETIIGAVPKATIVQTVEKY 138
Query: 182 L 182
L
Sbjct: 139 L 139
>emb|CAA55398.1| thioredoxin m [Chlamydomonas reinhardtii]
Length = 128
Score = 115 bits (287), Expect = 8e-25
Identities = 49/121 (40%), Positives = 78/121 (63%)
Query: 62 PHLPRSHKIRSLRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIA 121
P R+ RS+ A + D ++N +L+S +PVLV+F+A WCGPCR++ V+D+IA
Sbjct: 7 PAFARAAPRRSVVVRAEAGAVNDDTFKNVVLESSVPVLVDFWAPWCGPCRIIAPVVDEIA 66
Query: 122 KEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERV 181
EY +L C +NTD +A +Y I+++P +M+FK G+KC+ +IG +PK V +E+
Sbjct: 67 GEYKDKLKCVKLNTDESPNVASEYGIRSIPTIMVFKGGKKCETIIGAVPKATIVQTVEKY 126
Query: 182 L 182
L
Sbjct: 127 L 127
>ref|ZP_00110673.1| COG0526: Thiol-disulfide isomerase and thioredoxins [Nostoc
punctiforme PCC 73102]
Length = 107
Score = 112 bits (281), Expect = 4e-24
Identities = 47/104 (45%), Positives = 73/104 (70%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T ++ +L SD+PVLV+F+A WCGPCRMV V+D+I+++Y G++ VNTD +
Sbjct: 4 AAQVTDSSFKQEVLDSDVPVLVDFWAPWCGPCRMVAPVVDEISEQYKGQIKVVKVNTDEN 63
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
Q+A Y I+++P +M+FKDGQK D V+G +PK + +E+ L
Sbjct: 64 PQVASQYGIRSIPTLMIFKDGQKVDMVVGAVPKSTLASTLEKYL 107
>emb|CAE03864.2| OSJNBa0081C01.10 [Oryza sativa (japonica cultivar-group)]
gi|38346628|emb|CAD41211.2| OSJNBa0074L08.22 [Oryza
sativa (japonica cultivar-group)]
gi|50926654|ref|XP_473274.1| OSJNBa0074L08.22 [Oryza
sativa (japonica cultivar-group)]
Length = 180
Score = 112 bits (279), Expect = 7e-24
Identities = 42/102 (41%), Positives = 77/102 (75%)
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+TK W++ +++S++PVLV ++A+WCGPC+M+ V+ ++KEY G+L C+ +NTD + I
Sbjct: 78 VTKSTWQSLVMESELPVLVGYWATWCGPCKMIDPVVGKLSKEYEGKLKCYKLNTDENPDI 137
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLK 183
A Y ++++P +M+FK+G+K DAVIG +P+ + +IE+ ++
Sbjct: 138 ASQYGVRSIPTMMIFKNGEKKDAVIGAVPESTLIASIEKFVE 179
>emb|CAA44209.1| thioredoxin Ch2 [Chlamydomonas reinhardtii]
Length = 106
Score = 111 bits (278), Expect = 9e-24
Identities = 45/104 (43%), Positives = 72/104 (68%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A + D ++N +L+S +PVLV+F+A WCGPCR++ V+D+IA EY +L C +NTD
Sbjct: 2 AGAVNDDTFKNVVLESSVPVLVDFWAPWCGPCRIIAPVVDEIAGEYKDKLKCVKLNTDES 61
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+A +Y I+++P +M+FK G+KC+ +IG +PK V +E+ L
Sbjct: 62 PNVASEYGIRSIPTIMVFKGGKKCETIIGAVPKATIVQTVEKYL 105
>pdb|1DBY|A Chain A, Nmr Structures Of Chloroplast Thioredoxin M Ch2 From The
Green Alga Chlamydomonas Reinhardtii
Length = 107
Score = 111 bits (278), Expect = 9e-24
Identities = 45/104 (43%), Positives = 72/104 (68%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A + D ++N +L+S +PVLV+F+A WCGPCR++ V+D+IA EY +L C +NTD
Sbjct: 3 AGAVNDDTFKNVVLESSVPVLVDFWAPWCGPCRIIAPVVDEIAGEYKDKLKCVKLNTDES 62
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+A +Y I+++P +M+FK G+KC+ +IG +PK V +E+ L
Sbjct: 63 PNVASEYGIRSIPTIMVFKGGKKCETIIGAVPKATIVQTVEKYL 106
>emb|CAA06736.1| thioredoxin M [Oryza sativa] gi|11135471|sp|Q9ZP20|TRXM_ORYSA
Thioredoxin M-type, chloroplast precursor (TRX-M)
Length = 172
Score = 110 bits (274), Expect = 2e-23
Identities = 46/94 (48%), Positives = 69/94 (72%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
W++ +L S+ PVLVEF+A WCGPCRM+ VID++AKEY G++ C VNTD IA +Y
Sbjct: 75 WDSMVLGSEAPVLVEFWAPWCGPCRMIAPVIDELAKEYVGKIKCCKVNTDDSPNIATNYG 134
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
I+++P V++FK+G+K ++VIG +PK I++
Sbjct: 135 IRSIPTVLMFKNGEKKESVIGAVPKTTLATIIDK 168
>sp|P0A4L2|THIO1_ANASO Thioredoxin 1 (TRX-1) (Thioredoxin M)
gi|61248483|sp|P0A4L1|THIO1_ANASP Thioredoxin 1 (TRX-1)
(Thioredoxin M) gi|17135030|dbj|BAB77576.1| thioredoxin
[Nostoc sp. PCC 7120] gi|7427614|pir||TXAI thioredoxin 1
- Anabaena sp gi|17227548|ref|NP_484096.1| thioredoxin
[Nostoc sp. PCC 7120] gi|45510273|ref|ZP_00162605.1|
COG0526: Thiol-disulfide isomerase and thioredoxins
[Anabaena variabilis ATCC 29413]
gi|142118|gb|AAA22049.1| thioredoxin
Length = 107
Score = 109 bits (273), Expect = 3e-23
Identities = 47/104 (45%), Positives = 71/104 (68%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T ++ +L SD+PVLV+F+A WCGPCRMV V+D+IA++Y G++ VNTD +
Sbjct: 4 AAQVTDSTFKQEVLDSDVPVLVDFWAPWCGPCRMVAPVVDEIAQQYEGKIKVVKVNTDEN 63
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
Q+A Y I+++P +M+FK GQK D V+G +PK +E+ L
Sbjct: 64 PQVASQYGIRSIPTLMIFKGGQKVDMVVGAVPKTTLSQTLEKHL 107
>emb|CAE21302.1| Thioredoxin [Prochlorococcus marinus str. MIT 9313]
gi|33863398|ref|NP_894958.1| Thioredoxin
[Prochlorococcus marinus str. MIT 9313]
Length = 107
Score = 108 bits (271), Expect = 6e-23
Identities = 46/106 (43%), Positives = 71/106 (66%)
Query: 77 TRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTD 136
+ A +T +E +L+SD+PVLV+F+A WCGPCRMV ++D+IAKE+ ++ F +NTD
Sbjct: 2 SNAAAVTDASFEQDVLQSDVPVLVDFWAPWCGPCRMVAPIVDEIAKEFESKIKVFKLNTD 61
Query: 137 TDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+ +A Y I+++P +M+FK GQK D V+G +PK I + L
Sbjct: 62 ENPNVASQYGIRSIPTLMVFKGGQKVDTVVGAVPKATLSGTISKYL 107
>ref|NP_681601.1| thioredoxin [Thermosynechococcus elongatus BP-1]
gi|22294533|dbj|BAC08363.1| thioredoxin
[Thermosynechococcus elongatus BP-1]
Length = 107
Score = 108 bits (271), Expect = 6e-23
Identities = 47/93 (50%), Positives = 67/93 (71%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T +E +L SDIPVLV+F+A WCGPCRMV V+D+IA EY GR+ VNTD +
Sbjct: 4 ALSVTDATFEEEVLNSDIPVLVDFWAPWCGPCRMVAPVVDEIANEYQGRVKVVKVNTDEN 63
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPK 171
++A D+ I+++P +M+FK GQK D ++G +PK
Sbjct: 64 SKVATDFGIRSIPTLMIFKGGQKVDILVGAVPK 96
>emb|CAE07239.1| Thioredoxin [Synechococcus sp. WH 8102]
gi|33865258|ref|NP_896817.1| Thioredoxin [Synechococcus
sp. WH 8102]
Length = 107
Score = 108 bits (270), Expect = 7e-23
Identities = 46/104 (44%), Positives = 71/104 (68%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T +E +L+SD+PVLV+F+A WCGPCRMV ++++IAKE+ G++ F +NTD +
Sbjct: 4 AAAVTDASFEQDVLQSDVPVLVDFWAPWCGPCRMVAPIVEEIAKEFDGQIKVFKLNTDEN 63
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+A Y I+++P +M+FK GQK D V+G +PK I + L
Sbjct: 64 PNVASQYGIRSIPTLMVFKGGQKVDTVVGAVPKATLSGTISKYL 107
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.131 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 305,040,911
Number of Sequences: 2540612
Number of extensions: 12435879
Number of successful extensions: 178770
Number of sequences better than 10.0: 2806
Number of HSP's better than 10.0 without gapping: 2164
Number of HSP's successfully gapped in prelim test: 660
Number of HSP's that attempted gapping in prelim test: 147745
Number of HSP's gapped (non-prelim): 12463
length of query: 184
length of database: 863,360,394
effective HSP length: 120
effective length of query: 64
effective length of database: 558,486,954
effective search space: 35743165056
effective search space used: 35743165056
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0227.2


