

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0211.14
(280 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
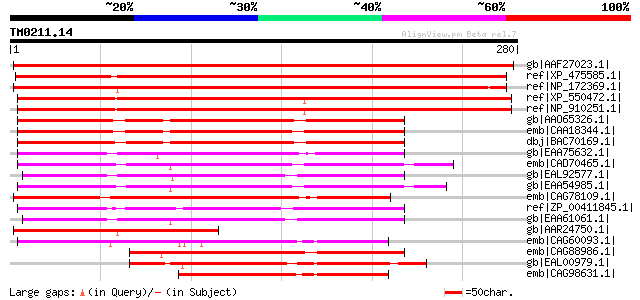
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF27023.1| unknown protein [Arabidopsis thaliana] gi|1522991... 422 e-117
ref|XP_475585.1| unknown protein [Oryza sativa (japonica cultiva... 391 e-107
ref|NP_172369.1| phosphoglycerate/bisphosphoglycerate mutase fam... 390 e-107
ref|XP_550472.1| hypothetical protein [Oryza sativa (japonica cu... 288 9e-77
ref|NP_910251.1| P0514G12.25 [Oryza sativa (japonica cultivar-gr... 288 9e-77
gb|AAO65326.1| unknown [Streptomyces murayamaensis] 196 6e-49
emb|CAA18344.1| conserved hypothetical protein SC4H2.29 [Strepto... 196 8e-49
dbj|BAC70169.1| hypothetical protein [Streptomyces avermitilis M... 190 3e-47
gb|EAA75632.1| hypothetical protein FG05987.1 [Gibberella zeae P... 185 1e-45
emb|CAD70465.1| conserved hypothetical protein [Neurospora crass... 184 2e-45
gb|EAL92577.1| phosphoglycerate mutase family domain protein [As... 180 3e-44
gb|EAA54985.1| hypothetical protein MG06642.4 [Magnaporthe grise... 179 8e-44
emb|CAG78109.1| unnamed protein product [Yarrowia lipolytica CLI... 179 1e-43
ref|ZP_00411845.1| Phosphoglycerate/bisphosphoglycerate mutase [... 172 1e-41
gb|EAA61061.1| hypothetical protein AN4983.2 [Aspergillus nidula... 170 4e-41
gb|AAR24750.1| At1g08940 [Arabidopsis thaliana] gi|38454096|gb|A... 160 5e-38
emb|CAG60093.1| unnamed protein product [Candida glabrata CBS138... 137 4e-31
emb|CAG88986.1| unnamed protein product [Debaryomyces hansenii C... 136 6e-31
gb|EAL00979.1| hypothetical protein CaO19.6747 [Candida albicans... 135 1e-30
emb|CAG98631.1| unnamed protein product [Kluyveromyces lactis NR... 120 3e-26
>gb|AAF27023.1| unknown protein [Arabidopsis thaliana] gi|15229917|ref|NP_187168.1|
phosphoglycerate/bisphosphoglycerate mutase family
protein [Arabidopsis thaliana]
Length = 316
Score = 422 bits (1084), Expect = e-117
Identities = 194/276 (70%), Positives = 232/276 (83%)
Query: 3 VLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPD 62
+LPKRIIL+RHGES+GNLDT+AYTTTPDH IQLT G+ QA+ AG RL +++++ SP+
Sbjct: 7 LLPKRIILVRHGESEGNLDTAAYTTTPDHKIQLTDSGLLQAQEAGARLHALISSNPSSPE 66
Query: 63 WRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRE 122
WRV FYVSPY RTRSTLRE+GRSFS++RVIGVREE RIREQDFGNFQV+ERM+ K+ RE
Sbjct: 67 WRVYFYVSPYDRTRSTLREIGRSFSRRRVIGVREECRIREQDFGNFQVKERMRATKKVRE 126
Query: 123 RFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIF 182
RFGRFFYRFPEGESAADV+DR+S F E+LWRDID+NRLH +PS++LN VIVSHGLTSR+F
Sbjct: 127 RFGRFFYRFPEGESAADVFDRVSSFLESLWRDIDMNRLHINPSHELNFVIVSHGLTSRVF 186
Query: 183 LMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWGLSPEMVADQKW 242
LMKWFKW+VEQFE LNN GN E RVME G GG+YSLA+HHTEEE+ WGLSPEM+ADQKW
Sbjct: 187 LMKWFKWSVEQFEGLNNPGNSEIRVMELGQGGDYSLAIHHTEEELATWGLSPEMIADQKW 246
Query: 243 RAGASRGAWNDQCSWYLDGFFDRLAEESDDEEASSS 278
RA A +G W + C WY FFD +A+ + E ++
Sbjct: 247 RANAHKGEWKEDCKWYFGDFFDHMADSDKECETEAT 282
>ref|XP_475585.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|46485803|gb|AAS98428.1| unknown protein [Oryza sativa
(japonica cultivar-group)] gi|46391117|gb|AAS90644.1|
unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 295
Score = 391 bits (1005), Expect = e-107
Identities = 181/271 (66%), Positives = 224/271 (81%), Gaps = 3/271 (1%)
Query: 4 LPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDW 63
LPKRIIL+RHGESQGNLD SAYTTTPD+ I LTP G+ QAR AG + V+++ + +W
Sbjct: 13 LPKRIILVRHGESQGNLDMSAYTTTPDYRIPLTPLGVDQARAAGKGILDVVSS---AANW 69
Query: 64 RVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRER 123
+V FYVSPY RTR+TLRE+G +F + RVIG REE R+REQDFGNFQVEERM+ +K+TR+R
Sbjct: 70 KVYFYVSPYERTRATLREIGAAFPRHRVIGAREECRVREQDFGNFQVEERMRAVKETRDR 129
Query: 124 FGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIFL 183
FGRFF+RFPEGESAADV+DR++ F E+LWRDID+ RL D S + NLVIVSHGLTSR+FL
Sbjct: 130 FGRFFFRFPEGESAADVFDRVASFLESLWRDIDMGRLEQDASCETNLVIVSHGLTSRVFL 189
Query: 184 MKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWGLSPEMVADQKWR 243
MKWFKWTV+QFE LNNF NCEFRVM+ G GEYSL +HHT+EE+ +WGLSPEM+ADQ+WR
Sbjct: 190 MKWFKWTVDQFERLNNFDNCEFRVMQLGPAGEYSLLIHHTKEELQRWGLSPEMIADQQWR 249
Query: 244 AGASRGAWNDQCSWYLDGFFDRLAEESDDEE 274
A A+R +W D+CS +L FFD E+ +D++
Sbjct: 250 ASANRRSWADECSSFLATFFDHWNEDDNDDD 280
>ref|NP_172369.1| phosphoglycerate/bisphosphoglycerate mutase family protein
[Arabidopsis thaliana] gi|25407069|pir||C86221
hypothetical protein [imported] - Arabidopsis thaliana
gi|2342686|gb|AAB70412.1| Similar to Saccharomyces
hypothetical protein YDR051c (gb|Z49209). ESTs
gb|T44436,gb|42252 come from this gene. [Arabidopsis
thaliana]
Length = 281
Score = 390 bits (1003), Expect = e-107
Identities = 186/276 (67%), Positives = 224/276 (80%), Gaps = 5/276 (1%)
Query: 3 VLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADG---C 59
+LPKRIILMRHGES GN+D AY TTPDH I LT +G +QAR AG ++R +++ C
Sbjct: 7 MLPKRIILMRHGESAGNIDAGAYATTPDHKIPLTEEGRAQAREAGKKMRALISTQSGGAC 66
Query: 60 SPDWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQ 119
+WRV FYVSPY RTR+TLREVG+ FS+KRVIGVREE RIREQDFGNFQVEERM+++K+
Sbjct: 67 GENWRVYFYVSPYERTRTTLREVGKGFSRKRVIGVREECRIREQDFGNFQVEERMRVVKE 126
Query: 120 TRERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTS 179
TRERFGRFFYRFPEGESAADVYDR+S F E++WRD+D+NR DPS++LNLVIVSHGLTS
Sbjct: 127 TRERFGRFFYRFPEGESAADVYDRVSSFLESMWRDVDMNRHQVDPSSELNLVIVSHGLTS 186
Query: 180 RIFLMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWGLSPEMVAD 239
R+FL KWFKWTV +FE LNNFGNCEFRVME G+ GEY+ A+HH+EEEML WG+S +M+ D
Sbjct: 187 RVFLTKWFKWTVAEFERLNNFGNCEFRVMELGASGEYTFAIHHSEEEMLDWGMSKDMIDD 246
Query: 240 QKWRAGASR-GAWNDQCSWYLDGFFDRLAEESDDEE 274
QK R R ND CS +L+ +FD L + +DDEE
Sbjct: 247 QKDRVDGCRVTTSNDSCSLHLNEYFD-LLDVTDDEE 281
>ref|XP_550472.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|55296173|dbj|BAD67891.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
gi|55296098|dbj|BAD67688.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 375
Score = 288 bits (738), Expect = 9e-77
Identities = 142/275 (51%), Positives = 187/275 (67%), Gaps = 3/275 (1%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P+RI+L+RHGES+GN+D +AYT PD I LTPQG A G RLR +++ G DW+
Sbjct: 89 PRRIVLVRHGESEGNVDEAAYTRVPDPRIGLTPQGWRDAEDCGRRLRHLLSTGG-GDDWK 147
Query: 65 VNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRERF 124
V FYVSPY RT TLR +GR+F +R+ GVREE R+REQDFGNFQ ++M++ K+ R R+
Sbjct: 148 VYFYVSPYRRTLETLRGLGRAFEARRIAGVREEPRLREQDFGNFQDRDKMRVEKEIRRRY 207
Query: 125 GRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLH--HDPSNDLNLVIVSHGLTSRIF 182
GRFFYRFP GESAADVYDRI+GF E L DID+ R + + D+N+V+VSHGLT R+F
Sbjct: 208 GRFFYRFPNGESAADVYDRITGFRETLRADIDIGRFQPPGERNPDMNVVLVSHGLTLRVF 267
Query: 183 LMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWGLSPEMVADQKW 242
LM+W+KWTV QFE L N N VM+ G+GG YSL VHH+ +E+ ++GL+ +M+ DQKW
Sbjct: 268 LMRWYKWTVSQFEGLANLSNGGALVMQTGAGGRYSLLVHHSVDELREFGLTDDMIEDQKW 327
Query: 243 RAGASRGAWNDQCSWYLDGFFDRLAEESDDEEASS 277
+ A G N FF D+ ++
Sbjct: 328 QMTARPGELNYNFITNGPSFFTHFTHHHHDKHKAA 362
>ref|NP_910251.1| P0514G12.25 [Oryza sativa (japonica cultivar-group)]
Length = 450
Score = 288 bits (738), Expect = 9e-77
Identities = 142/275 (51%), Positives = 187/275 (67%), Gaps = 3/275 (1%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P+RI+L+RHGES+GN+D +AYT PD I LTPQG A G RLR +++ G DW+
Sbjct: 164 PRRIVLVRHGESEGNVDEAAYTRVPDPRIGLTPQGWRDAEDCGRRLRHLLSTGG-GDDWK 222
Query: 65 VNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRERF 124
V FYVSPY RT TLR +GR+F +R+ GVREE R+REQDFGNFQ ++M++ K+ R R+
Sbjct: 223 VYFYVSPYRRTLETLRGLGRAFEARRIAGVREEPRLREQDFGNFQDRDKMRVEKEIRRRY 282
Query: 125 GRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLH--HDPSNDLNLVIVSHGLTSRIF 182
GRFFYRFP GESAADVYDRI+GF E L DID+ R + + D+N+V+VSHGLT R+F
Sbjct: 283 GRFFYRFPNGESAADVYDRITGFRETLRADIDIGRFQPPGERNPDMNVVLVSHGLTLRVF 342
Query: 183 LMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWGLSPEMVADQKW 242
LM+W+KWTV QFE L N N VM+ G+GG YSL VHH+ +E+ ++GL+ +M+ DQKW
Sbjct: 343 LMRWYKWTVSQFEGLANLSNGGALVMQTGAGGRYSLLVHHSVDELREFGLTDDMIEDQKW 402
Query: 243 RAGASRGAWNDQCSWYLDGFFDRLAEESDDEEASS 277
+ A G N FF D+ ++
Sbjct: 403 QMTARPGELNYNFITNGPSFFTHFTHHHHDKHKAA 437
>gb|AAO65326.1| unknown [Streptomyces murayamaensis]
Length = 219
Score = 196 bits (498), Expect = 6e-49
Identities = 98/214 (45%), Positives = 136/214 (62%), Gaps = 15/214 (7%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P+RI+L+RHGES+GN D + Y PDH+++LT G QAR G RLR + + R
Sbjct: 4 PRRIVLVRHGESEGNADDTVYEREPDHALRLTEPGRLQARATGERLREIFGRE------R 57
Query: 65 VNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRERF 124
V+ YVSPY RT T RE G ++ +REE R+REQD+GN+Q + +++ K R+ +
Sbjct: 58 VSVYVSPYRRTHETFREFGLD---PDLVRIREEPRLREQDWGNWQDRDDVRLQKAYRDAY 114
Query: 125 GRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIFLM 184
G FFYRF +GES ADVYDR+ F E+L R + P + N+++V+HGLT R+F M
Sbjct: 115 GHFFYRFAQGESGADVYDRVGSFLESLHRSFEA------PDHPPNVLLVTHGLTMRLFCM 168
Query: 185 KWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSL 218
+WF WTV FE L+N GN E R + G+ G Y+L
Sbjct: 169 RWFHWTVADFESLSNPGNAETRTLVLGTDGRYTL 202
>emb|CAA18344.1| conserved hypothetical protein SC4H2.29 [Streptomyces coelicolor
A3(2)] gi|7479557|pir||T35128 hypothetical protein
SC4H2.29 SC4H2.29 - Streptomyces coelicolor
gi|21224153|ref|NP_629932.1| hypothetical protein
SCO5808 [Streptomyces coelicolor A3(2)]
Length = 219
Score = 196 bits (497), Expect = 8e-49
Identities = 98/214 (45%), Positives = 139/214 (64%), Gaps = 15/214 (7%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P+RI+L+RHGES GN+D + Y PDH++ LT +G +QA G LR V ++ R
Sbjct: 4 PRRIVLVRHGESIGNVDDTVYEREPDHALALTDRGRAQAEETGEGLREVFGSE------R 57
Query: 65 VNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRERF 124
++ YVSPY RT TLR +I +REE R+REQD+GN+Q + +++ K R+ +
Sbjct: 58 ISVYVSPYRRTHETLRAFRLD---PDLIRIREEPRLREQDWGNWQDRDDVRLQKAYRDAY 114
Query: 125 GRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIFLM 184
G FFYRF +GES ADVYDR+ GF E+L+R + DP + N+++V+HGL R+F M
Sbjct: 115 GHFFYRFAQGESGADVYDRVGGFLESLFRSFE------DPDHPPNVLLVTHGLAMRLFCM 168
Query: 185 KWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSL 218
+WF WTV +FE L N GN E R++ G+ G+YSL
Sbjct: 169 RWFHWTVAEFESLANPGNAEMRMLVLGADGKYSL 202
>dbj|BAC70169.1| hypothetical protein [Streptomyces avermitilis MA-4680]
gi|29829000|ref|NP_823634.1| hypothetical protein
SAV2458 [Streptomyces avermitilis MA-4680]
Length = 219
Score = 190 bits (483), Expect = 3e-47
Identities = 97/214 (45%), Positives = 136/214 (63%), Gaps = 15/214 (7%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P+RI+L+RHGES GN D + Y PDH++ LT G QA G +LR + +G
Sbjct: 4 PRRIVLVRHGESAGNADDTVYEREPDHALPLTEVGWRQAEERGKQLRELFGQEG------ 57
Query: 65 VNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRERF 124
V+ YVSPY RT TLR ++ VREE R+REQD+GN+Q + +++ K R+ +
Sbjct: 58 VSVYVSPYRRTLETLRAFHLD---PDLVRVREEPRLREQDWGNWQDRDDVRLQKAYRDAY 114
Query: 125 GRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIFLM 184
G FFYRF +GES ADVYDR+ GF E+L+R + P + N++IV+HGL R+F M
Sbjct: 115 GHFFYRFAQGESGADVYDRVGGFLESLFRSFEA------PDHPPNVLIVTHGLAMRLFCM 168
Query: 185 KWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSL 218
+WF W+V +FE L+N GN E R++ G+ G+YSL
Sbjct: 169 RWFHWSVAEFESLSNPGNAEMRMLVLGNDGKYSL 202
>gb|EAA75632.1| hypothetical protein FG05987.1 [Gibberella zeae PH-1]
gi|46123219|ref|XP_386163.1| hypothetical protein
FG05987.1 [Gibberella zeae PH-1]
Length = 494
Score = 185 bits (470), Expect = 1e-45
Identities = 104/224 (46%), Positives = 135/224 (59%), Gaps = 21/224 (9%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P+ IIL+RHG+S+GN + + T PDH ++LTP+G SQA AG RLR ++ PD
Sbjct: 4 PRLIILVRHGQSEGNKNREIHQTVPDHRVKLTPEGWSQAHDAGRRLRSLL-----RPDDT 58
Query: 65 VNFYVSPYARTRSTLR---------EVGRSFSKKRVIGVREESRIREQDFGNFQ-VEERM 114
+ F+ SPY RTR T E S ++ I V EE R+REQDFGNFQ M
Sbjct: 59 LQFFTSPYRRTRETTEGILETLTSDETSPSPFRRNNIKVYEEPRLREQDFGNFQPCSAEM 118
Query: 115 KIIKQTRERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVS 174
+ + Q R +G FFYR P GESAAD YDR+SGF E+LWR + PS V+V+
Sbjct: 119 ERMWQERADYGHFFYRIPNGESAADAYDRVSGFNESLWRQFGEDDF---PS---VCVLVT 172
Query: 175 HGLTSRIFLMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSL 218
HGL SR+FLMKW+ +TVE FE L N +CEF +M + +Y L
Sbjct: 173 HGLMSRVFLMKWYHFTVEYFEDLRNINHCEFLIMRKQENQKYLL 216
>emb|CAD70465.1| conserved hypothetical protein [Neurospora crassa]
gi|32415611|ref|XP_328284.1| hypothetical protein
[Neurospora crassa] gi|28917700|gb|EAA27393.1|
hypothetical protein [Neurospora crassa]
Length = 570
Score = 184 bits (467), Expect = 2e-45
Identities = 106/252 (42%), Positives = 143/252 (56%), Gaps = 27/252 (10%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P+ IIL+RH +S+GN + + T PDH ++LT +G QA AG RLR+++ AD
Sbjct: 4 PRLIILIRHAQSEGNKNRDIHQTIPDHRVKLTDEGWQQAYDAGRRLRKLLRADDT----- 58
Query: 65 VNFYVSPYARTRSTLREVGRSFS---------KKRVIGVREESRIREQDFGNFQ-VEERM 114
+ F+ SPY RTR T + + + K+ I V EE R+REQDFGNFQ M
Sbjct: 59 IQFFTSPYRRTRETTEGILATLTSDDPEPSPFKRNHIKVYEEPRLREQDFGNFQPCSAEM 118
Query: 115 KIIKQTRERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVS 174
+ + Q R +G FFYR P GESAAD YDR+SGF E+LWR + D V+V+
Sbjct: 119 ERMWQERADYGHFFYRIPNGESAADAYDRVSGFNESLWRQFN------DDDFASVCVLVT 172
Query: 175 HGLTSRIFLMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQWG-LS 233
HGL SR+FLMKW+ ++VE FE L N +CEF +M + G+Y L E ++ W L
Sbjct: 173 HGLMSRVFLMKWYHFSVEYFEDLRNVNHCEFLIMRKNDSGKYIL-----ENKLRTWSELR 227
Query: 234 PEMVADQKWRAG 245
E A Q G
Sbjct: 228 KERAAQQALLTG 239
>gb|EAL92577.1| phosphoglycerate mutase family domain protein [Aspergillus
fumigatus Af293]
Length = 557
Score = 180 bits (457), Expect = 3e-44
Identities = 100/222 (45%), Positives = 133/222 (59%), Gaps = 22/222 (9%)
Query: 8 IILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWRVNF 67
IIL+RH +S+GN + + T PDH +QLTP+G QAR AG++LR ++ PD ++F
Sbjct: 2 IILIRHAQSEGNKNREIHQTIPDHRVQLTPEGHRQAREAGSKLRALL-----RPDDTIHF 56
Query: 68 YVSPYARTRSTLREVGRSFSK---------KRVIGVREESRIREQDFGNFQ-VEERMKII 117
+ SPY RTR T + S + + I V EE R+REQDFGNFQ M+ +
Sbjct: 57 FTSPYRRTRETTEGILESLTSDSPSPSPFPRHTIKVYEEPRLREQDFGNFQPCSAEMERM 116
Query: 118 KQTRERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGL 177
R +G FFYR P GESAAD YDRISGF E+LW RL + V+V+HGL
Sbjct: 117 WLERADYGHFFYRIPNGESAADAYDRISGFNESLW------RLFGEDDFASVCVLVTHGL 170
Query: 178 TSRIFLMKWFKWTVEQFEHLNNFGNCEFRVME-QGSGGEYSL 218
+R+FLMKW+ W+VE FE L N +CEF +M+ G+Y L
Sbjct: 171 MTRVFLMKWYHWSVEYFEDLRNINHCEFVIMKLNPDNGKYVL 212
>gb|EAA54985.1| hypothetical protein MG06642.4 [Magnaporthe grisea 70-15]
gi|39977515|ref|XP_370145.1| hypothetical protein
MG06642.4 [Magnaporthe grisea 70-15]
Length = 633
Score = 179 bits (454), Expect = 8e-44
Identities = 107/249 (42%), Positives = 139/249 (54%), Gaps = 28/249 (11%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P+ IIL+RH +S+GN + T PDH ++LT G QA AG RLR ++ AD
Sbjct: 4 PRLIILIRHAQSEGNKKREIHQTIPDHRVKLTQDGWQQAYEAGRRLRSLLRADDT----- 58
Query: 65 VNFYVSPYARTRSTLREVGRSFS---------KKRVIGVREESRIREQDFGNFQ-VEERM 114
+ F+ SPY RTR T + + + K+ I V EE R+REQDFGNFQ M
Sbjct: 59 LQFFTSPYRRTRETTEGILATLTADEPDPSPFKREKIKVYEEPRLREQDFGNFQPCSAEM 118
Query: 115 KIIKQTRERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVS 174
+ + Q R +G FFYR P GESAAD YDR+SGF E+LWR D V+V+
Sbjct: 119 ERMWQERADYGHFFYRIPNGESAADAYDRVSGFNESLWRQFG------DEDFASVCVLVT 172
Query: 175 HGLTSRIFLMKWFKWTVEQFEHLNNFGNCEFRVMEQGSG-GEYSLAVHHTEEEMLQWG-L 232
HGL SR+FLMKW+ ++VE FE L N +CEF +M Q G G+Y L E ++ W L
Sbjct: 173 HGLMSRVFLMKWYHFSVEYFEDLRNVNHCEFLIMRQREGTGKYDL-----ENKLRTWSEL 227
Query: 233 SPEMVADQK 241
E QK
Sbjct: 228 RRERAQHQK 236
>emb|CAG78109.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50555788|ref|XP_505302.1| hypothetical protein
[Yarrowia lipolytica]
Length = 289
Score = 179 bits (453), Expect = 1e-43
Identities = 100/210 (47%), Positives = 131/210 (61%), Gaps = 13/210 (6%)
Query: 3 VLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPD 62
+L + IIL+RHGES+GN D S T +H I+LT G QAR AG RL AD + D
Sbjct: 9 ILTQLIILIRHGESEGNCDKSVNRHTSNHRIKLTANGEEQARDAGKRL-----ADMVNKD 63
Query: 63 WRVNFYVSPYARTRSTLREVGRSFSKKRV-IGVREESRIREQDFGNFQVE-ERMKIIKQT 120
+ FY SPY RTR T + + +K + V EE R+REQDFGNFQ M I
Sbjct: 64 DTLLFYTSPYQRTRQTTQLICEGIEEKNISYKVHEEPRLREQDFGNFQASASEMNKIWND 123
Query: 121 RERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSR 180
R ++G FFYR P GESAADVYDR +GF E L+R + ++ PS LV+V+HG+ R
Sbjct: 124 RSQYGHFFYRIPNGESAADVYDRCAGFNETLFRQFNSDKF---PSV---LVLVAHGIWIR 177
Query: 181 IFLMKWFKWTVEQFEHLNNFGNCEFRVMEQ 210
+FLMKW++WT E+FE L N +CEF +M++
Sbjct: 178 VFLMKWYRWTYEKFESLRNLRHCEFIIMKR 207
>ref|ZP_00411845.1| Phosphoglycerate/bisphosphoglycerate mutase [Arthrobacter sp. FB24]
gi|66869802|gb|EAL97168.1|
Phosphoglycerate/bisphosphoglycerate mutase
[Arthrobacter sp. FB24]
Length = 263
Score = 172 bits (435), Expect = 1e-41
Identities = 97/214 (45%), Positives = 128/214 (59%), Gaps = 15/214 (7%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWR 64
P +II++RHG+S N DTS Y PD+ I LT G++QAR AG +RR + DG +
Sbjct: 41 PGKIIMIRHGQSAANADTSIYNRVPDYRIPLTELGVAQARAAGEEIRREL--DGR----Q 94
Query: 65 VNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMKIIKQTRERF 124
V YVSPY R TL + +R+I EE R+REQD+ NFQ+ ++ K+ R +
Sbjct: 95 VCVYVSPYLRAYQTLEALNLGPLVERII---EEPRLREQDWANFQIAGDIEDQKELRNAY 151
Query: 125 GRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIFLM 184
G FFYRF EGES +DVYDRIS F E L+R P+ N + V+HGLT R+F M
Sbjct: 152 GHFFYRFREGESGSDVYDRISSFMET------LHRHWSKPTYAPNALFVTHGLTMRLFCM 205
Query: 185 KWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSL 218
+WF W+VE FE LNN N E R + + G+Y L
Sbjct: 206 RWFHWSVEYFESLNNPENAEVRTLVRTDKGKYEL 239
>gb|EAA61061.1| hypothetical protein AN4983.2 [Aspergillus nidulans FGSC A4]
gi|67537626|ref|XP_662587.1| hypothetical protein
AN4983_2 [Aspergillus nidulans FGSC A4]
gi|49095318|ref|XP_409120.1| hypothetical protein
AN4983.2 [Aspergillus nidulans FGSC A4]
Length = 651
Score = 170 bits (431), Expect = 4e-41
Identities = 97/222 (43%), Positives = 130/222 (57%), Gaps = 22/222 (9%)
Query: 8 IILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADGCSPDWRVNF 67
IIL+RH +S+GN + + T PDH ++LTP+G QAR AG RLR ++ PD ++F
Sbjct: 82 IILIRHAQSEGNKNREIHQTIPDHRVKLTPEGHRQARDAGTRLRGLL-----RPDDTIHF 136
Query: 68 YVSPYARTRSTLREVGRSFS---------KKRVIGVREESRIREQDFGNFQ-VEERMKII 117
+ SPY RTR T + S + + I V EE R+REQDFGNFQ M+ +
Sbjct: 137 FTSPYRRTRETTEGILESLTADTPSPSPFPRHTIKVYEEPRLREQDFGNFQPCSAEMERM 196
Query: 118 KQTRERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGL 177
R +G FFYR P GESAAD YDR+SGF E++W RL + V+V+HGL
Sbjct: 197 WLERADYGHFFYRIPNGESAADAYDRVSGFNESMW------RLFGEKDFASVCVLVTHGL 250
Query: 178 TSRIFLMKWFKWTVEQFEHLNNFGNCEFRVME-QGSGGEYSL 218
+R+FLMKW+ VE FE L N +CEF +M+ G+Y L
Sbjct: 251 MTRVFLMKWYHSFVEYFEDLRNINHCEFVIMKLNEDSGKYVL 292
>gb|AAR24750.1| At1g08940 [Arabidopsis thaliana] gi|38454096|gb|AAR20742.1|
At1g08940 [Arabidopsis thaliana]
Length = 131
Score = 160 bits (404), Expect = 5e-38
Identities = 77/116 (66%), Positives = 92/116 (78%), Gaps = 3/116 (2%)
Query: 3 VLPKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVMAADG---C 59
+LPKRIILMRHGES GN+D AY TTPDH I LT +G +QAR AG ++R +++ C
Sbjct: 7 MLPKRIILMRHGESAGNIDAGAYATTPDHKIPLTEEGRAQAREAGKKMRALISTQSGGAC 66
Query: 60 SPDWRVNFYVSPYARTRSTLREVGRSFSKKRVIGVREESRIREQDFGNFQVEERMK 115
+WRV FYVSPY RTR+TLREVG+ FS+KRVIGVREE RIREQDFGNFQVEE+ +
Sbjct: 67 GENWRVYFYVSPYERTRTTLREVGKGFSRKRVIGVREECRIREQDFGNFQVEEKKR 122
>emb|CAG60093.1| unnamed protein product [Candida glabrata CBS138]
gi|50289457|ref|XP_447160.1| unnamed protein product
[Candida glabrata]
Length = 319
Score = 137 bits (344), Expect = 4e-31
Identities = 93/287 (32%), Positives = 127/287 (43%), Gaps = 85/287 (29%)
Query: 5 PKRIILMRHGESQGNLDTSAYTTTPDHSIQLTPQGISQARRAGNRLRRVM---------- 54
P+ I+L+RHGES+ N D TP+H I LTP G QA AG +L R++
Sbjct: 8 PRLIVLIRHGESESNKDKKINEVTPNHLIPLTPYGKKQAHNAGLKLLRLLNMADASVIEK 67
Query: 55 --------------------AADGCSPDWRVNFYVSPYARTRSTLREVGRSFSKKRVI-- 92
+G D + FY SPY RTR TL+ V + +
Sbjct: 68 LGEEYALPATSTNTLELRGYKPNGKHMDDNIIFYTSPYKRTRETLKGVLEVIDRYNELKS 127
Query: 93 GVR----------------------EESRIREQD-------------------------- 104
G+ E S I E D
Sbjct: 128 GINMCAEQTYNPYGKQKHAIWPHDLENSGIYENDESTHCGPEREGTYIRYRIIDEPRLRE 187
Query: 105 --FGNFQVEERMKIIKQTRERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHH 162
FGN+Q M+ + + R+ +G FF+RFPEGESAADVYDR++ F + L+R + H
Sbjct: 188 QDFGNYQEVSSMQDVMEKRKTYGHFFFRFPEGESAADVYDRVASFQDTLYRHFQFRQ--H 245
Query: 163 DPSNDLNLVIVSHGLTSRIFLMKWFKWTVEQFEHLNNFGNCEFRVME 209
D+ +V+V+HG+ SR+FLMKWF+WT E+FE N N VME
Sbjct: 246 TKGRDV-VVLVTHGIYSRVFLMKWFRWTFEEFESFTNVPNGSLMVME 291
>emb|CAG88986.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50428257|ref|XP_460654.1| unnamed protein product
[Debaryomyces hansenii]
Length = 271
Score = 136 bits (343), Expect = 6e-31
Identities = 71/156 (45%), Positives = 98/156 (62%), Gaps = 10/156 (6%)
Query: 67 FYVSPYARTRSTLREV--GRSFSKKRVIGVREESRIREQDFGNFQ-VEERMKIIKQTRER 123
FY SPY R R T + G + + V EE R+REQDFGNFQ E+M+ I Q R
Sbjct: 3 FYTSPYLRARQTCNNIIDGIKYVPGVMYKVHEEPRMREQDFGNFQSTSEQMEKIWQERAH 62
Query: 124 FGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRIFL 183
+G FFYR P GESAADVYDRI+ F E L+R + + L++V+HG+ +R+FL
Sbjct: 63 YGHFFYRIPHGESAADVYDRIASFNETLFRQFQQDNFPNI------LILVTHGIWARVFL 116
Query: 184 MKWFKWTVEQFEHLNNFGNCEFRVM-EQGSGGEYSL 218
MKWF+W+ E+FE L N +C++ +M +Q GG++ L
Sbjct: 117 MKWFRWSYEEFESLRNIPHCQYLIMKKQSEGGKFRL 152
>gb|EAL00979.1| hypothetical protein CaO19.6747 [Candida albicans SC5314]
gi|46441558|gb|EAL00854.1| hypothetical protein
CaO19.14039 [Candida albicans SC5314]
Length = 240
Score = 135 bits (340), Expect = 1e-30
Identities = 75/169 (44%), Positives = 105/169 (61%), Gaps = 17/169 (10%)
Query: 67 FYVSPYARTRSTLREVGRSFSKKRVIGV----REESRIREQDFGNFQVE-ERMKIIKQTR 121
F SPY RTR T + K V GV REE R+REQDFGNFQ + M++I + R
Sbjct: 3 FITSPYLRTRQTCNNIIEGI--KNVPGVEYKVREEPRMREQDFGNFQPSSDEMELIWKER 60
Query: 122 ERFGRFFYRFPEGESAADVYDRISGFFEALWRDIDLNRLHHDPSNDLNLVIVSHGLTSRI 181
++G FFYR P GESAADVYDR++ F E+L+R + D ++ LV+VSHG+ SR+
Sbjct: 61 AQYGHFFYRIPHGESAADVYDRVASFMESLFR-----QFRSDDFPNI-LVLVSHGIWSRV 114
Query: 182 FLMKWFKWTVEQFEHLNNFGNCEFRVMEQGSGGEYSLAVHHTEEEMLQW 230
F+MKWFKW+ E+FE L N +C++ VM++ + +H + +L W
Sbjct: 115 FIMKWFKWSYEEFESLKNVPHCKYLVMKK----DDETQKYHLKTRLLTW 159
>emb|CAG98631.1| unnamed protein product [Kluyveromyces lactis NRRL Y-1140]
gi|50311789|ref|XP_455923.1| unnamed protein product
[Kluyveromyces lactis]
Length = 313
Score = 120 bits (302), Expect = 3e-26
Identities = 62/116 (53%), Positives = 78/116 (66%), Gaps = 4/116 (3%)
Query: 94 VREESRIREQDFGNFQVEERMKIIKQTRERFGRFFYRFPEGESAADVYDRISGFFEALWR 153
V+EE RIREQDFGNFQ M + TR +G FF+RFP+GESAADVYDR SGF E+L+R
Sbjct: 184 VKEEPRIREQDFGNFQKTASMTEVMNTRANYGHFFFRFPQGESAADVYDRCSGFQESLFR 243
Query: 154 DIDLNRLHHDPSNDLNLVIVSHGLTSRIFLMKWFKWTVEQFEHLNNFGNCEFRVME 209
+ + D+ + IV+HG+ R+FLMKWF+WT E+FE L N N VME
Sbjct: 244 HFEKS---GGKKRDV-VAIVTHGIFLRVFLMKWFRWTYEEFESLTNVPNGSVIVME 295
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.136 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 496,735,812
Number of Sequences: 2540612
Number of extensions: 20565823
Number of successful extensions: 48267
Number of sequences better than 10.0: 303
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 276
Number of HSP's that attempted gapping in prelim test: 48005
Number of HSP's gapped (non-prelim): 325
length of query: 280
length of database: 863,360,394
effective HSP length: 126
effective length of query: 154
effective length of database: 543,243,282
effective search space: 83659465428
effective search space used: 83659465428
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0211.14


