

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0206b.3
(304 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
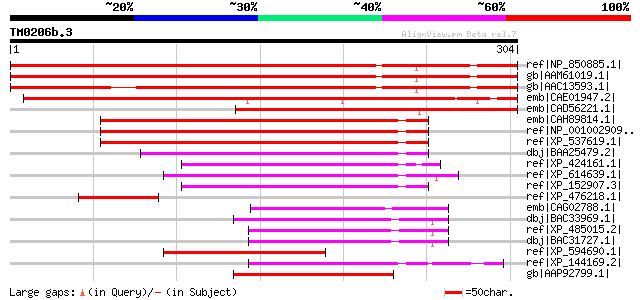
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_850885.1| D111/G-patch domain-containing protein [Arabido... 386 e-106
gb|AAM61019.1| unknown [Arabidopsis thaliana] 386 e-106
gb|AAC13593.1| contains similarity to Arabidopsis thaliana DNA-d... 358 1e-97
emb|CAE01947.2| OSJNBa0073L13.10 [Oryza sativa (japonica cultiva... 334 2e-90
emb|CAD56221.1| hypothetical protein [Cicer arietinum] 285 9e-76
emb|CAH89814.1| hypothetical protein [Pongo pygmaeus] 139 8e-32
ref|NP_001002909.1| hypothetical protein LOC23131 [Homo sapiens] 139 8e-32
ref|XP_537619.1| PREDICTED: similar to KIAA0553 protein [Canis f... 139 1e-31
dbj|BAA25479.2| KIAA0553 protein [Homo sapiens] 101 2e-20
ref|XP_424161.1| PREDICTED: similar to Hypothetical protein KIAA... 98 3e-19
ref|XP_614639.1| PREDICTED: similar to KIAA0553 protein, partial... 98 3e-19
ref|XP_152907.3| PREDICTED: hypothetical protein LOC237943 [Mus ... 96 1e-18
ref|XP_476218.1| unknown protein [Oryza sativa (japonica cultiva... 89 2e-16
emb|CAG02788.1| unnamed protein product [Tetraodon nigroviridis] 75 2e-12
dbj|BAC33969.1| unnamed protein product [Mus musculus] 75 3e-12
ref|XP_485015.2| PREDICTED: RIKEN cDNA C630007C17 gene [Mus musc... 74 4e-12
dbj|BAC31727.1| unnamed protein product [Mus musculus] 74 4e-12
ref|XP_594690.1| PREDICTED: similar to KIAA0553 protein, partial... 74 5e-12
ref|XP_144169.2| PREDICTED: similar to hypothetical protein FLJ3... 74 5e-12
gb|AAP92799.1| hypothetical protein [Homo sapiens] gi|34740327|r... 74 7e-12
>ref|NP_850885.1| D111/G-patch domain-containing protein [Arabidopsis thaliana]
gi|30690349|ref|NP_850884.1| D111/G-patch
domain-containing protein [Arabidopsis thaliana]
Length = 301
Score = 386 bits (992), Expect = e-106
Identities = 198/308 (64%), Positives = 244/308 (78%), Gaps = 11/308 (3%)
Query: 1 MDYRRSSGRQESGGRGRRQFKKEEAHQDSLIGDLAEDFRLPINHRPTENVDLENVEQASL 60
MD R + E+ R +R ++K++ +Q+S I LAE+FRLPI HR TENVDLE+VEQASL
Sbjct: 1 MDISRRESKTEASSREKRIYEKDQMNQESFIEGLAEEFRLPITHRVTENVDLEDVEQASL 60
Query: 61 DTQLTSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEEN 120
D +++SSN+GF+LLQKMGWKGKGLGK EQGI EPIKSG+RD RLGLGKQEEDD+FTAEEN
Sbjct: 61 DVKISSSNVGFRLLQKMGWKGKGLGKQEQGITEPIKSGIRDRRLGLGKQEEDDYFTAEEN 120
Query: 121 IQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHL 180
IQRKKLD+E+EETEE +KREVLAEREQKIQ++VKEIRKVFYC+LC+KQY+ MEFE HL
Sbjct: 121 IQRKKLDIEIEETEEIAKKREVLAEREQKIQSDVKEIRKVFYCELCSKQYRTVMEFEGHL 180
Query: 181 SSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESG 240
SSYDHNH+KRFK+MKEMHG S RD+R+KREQQRQERE+ K +ADA+KQ ++Q Q+
Sbjct: 181 SSYDHNHKKRFKEMKEMHGASGRDERKKREQQRQEREMTK---MADARKQHQMQQSQQEV 237
Query: 241 SE--PVSSETR-TATPLTDQEQRNALKFGFSSK-GSASKNVIGPKRQMVAKKQNIPVSSI 296
E PVS+ + T PL Q+QR LKFGFSSK G SK+ + KK + ++S+
Sbjct: 238 PENVPVSAPAKTTVAPLAVQDQRKTLKFGFSSKSGIISKS----QPTSSIKKPKVAIASV 293
Query: 297 FNNDSDEE 304
F NDSDE+
Sbjct: 294 FGNDSDED 301
>gb|AAM61019.1| unknown [Arabidopsis thaliana]
Length = 301
Score = 386 bits (992), Expect = e-106
Identities = 198/308 (64%), Positives = 244/308 (78%), Gaps = 11/308 (3%)
Query: 1 MDYRRSSGRQESGGRGRRQFKKEEAHQDSLIGDLAEDFRLPINHRPTENVDLENVEQASL 60
MD R + E+ R +R ++K++ +Q+S I LAE+FRLPI HR TENVDLE+VEQASL
Sbjct: 1 MDISRRESKTEASSREKRIYEKDQMNQESFIEGLAEEFRLPITHRVTENVDLEDVEQASL 60
Query: 61 DTQLTSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEEN 120
D +++SSN+GF+LLQKMGWKGKGLGK EQGI EPIKSG+RD RLGLGKQEEDD+FTAEEN
Sbjct: 61 DVKISSSNVGFRLLQKMGWKGKGLGKQEQGITEPIKSGIRDRRLGLGKQEEDDYFTAEEN 120
Query: 121 IQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHL 180
IQRKKLD+E+EETEE +KREVLAEREQKIQ++VKEIRKVFYC+LC+KQY+ M+FE HL
Sbjct: 121 IQRKKLDIEIEETEEIAKKREVLAEREQKIQSDVKEIRKVFYCELCSKQYRTVMKFEGHL 180
Query: 181 SSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESG 240
SSYDHNH+KRFK+MKEMHG S RDDR+KREQQRQERE+ K +ADA+KQ ++Q Q+
Sbjct: 181 SSYDHNHKKRFKEMKEMHGASGRDDRKKREQQRQEREMTK---MADARKQHQMQQSQQEV 237
Query: 241 SE--PVSSETR-TATPLTDQEQRNALKFGFSSK-GSASKNVIGPKRQMVAKKQNIPVSSI 296
E PVS+ + T PL Q+QR LKFGFSSK G SK+ + KK + ++S+
Sbjct: 238 PENVPVSAPAKTTVAPLAVQDQRKTLKFGFSSKSGIISKS----QPTSSIKKPKVAIASV 293
Query: 297 FNNDSDEE 304
F NDSDE+
Sbjct: 294 FGNDSDED 301
>gb|AAC13593.1| contains similarity to Arabidopsis thaliana
DNA-damage-repair/tolerance resistance protein DRT111
(SW:P42698) gi|7485939|pir||T01192 hypothetical protein
F21E10.7 - Arabidopsis thaliana
Length = 287
Score = 358 bits (919), Expect = 1e-97
Identities = 190/308 (61%), Positives = 231/308 (74%), Gaps = 25/308 (8%)
Query: 1 MDYRRSSGRQESGGRGRRQFKKEEAHQDSLIGDLAEDFRLPINHRPTENVDLENVEQASL 60
MD R + E+ R +R ++K++ +Q+S I LAE+FRLPI HR TENVDLE+VEQASL
Sbjct: 1 MDISRRESKTEASSREKRIYEKDQMNQESFIEGLAEEFRLPITHRVTENVDLEDVEQASL 60
Query: 61 DTQLTSSNIGFKLLQKMGWKGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEEN 120
D KMGWKGKGLGK EQGI EPIKSG+RD RLGLGKQEEDD+FTAEEN
Sbjct: 61 D--------------KMGWKGKGLGKQEQGITEPIKSGIRDRRLGLGKQEEDDYFTAEEN 106
Query: 121 IQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHL 180
IQRKKLD+E+EETEE +KREVLAEREQKIQ++VKEIRKVFYC+LC+KQY+ MEFE HL
Sbjct: 107 IQRKKLDIEIEETEEIAKKREVLAEREQKIQSDVKEIRKVFYCELCSKQYRTVMEFEGHL 166
Query: 181 SSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESG 240
SSYDHNH+KRFK+MKEMHG S RD+R+KREQQRQERE+ K +ADA+KQ ++Q Q+
Sbjct: 167 SSYDHNHKKRFKEMKEMHGASGRDERKKREQQRQEREMTK---MADARKQHQMQQSQQEV 223
Query: 241 SE--PVSSETR-TATPLTDQEQRNALKFGFSSK-GSASKNVIGPKRQMVAKKQNIPVSSI 296
E PVS+ + T PL Q+QR LKFGFSSK G SK+ + KK + ++S+
Sbjct: 224 PENVPVSAPAKTTVAPLAVQDQRKTLKFGFSSKSGIISKS----QPTSSIKKPKVAIASV 279
Query: 297 FNNDSDEE 304
F NDSDE+
Sbjct: 280 FGNDSDED 287
>emb|CAE01947.2| OSJNBa0073L13.10 [Oryza sativa (japonica cultivar-group)]
gi|50921349|ref|XP_471035.1| OSJNBa0073L13.10 [Oryza
sativa (japonica cultivar-group)]
Length = 345
Score = 334 bits (857), Expect = 2e-90
Identities = 181/346 (52%), Positives = 234/346 (67%), Gaps = 55/346 (15%)
Query: 9 RQESGGRGRRQFKKEEAHQDSLIGDLAEDFRLPINHRPTENVDLENVEQASLDTQLTSSN 68
R GG R ++ + D ++FRLP++HRPTEN++ E +EQAS+DTQLTSSN
Sbjct: 4 RDRGGGSSSRNPHGAPVEEEEELEDDFDEFRLPMSHRPTENLETEGLEQASVDTQLTSSN 63
Query: 69 IGFKLLQKMGWKGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDV 128
+GF+LLQKMGWKGKGLGK+EQGI EPIK+G+RD +LG+GKQE+DDFFT+E+N+QR+KL++
Sbjct: 64 VGFRLLQKMGWKGKGLGKNEQGITEPIKAGIRDAKLGVGKQEQDDFFTSEDNVQRRKLNI 123
Query: 129 ELEETEENVRKRE----------------------------------------------V 142
ELEETEE+++KRE V
Sbjct: 124 ELEETEEHIKKRELLALHIDICILFMQELLASALMILEHFGHFLFGGSFSCLVQIMTIQV 183
Query: 143 LAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMH--GG 200
+AEREQKI++EVKEI+KVF+C LCNKQYKLA EFE+HLSSYDHNHRKRFK+MKEM
Sbjct: 184 IAEREQKIRSEVKEIQKVFFCSLCNKQYKLAHEFESHLSSYDHNHRKRFKEMKEMQSSSS 243
Query: 201 SSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTATPLTDQEQR 260
SSRDDRQKREQQR+E+E+AKFAQ+ADA ++Q+ Q + S S + ++Q+QR
Sbjct: 244 SSRDDRQKREQQREEKELAKFAQLADAHRKQQQQKQEPSESSSERITMKNLPNPSNQDQR 303
Query: 261 NALKFGFSSKGSASKNVIG--PKRQMVAKKQNIPVSSIFNNDSDEE 304
LKFGF SK + SK +G K+ VA K +SS+F N+SDEE
Sbjct: 304 KTLKFGF-SKMAPSKAPVGNVSKKPKVATK----MSSVFGNESDEE 344
>emb|CAD56221.1| hypothetical protein [Cicer arietinum]
Length = 206
Score = 285 bits (730), Expect = 9e-76
Identities = 145/171 (84%), Positives = 156/171 (90%), Gaps = 2/171 (1%)
Query: 136 NVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMK 195
++ K +VLAEREQKIQTEVKEIRKVFYC+LCNKQYKLAMEFEAHLSSYDHNHRKRFKQMK
Sbjct: 36 HIMKIQVLAEREQKIQTEVKEIRKVFYCELCNKQYKLAMEFEAHLSSYDHNHRKRFKQMK 95
Query: 196 EMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPV--SSETRTATP 253
EMHG SSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQL QESGS PV SSE+RTAT
Sbjct: 96 EMHGSSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLQQESGSAPVSASSESRTATA 155
Query: 254 LTDQEQRNALKFGFSSKGSASKNVIGPKRQMVAKKQNIPVSSIFNNDSDEE 304
LTDQEQRN LKFGFSSKG+ASK+ I K+Q V KKQN+PV+SIF NDSD+E
Sbjct: 156 LTDQEQRNTLKFGFSSKGTASKSTISAKKQNVPKKQNVPVASIFGNDSDDE 206
>emb|CAH89814.1| hypothetical protein [Pongo pygmaeus]
Length = 678
Score = 139 bits (351), Expect = 8e-32
Identities = 71/198 (35%), Positives = 122/198 (60%), Gaps = 5/198 (2%)
Query: 55 VEQASLDTQLTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDD 113
+EQASLD + S NIG +LLQK GWK G+GLGK QG +PI ++ +G+G+ E +
Sbjct: 29 IEQASLDKPIESDNIGHRLLQKHGWKLGQGLGKSLQGRTDPIPIVVKYDVMGMGRMEMEL 88
Query: 114 FFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLA 173
+ + +R+ L+VE E+TEE +K + ++E+ I ++++R FYC+LC+KQY+
Sbjct: 89 DYAEDATERRRVLEVEKEDTEELRQKYKDYVDKEKAIAKALEDLRANFYCELCDKQYQKH 148
Query: 174 MEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRL 233
EF+ H++SYDH H++R K +K+ + R ++++++QE+ + + ++A+ +KQ
Sbjct: 149 QEFDNHINSYDHAHKQRLKDLKQREFARNVSSRSRKDEKKQEKALRRLHELAEQRKQAEC 208
Query: 234 QLLQESGSEPVSSETRTA 251
GS P+ T A
Sbjct: 209 ----APGSGPMFKPTTVA 222
>ref|NP_001002909.1| hypothetical protein LOC23131 [Homo sapiens]
Length = 1502
Score = 139 bits (351), Expect = 8e-32
Identities = 71/198 (35%), Positives = 122/198 (60%), Gaps = 5/198 (2%)
Query: 55 VEQASLDTQLTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDD 113
+EQASLD + S NIG +LLQK GWK G+GLGK QG +PI ++ +G+G+ E +
Sbjct: 29 IEQASLDKPIESDNIGHRLLQKHGWKLGQGLGKSLQGRTDPIPIVVKYDVMGMGRMEMEL 88
Query: 114 FFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLA 173
+ + +R+ L+VE E+TEE +K + ++E+ I ++++R FYC+LC+KQY+
Sbjct: 89 DYAEDATERRRVLEVEKEDTEELRQKYKDYVDKEKAIAKALEDLRANFYCELCDKQYQKH 148
Query: 174 MEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRL 233
EF+ H++SYDH H++R K +K+ + R ++++++QE+ + + ++A+ +KQ
Sbjct: 149 QEFDNHINSYDHAHKQRLKDLKQREFARNVSSRSRKDEKKQEKALRRLHELAEQRKQAEC 208
Query: 234 QLLQESGSEPVSSETRTA 251
GS P+ T A
Sbjct: 209 ----APGSGPMFKPTTVA 222
>ref|XP_537619.1| PREDICTED: similar to KIAA0553 protein [Canis familiaris]
Length = 1502
Score = 139 bits (350), Expect = 1e-31
Identities = 71/198 (35%), Positives = 122/198 (60%), Gaps = 5/198 (2%)
Query: 55 VEQASLDTQLTSSNIGFKLLQKMGWK-GKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDD 113
+EQASLD + S NIG +LLQK GWK G+GLGK QG +PI ++ +G+G+ E +
Sbjct: 29 IEQASLDKPIESDNIGHRLLQKHGWKLGQGLGKSLQGRTDPIPIVVKYDVMGMGRMEMEL 88
Query: 114 FFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLA 173
+ + +R+ L+VE E+TEE +K + ++E+ I ++++R FYC+LC+KQY+
Sbjct: 89 DYAEDATERRRVLEVEKEDTEELRQKYKDYVDKEKAIAKALEDLRANFYCELCDKQYQKH 148
Query: 174 MEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRL 233
EF+ H++SYDH H++R K +K+ + R ++++++QE+ + + ++A+ +KQ
Sbjct: 149 QEFDNHINSYDHAHKQRLKDLKQREFARNVSSRSRKDEKKQEKALRRLHELAEQRKQAEC 208
Query: 234 QLLQESGSEPVSSETRTA 251
GS P+ T A
Sbjct: 209 ----APGSGPMFRPTTVA 222
>dbj|BAA25479.2| KIAA0553 protein [Homo sapiens]
Length = 1450
Score = 101 bits (252), Expect = 2e-20
Identities = 51/173 (29%), Positives = 98/173 (56%), Gaps = 4/173 (2%)
Query: 79 WKGKGLGKDEQGIIEPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELEETEENVR 138
W G G G +PI ++ +G+G+ E + + + +R+ L+VE E+TEE +
Sbjct: 2 WTCCGDGNLSSGRTDPIPIVVKYDVMGMGRMEMELDYAEDATERRRVLEVEKEDTEELRQ 61
Query: 139 KREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMH 198
K + ++E+ I ++++R FYC+LC+KQY+ EF+ H++SYDH H++R K +K+
Sbjct: 62 KYKDYVDKEKAIAKALEDLRANFYCELCDKQYQKHQEFDNHINSYDHAHKQRLKDLKQRE 121
Query: 199 GGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTA 251
+ R ++++++QE+ + + ++A+ +KQ GS P+ T A
Sbjct: 122 FARNVSSRSRKDEKKQEKALRRLHELAEQRKQAEC----APGSGPMFKPTTVA 170
>ref|XP_424161.1| PREDICTED: similar to Hypothetical protein KIAA0553, partial
[Gallus gallus]
Length = 1156
Score = 98.2 bits (243), Expect = 3e-19
Identities = 48/155 (30%), Positives = 93/155 (59%), Gaps = 7/155 (4%)
Query: 104 LGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYC 163
+G+G+ E + + + +R+ L+VE E+TEE +K + ++E+ I ++++R FYC
Sbjct: 1 MGMGRMEMELDYAEDATERRRVLEVEKEDTEELRQKYKDYVDKEKAIAKALEDLRANFYC 60
Query: 164 DLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQ 223
+LC+KQY+ EF+ H++SYDH H++R K +K+ + R ++++++QER + + +
Sbjct: 61 ELCDKQYQKHQEFDNHINSYDHAHKQRLKDLKQREFARNVSSRSRKDERKQERALRRLHE 120
Query: 224 IADAQKQQRLQLLQESGSEPVSSETRTATPLTDQE 258
+A+ ++Q GS P+ RT T D+E
Sbjct: 121 LAEQRRQPEC----APGSGPM---FRTTTVAVDEE 148
>ref|XP_614639.1| PREDICTED: similar to KIAA0553 protein, partial [Bos taurus]
Length = 1040
Score = 97.8 bits (242), Expect = 3e-19
Identities = 51/180 (28%), Positives = 102/180 (56%), Gaps = 7/180 (3%)
Query: 93 EPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQT 152
+PI ++ +G+G+ E + + + +R+ L+VE E+TEE +K + ++E+ I
Sbjct: 3 DPIPIVVKYDVMGMGRMEMELDYAEDATERRRVLEVEKEDTEELRQKYKDYVDKEKAIAK 62
Query: 153 EVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQ 212
++++R FYC+LC+KQY+ EF+ H++SYDH H++R K +K+ + R +++++
Sbjct: 63 ALEDLRANFYCELCDKQYQKHQEFDNHINSYDHAHKQRLKDLKQREFARNVSSRSRKDEK 122
Query: 213 RQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTATPL---TDQEQRNALKFGFSS 269
+QE+ + + ++A+ +KQ GS P+ T A D++ +A G S+
Sbjct: 123 KQEKALRRLHELAEQRKQAEC----APGSGPMFRPTTVAVDEEAGDDEKDESATNSGTST 178
>ref|XP_152907.3| PREDICTED: hypothetical protein LOC237943 [Mus musculus]
Length = 1427
Score = 96.3 bits (238), Expect = 1e-18
Identities = 45/148 (30%), Positives = 89/148 (59%), Gaps = 4/148 (2%)
Query: 104 LGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQTEVKEIRKVFYC 163
+G+G+ E + + + +R+ L+VE E+TEE +K + ++E+ I ++++R FYC
Sbjct: 1 MGMGRMEMELDYAEDATERRRVLEVEKEDTEELRQKYKDYVDKEKAIAKALEDLRANFYC 60
Query: 164 DLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSRDDRQKREQQRQEREIAKFAQ 223
+LC+KQY+ EF+ H++SYDH H++R K +K+ + R ++++++QE+ + + +
Sbjct: 61 ELCDKQYQKHQEFDNHINSYDHAHKQRLKDLKQREFARNVSSRSRKDEKKQEKALRRLHE 120
Query: 224 IADAQKQQRLQLLQESGSEPVSSETRTA 251
+A+ +KQ GS P+ T A
Sbjct: 121 LAEQRKQAEC----APGSGPMFRPTTVA 144
>ref|XP_476218.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|51038033|gb|AAT93837.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 70
Score = 89.0 bits (219), Expect = 2e-16
Identities = 40/48 (83%), Positives = 46/48 (95%)
Query: 42 INHRPTENVDLENVEQASLDTQLTSSNIGFKLLQKMGWKGKGLGKDEQ 89
+NHRPTEN+D E +EQAS+DTQLTSSNIGF+LLQKMGWKGKGLGK+EQ
Sbjct: 1 MNHRPTENLDTEGLEQASVDTQLTSSNIGFRLLQKMGWKGKGLGKNEQ 48
>emb|CAG02788.1| unnamed protein product [Tetraodon nigroviridis]
Length = 1355
Score = 75.5 bits (184), Expect = 2e-12
Identities = 36/119 (30%), Positives = 68/119 (56%), Gaps = 4/119 (3%)
Query: 145 EREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSRD 204
E+E+ I ++++R FYC+LC+KQY EF+ H++SYDH H++R K++K+ +
Sbjct: 4 EKEKAIAKALEDLRANFYCELCDKQYTKHQEFDNHINSYDHAHKQRLKELKQREFARNVS 63
Query: 205 DRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTATPLTDQEQRNAL 263
R +++ ++QE+ + + ++A +QR Q GS P+ T A E +++
Sbjct: 64 SRSRKDGKKQEKMLRRLHELA----EQRKQFDCTPGSGPMFKTTTVAVDGEKMEDNDSV 118
>dbj|BAC33969.1| unnamed protein product [Mus musculus]
Length = 585
Score = 74.7 bits (182), Expect = 3e-12
Identities = 37/131 (28%), Positives = 74/131 (56%), Gaps = 6/131 (4%)
Query: 135 ENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQM 194
+N K AE+E I +++++ FYC+LC+KQY EF+ H++SYDH H++R K++
Sbjct: 31 KNGNKTLDYAEKENTIAKALEDLKANFYCELCDKQYYKHQEFDNHINSYDHAHKQRLKEL 90
Query: 195 KEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTAT-- 252
K+ + + ++++++QE+ + + ++A+ +K + + GS P+ T
Sbjct: 91 KQREFARNVASKSRKDERKQEKALQRLHKLAELRK----ETVCAPGSGPMFKSTTVTVRE 146
Query: 253 PLTDQEQRNAL 263
L D QR ++
Sbjct: 147 NLNDVSQRESV 157
>ref|XP_485015.2| PREDICTED: RIKEN cDNA C630007C17 gene [Mus musculus]
Length = 1180
Score = 74.3 bits (181), Expect = 4e-12
Identities = 35/122 (28%), Positives = 71/122 (57%), Gaps = 6/122 (4%)
Query: 144 AEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSR 203
AE+E I +++++ FYC+LC+KQY EF+ H++SYDH H++R K++K+ +
Sbjct: 20 AEKENTIAKALEDLKANFYCELCDKQYYKHQEFDNHINSYDHAHKQRLKELKQREFARNV 79
Query: 204 DDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTAT--PLTDQEQRN 261
+ ++++++QE+ + + ++A+ +K + + GS P+ T L D QR
Sbjct: 80 ASKSRKDERKQEKALQRLHKLAELRK----ETVCAPGSGPMFKSTTVTVRENLNDVSQRE 135
Query: 262 AL 263
++
Sbjct: 136 SV 137
>dbj|BAC31727.1| unnamed protein product [Mus musculus]
Length = 316
Score = 74.3 bits (181), Expect = 4e-12
Identities = 35/122 (28%), Positives = 71/122 (57%), Gaps = 6/122 (4%)
Query: 144 AEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSR 203
AE+E I +++++ FYC+LC+KQY EF+ H++SYDH H++R K++K+ +
Sbjct: 6 AEKENTIAKALEDLKANFYCELCDKQYYKHQEFDNHINSYDHAHKQRLKELKQREFARNV 65
Query: 204 DDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTAT--PLTDQEQRN 261
+ ++++++QE+ + + ++A+ +K + + GS P+ T L D QR
Sbjct: 66 ASKSRKDERKQEKALQRLHKLAELRK----ETVCAPGSGPMFKSTTVTVRENLNDVSQRE 121
Query: 262 AL 263
++
Sbjct: 122 SV 123
>ref|XP_594690.1| PREDICTED: similar to KIAA0553 protein, partial [Bos taurus]
Length = 99
Score = 73.9 bits (180), Expect = 5e-12
Identities = 33/97 (34%), Positives = 63/97 (64%)
Query: 93 EPIKSGMRDPRLGLGKQEEDDFFTAEENIQRKKLDVELEETEENVRKREVLAEREQKIQT 152
+PI ++ +G+G+ E + + + +R+ L+VE E+TEE +K + ++E+ I
Sbjct: 3 DPIPIVVKYDVMGMGRMEMELDYAEDATERRRVLEVEKEDTEELRQKYKDYVDKEKAIAK 62
Query: 153 EVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRK 189
++++R FYC+LC+KQY+ EF+ H++SYDH H++
Sbjct: 63 ALEDLRANFYCELCDKQYQKHQEFDNHINSYDHAHKQ 99
>ref|XP_144169.2| PREDICTED: similar to hypothetical protein FLJ32110 [Mus musculus]
Length = 1376
Score = 73.9 bits (180), Expect = 5e-12
Identities = 43/153 (28%), Positives = 79/153 (51%), Gaps = 13/153 (8%)
Query: 144 AEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQMKEMHGGSSR 203
AE+E+ +++ + FYC+LC+KQY EF+ H++SYDH H++R K++K +
Sbjct: 72 AEKEKSTAKSLEDAKANFYCELCDKQYNKHQEFDNHINSYDHAHKQRLKELKHREFARNV 131
Query: 204 DDRQKREQQRQEREIAKFAQIADAQKQQRLQLLQESGSEPVSSETRTATPLTDQEQRNAL 263
+ +++++QE+ + + Q+A+ ++Q SG+ P R A + Q Q+
Sbjct: 132 ASKSWKDEKKQEKALKRLHQLAELRQQSEC----VSGNGPAYKTPRIA--MEKQLQQGIF 185
Query: 264 KFGFSSKGSASKNVIGPKRQMVAKKQNIPVSSI 296
K S K+ V K +N+P SS+
Sbjct: 186 PVKNGRKVSCMKSA-------VFKGKNVPRSSM 211
>gb|AAP92799.1| hypothetical protein [Homo sapiens] gi|34740327|ref|NP_919226.1|
hypothetical protein LOC91752 [Homo sapiens]
Length = 1209
Score = 73.6 bits (179), Expect = 7e-12
Identities = 29/96 (30%), Positives = 62/96 (64%)
Query: 135 ENVRKREVLAEREQKIQTEVKEIRKVFYCDLCNKQYKLAMEFEAHLSSYDHNHRKRFKQM 194
+N K AE+E I +++++ FYC+LC+KQY EF+ H++SYDH H++R K++
Sbjct: 31 KNGNKTLDYAEKENTIAKALEDLKANFYCELCDKQYYKHQEFDNHINSYDHAHKQRLKEL 90
Query: 195 KEMHGGSSRDDRQKREQQRQEREIAKFAQIADAQKQ 230
K+ + + ++++++QE+ + + ++A+ +K+
Sbjct: 91 KQREFARNVASKSRKDERKQEKALQRLHKLAELRKE 126
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.311 0.130 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 483,997,490
Number of Sequences: 2540612
Number of extensions: 20213486
Number of successful extensions: 126424
Number of sequences better than 10.0: 2515
Number of HSP's better than 10.0 without gapping: 214
Number of HSP's successfully gapped in prelim test: 2387
Number of HSP's that attempted gapping in prelim test: 114049
Number of HSP's gapped (non-prelim): 7132
length of query: 304
length of database: 863,360,394
effective HSP length: 127
effective length of query: 177
effective length of database: 540,702,670
effective search space: 95704372590
effective search space used: 95704372590
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0206b.3


