

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0212.5
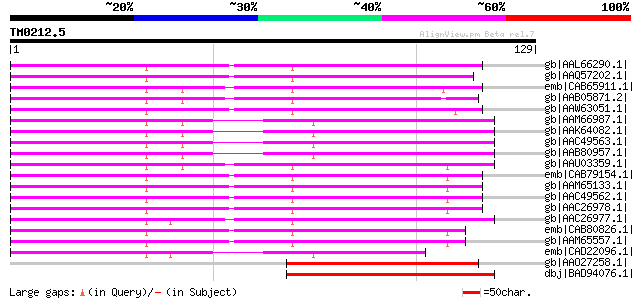
(129 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL66290.1| adenosine 5'-phosphosulfate reductase [Glycine max] 105 2e-22
gb|AAQ57202.1| adenosine 5' phosphosulfate reductase [Populus al... 97 7e-20
emb|CAB65911.1| adenosine 5'-phosphosulphate reductase [Lemna mi... 97 1e-19
gb|AAB05871.2| PAPS-reductase-like protein [Catharanthus roseus] 95 3e-19
gb|AAW63051.1| adenosine 5'-phosphosulfate reductase 1 [Zea mays] 94 1e-18
gb|AAM66987.1| 5'-adenylylphosphosulfate reductase, putative [Ar... 93 1e-18
gb|AAK64082.1| putative 5'-adenylylphosphosulfate reductase [Ara... 93 1e-18
gb|AAC49563.1| PRH43 [Arabidopsis thaliana] 93 1e-18
gb|AAB80957.1| adenosine-5'-phosphosulfate reductase [Arabidopsi... 93 1e-18
gb|AAU03359.1| adenylyl-sulfate reductase [Lycopersicon esculentum] 93 2e-18
emb|CAB79154.1| PRH26 protein [Arabidopsis thaliana] gi|2961344|... 91 5e-18
gb|AAM65133.1| PRH26 protein [Arabidopsis thaliana] 91 5e-18
gb|AAC49562.1| PRH26 [Arabidopsis thaliana] 91 5e-18
gb|AAC26978.1| 5'-adenylylphosphosulfate reductase [Arabidopsis ... 91 5e-18
gb|AAC26977.1| 5'-adenylylphosphosulfate reductase [Arabidopsis ... 91 8e-18
emb|CAB80826.1| 5'-adenylylsulfate reductase [Arabidopsis thalia... 90 1e-17
gb|AAM65557.1| 5-adenylylsulfate reductase [Arabidopsis thaliana] 90 1e-17
emb|CAD22096.1| adenosine 5' phosphosulfate reductase [Physcomit... 86 2e-16
gb|AAO27258.1| putative adenosine 5'-phosphosulphate reductase [... 75 5e-13
dbj|BAD94076.1| putative adenosine-5'-phosphosulfate reductase [... 64 1e-09
>gb|AAL66290.1| adenosine 5'-phosphosulfate reductase [Glycine max]
Length = 470
Score = 105 bits (262), Expect = 2e-22
Identities = 66/143 (46%), Positives = 78/143 (54%), Gaps = 28/143 (19%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 202 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGIGSLVKWNP 261
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 262 VANVNGLDIWSFLRT-MDVPVNSLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKAK 320
Query: 94 DCGLHKGNVKQDVGAQLNGNGAA 116
+CGLHKGN+K + AQLNGNGA+
Sbjct: 321 ECGLHKGNIKHEDAAQLNGNGAS 343
>gb|AAQ57202.1| adenosine 5' phosphosulfate reductase [Populus alba x Populus
tremula]
Length = 465
Score = 97.4 bits (241), Expect = 7e-20
Identities = 63/141 (44%), Positives = 75/141 (52%), Gaps = 28/141 (19%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +L+KWN
Sbjct: 195 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGAGSLIKWNP 254
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
+A + L M + ++ + + P VL QHEREG WWEDA AK
Sbjct: 255 MANVEGQDVWKFLRT-MDVPVNSLHSKGYISIGCEPCTRPVLPGQHEREGRWWWEDATAK 313
Query: 94 DCGLHKGNVKQDVGAQLNGNG 114
+CGLHKGN+KQ AQLNGNG
Sbjct: 314 ECGLHKGNLKQGDAAQLNGNG 334
>emb|CAB65911.1| adenosine 5'-phosphosulphate reductase [Lemna minor]
Length = 459
Score = 96.7 bits (239), Expect = 1e-19
Identities = 64/146 (43%), Positives = 77/146 (51%), Gaps = 32/146 (21%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR L+G+ I GQ KDQSPGT +L+KWN
Sbjct: 199 CCRVRKVRPLRRALRGLRAWITGQRKDQSPGTRASVPTVQVDPSFEGFEGGTGSLIKWNP 258
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
VA + I+ M + ++ + + P VL QHEREG WWEDAKA
Sbjct: 259 VANVDGQDIWRFLRT--MAVPVNSLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKA 316
Query: 93 KDCGLHKGNVKQD--VGAQLNGNGAA 116
K+CGLHKGN+KQD V NGNG A
Sbjct: 317 KECGLHKGNIKQDESVPPSSNGNGTA 342
>gb|AAB05871.2| PAPS-reductase-like protein [Catharanthus roseus]
Length = 464
Score = 95.1 bits (235), Expect = 3e-19
Identities = 63/143 (44%), Positives = 76/143 (53%), Gaps = 31/143 (21%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 197 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGMDGGVGSLVKWNP 256
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
VA + I+ M + ++ + + P VL QHEREG WWEDAKA
Sbjct: 257 VANVEGKDIWNFLRA--MDVPVNTLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKA 314
Query: 93 KDCGLHKGNVKQDVGAQLNGNGA 115
K+CGLHKGN+K++ NGNGA
Sbjct: 315 KECGLHKGNIKEET-LNNNGNGA 336
>gb|AAW63051.1| adenosine 5'-phosphosulfate reductase 1 [Zea mays]
Length = 461
Score = 93.6 bits (231), Expect = 1e-18
Identities = 64/147 (43%), Positives = 75/147 (50%), Gaps = 32/147 (21%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 195 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRASIPIVQVDPSFEGLDGGAGSLVKWNP 254
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 255 VANVDGKDIWTFLRT-MDVPVNTLHAQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKAK 313
Query: 94 DCGLHKGNVKQDVGA----QLNGNGAA 116
+CGLHKGN+ +D A NGNG+A
Sbjct: 314 ECGLHKGNIDKDAQAAAPRSANGNGSA 340
>gb|AAM66987.1| 5'-adenylylphosphosulfate reductase, putative [Arabidopsis
thaliana] gi|2098778|gb|AAB57688.1| APS reductase
[Arabidopsis thaliana]
Length = 455
Score = 93.2 bits (230), Expect = 1e-18
Identities = 62/157 (39%), Positives = 77/157 (48%), Gaps = 50/157 (31%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 195 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVDPVFEGLDGGVGSLVKWNP 254
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHERE 82
+A + A ++ +R P+ AL VL QHERE
Sbjct: 255 LANVEGADVWNF------------LRTMDVPVNALHAQGYVSIGCEPCTRPVLPGQHERE 302
Query: 83 GNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQV 119
G WWEDAKAK+CGLHKGN+K++ GA + A ++
Sbjct: 303 GRWWWEDAKAKECGLHKGNIKEEDGAADSKPAAVQEI 339
>gb|AAK64082.1| putative 5'-adenylylphosphosulfate reductase [Arabidopsis thaliana]
gi|13430560|gb|AAK25902.1| putative
5'-adenylylphosphosulfate reductase [Arabidopsis
thaliana] gi|15220723|ref|NP_176409.1|
5'-adenylylsulfate reductase 2, chloroplast (APR2)
(APSR) / adenosine 5'-phosphosulfate 5'-adenylylsulfate
(APS) sulfotransferase 2 /
3'-phosphoadenosine-5'-phosphosulfate (PAPS) reductase
homolog 43 (PRH-43) [Arabidopsis thaliana]
gi|2738758|gb|AAC26980.1| 5'-adenylylsulfate reductase
[Arabidopsis thaliana] gi|25288823|pir||C96648
hypothetical protein F19K23.11 [imported] - Arabidopsis
thaliana gi|2160142|gb|AAB60764.1| Strong similarity to
Arabidopsis APR2 (gb|U56921). [Arabidopsis thaliana]
gi|24211447|sp|P92981|APR2_ARATH 5'-adenylylsulfate
reductase 2, chloroplast precursor (Adenosine
5'-phosphosulfate 5'-adenylylsulfate sulfotransferase 2)
(APS sulfotransferase 2) (Thioredoxin independent APS
reductase 2) (3'-phosphoadenosine-5'-phosphosulfate
reductase homolog 43) (PAPS reductase homolog 43)
(Prh-43)
Length = 454
Score = 93.2 bits (230), Expect = 1e-18
Identities = 62/157 (39%), Positives = 77/157 (48%), Gaps = 50/157 (31%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHERE 82
+A + A ++ +R P+ AL VL QHERE
Sbjct: 254 LANVEGADVWNF------------LRTMDVPVNALHAQGYVSIGCEPCTRPVLPGQHERE 301
Query: 83 GNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQV 119
G WWEDAKAK+CGLHKGN+K++ GA + A ++
Sbjct: 302 GRWWWEDAKAKECGLHKGNIKEEDGAADSKPAAVQEI 338
>gb|AAC49563.1| PRH43 [Arabidopsis thaliana]
Length = 453
Score = 93.2 bits (230), Expect = 1e-18
Identities = 62/157 (39%), Positives = 77/157 (48%), Gaps = 50/157 (31%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 193 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVDPVFEGLDGGVGSLVKWNP 252
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHERE 82
+A + A ++ +R P+ AL VL QHERE
Sbjct: 253 LANVEGADVWNF------------LRTMDVPVNALHAQGYVSIGCEPCTRPVLPGQHERE 300
Query: 83 GNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQV 119
G WWEDAKAK+CGLHKGN+K++ GA + A ++
Sbjct: 301 GRWWWEDAKAKECGLHKGNIKEEDGAADSKPAAVQEI 337
>gb|AAB80957.1| adenosine-5'-phosphosulfate reductase [Arabidopsis thaliana]
Length = 454
Score = 93.2 bits (230), Expect = 1e-18
Identities = 62/157 (39%), Positives = 77/157 (48%), Gaps = 50/157 (31%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHERE 82
+A + A ++ +R P+ AL VL QHERE
Sbjct: 254 LANVEGADVWNF------------LRTMDVPVNALHAQGYVSIGCEPCTRPVLPGQHERE 301
Query: 83 GNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQV 119
G WWEDAKAK+CGLHKGN+K++ GA + A ++
Sbjct: 302 GRWWWEDAKAKECGLHKGNIKEEDGAADSKPAAVQEI 338
>gb|AAU03359.1| adenylyl-sulfate reductase [Lycopersicon esculentum]
Length = 461
Score = 92.8 bits (229), Expect = 2e-18
Identities = 62/150 (41%), Positives = 75/150 (49%), Gaps = 33/150 (22%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 198 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPIVQVDPSFEGLDGGAGSLVKWNP 257
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
VA + I+ M + ++ + + P VL QHEREG WWEDAKA
Sbjct: 258 VANVDGKDIWNFLRA--MNVPVNSLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKA 315
Query: 93 KDCGLHKGNVKQDV---GAQLNGNGAAMQV 119
K+CGLHKGN+K + AQ NG +
Sbjct: 316 KECGLHKGNIKDETVNGAAQTNGTATVADI 345
>emb|CAB79154.1| PRH26 protein [Arabidopsis thaliana] gi|2961344|emb|CAA18102.1|
PRH26 protein [Arabidopsis thaliana]
gi|21655305|gb|AAM65364.1| AT4g21990/F1N20_90
[Arabidopsis thaliana] gi|20259942|gb|AAM13318.1| PRH26
protein [Arabidopsis thaliana]
gi|17065178|gb|AAL32743.1| PRH26 protein [Arabidopsis
thaliana] gi|15234684|ref|NP_193930.1|
5'-adenylylsulfate reductase (APR3) / PAPS reductase
homolog (PRH26) [Arabidopsis thaliana]
gi|16226614|gb|AAL16214.1| AT4g21990/F1N20_90
[Arabidopsis thaliana] gi|15809816|gb|AAL06836.1|
AT4g21990/F1N20_90 [Arabidopsis thaliana]
gi|2738760|gb|AAC26981.1| 5'-adenylylsulfate reductase
[Arabidopsis thaliana] gi|11358631|pir||T49106 PRH26
protein - Arabidopsis thaliana
gi|24211446|sp|P92980|APR3_ARATH 5'-adenylylsulfate
reductase 3, chloroplast precursor (Adenosine
5'-phosphosulfate 5'-adenylylsulfate sulfotransferase 3)
(APS sulfotransferase 3) (Thioredoxin independent APS
reductase 3) (3'-phosphoadenosine-5'-phosphosulfate
reductase homolog 26) (PAPS reductase homolog 26)
(Prh-26)
Length = 458
Score = 91.3 bits (225), Expect = 5e-18
Identities = 62/145 (42%), Positives = 75/145 (50%), Gaps = 30/145 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR+RKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 254 VANVEGNDVWNFLRT-MDVPVNTLHAAGYVSIGCEPCTRAVLPGQHEREGRWWWEDAKAK 312
Query: 94 DCGLHKGNVKQDV--GAQLNGNGAA 116
+CGLHKGN+K++ A N NG A
Sbjct: 313 ECGLHKGNIKENTNGNATANVNGTA 337
>gb|AAM65133.1| PRH26 protein [Arabidopsis thaliana]
Length = 458
Score = 91.3 bits (225), Expect = 5e-18
Identities = 62/145 (42%), Positives = 75/145 (50%), Gaps = 30/145 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR+RKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 254 VANVEGNDVWNFLRT-MDVPVNTLHAAGYVSIGCEPCTRAVLPGQHEREGRWWWEDAKAK 312
Query: 94 DCGLHKGNVKQDV--GAQLNGNGAA 116
+CGLHKGN+K++ A N NG A
Sbjct: 313 ECGLHKGNIKENTNGNATANVNGTA 337
>gb|AAC49562.1| PRH26 [Arabidopsis thaliana]
Length = 458
Score = 91.3 bits (225), Expect = 5e-18
Identities = 62/145 (42%), Positives = 75/145 (50%), Gaps = 30/145 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR+RKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 254 VANVEGNDVWNFLRT-MDVPVNTLHAAGYVSIGCEPCTRAVLPGQHEREGRWWWEDAKAK 312
Query: 94 DCGLHKGNVKQDV--GAQLNGNGAA 116
+CGLHKGN+K++ A N NG A
Sbjct: 313 ECGLHKGNIKENTNGNATANVNGTA 337
>gb|AAC26978.1| 5'-adenylylphosphosulfate reductase [Arabidopsis thaliana]
gi|2129519|pir||S71243 phosphoadenylyl-sulfate reductase
(thioredoxin) (EC 1.8.4.8) APR3 precursor - Arabidopsis
thaliana (fragment)
Length = 422
Score = 91.3 bits (225), Expect = 5e-18
Identities = 62/145 (42%), Positives = 75/145 (50%), Gaps = 30/145 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR+RKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 167 CCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 226
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 227 VANVEGNDVWNFLRT-MDVPVNTLHAAGYVSIGCEPCTRAVLPGQHEREGRWWWEDAKAK 285
Query: 94 DCGLHKGNVKQDV--GAQLNGNGAA 116
+CGLHKGN+K++ A N NG A
Sbjct: 286 ECGLHKGNIKENTNGNATANVNGTA 310
>gb|AAC26977.1| 5'-adenylylphosphosulfate reductase [Arabidopsis thaliana]
gi|2129518|pir||S71242 phosphoadenylyl-sulfate reductase
(thioredoxin) (EC 1.8.4.8) APR2 - Arabidopsis thaliana
Length = 406
Score = 90.5 bits (223), Expect = 8e-18
Identities = 59/147 (40%), Positives = 77/147 (52%), Gaps = 30/147 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWN- 38
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 146 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVDPVFEGLDGGVGSLVKWNP 205
Query: 39 LVAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
L + A ++ M + ++ + + P + VL QHEREG WWEDAKA
Sbjct: 206 LANVEGADVWNFLRT--MDVPVNALHAQGYVSIGCEPCTSPVLPGQHEREGRWWWEDAKA 263
Query: 93 KDCGLHKGNVKQDVGAQLNGNGAAMQV 119
K+CGLHKGN+ ++ GA + A ++
Sbjct: 264 KECGLHKGNIFEEDGAADSKPAAVQEI 290
>emb|CAB80826.1| 5'-adenylylsulfate reductase [Arabidopsis thaliana]
gi|27363218|gb|AAO11528.1| At4g04610/F4H6_13
[Arabidopsis thaliana] gi|15983416|gb|AAL11576.1|
AT4g04610/F4H6_13 [Arabidopsis thaliana]
gi|4773905|gb|AAD29775.1| 5'-adenylylsulfate reductase
[Arabidopsis thaliana] gi|2738756|gb|AAC26979.1|
5'-adenylylsulfate reductase [Arabidopsis thaliana]
gi|15233591|ref|NP_192370.1| 5'-adenylylsulfate
reductase (APR1) / PAPS reductase homolog (PRH19)
[Arabidopsis thaliana] gi|25288822|pir||B85058
5'-adenylylsulfate reductase [imported] - Arabidopsis
thaliana gi|24211445|sp|P92979|APR1_ARATH
5'-adenylylsulfate reductase 1, chloroplast precursor
(Adenosine 5'-phosphosulfate 5'-adenylylsulfate
sulfotransferase 1) (APS sulfotransferase 1)
(Thioredoxin independent APS reductase 1)
(3'-phosphoadenosine-5'-phosphosulfate reductase homolog
19) (PAPS reductase homolog 19) (Prh-19)
Length = 465
Score = 90.1 bits (222), Expect = 1e-17
Identities = 62/140 (44%), Positives = 75/140 (53%), Gaps = 29/140 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 202 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 261
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 262 VANVEGNDVWNFLRT-MDVPVNTLHAAGYISIGCEPCTKAVLPGQHEREGRWWWEDAKAK 320
Query: 94 DCGLHKGNVKQDV-GAQLNG 112
+CGLHKGNVK++ A++NG
Sbjct: 321 ECGLHKGNVKENSDDAKVNG 340
>gb|AAM65557.1| 5-adenylylsulfate reductase [Arabidopsis thaliana]
Length = 465
Score = 90.1 bits (222), Expect = 1e-17
Identities = 62/140 (44%), Positives = 75/140 (53%), Gaps = 29/140 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 202 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 261
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 262 VANVEGNDVWNFLRT-MDVPVNTLHAAGYISIGCEPCTKAVLPGQHEREGRWWWEDAKAK 320
Query: 94 DCGLHKGNVKQDV-GAQLNG 112
+CGLHKGNVK++ A++NG
Sbjct: 321 ECGLHKGNVKENSDDAKVNG 340
>emb|CAD22096.1| adenosine 5' phosphosulfate reductase [Physcomitrella patens]
Length = 465
Score = 85.9 bits (211), Expect = 2e-16
Identities = 58/140 (41%), Positives = 67/140 (47%), Gaps = 50/140 (35%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWN- 38
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 203 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRANVPVVQVDPAFEGLDGGPGSLVKWNP 262
Query: 39 LVAISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHERE 82
L +S ++ +R P+ AL VL QHERE
Sbjct: 263 LSNVSGTAVWSF------------LRTMDVPVNALHFKGYVSIGCEPCTRAVLPGQHERE 310
Query: 83 GNRWWEDAKAKDCGLHKGNV 102
G WWEDAKAK+CGLHKGN+
Sbjct: 311 GRWWWEDAKAKECGLHKGNL 330
>gb|AAO27258.1| putative adenosine 5'-phosphosulphate reductase [Pisum sativum]
Length = 192
Score = 74.7 bits (182), Expect = 5e-13
Identities = 33/47 (70%), Positives = 36/47 (76%)
Query: 69 PLEALVLREQHEREGNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGA 115
P VL QHEREG WWEDAKAK+CGLHKGN+KQ+ A LNGNGA
Sbjct: 17 PCTRAVLPGQHEREGRWWWEDAKAKECGLHKGNIKQEDDAHLNGNGA 63
>dbj|BAD94076.1| putative adenosine-5'-phosphosulfate reductase [Arabidopsis
thaliana]
Length = 186
Score = 63.5 bits (153), Expect = 1e-09
Identities = 28/51 (54%), Positives = 35/51 (67%)
Query: 69 PLEALVLREQHEREGNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQV 119
P VL QHEREG WWEDAKAK+CGLHKGN+K++ GA + A ++
Sbjct: 20 PCTRPVLPGQHEREGRWWWEDAKAKECGLHKGNIKEEDGAADSKPAAVQEI 70
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.344 0.153 0.534
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 194,756,831
Number of Sequences: 2540612
Number of extensions: 6865819
Number of successful extensions: 23152
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 89
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 22959
Number of HSP's gapped (non-prelim): 167
length of query: 129
length of database: 863,360,394
effective HSP length: 105
effective length of query: 24
effective length of database: 596,596,134
effective search space: 14318307216
effective search space used: 14318307216
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0212.5


