

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
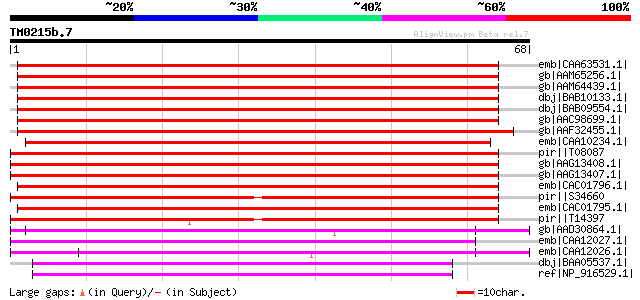
Query= TM0215b.7
(68 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA63531.1| pollen coat protein [Brassica oleracea] gi|74885... 91 1e-17
gb|AAM65256.1| pollen coat-like protein [Arabidopsis thaliana] 89 4e-17
gb|AAM64439.1| ABA-inducible protein-like protein [Arabidopsis t... 87 9e-17
dbj|BAB10133.1| pollen coat protein-like [Arabidopsis thaliana] ... 87 1e-16
dbj|BAB09554.1| ABA-inducible protein-like [Arabidopsis thaliana... 86 3e-16
gb|AAC98699.1| pollen coat protein homolog [Brassica rapa] gi|74... 86 3e-16
gb|AAF32455.1| unknown protein [Arabidopsis thaliana] gi|1865534... 73 2e-12
emb|CAA10234.1| ABA-inducible protein [Fagus sylvatica] 73 2e-12
pir||T08087 cold-induced protein kin1 - rape gi|167146|gb|AAA329... 60 1e-08
gb|AAG13408.1| BN28b [Brassica napus] 60 1e-08
gb|AAG13407.1| BN28a [Brassica napus] 60 1e-08
emb|CAC01796.1| cold-regulated protein COR6.6 (KIN2) [Arabidopsi... 55 5e-07
pir||S34660 probable cold-regulated protein kin1 - turnip gi|395... 55 6e-07
emb|CAC01795.1| cold and ABA inducible protein kin1 [Arabidopsis... 52 4e-06
pir||T14397 probable cold-induced protein kin - turnip gi|120926... 51 7e-06
gb|AAD30864.1| seed maturation protein PM30 [Glycine max] 51 9e-06
emb|CAA12027.1| LEA PROTEIN [Cicer arietinum] gi|24418489|sp|O49... 50 2e-05
emb|CAA12026.1| LEA protein [Cicer arietinum] gi|24418488|sp|O49... 45 4e-04
dbj|BAA05537.1| WSI18 protein induced by water stress [Oryza sat... 45 5e-04
ref|NP_916529.1| WSI18 protein [Oryza sativa (japonica cultivar-... 45 5e-04
>emb|CAA63531.1| pollen coat protein [Brassica oleracea] gi|7488569|pir||T14467
pollen coat protein - wild cabbage
Length = 67
Score = 90.5 bits (223), Expect = 1e-17
Identities = 43/63 (68%), Positives = 53/63 (83%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+SQ MS+NAGQAKGQ +EK SNLMD ASNAAQSAKE++Q GQQ+K AQGA++A+K T
Sbjct: 4 NSQSMSFNAGQAKGQTQEKASNLMDKASNAAQSAKESLQEGGQQLKQKAQGASEAIKEKT 63
Query: 62 GMN 64
G+N
Sbjct: 64 GLN 66
>gb|AAM65256.1| pollen coat-like protein [Arabidopsis thaliana]
Length = 67
Score = 88.6 bits (218), Expect = 4e-17
Identities = 43/63 (68%), Positives = 52/63 (82%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+SQ +S+ AGQAKGQ +EK S +MD ASNAAQSAKE++Q GQQIK AQGA ++VKNAT
Sbjct: 4 NSQNISFQAGQAKGQTQEKASTMMDKASNAAQSAKESLQETGQQIKEKAQGATESVKNAT 63
Query: 62 GMN 64
GMN
Sbjct: 64 GMN 66
>gb|AAM64439.1| ABA-inducible protein-like protein [Arabidopsis thaliana]
Length = 67
Score = 87.4 bits (215), Expect = 9e-17
Identities = 41/63 (65%), Positives = 52/63 (82%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+SQ MS+NAGQAKGQ +EK SNLMD ASNAAQSAKE++Q GQQ+K AQGA++ +K T
Sbjct: 4 NSQSMSFNAGQAKGQTQEKASNLMDKASNAAQSAKESIQEGGQQLKQKAQGASETIKEKT 63
Query: 62 GMN 64
G++
Sbjct: 64 GIS 66
>dbj|BAB10133.1| pollen coat protein-like [Arabidopsis thaliana]
gi|15240998|ref|NP_198692.1| expressed protein
[Arabidopsis thaliana]
Length = 67
Score = 87.0 bits (214), Expect = 1e-16
Identities = 42/63 (66%), Positives = 52/63 (81%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+SQ +S+ AGQAKGQ +EK S +MD ASNAAQSAKE+++ GQQIK AQGA ++VKNAT
Sbjct: 4 NSQNISFQAGQAKGQTQEKASTMMDKASNAAQSAKESLKETGQQIKEKAQGATESVKNAT 63
Query: 62 GMN 64
GMN
Sbjct: 64 GMN 66
>dbj|BAB09554.1| ABA-inducible protein-like [Arabidopsis thaliana]
gi|15238849|ref|NP_200193.1| expressed protein
[Arabidopsis thaliana]
Length = 67
Score = 85.9 bits (211), Expect = 3e-16
Identities = 40/63 (63%), Positives = 52/63 (82%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+SQ MS+NAGQAKGQ +EK SNL+D ASNAAQSAKE++Q GQQ+K AQGA++ +K T
Sbjct: 4 NSQSMSFNAGQAKGQTQEKASNLIDKASNAAQSAKESIQEGGQQLKQKAQGASETIKEKT 63
Query: 62 GMN 64
G++
Sbjct: 64 GIS 66
>gb|AAC98699.1| pollen coat protein homolog [Brassica rapa]
gi|7488528|pir||T14410 pollen coat protein homolog -
turnip
Length = 67
Score = 85.9 bits (211), Expect = 3e-16
Identities = 41/63 (65%), Positives = 50/63 (79%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+SQ S+NAGQAKGQ +EK SNLMD ASNAAQSAKE++ GQQ+K AQGA +A+K T
Sbjct: 4 NSQNWSFNAGQAKGQTQEKASNLMDKASNAAQSAKESLSEGGQQLKQKAQGATEAIKEKT 63
Query: 62 GMN 64
G+N
Sbjct: 64 GLN 66
>gb|AAF32455.1| unknown protein [Arabidopsis thaliana] gi|18655343|gb|AAL76127.1|
AT3g02480/F16B3_11 [Arabidopsis thaliana]
gi|14335128|gb|AAK59844.1| AT3g02480/F16B3_11
[Arabidopsis thaliana] gi|18396202|ref|NP_566173.1|
ABA-responsive protein-related [Arabidopsis thaliana]
Length = 68
Score = 73.2 bits (178), Expect = 2e-12
Identities = 35/65 (53%), Positives = 46/65 (69%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
+ Q SY AGQA GQ +EK +MD A +AA SA++++Q GQQ+K AQGAAD VK+ T
Sbjct: 3 NKQNASYQAGQATGQTKEKAGGMMDKAKDAAASAQDSLQQTGQQMKEKAQGAADVVKDKT 62
Query: 62 GMNNN 66
GMN +
Sbjct: 63 GMNKS 67
>emb|CAA10234.1| ABA-inducible protein [Fagus sylvatica]
Length = 68
Score = 73.2 bits (178), Expect = 2e-12
Identities = 36/61 (59%), Positives = 44/61 (72%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNATG 62
SQ + AGQAKGQ +EK SNLM+ A NA QSAKET AG+Q+ A AQGAA+ K+A G
Sbjct: 4 SQNTCFQAGQAKGQTQEKASNLMEKAGNAGQSAKETCLEAGKQVPAKAQGAANTAKDAVG 63
Query: 63 M 63
+
Sbjct: 64 L 64
>pir||T08087 cold-induced protein kin1 - rape gi|167146|gb|AAA32993.1| kin1
gi|384330|prf||1905417A bn28 gene
Length = 65
Score = 60.5 bits (145), Expect = 1e-08
Identities = 32/64 (50%), Positives = 41/64 (64%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M K S+ AGQA G+AEEK + LMD +AA +A + Q AGQ+I +A GA + VK
Sbjct: 1 MADNKQSFQAGQASGRAEEKGNVLMDKVKDAATAAGASAQTAGQKITEAAGGAVNLVKEK 60
Query: 61 TGMN 64
TGMN
Sbjct: 61 TGMN 64
>gb|AAG13408.1| BN28b [Brassica napus]
Length = 65
Score = 60.1 bits (144), Expect = 1e-08
Identities = 31/64 (48%), Positives = 41/64 (63%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M K S+ AGQ G+AEEK++ LMD +AA SA + Q AGQ+I +A GA + VK
Sbjct: 1 MADNKQSFQAGQTAGRAEEKSNVLMDKVKDAATSAGASAQTAGQKISETAGGAVNVVKEK 60
Query: 61 TGMN 64
TG+N
Sbjct: 61 TGIN 64
>gb|AAG13407.1| BN28a [Brassica napus]
Length = 65
Score = 60.1 bits (144), Expect = 1e-08
Identities = 32/64 (50%), Positives = 41/64 (64%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M K S+ AGQA G+AEEK + LMD +AA +A + Q AGQ+I +A GA + VK
Sbjct: 1 MADNKQSFQAGQAAGRAEEKGNVLMDKVKDAATAAGASAQTAGQKITEAAGGAVNLVKEK 60
Query: 61 TGMN 64
TGMN
Sbjct: 61 TGMN 64
>emb|CAC01796.1| cold-regulated protein COR6.6 (KIN2) [Arabidopsis thaliana]
gi|21386921|gb|AAM47864.1| cold-regulated protein
COR6.6 (KIN2) [Arabidopsis thaliana]
gi|16230|emb|CAA38894.1| cor6.6 [Arabidopsis thaliana]
gi|18252153|gb|AAL61909.1| cold-regulated protein
COR6.6 (KIN2) [Arabidopsis thaliana]
gi|15237236|ref|NP_197101.1| stress-responsive protein
(KIN2) / stress-induced protein (KIN2) /
cold-responsive protein (COR6.6) / cold-regulated
protein (COR6.6) [Arabidopsis thaliana]
gi|1084343|pir||S22529 cold-regulated protein kin2
(COR6.6) - Arabidopsis thaliana
gi|399298|sp|P31169|KIN2_ARATH Stress-induced KIN2
protein (Cold-induced COR6.6 protein)
Length = 66
Score = 55.1 bits (131), Expect = 5e-07
Identities = 27/63 (42%), Positives = 43/63 (67%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
++ K ++ AGQA G+AEEK++ L+D A +AA +A + Q AG+ I +A G + VK+ T
Sbjct: 3 ETNKNAFQAGQAAGKAEEKSNVLLDKAKDAAAAAGASAQQAGKSISDAAVGGVNFVKDKT 62
Query: 62 GMN 64
G+N
Sbjct: 63 GLN 65
>pir||S34660 probable cold-regulated protein kin1 - turnip
gi|395075|emb|CAA80862.1| kin1 [Brassica rapa]
gi|347060|gb|AAA33011.1| kin1
Length = 64
Score = 54.7 bits (130), Expect = 6e-07
Identities = 31/64 (48%), Positives = 41/64 (63%), Gaps = 1/64 (1%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M K S+ AGQA G+AEEK + LMD +AA +A + + AGQ+I +A GA + VK
Sbjct: 1 MADNKQSFQAGQAAGRAEEKGNVLMDKVKDAA-TAGASAETAGQKITEAAGGAVNLVKEK 59
Query: 61 TGMN 64
TGMN
Sbjct: 60 TGMN 63
>emb|CAC01795.1| cold and ABA inducible protein kin1 [Arabidopsis thaliana]
gi|21592351|gb|AAM64302.1| cold and ABA inducible
protein kin1 [Arabidopsis thaliana]
gi|21387005|gb|AAM47906.1| cold and ABA inducible
protein kin1 [Arabidopsis thaliana]
gi|296033|emb|CAA35838.1| kin1 [Arabidopsis thaliana]
gi|17065546|gb|AAL32927.1| cold and ABA inducible
protein kin1 [Arabidopsis thaliana]
gi|15237228|ref|NP_197100.1| stress-responsive protein
(KIN1) / stress-induced protein (KIN1) [Arabidopsis
thaliana] gi|125408|sp|P18612|KIN1_ARATH Stress-induced
KIN1 protein
Length = 66
Score = 52.0 bits (123), Expect = 4e-06
Identities = 25/63 (39%), Positives = 40/63 (62%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNAT 61
++ K ++ AGQ G+AEEK++ L+D A +AA A Q AG+ + +A G + VK+ T
Sbjct: 3 ETNKNAFQAGQTAGKAEEKSNVLLDKAKDAAAGAGAGAQQAGKSVSDAAAGGVNFVKDKT 62
Query: 62 GMN 64
G+N
Sbjct: 63 GLN 65
>pir||T14397 probable cold-induced protein kin - turnip
gi|1209262|gb|AAA91051.1| kin gene product
Length = 65
Score = 51.2 bits (121), Expect = 7e-06
Identities = 32/65 (49%), Positives = 41/65 (62%), Gaps = 2/65 (3%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTS-NLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKN 59
M K S+ AGQA G+AEEK + LMD +AA +A +Q AGQ+I +A GA + VK
Sbjct: 1 MADNKQSFQAGQAAGRAEEKGNVLLMDKVKDAA-TAAGALQTAGQKITEAAGGAVNLVKE 59
Query: 60 ATGMN 64
TGMN
Sbjct: 60 KTGMN 64
>gb|AAD30864.1| seed maturation protein PM30 [Glycine max]
Length = 140
Score = 50.8 bits (120), Expect = 9e-06
Identities = 27/61 (44%), Positives = 32/61 (52%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M S + SY AGQ KG+ EEKT+ M AQ+AKE Q Q K Q A A K+
Sbjct: 1 MASHRQSYEAGQTKGRTEEKTNQTMGNIGEKAQAAKEKTQEMAQAAKEKTQQTAQAAKDK 60
Query: 61 T 61
T
Sbjct: 61 T 61
Score = 46.2 bits (108), Expect = 2e-04
Identities = 24/70 (34%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQG----AGQQIKASAQGAADAVK 58
SQ Q G A++KTS + +AQS K+ QG G+++K +AQGA +AVK
Sbjct: 65 SQAAKEKTQQNTGAAQQKTSEMGQSTKESAQSGKDNTQGFLQQTGEKVKGAAQGATEAVK 124
Query: 59 NATGMNNNSK 68
G+ + +
Sbjct: 125 QTLGLGEHDQ 134
>emb|CAA12027.1| LEA PROTEIN [Cicer arietinum] gi|24418489|sp|O49817|LEA2_CICAR
Late embryogenesis abundant protein 2 (CapLEA-2)
Length = 155
Score = 49.7 bits (117), Expect = 2e-05
Identities = 26/61 (42%), Positives = 33/61 (53%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M S SY AG+AKG+ EEKT+ ++ + AQ+AKE Q A Q K A A K
Sbjct: 1 MTSHDQSYRAGEAKGRTEEKTNQMIGNIEDKAQAAKEKAQQAAQTAKDKTSQTAQAAKEK 60
Query: 61 T 61
T
Sbjct: 61 T 61
Score = 43.5 bits (101), Expect = 0.001
Identities = 25/70 (35%), Positives = 35/70 (49%), Gaps = 4/70 (5%)
Query: 3 SQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKET----VQGAGQQIKASAQGAADAVK 58
+Q A G+A+EK S + AQS K+ +Q G++ K AQGA DAVK
Sbjct: 76 TQATKEKAQDTTGRAKEKGSEMGQSTKETAQSGKDNSAGFLQQTGEKAKGMAQGATDAVK 135
Query: 59 NATGMNNNSK 68
GM N+ +
Sbjct: 136 QTFGMTNDDQ 145
>emb|CAA12026.1| LEA protein [Cicer arietinum] gi|24418488|sp|O49816|LEA1_CICAR
Late embryogenesis abundant protein 1 (CapLEA-1)
Length = 177
Score = 45.4 bits (106), Expect = 4e-04
Identities = 24/61 (39%), Positives = 31/61 (50%)
Query: 1 MDSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVKNA 60
M S SY AG+ G+ EEKT+ ++ + AQ+AKE Q A Q K A A K
Sbjct: 1 MASHDQSYKAGETMGRTEEKTNQMIGNIEDKAQAAKEKAQQAAQTAKDKTSQTAQAAKEK 60
Query: 61 T 61
T
Sbjct: 61 T 61
Score = 44.7 bits (104), Expect = 6e-04
Identities = 25/63 (39%), Positives = 33/63 (51%), Gaps = 4/63 (6%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKET----VQGAGQQIKASAQGAADAVKNATGMNN 65
A G+A EK S + AQS K+ +Q G+++K AQGA DAVK GM N
Sbjct: 105 AQDTTGRAREKGSEMGQSTKETAQSGKDNSAGFLQQTGEKVKGMAQGATDAVKQTFGMAN 164
Query: 66 NSK 68
+ K
Sbjct: 165 DDK 167
>dbj|BAA05537.1| WSI18 protein induced by water stress [Oryza sativa (japonica
cultivar-group)] gi|1076757|pir||S52642 WSI18 protein -
rice
Length = 214
Score = 45.1 bits (105), Expect = 5e-04
Identities = 21/55 (38%), Positives = 31/55 (56%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVK 58
++ SY+AG+ K +AEEKT +M A A+ AK+T A + GA +A K
Sbjct: 6 ERASYHAGETKARAEEKTGRMMGTAQEKAREAKDTASDAAGRAMGRGHGAKEATK 60
Score = 37.7 bits (86), Expect = 0.079
Identities = 25/70 (35%), Positives = 34/70 (47%), Gaps = 11/70 (15%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKET-----------VQGAGQQIKASAQGAADAVK 58
AGQ + A+ AAQ AKET +Q AG+Q+K+ A GA DAV
Sbjct: 124 AGQTGSYIGQTAEAAKQKAAGAAQYAKETAIAGKDKTGAVLQQAGEQVKSVAVGAKDAVM 183
Query: 59 NATGMNNNSK 68
GM+ ++K
Sbjct: 184 YTLGMSGDNK 193
Score = 31.2 bits (69), Expect = 7.4
Identities = 21/68 (30%), Positives = 33/68 (47%), Gaps = 1/68 (1%)
Query: 2 DSQKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQ-GAGQQIKASAQGAADAVKNA 60
D+ + + G+A G +K + D A++ AQSA + + GAGQ Q A A + A
Sbjct: 83 DATGRAMDKGRAAGATRDKAYDAKDRAADTAQSAADRARDGAGQTGSYIGQTAEAAKQKA 142
Query: 61 TGMNNNSK 68
G +K
Sbjct: 143 AGAAQYAK 150
>ref|NP_916529.1| WSI18 protein [Oryza sativa (japonica cultivar-group)]
gi|19571082|dbj|BAB86507.1| WSI18 protein induced by
water stress [Oryza sativa (japonica cultivar-group)]
gi|13872923|dbj|BAB44029.1| WSI18 protein induced by
water stress [Oryza sativa (japonica cultivar-group)]
Length = 215
Score = 45.1 bits (105), Expect = 5e-04
Identities = 21/55 (38%), Positives = 31/55 (56%)
Query: 4 QKMSYNAGQAKGQAEEKTSNLMDMASNAAQSAKETVQGAGQQIKASAQGAADAVK 58
++ SY+AG+ K +AEEKT +M A A+ AK+T A + GA +A K
Sbjct: 6 ERASYHAGETKARAEEKTGRMMGTAQEKAREAKDTASDAAGRAMGRGHGAKEATK 60
Score = 37.7 bits (86), Expect = 0.079
Identities = 25/70 (35%), Positives = 34/70 (47%), Gaps = 11/70 (15%)
Query: 10 AGQAKGQAEEKTSNLMDMASNAAQSAKET-----------VQGAGQQIKASAQGAADAVK 58
AGQ + A+ AAQ AKET +Q AG+Q+K+ A GA DAV
Sbjct: 125 AGQTGSYIGQTAEAAKQKAAGAAQYAKETAIAGKDKTGAVLQQAGEQVKSVAVGAKDAVM 184
Query: 59 NATGMNNNSK 68
GM+ ++K
Sbjct: 185 YTLGMSGDNK 194
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.295 0.108 0.262
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 82,128,483
Number of Sequences: 2540612
Number of extensions: 1966254
Number of successful extensions: 6198
Number of sequences better than 10.0: 162
Number of HSP's better than 10.0 without gapping: 118
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 5852
Number of HSP's gapped (non-prelim): 351
length of query: 68
length of database: 863,360,394
effective HSP length: 44
effective length of query: 24
effective length of database: 751,573,466
effective search space: 18037763184
effective search space used: 18037763184
T: 11
A: 40
X1: 17 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (21.9 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0215b.7


