

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0212.10
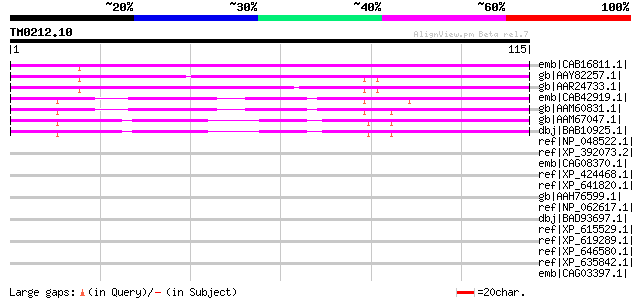
(115 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB16811.1| putative protein [Arabidopsis thaliana] gi|72705... 93 2e-18
gb|AAY82257.1| hypothetical protein At2g18200 [Arabidopsis thali... 84 6e-16
gb|AAR24733.1| At2g18210 [Arabidopsis thaliana] gi|38603892|gb|A... 84 1e-15
emb|CAB42919.1| hypothetical protein [Arabidopsis thaliana] gi|2... 83 1e-15
gb|AAM60831.1| unknown [Arabidopsis thaliana] 83 1e-15
gb|AAM67047.1| unknown [Arabidopsis thaliana] 77 8e-14
dbj|BAB10925.1| unnamed protein product [Arabidopsis thaliana] g... 76 2e-13
ref|NP_048522.1| a174L [Paramecium bursaria Chlorella virus 1] g... 33 1.3
ref|XP_392073.2| PREDICTED: similar to Colorectal mutant cancer ... 33 2.1
emb|CAG08370.1| unnamed protein product [Tetraodon nigroviridis] 32 2.8
ref|XP_424468.1| PREDICTED: similar to C1orf25 [Gallus gallus] 32 3.6
ref|XP_641820.1| hypothetical protein DDB0204704 [Dictyostelium ... 32 3.6
gb|AAH76599.1| Tera protein [Mus musculus] gi|26338165|dbj|BAC32... 32 4.8
ref|NP_062617.1| teratocarcinoma expressed, serine rich [Mus mus... 32 4.8
dbj|BAD93697.1| vitellogenin [Gambusia affinis] 32 4.8
ref|XP_615529.1| PREDICTED: similar to a disintegrin-like and me... 31 6.2
ref|XP_619289.1| PREDICTED: similar to Tera [Mus musculus] 31 8.1
ref|XP_646580.1| hypothetical protein DDB0190842 [Dictyostelium ... 31 8.1
ref|XP_635842.1| hypothetical protein DDB0219539 [Dictyostelium ... 31 8.1
emb|CAG03397.1| unnamed protein product [Tetraodon nigroviridis] 31 8.1
>emb|CAB16811.1| putative protein [Arabidopsis thaliana] gi|7270598|emb|CAB80316.1|
putative protein [Arabidopsis thaliana]
gi|28827518|gb|AAO50603.1| unknown protein [Arabidopsis
thaliana] gi|28393196|gb|AAO42028.1| unknown protein
[Arabidopsis thaliana] gi|15234426|ref|NP_195368.1|
expressed protein [Arabidopsis thaliana]
gi|25334980|pir||H85430 hypothetical protein AT4g36500
[imported] - Arabidopsis thaliana
Length = 122
Score = 92.8 bits (229), Expect = 2e-18
Identities = 52/120 (43%), Positives = 69/120 (57%), Gaps = 5/120 (4%)
Query: 1 MLRSFTTRRYERLG-----KETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQ 55
MLRS +TR R G + ++ +LL +RSTS+P A S L
Sbjct: 3 MLRSLSTRTRSRRGGYERVSDDSTFSLLGAKLRRSTSVPYYAPSIKLGAGGVPTILEELP 62
Query: 56 RNPTKKANNSSKSTHPLFSFLDFRRKKKTTARPEFTRYLEYLKEGGMWDLNSNKPVIHYK 115
R +KK + K +HP+FS ++KK TT +PEF+RYLEYLKEGGMWD +N PVI+YK
Sbjct: 63 RQKSKKVKPTGKFSHPIFSLFYGKKKKSTTTKPEFSRYLEYLKEGGMWDARANAPVIYYK 122
>gb|AAY82257.1| hypothetical protein At2g18200 [Arabidopsis thaliana]
Length = 124
Score = 84.3 bits (207), Expect = 6e-16
Identities = 55/123 (44%), Positives = 71/123 (57%), Gaps = 9/123 (7%)
Query: 1 MLRSFTTRRYERLG-----KETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQ 55
MLR+ +TR R G + ++ +LL +RSTS+P A S R L
Sbjct: 3 MLRNLSTRTRSRRGGYERVSDDSTFSLLGAKLRRSTSVPYYAPS-IRLGGDXPVILEKLP 61
Query: 56 RNPTKKANNSSKSTHPLFSFLD-FRR--KKKTTARPEFTRYLEYLKEGGMWDLNSNKPVI 112
R K +SK +HP+FS D +RR KKK TA+PEF+RY EYLKE GMWDL SN PVI
Sbjct: 62 RQKPTKTVVTSKLSHPIFSLFDGYRRHNKKKATAKPEFSRYHEYLKESGMWDLRSNSPVI 121
Query: 113 HYK 115
++K
Sbjct: 122 YFK 124
>gb|AAR24733.1| At2g18210 [Arabidopsis thaliana] gi|38603892|gb|AAR24691.1|
At2g18210 [Arabidopsis thaliana]
gi|20198202|gb|AAM15456.1| unknown protein [Arabidopsis
thaliana] gi|4309745|gb|AAD15515.1| unknown protein
[Arabidopsis thaliana] gi|15224130|ref|NP_179413.1|
expressed protein [Arabidopsis thaliana]
gi|25334982|pir||F84561 hypothetical protein At2g18210
[imported] - Arabidopsis thaliana
Length = 124
Score = 83.6 bits (205), Expect = 1e-15
Identities = 54/123 (43%), Positives = 72/123 (57%), Gaps = 9/123 (7%)
Query: 1 MLRSFTTRRYERLG-----KETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQ 55
MLR+ +TR R G + ++ +LL +RSTS+P A S +
Sbjct: 3 MLRNLSTRTRSRRGGYERVSDDSTFSLLGAKLRRSTSVPYYAPSIRLGGDFPVILEKLPR 62
Query: 56 RNPTKKANNSSKSTHPLFSFLD-FRR--KKKTTARPEFTRYLEYLKEGGMWDLNSNKPVI 112
+ PTK +SK +HP+FS D +RR KKK TA+PEF+RY EYLKE GMWDL SN PVI
Sbjct: 63 QKPTKTVV-TSKLSHPIFSLFDGYRRHNKKKATAKPEFSRYHEYLKESGMWDLRSNSPVI 121
Query: 113 HYK 115
++K
Sbjct: 122 YFK 124
>emb|CAB42919.1| hypothetical protein [Arabidopsis thaliana]
gi|26450545|dbj|BAC42385.1| unknown protein [Arabidopsis
thaliana] gi|18409206|ref|NP_566940.1| expressed protein
[Arabidopsis thaliana] gi|7459217|pir||T08411
hypothetical protein F18B3.180 - Arabidopsis thaliana
Length = 114
Score = 83.2 bits (204), Expect = 1e-15
Identities = 52/129 (40%), Positives = 71/129 (54%), Gaps = 29/129 (22%)
Query: 1 MLRSFTTRR----YERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQR 56
M R+ +TR+ YE+LG E A KR +S+P+ +R +++
Sbjct: 1 MFRAMSTRKIHGGYEKLGDEEAR-------LKRVSSVPASVYGHSRNPVQ------EVKK 47
Query: 57 NPTKKANNSSKSTHPLFSFLD--FRRKKKTTAR--------PEFTRYLEYLKEGGMWDLN 106
PT K S HPLFSF D F+RKKK TA+ PEF RY+EY++EGG+WD +
Sbjct: 48 TPTAKPTGGS--VHPLFSFFDVHFQRKKKNTAKKKSLATAKPEFARYMEYVREGGVWDPS 105
Query: 107 SNKPVIHYK 115
SN PVIHY+
Sbjct: 106 SNAPVIHYR 114
>gb|AAM60831.1| unknown [Arabidopsis thaliana]
Length = 114
Score = 83.2 bits (204), Expect = 1e-15
Identities = 52/129 (40%), Positives = 71/129 (54%), Gaps = 29/129 (22%)
Query: 1 MLRSFTTRR----YERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQR 56
M R+ +TR+ YE+LG E A KR +S+P+ +R +++
Sbjct: 1 MFRAMSTRKVHGGYEKLGDEEAR-------LKRVSSVPASVYGHSRNPVQ------EVKK 47
Query: 57 NPTKKANNSSKSTHPLFSFLD--FRRKKK--------TTARPEFTRYLEYLKEGGMWDLN 106
PT K S HPLFSF D F+RKKK TA+PEF RY+EY++EGG+WD +
Sbjct: 48 TPTAKPTGGS--VHPLFSFFDVHFQRKKKKTTKKKSLATAKPEFARYMEYVREGGVWDPS 105
Query: 107 SNKPVIHYK 115
SN PVIHY+
Sbjct: 106 SNAPVIHYR 114
>gb|AAM67047.1| unknown [Arabidopsis thaliana]
Length = 109
Score = 77.4 bits (189), Expect = 8e-14
Identities = 50/125 (40%), Positives = 70/125 (56%), Gaps = 26/125 (20%)
Query: 1 MLRSFTTRR----YERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQR 56
M+R+ +TR+ Y++LG E + A L E + S+P+ + K+
Sbjct: 1 MIRALSTRKVRGSYKKLGDEEEAGAGLLE--VKPESVPTNPHGQSPKV-----------N 47
Query: 57 NPTKKANNSSKSTHPLFSFLDF---RRKKK---TTARPEFTRYLEYLKEGGMWDLNSNKP 110
P +K S+ HPLFSF D R+KKK TTA+PEF RYLEY+KEGG+WD SN P
Sbjct: 48 KPVEKTRGSA---HPLFSFFDMSLKRKKKKKSTTTAKPEFARYLEYVKEGGVWDNTSNGP 104
Query: 111 VIHYK 115
I+Y+
Sbjct: 105 AIYYR 109
>dbj|BAB10925.1| unnamed protein product [Arabidopsis thaliana]
gi|26450203|dbj|BAC42220.1| unknown protein [Arabidopsis
thaliana] gi|28827370|gb|AAO50529.1| unknown protein
[Arabidopsis thaliana] gi|15239976|ref|NP_201450.1|
expressed protein [Arabidopsis thaliana]
Length = 109
Score = 75.9 bits (185), Expect = 2e-13
Identities = 49/125 (39%), Positives = 70/125 (55%), Gaps = 26/125 (20%)
Query: 1 MLRSFTTRR----YERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQR 56
M+R+ +TR+ Y++LG E + A L E + S+P+ + K+
Sbjct: 1 MIRALSTRKVRGSYKKLGDEEEAGAGLLE--VKPESVPTNPHGQSPKV-----------N 47
Query: 57 NPTKKANNSSKSTHPLFSFLDF---RRKKK---TTARPEFTRYLEYLKEGGMWDLNSNKP 110
P +K S+ HPLFSF + R+KKK TTA+PEF RYLEY+KEGG+WD SN P
Sbjct: 48 KPVEKTRGSA---HPLFSFFEMSLKRKKKKKSTTTAKPEFARYLEYVKEGGVWDNTSNGP 104
Query: 111 VIHYK 115
I+Y+
Sbjct: 105 AIYYR 109
>ref|NP_048522.1| a174L [Paramecium bursaria Chlorella virus 1]
gi|1181337|gb|AAC96542.1| a174L [Paramecium bursaria
Chlorella virus 1] gi|7461320|pir||T17665 hypothetical
protein a174L - Chlorella virus PBCV-1
Length = 65
Score = 33.5 bits (75), Expect = 1.3
Identities = 19/44 (43%), Positives = 23/44 (52%), Gaps = 3/44 (6%)
Query: 28 KRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSKSTHP 71
KR S P RASS RK A F + P KK NN++ + HP
Sbjct: 8 KRRASSPKRASSPKRKGVMALF---KKKSAPKKKTNNNNNNNHP 48
>ref|XP_392073.2| PREDICTED: similar to Colorectal mutant cancer protein (MCC
protein) [Apis mellifera]
Length = 1941
Score = 32.7 bits (73), Expect = 2.1
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 4/76 (5%)
Query: 11 ERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTK---KANNSSK 67
E +G+E +S++L +G +RS LP+ ++ S + AT + +++ A SK
Sbjct: 564 EGIGRERETSSILYDGVQRS-QLPAASTLSQPTLLDATTTSSKIEQTSYACLGVAMGRSK 622
Query: 68 STHPLFSFLDFRRKKK 83
ST P S + F+ K+K
Sbjct: 623 STEPEESVMGFKDKQK 638
>emb|CAG08370.1| unnamed protein product [Tetraodon nigroviridis]
Length = 238
Score = 32.3 bits (72), Expect = 2.8
Identities = 21/73 (28%), Positives = 35/73 (47%), Gaps = 3/73 (4%)
Query: 8 RRYERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATF---GNINLQRNPTKKANN 64
RR E L A + + + SL A+SS M H++ G+ + Q + T +++
Sbjct: 162 RREEELANMAAQTHIRAPASPDAESLTPPAASSRPTMLHSSVTLQGSFSAQTSFTGESST 221
Query: 65 SSKSTHPLFSFLD 77
+ HP FSFL+
Sbjct: 222 QVRHVHPFFSFLN 234
>ref|XP_424468.1| PREDICTED: similar to C1orf25 [Gallus gallus]
Length = 908
Score = 32.0 bits (71), Expect = 3.6
Identities = 15/51 (29%), Positives = 23/51 (44%)
Query: 51 NINLQRNPTKKANNSSKSTHPLFSFLDFRRKKKTTARPEFTRYLEYLKEGG 101
N + RN TK+ + + HP F + R K P+ ++L L E G
Sbjct: 766 NNEVMRNATKRQKTDNSAEHPAFYYNIHRHSIKGMNMPKLNKFLNCLSEAG 816
>ref|XP_641820.1| hypothetical protein DDB0204704 [Dictyostelium discoideum]
gi|60469866|gb|EAL67852.1| hypothetical protein
DDB0204704 [Dictyostelium discoideum]
Length = 1317
Score = 32.0 bits (71), Expect = 3.6
Identities = 18/55 (32%), Positives = 27/55 (48%), Gaps = 5/55 (9%)
Query: 17 TASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSKSTHP 71
T SS + + R T +PS+A SS F N N+ N +NN++K+ P
Sbjct: 371 TTSSLSNTKSYIRPTPMPSKAQSS-----RTNFNNNNINNNYYNNSNNNNKNRQP 420
>gb|AAH76599.1| Tera protein [Mus musculus] gi|26338165|dbj|BAC32768.1| unnamed
protein product [Mus musculus]
Length = 221
Score = 31.6 bits (70), Expect = 4.8
Identities = 20/61 (32%), Positives = 34/61 (54%), Gaps = 2/61 (3%)
Query: 23 LQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSKSTHPLFSFLDFRRKK 82
LQ+ FKR S +SSA + N + + + T+ A++S+++ P+FSFLD K
Sbjct: 111 LQKEFKRHNSDAHSTTSSASPAQSPCYSNQSDEGSDTEMASSSNRT--PVFSFLDLTYWK 168
Query: 83 K 83
+
Sbjct: 169 R 169
>ref|NP_062617.1| teratocarcinoma expressed, serine rich [Mus musculus]
gi|1575505|gb|AAB09547.1| Tera
Length = 277
Score = 31.6 bits (70), Expect = 4.8
Identities = 20/61 (32%), Positives = 34/61 (54%), Gaps = 2/61 (3%)
Query: 23 LQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSKSTHPLFSFLDFRRKK 82
LQ+ FKR S +SSA + N + + + T+ A++S+++ P+FSFLD K
Sbjct: 167 LQKEFKRHNSDAHSTTSSASPAQSPCYSNQSDEGSDTEMASSSNRT--PVFSFLDLTYWK 224
Query: 83 K 83
+
Sbjct: 225 R 225
>dbj|BAD93697.1| vitellogenin [Gambusia affinis]
Length = 1695
Score = 31.6 bits (70), Expect = 4.8
Identities = 25/87 (28%), Positives = 39/87 (44%), Gaps = 3/87 (3%)
Query: 3 RSFTTRRYERLGKETASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKA 62
RS + R + SSA +S SR+SSS K F ++ + NP+K A
Sbjct: 1155 RSRSGRSRSSSSSDRTSSASSIASLFIDSSSSSRSSSSRSKQVTERFQRLHNKENPSKSA 1214
Query: 63 NNS---SKSTHPLFSFLDFRRKKKTTA 86
+ S S S +F+ F R+++ A
Sbjct: 1215 SRSRSISASVEAIFNQKKFLREEEVVA 1241
>ref|XP_615529.1| PREDICTED: similar to a disintegrin-like and metalloprotease
(reprolysin type) with thrombospondin type 1 motif, 3
proprotein, partial [Bos taurus]
Length = 399
Score = 31.2 bits (69), Expect = 6.2
Identities = 14/45 (31%), Positives = 24/45 (53%)
Query: 40 SARKMAHATFGNINLQRNPTKKANNSSKSTHPLFSFLDFRRKKKT 84
S KM H +F +N + P ++ N + THPL++ ++ KT
Sbjct: 68 SDNKMVHRSFCEVNKKPKPIRRMCNIQECTHPLWAAEEWEHCTKT 112
>ref|XP_619289.1| PREDICTED: similar to Tera [Mus musculus]
Length = 177
Score = 30.8 bits (68), Expect = 8.1
Identities = 20/61 (32%), Positives = 34/61 (54%), Gaps = 2/61 (3%)
Query: 23 LQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNSSKSTHPLFSFLDFRRKK 82
LQ+ FKR S +SSA + N + + + T+ A++S+++ P+FSFLD K
Sbjct: 67 LQKEFKRHNSDAHSTTSSASPGQSPCYSNQSDEGSDTEMASSSNRT--PVFSFLDLTYWK 124
Query: 83 K 83
+
Sbjct: 125 R 125
>ref|XP_646580.1| hypothetical protein DDB0190842 [Dictyostelium discoideum]
gi|60474488|gb|EAL72425.1| hypothetical protein
DDB0190842 [Dictyostelium discoideum]
Length = 889
Score = 30.8 bits (68), Expect = 8.1
Identities = 14/48 (29%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query: 52 INLQRNPTKKANNSSKSTHPLFSFLDFRRKKKTTARPEFTRYL-EYLK 98
++++ N NN + + + +F+F+DF K K F++Y+ E LK
Sbjct: 655 LSIENNNNNNNNNDNNNKNEIFTFIDFSNKFKEIGNIFFSKYIREQLK 702
>ref|XP_635842.1| hypothetical protein DDB0219539 [Dictyostelium discoideum]
gi|60464186|gb|EAL62346.1| hypothetical protein
DDB0219539 [Dictyostelium discoideum]
Length = 936
Score = 30.8 bits (68), Expect = 8.1
Identities = 18/48 (37%), Positives = 30/48 (62%), Gaps = 2/48 (4%)
Query: 18 ASSALLQEGFKRSTSLPSRASSSARKMAHATFGNINLQR-NPTKKANN 64
A LL+EG+ + + S +SSS+ + ++ +IN+Q+ NPT K NN
Sbjct: 206 ALKILLEEGYYQFEQISSPSSSSSSSSSSSS-SSINIQKTNPTSKINN 252
>emb|CAG03397.1| unnamed protein product [Tetraodon nigroviridis]
Length = 209
Score = 30.8 bits (68), Expect = 8.1
Identities = 22/78 (28%), Positives = 41/78 (52%), Gaps = 4/78 (5%)
Query: 8 RRYERLGKETASSAL--LQEGFKRSTSLPSRASSSARKMAHATFGNINLQRNPTKKANNS 65
++ + + K+ S + LQ+ KRS S +SSA ++ +++ + T+ + S
Sbjct: 93 KKIKSISKKARPSQISRLQKELKRSNSDAHSTTSSASPAQSPSYSHLSDDGSDTELSPGS 152
Query: 66 SKSTHPLFSFLDFRRKKK 83
S+S P+FSFLD K+
Sbjct: 153 SRS--PVFSFLDLTYWKR 168
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.127 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 182,114,003
Number of Sequences: 2540612
Number of extensions: 6128725
Number of successful extensions: 18897
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 18873
Number of HSP's gapped (non-prelim): 23
length of query: 115
length of database: 863,360,394
effective HSP length: 91
effective length of query: 24
effective length of database: 632,164,702
effective search space: 15171952848
effective search space used: 15171952848
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0212.10


