

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0223.2
(365 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
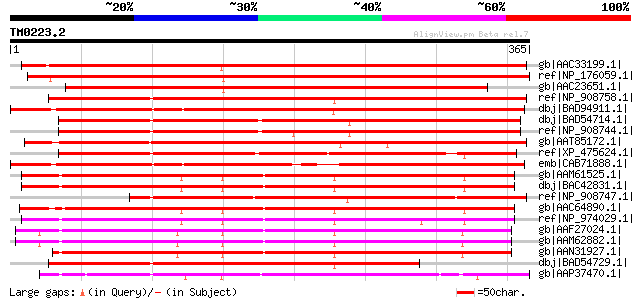
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC33199.1| Similar to nodulins and lipase [Arabidopsis thali... 479 e-134
ref|NP_176059.1| GDSL-motif lipase/hydrolase family protein [Ara... 459 e-128
gb|AAC23651.1| lipase homolog [Arabidopsis thaliana] gi|25336281... 405 e-111
ref|NP_908758.1| putative lipase homolog [Oryza sativa (japonica... 382 e-104
dbj|BAD94911.1| putative protein [Arabidopsis thaliana] gi|45773... 361 2e-98
dbj|BAD54714.1| putative early nodule-specific protein ENOD8 [Or... 353 5e-96
ref|NP_908744.1| putative lipase homolog [Oryza sativa (japonica... 344 2e-93
gb|AAT85172.1| hypothetical protein [Oryza sativa (japonica cult... 338 2e-91
ref|XP_475624.1| putative GDSL-like lipase/acylhydrolase [Oryza ... 318 1e-85
emb|CAB71888.1| putative protein [Arabidopsis thaliana] gi|15228... 314 3e-84
gb|AAM61525.1| early nodule-specific protein, putative [Arabidop... 307 3e-82
dbj|BAC42831.1| unknown protein [Arabidopsis thaliana] gi|289735... 307 3e-82
ref|NP_908747.1| putative lipase homolog [Oryza sativa (japonica... 306 6e-82
gb|AAC64890.1| Similar to nodulins and lipase homolog F14J9.5 gi... 298 2e-79
ref|NP_974029.1| GDSL-motif lipase/hydrolase family protein [Ara... 293 6e-78
gb|AAF27024.1| putative nodulin [Arabidopsis thaliana] gi|158102... 278 1e-73
gb|AAM62882.1| putative nodulin [Arabidopsis thaliana] 278 1e-73
gb|AAN31927.1| putative nodulin [Arabidopsis thaliana] 278 2e-73
dbj|BAD54729.1| putative lipase homolog [Oryza sativa (japonica ... 273 8e-72
gb|AAP37470.1| ENSP-like protein [Hevea brasiliensis] gi|5131578... 272 1e-71
>gb|AAC33199.1| Similar to nodulins and lipase [Arabidopsis thaliana]
gi|28393967|gb|AAO42391.1| putative lipase [Arabidopsis
thaliana] gi|27754509|gb|AAO22702.1| putative lipase
[Arabidopsis thaliana] gi|15217506|ref|NP_172410.1|
GDSL-motif lipase/hydrolase family protein [Arabidopsis
thaliana] gi|25336317|pir||B86227 hypothetical protein
[imported] - Arabidopsis thaliana
Length = 370
Score = 479 bits (1233), Expect = e-134
Identities = 228/358 (63%), Positives = 279/358 (77%), Gaps = 4/358 (1%)
Query: 9 LHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHR 68
L +L F+ I + AV C VP V+F FGDSNSDTGGL +GLG+ I LPNGR+FF R
Sbjct: 12 LLVLLPFILILRQNLAVA-GGCQVPPVIFNFGDSNSDTGGLVAGLGYSIGLPNGRSFFQR 70
Query: 69 STGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVPFSLNI 128
STGRLSDGRLVID LCQSLNT L PYLDSL GS F NGANFA+VGSSTLP+YVPF+LNI
Sbjct: 71 STGRLSDGRLVIDFLCQSLNTSLLNPYLDSLVGSKFQNGANFAIVGSSTLPRYVPFALNI 130
Query: 129 QVMQFLHFKARTLELVSAG---AKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVI 185
Q+MQFLHFK+R LEL S + +I + GFR ALY+IDIGQND+ADSF+K L+Y +V+
Sbjct: 131 QLMQFLHFKSRALELASISDPLKEMMIGESGFRNALYMIDIGQNDIADSFSKGLSYSRVV 190
Query: 186 KKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSYNSA 245
K IP VI+EI++A+K LY+EG RKFWVHNTGPLGCLP+ L++ K D GCL++YN+A
Sbjct: 191 KLIPNVISEIKSAIKILYDEGGRKFWVHNTGPLGCLPQKLSMVHSKGFDKHGCLATYNAA 250
Query: 246 ARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPY 305
A+LFNE L H + LRT LK+A +VYVDIYAIK+DLIAN+ YGF PL CCG+GGPPY
Sbjct: 251 AKLFNEGLDHMCRDLRTELKEANIVYVDIYAIKYDLIANSNNYGFEKPLMACCGYGGPPY 310
Query: 306 NFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFC 363
N+++ +TCG G + CDEGSR+++WDG H+TE AN +A K+LS +STP TPF FFC
Sbjct: 311 NYNVNITCGNGGSKSCDEGSRFISWDGIHYTETANAIVAMKVLSMQHSTPPTPFHFFC 368
>ref|NP_176059.1| GDSL-motif lipase/hydrolase family protein [Arabidopsis thaliana]
gi|25404142|pir||D96608 hypothetical protein F25P12.90
[imported] - Arabidopsis thaliana
gi|9954747|gb|AAG09098.1| Similar to nodulins
[Arabidopsis thaliana]
Length = 373
Score = 459 bits (1180), Expect = e-128
Identities = 216/363 (59%), Positives = 274/363 (74%), Gaps = 10/363 (2%)
Query: 13 FLFLWITSSSAAVMM-------SKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTF 65
F F +IT S A+++ + C V+F FGDSNSDTGGL +GLG+PI PNGR F
Sbjct: 11 FSFFFITLVSLALLILRQPSRAASCTARPVIFNFGDSNSDTGGLVAGLGYPIGFPNGRLF 70
Query: 66 FHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVPFS 125
F RSTGRLSDGRL+ID LCQSLNT L PYLDSL + F NGANFA+ GS TLPK VPFS
Sbjct: 71 FRRSTGRLSDGRLLIDFLCQSLNTSLLRPYLDSLGRTRFQNGANFAIAGSPTLPKNVPFS 130
Query: 126 LNIQVMQFLHFKARTLELVSAGAK---NVINDEGFRAALYLIDIGQNDLADSFAKNLTYV 182
LNIQV QF HFK+R+LEL S+ I++ GF+ ALY+IDIGQND+A SFA+ +Y
Sbjct: 131 LNIQVKQFSHFKSRSLELASSSNSLKGMFISNNGFKNALYMIDIGQNDIARSFARGNSYS 190
Query: 183 QVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSY 242
Q +K IP +ITEI++++K LY+EG R+FW+HNTGPLGCLP+ L++ + KDLD GCL SY
Sbjct: 191 QTVKLIPQIITEIKSSIKRLYDEGGRRFWIHNTGPLGCLPQKLSMVKSKDLDQHGCLVSY 250
Query: 243 NSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGG 302
NSAA LFN+ L H ++LRT L+DAT++Y+DIYAIK+ LIAN+ +YGF +PL CCG+GG
Sbjct: 251 NSAATLFNQGLDHMCEELRTELRDATIIYIDIYAIKYSLIANSNQYGFKSPLMACCGYGG 310
Query: 303 PPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFF 362
PYN+++++TCG G VC+EGSR+++WDG H+TE AN +A K+LS YS P TPF FF
Sbjct: 311 TPYNYNVKITCGHKGSNVCEEGSRFISWDGIHYTETANAIVAMKVLSMHYSKPPTPFHFF 370
Query: 363 CHR 365
C R
Sbjct: 371 CRR 373
>gb|AAC23651.1| lipase homolog [Arabidopsis thaliana] gi|25336281|pir||T52366
lipase-like protein Lip-4 [imported] - Arabidopsis
thaliana (fragment)
Length = 301
Score = 405 bits (1040), Expect = e-111
Identities = 188/300 (62%), Positives = 237/300 (78%), Gaps = 3/300 (1%)
Query: 40 GDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSL 99
GDSNSDTGGL +GLG+PI PNGR FF RSTGRLSDGRL+ID LCQSLNT L PYLDSL
Sbjct: 1 GDSNSDTGGLVAGLGYPIGFPNGRLFFRRSTGRLSDGRLLIDFLCQSLNTSLLRPYLDSL 60
Query: 100 SGSSFTNGANFAVVGSSTLPKYVPFSLNIQVMQFLHFKARTLELVSAGAK---NVINDEG 156
+ F N ANFA+ GSSTLPK VPFSLNIQV QF HFK+R+LEL S+ I++ G
Sbjct: 61 GRTRFQNVANFAIAGSSTLPKNVPFSLNIQVKQFSHFKSRSLELASSSNSLKGMFISNNG 120
Query: 157 FRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTG 216
F+ ALY+IDIGQND+A SFA+ +Y Q +K IP +ITEI++++K LY+E R+FW+HNTG
Sbjct: 121 FKNALYMIDIGQNDIALSFARGNSYSQTVKLIPQIITEIKSSIKRLYDEEGRRFWIHNTG 180
Query: 217 PLGCLPKILALAQKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYA 276
PLGCLP+ L++ + KDLD GCL SYNSAA LFN+ L H ++LRT L+DAT++Y+DIYA
Sbjct: 181 PLGCLPQKLSMVKSKDLDQLGCLVSYNSAATLFNQGLDHMCEELRTELRDATIIYIDIYA 240
Query: 277 IKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHT 336
IK+ LIAN+ +YGF +PL CCG+GG PYN+++++TCG G VC EGSR+++WDG H+T
Sbjct: 241 IKYSLIANSNQYGFKSPLMACCGYGGTPYNYNVKITCGHKGSNVCKEGSRFISWDGIHYT 300
>ref|NP_908758.1| putative lipase homolog [Oryza sativa (japonica cultivar-group)]
Length = 384
Score = 382 bits (980), Expect = e-104
Identities = 184/340 (54%), Positives = 246/340 (72%), Gaps = 5/340 (1%)
Query: 28 SKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVIDLLCQSL 87
+ C V+F FGDSN+DTGG+ +G+G+ LP GR FF R+TGRL DGRLVID LC+SL
Sbjct: 44 ASCARRPVVFAFGDSNTDTGGIAAGMGYYFPLPEGRAFFRRATGRLCDGRLVIDHLCESL 103
Query: 88 NTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVPFSLNIQVMQFLHFKARTLELVSAG 147
N +L+PYL+ L G+ FTNGANFA+ G++T P+ FSL+IQV QF+HFK R+LEL S G
Sbjct: 104 NMSYLSPYLEPL-GTDFTNGANFAISGAATAPRNAAFSLHIQVQQFIHFKQRSLELASRG 162
Query: 148 AKNVINDEGFRAALYLIDIGQNDLADSF-AKNLTYVQVIK-KIPTVITEIENAVKSLYNE 205
++ +GFR ALYLIDIGQNDL+ +F A L Y V++ + P +++EI++A++SLY
Sbjct: 163 EAVPVDADGFRNALYLIDIGQNDLSAAFSAGGLPYDDVVRQRFPAILSEIKDAIQSLYYN 222
Query: 206 GARKFWVHNTGPLGCLPKILAL--AQKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTR 263
GA+ W+H TGPLGCLP+ LA+ A DLD GCL + N+ A FN L +L ++
Sbjct: 223 GAKNLWIHGTGPLGCLPQKLAVPRADDGDLDPSGCLKTLNAGAYEFNSQLSSICDQLSSQ 282
Query: 264 LKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDE 323
L+ AT+V+ DI AIK+DLIAN + YGF PL CCG GGPPYN+D V+C GY+VC++
Sbjct: 283 LRGATIVFTDILAIKYDLIANHSSYGFEEPLMACCGHGGPPYNYDFNVSCLGAGYRVCED 342
Query: 324 GSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFC 363
GS++V+WDG H+T+AAN +A KILS DYS P+ PF +FC
Sbjct: 343 GSKFVSWDGVHYTDAANAVVAGKILSADYSRPKLPFSYFC 382
>dbj|BAD94911.1| putative protein [Arabidopsis thaliana] gi|45773932|gb|AAS76770.1|
At3g62280 [Arabidopsis thaliana]
Length = 365
Score = 361 bits (927), Expect = 2e-98
Identities = 182/365 (49%), Positives = 247/365 (66%), Gaps = 8/365 (2%)
Query: 1 MSSALAFSLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLP 60
+SS F L L L ++S + +K +L FGDSNSDTGG+ +G+G PI LP
Sbjct: 5 VSSLQCFFLVLCLSLLVCSNSETSYKSNK---KPILINFGDSNSDTGGVLAGVGLPIGLP 61
Query: 61 NGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPK 120
+G TFFHR TGRL DGRL++D C+ L +L+PYLDSLS +F G NFAV G++ LP
Sbjct: 62 HGITFFHRGTGRLGDGRLIVDFYCEHLKMTYLSPYLDSLS-PNFKRGVNFAVSGATALPI 120
Query: 121 YVPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADS-FAKNL 179
+ F L IQ+ QF+HFK R+ EL+S+G +++I+D GFR ALY+IDIGQNDL + + NL
Sbjct: 121 F-SFPLAIQIRQFVHFKNRSQELISSGRRDLIDDNGFRNALYMIDIGQNDLLLALYDSNL 179
Query: 180 TYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILA--LAQKKDLDLFG 237
TY V++KIP+++ EI+ A++++Y G RKFWVHNTGPLGC PK LA L DLD G
Sbjct: 180 TYAPVVEKIPSMLLEIKKAIQTVYLYGGRKFWVHNTGPLGCAPKELAIHLHNDSDLDPIG 239
Query: 238 CLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVC 297
C +N A+ FN+ L +LR++ KDATLVYVDIY+IK+ L A+ YGF +PL C
Sbjct: 240 CFRVHNEVAKAFNKGLLSLCNELRSQFKDATLVYVDIYSIKYKLSADFKLYGFVDPLMAC 299
Query: 298 CGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRT 357
CG+GG P N+D + TCGQPG +C + ++ + WDG H+TEAAN F+ +L+ YS P+
Sbjct: 300 CGYGGRPNNYDRKATCGQPGSTICRDVTKAIVWDGVHYTEAANRFVVDAVLTNRYSYPKN 359
Query: 358 PFEFF 362
+ F
Sbjct: 360 SLDRF 364
>dbj|BAD54714.1| putative early nodule-specific protein ENOD8 [Oryza sativa
(japonica cultivar-group)]
Length = 436
Score = 353 bits (906), Expect = 5e-96
Identities = 170/327 (51%), Positives = 232/327 (69%), Gaps = 5/327 (1%)
Query: 35 VLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTP 94
V+F FGDSNSDTGG+ + +G I LP GRT+F R TGR+SDGRLVID +C+SLNT L+P
Sbjct: 100 VIFNFGDSNSDTGGMAAAMGLNIALPEGRTYFRRPTGRISDGRLVIDFICESLNTPHLSP 159
Query: 95 YLDSLSGSSFTNGANFAVVGSSTLPKYVPFSLNIQVMQFLHFKARTLELVSAGAKNVIND 154
YL SL GS F+NG NFA+ GS+ P FSL++Q+ QFL+F+ R++EL++ G + I+
Sbjct: 160 YLKSL-GSDFSNGVNFAIGGSTATPGGSTFSLDVQLHQFLYFRTRSIELINQGVRTPIDR 218
Query: 155 EGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHN 214
+GFR A+Y IDIGQNDLA NL Y QV+ KIPT++ I+ +++LY G RKFWVH
Sbjct: 219 DGFRNAIYTIDIGQNDLAAYM--NLPYDQVLAKIPTIVAHIKYTIEALYGHGGRKFWVHG 276
Query: 215 TGPLGCLPKILALAQKKDLDLFG--CLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYV 272
TG LGCLP+ L++ + D DL G CL +YN+AAR FN L + ++LR R+ DA +V+
Sbjct: 277 TGALGCLPQKLSIPRDDDSDLDGNGCLKTYNAAAREFNAQLGAACRRLRQRMADAAVVFT 336
Query: 273 DIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDG 332
D+YA K+DL+AN T +G PL CCG GGPPYN++ C ++CD G+R+ +WDG
Sbjct: 337 DVYAAKYDLVANHTLHGIERPLMACCGNGGPPYNYNHFKMCMSAEMELCDMGARFASWDG 396
Query: 333 THHTEAANTFIASKILSTDYSTPRTPF 359
H+TEAAN +A+++L+ +YSTP F
Sbjct: 397 VHYTEAANAIVAARVLTGEYSTPPVRF 423
>ref|NP_908744.1| putative lipase homolog [Oryza sativa (japonica cultivar-group)]
Length = 443
Score = 344 bits (883), Expect = 2e-93
Identities = 170/334 (50%), Positives = 231/334 (68%), Gaps = 12/334 (3%)
Query: 35 VLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTP 94
V+F FGDSNSDTGG+ + +G I LP GRT+F R TGR+SDGRLVID +C+SLNT L+P
Sbjct: 100 VIFNFGDSNSDTGGMAAAMGLNIALPEGRTYFRRPTGRISDGRLVIDFICESLNTPHLSP 159
Query: 95 YLDSLSGSSFTNGANFAVVGSSTLPKYVPFSLNIQVMQFLHFKARTLELVSAGAKNVIND 154
YL SL GS F+NG NFA+ GS+ P FSL++Q+ QFL+F+ R++EL++ G + I+
Sbjct: 160 YLKSL-GSDFSNGVNFAIGGSTATPGGSTFSLDVQLHQFLYFRTRSIELINQGVRTPIDR 218
Query: 155 EGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIENA-------VKSLYNEGA 207
+GFR A+Y IDIGQNDLA NL Y QV+ KIPT++ I +++LY G
Sbjct: 219 DGFRNAIYTIDIGQNDLAAYM--NLPYDQVLAKIPTIVAHINLMTQLTSWWMQALYGHGG 276
Query: 208 RKFWVHNTGPLGCLPKILALAQKKDLDLFG--CLSSYNSAARLFNEALYHSSQKLRTRLK 265
RKFWVH TG LGCLP+ L++ + D DL G CL +YN+AAR FN L + ++LR R+
Sbjct: 277 RKFWVHGTGALGCLPQKLSIPRDDDSDLDGNGCLKTYNAAAREFNAQLGAACRRLRQRMA 336
Query: 266 DATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCDEGS 325
DA +V+ D+YA K+DL+AN T +G PL CCG GGPPYN++ C ++CD G+
Sbjct: 337 DAAVVFTDVYAAKYDLVANHTLHGIERPLMACCGNGGPPYNYNHFKMCMSAEMELCDMGA 396
Query: 326 RYVNWDGTHHTEAANTFIASKILSTDYSTPRTPF 359
R+ +WDG H+TEAAN +A+++L+ +YSTP F
Sbjct: 397 RFASWDGVHYTEAANAIVAARVLTGEYSTPPVRF 430
>gb|AAT85172.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 363
Score = 338 bits (867), Expect = 2e-91
Identities = 178/360 (49%), Positives = 231/360 (63%), Gaps = 12/360 (3%)
Query: 11 LLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRST 70
LL L L + + +C V+F FGDSNSDTG L + GF + P GR FFHR T
Sbjct: 8 LLLLMLLQCGGLPGIAVGRC----VVFNFGDSNSDTGSLPAAFGFYLGPPAGRRFFHRQT 63
Query: 71 GRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKY-VPFSLNIQ 129
GR SDGRL ID + + L +L+PY++S SGS FT+G NFAV G++ K +P L+ Q
Sbjct: 64 GRWSDGRLYIDFIAEKLKISYLSPYMES-SGSDFTSGVNFAVAGAAVTQKSAIPLGLDTQ 122
Query: 130 VMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIP 189
V QFLHFK RT EL GA ++I + FR A+Y IDIGQND+ +F NLT +V +++
Sbjct: 123 VNQFLHFKNRTRELRPRGAGSMIAESEFRDAVYAIDIGQNDITLAFLANLTLPEVERELA 182
Query: 190 TVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKK--DLDLFGCLSSYNSAAR 247
+ +AV++L GARKFWV+NTGP+GCLP+ LAL QK +LD GCL+ YN+AAR
Sbjct: 183 ASAAMVADAVRALRASGARKFWVYNTGPIGCLPQTLALRQKPGDELDAAGCLAEYNAAAR 242
Query: 248 LFNEALYHSSQKLRTRL----KDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGP 303
FN L + ++L L AT+V D+YAIK++L AN ++YGF PL CCG GGP
Sbjct: 243 SFNAELAAACRRLAAELGGGEDGATVVCTDMYAIKYELFANHSRYGFERPLMACCGHGGP 302
Query: 304 PYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFC 363
PYN+ TCGQP C EG R+V WDG H+TE AN +A KILS D+S+PRT + C
Sbjct: 303 PYNYANLKTCGQPTATACPEGERHVIWDGVHYTEDANAIVARKILSGDFSSPRTKLKALC 362
>ref|XP_475624.1| putative GDSL-like lipase/acylhydrolase [Oryza sativa (japonica
cultivar-group)] gi|55168050|gb|AAV43918.1| putative
GDSL lipase/acylhydrolase [Oryza sativa (japonica
cultivar-group)]
Length = 431
Score = 318 bits (816), Expect = 1e-85
Identities = 161/326 (49%), Positives = 219/326 (66%), Gaps = 15/326 (4%)
Query: 35 VLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTP 94
VLF FGDSNSDTGG+ + G I P GRT+FH TGRLSDGR++ID +C+SLNTR L P
Sbjct: 100 VLFNFGDSNSDTGGVAAAGGIRIMPPEGRTYFHHPTGRLSDGRVIIDFICESLNTRELNP 159
Query: 95 YLDSLSGSSFTNGANFAVVGSSTLPKYVPFSLNIQVMQFLHFKARTLELVSAGAKNVIND 154
YL S+ GS ++NG NFA+ GS+ P+SLN+QV QF++FK R+LEL G K ++
Sbjct: 160 YLKSI-GSDYSNGVNFAMAGSTVSHGVSPYSLNVQVDQFVYFKHRSLELFERGQKGPVSK 218
Query: 155 EGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHN 214
EGF ALY++DIG ND+A + KK +++EI++A++ LY+ GARKFW+H
Sbjct: 219 EGFENALYMMDIGHNDVAG--VMHTPSDNWDKKFSKIVSEIKDAIRILYDNGARKFWIHG 276
Query: 215 TGPLGCLPKILALAQKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDI 274
TG LGCLP L + +K + D GCL++YN AAR FN+ L H ++R +LK+AT+VY D+
Sbjct: 277 TGALGCLPA-LVVQEKGEHDAHGCLANYNKAARQFNKKLSHLCDEMRLQLKNATVVYTDM 335
Query: 275 YAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGY----QVCDEGSRYVNW 330
+AIK+D +AN TKYG PL VCCG GGPPYNF +PG +C+ GS+ ++W
Sbjct: 336 FAIKYDFVANHTKYGIKWPLMVCCGNGGPPYNF-------KPGKFGCDDLCEPGSKVLSW 388
Query: 331 DGTHHTEAANTFIASKILSTDYSTPR 356
DG H T+ + A +S +YS P+
Sbjct: 389 DGVHFTDFGSGLAAKLAMSGEYSKPK 414
>emb|CAB71888.1| putative protein [Arabidopsis thaliana]
gi|15228721|ref|NP_191787.1| GDSL-motif lipase/hydrolase
family protein [Arabidopsis thaliana]
gi|11358071|pir||T48020 hypothetical protein T17J13.240
- Arabidopsis thaliana
Length = 343
Score = 314 bits (804), Expect = 3e-84
Identities = 165/363 (45%), Positives = 229/363 (62%), Gaps = 26/363 (7%)
Query: 1 MSSALAFSLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLP 60
+SS F L L L ++S + +K +L FGDSNSDTGG+ +G+G PI LP
Sbjct: 5 VSSLQCFFLVLCLSLLVCSNSETSYKSNK---KPILINFGDSNSDTGGVLAGVGLPIGLP 61
Query: 61 NGRTFFHRSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPK 120
+G TFFHR TGRL DGRL++D C+ L +L+PYLDSLS +F G NFAV G++ LP
Sbjct: 62 HGITFFHRGTGRLGDGRLIVDFYCEHLKMTYLSPYLDSLS-PNFKRGVNFAVSGATALPI 120
Query: 121 YVPFSLNIQVMQFLHFKARTLELVSAGAKNVINDEGFRAALYLIDIGQNDLADS-FAKNL 179
+ F L IQ+ QF+HFK R+ EL+S+G +++I+D GFR ALY+IDIGQNDL + + NL
Sbjct: 121 F-SFPLAIQIRQFVHFKNRSQELISSGRRDLIDDNGFRNALYMIDIGQNDLLLALYDSNL 179
Query: 180 TYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCL 239
TY V++KIP+++ EI+ A+ +G +HN DLD GC
Sbjct: 180 TYAPVVEKIPSMLLEIKKAI-----QGELAIHLHNDS---------------DLDPIGCF 219
Query: 240 SSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCG 299
+N A+ FN+ L +LR++ KDATLVYVDIY+IK+ L A+ YGF +PL CCG
Sbjct: 220 RVHNEVAKAFNKGLLSLCNELRSQFKDATLVYVDIYSIKYKLSADFKLYGFVDPLMACCG 279
Query: 300 FGGPPYNFDLRVTCGQPGYQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPF 359
+GG P N+D + TCGQPG +C + ++ + WDG H+TEAAN F+ +L+ YS P+
Sbjct: 280 YGGRPNNYDRKATCGQPGSTICRDVTKAIVWDGVHYTEAANRFVVDAVLTNRYSYPKNSL 339
Query: 360 EFF 362
+ F
Sbjct: 340 DRF 342
>gb|AAM61525.1| early nodule-specific protein, putative [Arabidopsis thaliana]
Length = 377
Score = 307 bits (787), Expect = 3e-82
Identities = 163/365 (44%), Positives = 223/365 (60%), Gaps = 20/365 (5%)
Query: 9 LHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHR 68
+ L ++ L+ SS + P+ F FGDSNSDTG L +GLG ++LPNG+ F
Sbjct: 1 MKLFYVILFFISSLQISNSIDFNYPSA-FNFGDSNSDTGDLVAGLGIRLDLPNGQNSFKT 59
Query: 69 STGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLP----KYVPF 124
S+ R DGRLVID L ++ FL PYLDSL +F G NFA GS+ LP PF
Sbjct: 60 SSQRFCDGRLVIDFLMDEMDLPFLNPYLDSLGLPNFKKGCNFAAAGSTILPANPTSVSPF 119
Query: 125 SLNIQVMQFLHFKARTLELVSAGA----KNVINDEGFRAALYLIDIGQNDLADSFAKNLT 180
S ++Q+ QF+ FK+R +EL+S K + + + LY+IDIGQND+A +F T
Sbjct: 120 SFDLQISQFIRFKSRAIELLSKTGRKYEKYLPPIDYYSKGLYMIDIGQNDIAGAFYSK-T 178
Query: 181 YVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILAL--AQKKDLDLFGC 238
QV+ IP+++ E +K LY EG R W+HNTGPLGCL + +A LD FGC
Sbjct: 179 LDQVLASIPSILETFEAGLKRLYEEGGRNIWIHNTGPLGCLAQNIAKFGTDSTKLDEFGC 238
Query: 239 LSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCC 298
+SS+N AA+LFN L+ S K + + DA + YVDI++IK +LIAN +++GF PL CC
Sbjct: 239 VSSHNQAAKLFNLQLHAMSNKFQAQYPDANVTYVDIFSIKSNLIANYSRFGFEKPLMACC 298
Query: 299 GFGGPPYNFDLRVTCGQPGY--------QVCDEGSRYVNWDGTHHTEAANTFIASKILST 350
G GG P N+D R+TCGQ + C++ S Y+NWDG H+TEAAN F++S+IL+
Sbjct: 299 GVGGAPLNYDSRITCGQTKVLDGISVTAKACNDSSEYINWDGIHYTEAANEFVSSQILTG 358
Query: 351 DYSTP 355
YS P
Sbjct: 359 KYSDP 363
>dbj|BAC42831.1| unknown protein [Arabidopsis thaliana] gi|28973587|gb|AAO64118.1|
putative early nodule-specific protein [Arabidopsis
thaliana] gi|18405064|ref|NP_564668.1| GDSL-motif
lipase/hydrolase family protein [Arabidopsis thaliana]
Length = 382
Score = 307 bits (787), Expect = 3e-82
Identities = 163/365 (44%), Positives = 223/365 (60%), Gaps = 20/365 (5%)
Query: 9 LHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHR 68
+ L ++ L+ SS + P+ F FGDSNSDTG L +GLG ++LPNG+ F
Sbjct: 6 MKLFYVILFFISSLQISNSIDFNYPSA-FNFGDSNSDTGDLVAGLGIRLDLPNGQNSFKT 64
Query: 69 STGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLP----KYVPF 124
S+ R DGRLVID L ++ FL PYLDSL +F G NFA GS+ LP PF
Sbjct: 65 SSQRFCDGRLVIDFLMDEMDLPFLNPYLDSLGLPNFKKGCNFAAAGSTILPANPTSVSPF 124
Query: 125 SLNIQVMQFLHFKARTLELVSAGA----KNVINDEGFRAALYLIDIGQNDLADSFAKNLT 180
S ++Q+ QF+ FK+R +EL+S K + + + LY+IDIGQND+A +F T
Sbjct: 125 SFDLQISQFIRFKSRAIELLSKTGRKYEKYLPPIDYYSKGLYMIDIGQNDIAGAFYSK-T 183
Query: 181 YVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILAL--AQKKDLDLFGC 238
QV+ IP+++ E +K LY EG R W+HNTGPLGCL + +A LD FGC
Sbjct: 184 LDQVLASIPSILETFEAGLKRLYEEGGRNIWIHNTGPLGCLAQNIAKFGTDSTKLDEFGC 243
Query: 239 LSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCC 298
+SS+N AA+LFN L+ S K + + DA + YVDI++IK +LIAN +++GF PL CC
Sbjct: 244 VSSHNQAAKLFNLQLHAMSNKFQAQYPDANVTYVDIFSIKSNLIANYSRFGFEKPLMACC 303
Query: 299 GFGGPPYNFDLRVTCGQPGY--------QVCDEGSRYVNWDGTHHTEAANTFIASKILST 350
G GG P N+D R+TCGQ + C++ S Y+NWDG H+TEAAN F++S+IL+
Sbjct: 304 GVGGAPLNYDSRITCGQTKVLDGISVTAKACNDSSEYINWDGIHYTEAANEFVSSQILTG 363
Query: 351 DYSTP 355
YS P
Sbjct: 364 KYSDP 368
>ref|NP_908747.1| putative lipase homolog [Oryza sativa (japonica cultivar-group)]
Length = 284
Score = 306 bits (784), Expect = 6e-82
Identities = 148/281 (52%), Positives = 205/281 (72%), Gaps = 5/281 (1%)
Query: 85 QSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVPFSLNIQVMQFLHFKARTLELV 144
+SLN +L+PYL +L GS ++NGANFA+ GS+TLP+ FSL+IQV QFL F+ R+LEL+
Sbjct: 5 ESLNIGYLSPYLKAL-GSDYSNGANFAIAGSATLPRDTLFSLHIQVKQFLFFRDRSLELI 63
Query: 145 SAGAKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSLYN 204
S G ++ EGFR ALY+IDIGQND+ ++ L+Y QV+ + P ++ EI++A+++LY+
Sbjct: 64 SQGLPGPVDAEGFRNALYMIDIGQNDV-NALLSYLSYDQVVARFPPILDEIKDAIQTLYD 122
Query: 205 EGARKFWVHNTGPLGCLPKILALAQKKDLDLF--GCLSSYNSAARLFNEALYHSSQKLRT 262
G+R FWVH TG LGCLP+ L++ +K D DL GCL +YN AA FN AL +L T
Sbjct: 123 NGSRNFWVHGTGALGCLPQKLSIPRKNDSDLDSNGCLKTYNRAAVTFNAALGSLCDQLST 182
Query: 263 RLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPGYQVCD 322
++KDAT+VY D++ +K+DLIAN TKYGF PL CCG+GGPPYN+++ + C Q CD
Sbjct: 183 QMKDATIVYTDLFPLKYDLIANRTKYGFDKPLMTCCGYGGPPYNYNITIGC-QDKNASCD 241
Query: 323 EGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFC 363
+GS++V+WDG H TEAAN +A ILS+DYS P+ F+ FC
Sbjct: 242 DGSKFVSWDGVHLTEAANAIVAKGILSSDYSRPKIKFDQFC 282
>gb|AAC64890.1| Similar to nodulins and lipase homolog F14J9.5 gi|3482914 from
Arabidopsis thaliana BAC gb|AC003970. Alternate first
exon from 72258 to 72509 gi|25405771|pir||A96590
hypothetical protein T22H22.20 [imported] - Arabidopsis
thaliana
Length = 383
Score = 298 bits (763), Expect = 2e-79
Identities = 160/366 (43%), Positives = 223/366 (60%), Gaps = 23/366 (6%)
Query: 8 SLHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFH 67
S+ L L SS ++++ PA++ FGDSNSDTG L S +N P G+T+F+
Sbjct: 9 SIVLFILISLFLPSSFSIILK---YPAIIN-FGDSNSDTGNLISAGIENVNPPYGQTYFN 64
Query: 68 RSTGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLP----KYVP 123
+GR DGRL++D L ++ FL PYLDSL +F G NFA GS+ LP P
Sbjct: 65 LPSGRYCDGRLIVDFLLDEMDLPFLNPYLDSLGLPNFKKGCNFAAAGSTILPANPTSVSP 124
Query: 124 FSLNIQVMQFLHFKARTLELVSAGA----KNVINDEGFRAALYLIDIGQNDLADSFAKNL 179
FS ++Q+ QF+ FK+R +EL+S K + + + LY+IDIGQND+A +F
Sbjct: 125 FSFDLQISQFIRFKSRAIELLSKTGRKYEKYLPPIDYYSKGLYMIDIGQNDIAGAFYSK- 183
Query: 180 TYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILAL--AQKKDLDLFG 237
T QV+ IP+++ E +K LY EG R W+HNTGPLGCL + +A LD FG
Sbjct: 184 TLDQVLASIPSILETFEAGLKRLYEEGGRNIWIHNTGPLGCLAQNIAKFGTDSTKLDEFG 243
Query: 238 CLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVC 297
C+SS+N AA+LFN L+ S K + + DA + YVDI++IK +LIAN +++GF PL C
Sbjct: 244 CVSSHNQAAKLFNLQLHAMSNKFQAQYPDANVTYVDIFSIKSNLIANYSRFGFEKPLMAC 303
Query: 298 CGFGGPPYNFDLRVTCGQPGY--------QVCDEGSRYVNWDGTHHTEAANTFIASKILS 349
CG GG P N+D R+TCGQ + C++ S Y+NWDG H+TEAAN F++S+IL+
Sbjct: 304 CGVGGAPLNYDSRITCGQTKVLDGISVTAKACNDSSEYINWDGIHYTEAANEFVSSQILT 363
Query: 350 TDYSTP 355
YS P
Sbjct: 364 GKYSDP 369
>ref|NP_974029.1| GDSL-motif lipase/hydrolase family protein [Arabidopsis thaliana]
Length = 408
Score = 293 bits (750), Expect = 6e-78
Identities = 163/391 (41%), Positives = 223/391 (56%), Gaps = 46/391 (11%)
Query: 9 LHLLFLFLWITSSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHR 68
+ L ++ L+ SS + P+ F FGDSNSDTG L +GLG ++LPNG+ F
Sbjct: 6 MKLFYVILFFISSLQISNSIDFNYPSA-FNFGDSNSDTGDLVAGLGIRLDLPNGQNSFKT 64
Query: 69 STGRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSSTLP----KYVPF 124
S+ R DGRLVID L ++ FL PYLDSL +F G NFA GS+ LP PF
Sbjct: 65 SSQRFCDGRLVIDFLMDEMDLPFLNPYLDSLGLPNFKKGCNFAAAGSTILPANPTSVSPF 124
Query: 125 SLNIQVMQFLHFKARTLELVSAGA----KNVINDEGFRAALYLIDIGQNDLADSFAKNLT 180
S ++Q+ QF+ FK+R +EL+S K + + + LY+IDIGQND+A +F T
Sbjct: 125 SFDLQISQFIRFKSRAIELLSKTGRKYEKYLPPIDYYSKGLYMIDIGQNDIAGAFYSK-T 183
Query: 181 YVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILAL--AQKKDLDLFGC 238
QV+ IP+++ E +K LY EG R W+HNTGPLGCL + +A LD FGC
Sbjct: 184 LDQVLASIPSILETFEAGLKRLYEEGGRNIWIHNTGPLGCLAQNIAKFGTDSTKLDEFGC 243
Query: 239 LSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKY---------- 288
+SS+N AA+LFN L+ S K + + DA + YVDI++IK +LIAN +++
Sbjct: 244 VSSHNQAAKLFNLQLHAMSNKFQAQYPDANVTYVDIFSIKSNLIANYSRFGKHFTKPLID 303
Query: 289 ----------------GFSNPLTVCCGFGGPPYNFDLRVTCGQPGY--------QVCDEG 324
GF PL CCG GG P N+D R+TCGQ + C++
Sbjct: 304 LNHLENVGYNKILNVLGFEKPLMACCGVGGAPLNYDSRITCGQTKVLDGISVTAKACNDS 363
Query: 325 SRYVNWDGTHHTEAANTFIASKILSTDYSTP 355
S Y+NWDG H+TEAAN F++S+IL+ YS P
Sbjct: 364 SEYINWDGIHYTEAANEFVSSQILTGKYSDP 394
>gb|AAF27024.1| putative nodulin [Arabidopsis thaliana] gi|15810237|gb|AAL07236.1|
putative nodulin protein [Arabidopsis thaliana]
gi|15229919|ref|NP_187169.1| GDSL-motif lipase/hydrolase
family protein [Arabidopsis thaliana]
Length = 379
Score = 278 bits (712), Expect = 1e-73
Identities = 157/370 (42%), Positives = 222/370 (59%), Gaps = 23/370 (6%)
Query: 5 LAFSLHLLFLFLWIT---SSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPN 61
L +L L LF+ I+ S A D PAV F FGDSNSDTG L+SGLGF
Sbjct: 4 LFHTLLRLLLFVAISHTLSPLAGSFRISNDFPAV-FNFGDSNSDTGELSSGLGFLPQPSY 62
Query: 62 GRTFFHRST-GRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSS---- 116
TFF T GR +GRL++D L ++++ +L PYLDS+S ++ G NFA S+
Sbjct: 63 EITFFRSPTSGRFCNGRLIVDFLMEAIDRPYLRPYLDSISRQTYRRGCNFAAAASTIQKA 122
Query: 117 TLPKYVPFSLNIQVMQFLHFKARTLELVSAGA---KNVINDEGFRAALYLIDIGQNDLAD 173
Y PF +QV QF+ FK++ L+L+ + + ++ F LY+ DIGQND+A
Sbjct: 123 NAASYSPFGFGVQVSQFITFKSKVLQLIQQDEELQRYLPSEYFFSNGLYMFDIGQNDIAG 182
Query: 174 SFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILAL--AQKK 231
+F T QV+ +P ++ ++ +K LY EGAR +W+HNTGPLGCL +++++ K
Sbjct: 183 AFYTK-TVDQVLALVPIILDIFQDGIKRLYAEGARNYWIHNTGPLGCLAQVVSIFGEDKS 241
Query: 232 DLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFS 291
LD FGC+S +N AA+LFN L+ +KL + ++ YVDI++IK DLI N +KYGF
Sbjct: 242 KLDEFGCVSDHNQAAKLFNLQLHGLFKKLPQQYPNSRFTYVDIFSIKSDLILNHSKYGFD 301
Query: 292 NPLTVCCGFGGPPYNFDLRVTCGQPG--------YQVCDEGSRYVNWDGTHHTEAANTFI 343
+ + VCCG GGPP N+D +V CG+ + C + S+YVNWDG H+TEAAN F+
Sbjct: 302 HSIMVCCGTGGPPLNYDDQVGCGKTARSNGTIITAKPCYDSSKYVNWDGIHYTEAANRFV 361
Query: 344 ASKILSTDYS 353
A IL+ YS
Sbjct: 362 ALHILTGKYS 371
>gb|AAM62882.1| putative nodulin [Arabidopsis thaliana]
Length = 379
Score = 278 bits (712), Expect = 1e-73
Identities = 157/370 (42%), Positives = 222/370 (59%), Gaps = 23/370 (6%)
Query: 5 LAFSLHLLFLFLWIT---SSSAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPN 61
L +L L LF+ I+ S A D PAV F FGDSNSDTG L+SGLGF
Sbjct: 4 LFHTLLRLLLFVAISHTLSPLAGSFRISNDFPAV-FNFGDSNSDTGELSSGLGFLPQPSY 62
Query: 62 GRTFFHRST-GRLSDGRLVIDLLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSS---- 116
TFF T GR +GRL++D L ++++ +L PYLDS+S ++ G NFA S+
Sbjct: 63 EITFFRSPTSGRFCNGRLIVDFLMEAIDRPYLRPYLDSISRQTYRRGCNFAAAASTIQKA 122
Query: 117 TLPKYVPFSLNIQVMQFLHFKARTLELVSAGA---KNVINDEGFRAALYLIDIGQNDLAD 173
Y PF +QV QF+ FK++ L+L+ + + ++ F LY+ DIGQND+A
Sbjct: 123 NAASYSPFGFGVQVSQFITFKSKVLQLIQQDEELQRYLPSEYFFSNGLYMFDIGQNDIAG 182
Query: 174 SFAKNLTYVQVIKKIPTVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILAL--AQKK 231
+F T QV+ +P ++ ++ +K LY EGAR +W+HNTGPLGCL +++++ K
Sbjct: 183 AFYTK-TLDQVLALVPIILDIFQDGIKRLYAEGARNYWIHNTGPLGCLAQVVSIFGKDKS 241
Query: 232 DLDLFGCLSSYNSAARLFNEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFS 291
LD FGC+S +N AA+LFN L+ +KL + ++ YVDI++IK DLI N +KYGF
Sbjct: 242 KLDEFGCVSDHNQAAKLFNLQLHGLFKKLPQQYPNSRFTYVDIFSIKSDLILNHSKYGFD 301
Query: 292 NPLTVCCGFGGPPYNFDLRVTCGQPG--------YQVCDEGSRYVNWDGTHHTEAANTFI 343
+ + VCCG GGPP N+D +V CG+ + C + S+YVNWDG H+TEAAN F+
Sbjct: 302 HSIMVCCGTGGPPLNYDDQVGCGKTARSNGTIITAKPCYDSSKYVNWDGIHYTEAANRFV 361
Query: 344 ASKILSTDYS 353
A IL+ YS
Sbjct: 362 ALHILTGKYS 371
>gb|AAN31927.1| putative nodulin [Arabidopsis thaliana]
Length = 355
Score = 278 bits (711), Expect = 2e-73
Identities = 149/341 (43%), Positives = 211/341 (61%), Gaps = 20/341 (5%)
Query: 31 DVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRST-GRLSDGRLVIDLLCQSLNT 89
D PAV F FGDSNSDTG L+SGLGF TFF T GR +GRL++D L ++++
Sbjct: 9 DFPAV-FNFGDSNSDTGELSSGLGFLPQPSYEITFFRSPTSGRFCNGRLIVDFLMEAIDR 67
Query: 90 RFLTPYLDSLSGSSFTNGANFAVVGSS----TLPKYVPFSLNIQVMQFLHFKARTLELVS 145
+L PYLDS+S ++ G NFA S+ Y PF +QV QF+ FK++ L+L+
Sbjct: 68 PYLRPYLDSISRQTYRRGCNFAAAASTIQKANAASYSPFGFGVQVSQFITFKSKVLQLIQ 127
Query: 146 AGA---KNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIPTVITEIENAVKSL 202
+ + ++ F LY+ DIGQND+A +F T QV+ +P ++ ++ +K L
Sbjct: 128 QDEELQRYLPSEYFFSNGLYMFDIGQNDIAGAFYTK-TVDQVLALVPIILDIFQDGIKRL 186
Query: 203 YNEGARKFWVHNTGPLGCLPKILAL--AQKKDLDLFGCLSSYNSAARLFNEALYHSSQKL 260
Y EGAR +W+HNTGPLGCL +++++ K LD FGC+S +N AA+LFN L+ +KL
Sbjct: 187 YAEGARNYWIHNTGPLGCLAQVVSIFGEDKSKLDEFGCVSDHNQAAKLFNLQLHGLFKKL 246
Query: 261 RTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQPG--- 317
+ ++ YVDI++IK DLI N +KYGF + + VCCG GGPP N+D +V CG+
Sbjct: 247 PQQYPNSRFTYVDIFSIKSDLILNHSKYGFDHSIMVCCGTGGPPLNYDDQVGCGKTARSN 306
Query: 318 -----YQVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYS 353
+ C + S+YVNWDG H+TEAAN F+A IL+ YS
Sbjct: 307 GTIITAKPCYDSSKYVNWDGIHYTEAANRFVALHILTGKYS 347
>dbj|BAD54729.1| putative lipase homolog [Oryza sativa (japonica cultivar-group)]
Length = 417
Score = 273 bits (697), Expect = 8e-72
Identities = 140/265 (52%), Positives = 189/265 (70%), Gaps = 5/265 (1%)
Query: 28 SKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVIDLLCQSL 87
+ C V+F FGDSN+DTGG+ +G+G+ LP GR FF R+TGRL DGRLVID LC+SL
Sbjct: 44 ASCARRPVVFAFGDSNTDTGGIAAGMGYYFPLPEGRAFFRRATGRLCDGRLVIDHLCESL 103
Query: 88 NTRFLTPYLDSLSGSSFTNGANFAVVGSSTLPKYVPFSLNIQVMQFLHFKARTLELVSAG 147
N +L+PYL+ L G+ FTNGANFA+ G++T P+ FSL+IQV QF+HFK R+LEL S G
Sbjct: 104 NMSYLSPYLEPL-GTDFTNGANFAISGAATAPRNAAFSLHIQVQQFIHFKQRSLELASRG 162
Query: 148 AKNVINDEGFRAALYLIDIGQNDLADSF-AKNLTYVQVIK-KIPTVITEIENAVKSLYNE 205
++ +GFR ALYLIDIGQNDL+ +F A L Y V++ + P +++EI++A++SLY
Sbjct: 163 EAVPVDADGFRNALYLIDIGQNDLSAAFSAGGLPYDDVVRQRFPAILSEIKDAIQSLYYN 222
Query: 206 GARKFWVHNTGPLGCLPKILAL--AQKKDLDLFGCLSSYNSAARLFNEALYHSSQKLRTR 263
GA+ W+H TGPLGCLP+ LA+ A DLD GCL + N+ A FN L +L ++
Sbjct: 223 GAKNLWIHGTGPLGCLPQKLAVPRADDGDLDPSGCLKTLNAGAYEFNSQLSSICDQLSSQ 282
Query: 264 LKDATLVYVDIYAIKHDLIANATKY 288
L+ AT+V+ DI AIK+DLIAN + Y
Sbjct: 283 LRGATIVFTDILAIKYDLIANHSSY 307
>gb|AAP37470.1| ENSP-like protein [Hevea brasiliensis]
gi|51315784|sp|Q7Y1X1|EST_HEVBR Esterase precursor
(Early nodule-specific protein homolog) (Latex allergen
Hev b 13)
Length = 391
Score = 272 bits (696), Expect = 1e-71
Identities = 157/367 (42%), Positives = 207/367 (55%), Gaps = 30/367 (8%)
Query: 22 SAAVMMSKCDVPAVLFVFGDSNSDTGGLTSGLGFPINLPNGRTFFHRSTGRLSDGRLVID 81
S A CD PA+ F FGDSNSDTGG + +P+N P G TFFHRSTGR SDGRL+ID
Sbjct: 22 SLAYASETCDFPAI-FNFGDSNSDTGGKAAAF-YPLNPPYGETFFHRSTGRYSDGRLIID 79
Query: 82 LLCQSLNTRFLTPYLDSLSGSSFTNGANFAVVGSST-LPKYV--------PFSLNIQVMQ 132
+ +S N +L+PYL SL GS+F +GA+FA GS+ LP + PF L++Q Q
Sbjct: 80 FIAESFNLPYLSPYLSSL-GSNFKHGADFATAGSTIKLPTTIIPAHGGFSPFYLDVQYSQ 138
Query: 133 FLHFKARTLELVSAG---AKNVINDEGFRAALYLIDIGQNDLADSFAKNLTYVQVIKKIP 189
F F R+ + G A+ V + F ALY DIGQNDL + F NLT +V +P
Sbjct: 139 FRQFIPRSQFIRETGGIFAELVPEEYYFEKALYTFDIGQNDLTEGFL-NLTVEEVNATVP 197
Query: 190 TVITEIENAVKSLYNEGARKFWVHNTGPLGCLPKILALAQKKDLDLFGCLSSYNSAARLF 249
++ VK +Y+ GAR FW+HNTGP+GCL IL + D GC +YN A+ F
Sbjct: 198 DLVNSFSANVKKIYDLGARTFWIHNTGPIGCLSFILTYFPWAEKDSAGCAKAYNEVAQHF 257
Query: 250 NEALYHSSQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDL 309
N L +LR L AT V+VDIY++K+ L + K+GF PL CCG+GG YNF +
Sbjct: 258 NHKLKEIVAQLRKDLPLATFVHVDIYSVKYSLFSEPEKHGFEFPLITCCGYGG-KYNFSV 316
Query: 310 RVTCGQPGYQVCDEGSRY-----------VNWDGTHHTEAANTFIASKILSTDYSTPRTP 358
CG D+G++ VNWDG H+TEAAN + +I + +S P P
Sbjct: 317 TAPCGDT--VTADDGTKIVVGSCACPSVRVNWDGAHYTEAANEYFFDQISTGAFSDPPVP 374
Query: 359 FEFFCHR 365
CH+
Sbjct: 375 LNMACHK 381
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 625,197,820
Number of Sequences: 2540612
Number of extensions: 26641756
Number of successful extensions: 55311
Number of sequences better than 10.0: 424
Number of HSP's better than 10.0 without gapping: 322
Number of HSP's successfully gapped in prelim test: 102
Number of HSP's that attempted gapping in prelim test: 53617
Number of HSP's gapped (non-prelim): 480
length of query: 365
length of database: 863,360,394
effective HSP length: 129
effective length of query: 236
effective length of database: 535,621,446
effective search space: 126406661256
effective search space used: 126406661256
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0223.2


