

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0204.4
(368 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
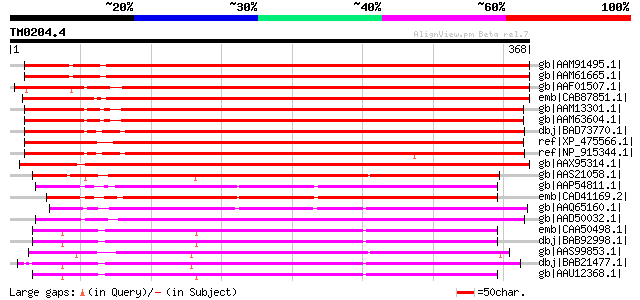
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM91495.1| AT5g05600/MOP10_14 [Arabidopsis thaliana] gi|1017... 514 e-144
gb|AAM61665.1| leucoanthocyanidin dioxygenase-like protein [Arab... 509 e-143
gb|AAF01507.1| putative leucoanthocyanidin dioxygenase [Arabidop... 506 e-142
emb|CAB87851.1| leucoanthocyanidin dioxygenase-like protein [Ara... 500 e-140
gb|AAM13301.1| putative anthocyanidin synthase [Arabidopsis thal... 439 e-122
gb|AAM63604.1| putative anthocyanidin synthase [Arabidopsis thal... 438 e-121
dbj|BAD73770.1| putative anthocyanidin synthase [Oryza sativa (j... 427 e-118
ref|XP_475566.1| putative leucoanthocyanidin dioxygenase (EC 1.1... 426 e-118
ref|NP_915344.1| leucoanthocyanidin dioxygenase-like protein [Or... 417 e-115
gb|AAX95314.1| leucoanthocyanidin dioxygenase-like protein [Oryz... 389 e-107
gb|AAS21058.1| flavonol synthase [Ginkgo biloba] 259 9e-68
gb|AAP54811.1| unknown protein [Oryza sativa (japonica cultivar-... 243 5e-63
emb|CAD41169.2| OSJNBa0064M23.14 [Oryza sativa (japonica cultiva... 242 1e-62
gb|AAQ65160.1| At4g10500 [Arabidopsis thaliana] gi|7267747|emb|C... 238 2e-61
gb|AAD50032.1| SRG1 Protein [Arabidopsis thaliana] gi|479047|emb... 237 4e-61
emb|CAA50498.1| anthocyanidin hydroxylase [Malus sp.] gi|4588783... 236 1e-60
dbj|BAB92998.1| anthocyanidin synthase [Malus x domestica] 235 1e-60
gb|AAS99853.1| anthocyanidin synthase [Allium cepa] 234 2e-60
dbj|BAB21477.1| anthocyanidin synthase [Torenia fournieri] 234 2e-60
gb|AAU12368.1| anthocyanidin synthase [Fragaria x ananassa] 234 3e-60
>gb|AAM91495.1| AT5g05600/MOP10_14 [Arabidopsis thaliana]
gi|10178137|dbj|BAB11549.1| leucoanthocyanidin
dioxygenase-like protein [Arabidopsis thaliana]
gi|15239179|ref|NP_196179.1| oxidoreductase, 2OG-Fe(II)
oxygenase family protein [Arabidopsis thaliana]
gi|14532538|gb|AAK63997.1| AT5g05600/MOP10_14
[Arabidopsis thaliana]
Length = 371
Score = 514 bits (1325), Expect = e-144
Identities = 248/358 (69%), Positives = 289/358 (80%), Gaps = 6/358 (1%)
Query: 11 WPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPD 70
WPEPIVRVQSL+E+ + S+P+RYIKP + RP+ ++ + A NIPIIDL GL +
Sbjct: 20 WPEPIVRVQSLAESNLSSLPDRYIKPASLRPT--TTEDAPTATNIPIIDLEGLF----SE 73
Query: 71 HPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPK 130
+ + +ISEAC+ WGFFQ+VNHGV P+LMD ARE WR+FFH+P+ K+ Y+NSP+
Sbjct: 74 EGLSDDVIMARISEACRGWGFFQVVNHGVKPELMDAARENWREFFHMPVNAKETYSNSPR 133
Query: 131 TYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLC 190
TYEGYGSRLGVEKGA LDWSDYYFLH LP LKD+NKWP+ PP+ REV DEYG ELVKL
Sbjct: 134 TYEGYGSRLGVEKGASLDWSDYYFLHLLPHHLKDFNKWPSFPPTIREVIDEYGEELVKLS 193
Query: 191 GRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLL 250
GR+M+VLS NL L+ED Q FGGE+IGACLRVN+YPKCP+PEL LGLS HSDPGGMT+L
Sbjct: 194 GRIMRVLSTNLGLKEDKFQEAFGGENIGACLRVNYYPKCPRPELALGLSPHSDPGGMTIL 253
Query: 251 LPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVS 310
LPDDQV GLQVRK D WITVKP PHAFIVNIGDQIQ+LSN+ YKSVEHRVIVNS+KERVS
Sbjct: 254 LPDDQVFGLQVRKDDTWITVKPHPHAFIVNIGDQIQILSNSTYKSVEHRVIVNSDKERVS 313
Query: 311 LAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 368
LAFFYNP+ DIPI+P +ELV P LY MTFD+YR FIR +GP GKSHVES SPR
Sbjct: 314 LAFFYNPKSDIPIQPLQELVSTHNPPLYPPMTFDQYRLFIRTQGPQGKSHVESHISPR 371
>gb|AAM61665.1| leucoanthocyanidin dioxygenase-like protein [Arabidopsis thaliana]
Length = 355
Score = 509 bits (1312), Expect = e-143
Identities = 246/358 (68%), Positives = 287/358 (79%), Gaps = 6/358 (1%)
Query: 11 WPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPD 70
WPEPIVRVQSL+E+ + S+P+RYIKP + RP+ ++ + A NIPIIDL GL +
Sbjct: 4 WPEPIVRVQSLAESNLSSLPDRYIKPASLRPT--TTEDAPTATNIPIIDLEGLF----SE 57
Query: 71 HPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPK 130
+ + +ISEAC+ WGFFQ+VNHGV P+LMD AR WR+FFH+P+ K+ Y+NSP+
Sbjct: 58 EGLSDDVIMARISEACRGWGFFQVVNHGVKPELMDAARXNWREFFHMPVNAKETYSNSPR 117
Query: 131 TYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLC 190
TYEGYGSRLGVEKGA LDWSDYYFLH LP LKD+NKWP+ PP+ REV DEYG ELVKL
Sbjct: 118 TYEGYGSRLGVEKGASLDWSDYYFLHLLPHHLKDFNKWPSFPPTIREVIDEYGEELVKLS 177
Query: 191 GRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLL 250
GR+M+VLS NL L+ED Q FGGE+IGACLRVN+YPKCP+P L LGLS HSDPGGMT+L
Sbjct: 178 GRIMRVLSTNLGLKEDKFQEAFGGENIGACLRVNYYPKCPRPVLALGLSPHSDPGGMTIL 237
Query: 251 LPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVS 310
LPDDQV GLQVRK D WITVKP PHAFIVNIGDQIQ+LSN+ YKSVEHRVIVNS+KERVS
Sbjct: 238 LPDDQVFGLQVRKDDTWITVKPHPHAFIVNIGDQIQILSNSTYKSVEHRVIVNSDKERVS 297
Query: 311 LAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSPR 368
LAFFYNP+ DIPI+P +ELV P LY MTFD+YR FIR +GP GKSHVES SPR
Sbjct: 298 LAFFYNPKSDIPIQPLQELVSTHNPPLYPPMTFDQYRLFIRTQGPQGKSHVESHISPR 355
>gb|AAF01507.1| putative leucoanthocyanidin dioxygenase [Arabidopsis thaliana]
gi|12321884|gb|AAG50980.1| leucoanthocyanidin
dioxygenase, putative; 41415-43854 [Arabidopsis
thaliana] gi|15229694|ref|NP_187728.1| oxidoreductase,
2OG-Fe(II) oxygenase family protein [Arabidopsis
thaliana]
Length = 400
Score = 506 bits (1304), Expect = e-142
Identities = 249/370 (67%), Positives = 294/370 (79%), Gaps = 14/370 (3%)
Query: 4 KNISPLN--WPEPIVRVQSLSETCIDSIPERYIKPLTDRPS---INSSSSSSDANNIPII 58
KN S L WPEPIVRVQSL+E+ + S+P+RYIKP + RP I+ +D N IPII
Sbjct: 40 KNKSGLGEKWPEPIVRVQSLAESNLTSLPDRYIKPPSQRPQTTIIDHQPEVADIN-IPII 98
Query: 59 DLRGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLP 118
DL L + D ++ISEAC+EWGFFQ++NHGV P+LMD ARETW+ FF+LP
Sbjct: 99 DLDSLFSGNEDDK--------KRISEACREWGFFQVINHGVKPELMDAARETWKSFFNLP 150
Query: 119 MELKQQYANSPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREV 178
+E K+ Y+NSP+TYEGYGSRLGVEKGAILDW+DYY+LH+LPL+LKD+NKWP+LP + RE+
Sbjct: 151 VEAKEVYSNSPRTYEGYGSRLGVEKGAILDWNDYYYLHFLPLALKDFNKWPSLPSNIREM 210
Query: 179 FDEYGRELVKLCGRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGL 238
DEYG+ELVKL GRLM +LS NL L + LQ FGGED+GACLRVN+YPKCP+PEL LGL
Sbjct: 211 NDEYGKELVKLGGRLMTILSSNLGLRAEQLQEAFGGEDVGACLRVNYYPKCPQPELALGL 270
Query: 239 SSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEH 298
S HSDPGGMT+LLPDDQV GLQVR GD WITV P+ HAFIVNIGDQIQ+LSN+ YKSVEH
Sbjct: 271 SPHSDPGGMTILLPDDQVVGLQVRHGDTWITVNPLRHAFIVNIGDQIQILSNSKYKSVEH 330
Query: 299 RVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGK 358
RVIVNS KERVSLAFFYNP+ DIPI+P ++LV P LY MTFD+YR FIR +GP GK
Sbjct: 331 RVIVNSEKERVSLAFFYNPKSDIPIQPMQQLVTSTMPPLYPPMTFDQYRLFIRTQGPRGK 390
Query: 359 SHVESLKSPR 368
SHVES SPR
Sbjct: 391 SHVESHISPR 400
>emb|CAB87851.1| leucoanthocyanidin dioxygenase-like protein [Arabidopsis thaliana]
gi|12043537|emb|CAC19787.1| putative leucoanthocyanidin
dioxygenase [Arabidopsis thaliana]
gi|15228785|ref|NP_191156.1| oxidoreductase, 2OG-Fe(II)
oxygenase family protein [Arabidopsis thaliana]
gi|11255840|pir||T49209 leucoanthocyanidin
dioxygenase-like protein - Arabidopsis thaliana
Length = 363
Score = 500 bits (1288), Expect = e-140
Identities = 242/362 (66%), Positives = 293/362 (80%), Gaps = 7/362 (1%)
Query: 10 NWPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSS-DANNIPIIDLRGLLYHGD 68
+WPEPIVRVQSLSE+ + +IP RY+KPL+ RP+I + + IPIIDL G LY
Sbjct: 6 DWPEPIVRVQSLSESNLGAIPNRYVKPLSQRPNITPHNKHNPQTTTIPIIDL-GRLY--- 61
Query: 69 PDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANS 128
D +A TL +IS+AC+E GFFQ+VNHG+SP LMD A+ TWR+FF+LPMELK +ANS
Sbjct: 62 TDDLTLQAKTLDEISKACRELGFFQVVNHGMSPQLMDQAKATWREFFNLPMELKNMHANS 121
Query: 129 PKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVK 188
PKTYEGYGSRLGVEKGAILDWSDYY+LHY P SLKDY KWP+LP CRE+ ++Y +E+VK
Sbjct: 122 PKTYEGYGSRLGVEKGAILDWSDYYYLHYQPSSLKDYTKWPSLPLHCREILEDYCKEMVK 181
Query: 189 LCGRLMKVLSLNLELEEDFLQNGFGG-EDIGACLRVNFYPKCPKPELTLGLSSHSDPGGM 247
LC LMK+LS NL L+ED LQN FGG E+ G CLRVN+YPKCP+PELTLG+S HSDPGG+
Sbjct: 182 LCENLMKILSKNLGLQEDRLQNAFGGKEESGGCLRVNYYPKCPQPELTLGISPHSDPGGL 241
Query: 248 TLLLPDDQVTGLQVRKGDN-WITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNK 306
T+LLPD+QV LQVR D+ WITV+P PHAFIVN+GDQIQ+LSN+IYKSVEHRVIVN
Sbjct: 242 TILLPDEQVASLQVRGSDDAWITVEPAPHAFIVNMGDQIQMLSNSIYKSVEHRVIVNPEN 301
Query: 307 ERVSLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKS 366
ER+SLAFFYNP+G++PIEP KELV D PALY++ T+D YR+FIR +GP K H++ LKS
Sbjct: 302 ERLSLAFFYNPKGNVPIEPLKELVTVDSPALYSSTTYDRYRQFIRTQGPRSKCHIDELKS 361
Query: 367 PR 368
PR
Sbjct: 362 PR 363
>gb|AAM13301.1| putative anthocyanidin synthase [Arabidopsis thaliana]
gi|3335372|gb|AAC27173.1| putative anthocyanidin
synthase [Arabidopsis thaliana]
gi|17065134|gb|AAL32721.1| putative anthocyanidin
synthase [Arabidopsis thaliana]
gi|15224472|ref|NP_181359.1| oxidoreductase, 2OG-Fe(II)
oxygenase family protein [Arabidopsis thaliana]
gi|7433241|pir||T01256 probable anthocyanidin synthase
[imported] - Arabidopsis thaliana
Length = 353
Score = 439 bits (1129), Expect = e-122
Identities = 211/355 (59%), Positives = 269/355 (75%), Gaps = 11/355 (3%)
Query: 11 WPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPD 70
WPEPIV VQSLS+T + ++P RY+KP RP N++ S + IP++D+ + G P+
Sbjct: 5 WPEPIVSVQSLSQTGVPTVPNRYVKPAHQRPVFNTTQSDAGIE-IPVLDMNDVW--GKPE 61
Query: 71 HPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPK 130
LR + AC+EWGFFQ+VNHGV+ LM+ R WR+FF LP+E K++YANSP
Sbjct: 62 G-------LRLVRSACEEWGFFQMVNHGVTHSLMERVRGAWREFFELPLEEKRKYANSPD 114
Query: 131 TYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLC 190
TYEGYGSRLGV K A LDWSDY+FL+YLP S+++ +KWP+ PP RE+ ++YG E+ KLC
Sbjct: 115 TYEGYGSRLGVVKDAKLDWSDYFFLNYLPSSIRNPSKWPSQPPKIRELIEKYGEEVRKLC 174
Query: 191 GRLMKVLSLNLELEEDFLQNGFGGED-IGACLRVNFYPKCPKPELTLGLSSHSDPGGMTL 249
RL + LS +L L+ + L GG D +GA LR NFYPKCP+P+LTLGLSSHSDPGG+T+
Sbjct: 175 ERLTETLSESLGLKPNKLMQALGGGDKVGASLRTNFYPKCPQPQLTLGLSSHSDPGGITI 234
Query: 250 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 309
LLPD++V GLQVR+GD W+T+K VP+A IVNIGDQ+Q+LSN IYKSVEH+VIVNS ERV
Sbjct: 235 LLPDEKVAGLQVRRGDGWVTIKSVPNALIVNIGDQLQILSNGIYKSVEHQVIVNSGMERV 294
Query: 310 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESL 364
SLAFFYNPR DIP+ P +ELV ++PALY + FDEYR IR KGPCGK+ V+SL
Sbjct: 295 SLAFFYNPRSDIPVGPIEELVTANRPALYKPIRFDEYRSLIRQKGPCGKNQVDSL 349
>gb|AAM63604.1| putative anthocyanidin synthase [Arabidopsis thaliana]
Length = 353
Score = 438 bits (1126), Expect = e-121
Identities = 210/355 (59%), Positives = 269/355 (75%), Gaps = 11/355 (3%)
Query: 11 WPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPD 70
WPEPIV VQSLS+T + ++P RY+KP RP N++ S + IP++D+ + G P+
Sbjct: 5 WPEPIVSVQSLSQTGVPTVPNRYVKPAHQRPVFNTTQSDAGIE-IPVLDMNDVW--GKPE 61
Query: 71 HPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPK 130
LR + AC+EWGFFQ+VNHGV+ LM+ R WR+FF LP+E K++YANSP
Sbjct: 62 G-------LRLVRSACEEWGFFQMVNHGVTHSLMERVRGAWREFFELPLEEKRKYANSPD 114
Query: 131 TYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLC 190
TYEGYGSRLGV + A LDWSDY+FL+YLP S+++ +KWP+ PP RE+ ++YG E+ KLC
Sbjct: 115 TYEGYGSRLGVVRDAKLDWSDYFFLNYLPSSIRNPSKWPSQPPKIRELIEKYGEEVRKLC 174
Query: 191 GRLMKVLSLNLELEEDFLQNGFGGED-IGACLRVNFYPKCPKPELTLGLSSHSDPGGMTL 249
RL + LS +L L+ + L GG D +GA LR NFYPKCP+P+LTLGLSSHSDPGG+T+
Sbjct: 175 ERLTETLSESLGLKPNKLMQALGGGDKVGASLRTNFYPKCPQPQLTLGLSSHSDPGGITI 234
Query: 250 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 309
LLPD++V GLQVR+GD W+T+K VP+A IVNIGDQ+Q+LSN IYKSVEH+VIVNS ERV
Sbjct: 235 LLPDEKVAGLQVRRGDGWVTIKSVPNALIVNIGDQLQILSNGIYKSVEHQVIVNSGMERV 294
Query: 310 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESL 364
SLAFFYNPR DIP+ P +ELV ++PALY + FDEYR IR KGPCGK+ V+SL
Sbjct: 295 SLAFFYNPRSDIPVGPIEELVTANRPALYKPIRFDEYRSLIRQKGPCGKNQVDSL 349
>dbj|BAD73770.1| putative anthocyanidin synthase [Oryza sativa (japonica
cultivar-group)]
Length = 366
Score = 427 bits (1098), Expect = e-118
Identities = 209/358 (58%), Positives = 273/358 (75%), Gaps = 11/358 (3%)
Query: 11 WPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPD 70
WPEP+VRVQ+L+++ +++IP Y+KP DRP+ + +SS A+ IP++DL +G D
Sbjct: 8 WPEPVVRVQALADSGLEAIPRCYVKPPCDRPAPEADDASSGAS-IPVVDLG----NGGDD 62
Query: 71 HPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPK 130
A ++ AC+ WGFFQ+VNHGV P+LM ARE W FF LP++ KQ+YANSP+
Sbjct: 63 EGGQLAEA---VAAACRGWGFFQVVNHGVRPELMRAAREAWHGFFRLPLQEKQKYANSPR 119
Query: 131 TYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNK-WPALPPSCREVFDEYGRELVKL 189
TYEGYGSRLGVEKGAILDW DYYFL P + K K WPA P C+EV +EYGRE++KL
Sbjct: 120 TYEGYGSRLGVEKGAILDWGDYYFLVLSPDAAKSPAKYWPANPGICKEVSEEYGREVIKL 179
Query: 190 CGRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTL 249
C RLM++LS +L L+E Q FGG D GA LR N+YP+CP+P+LTLGLS+HSDPG +T+
Sbjct: 180 CERLMRLLSASLGLDETRFQEAFGGADCGAGLRANYYPRCPQPDLTLGLSAHSDPGILTV 239
Query: 250 LLPDDQVTGLQVRKGD-NWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKER 308
LL DD V GLQVR+ D +W+TV+P+P AFIVN+GDQI++LSN++YKSVEHRVIVN+ +ER
Sbjct: 240 LLADDHVRGLQVRRRDGHWVTVQPLPDAFIVNVGDQIEILSNSMYKSVEHRVIVNAEEER 299
Query: 309 VSLAFFYNPRGDIPIEPAKELVKEDKPAL-YTAMTFDEYRRFIRMKGPCGKSHVESLK 365
+SLA FYNPRGD+P+ PA ELV ++P+L Y MTFDEYR ++R GP GK+ +E+LK
Sbjct: 300 ISLALFYNPRGDVPVAPAPELVTPERPSLYYRPMTFDEYRVYVRKNGPKGKAQLEALK 357
>ref|XP_475566.1| putative leucoanthocyanidin dioxygenase (EC 1.14.11.-) [Oryza
sativa (japonica cultivar-group)]
gi|46391159|gb|AAS90686.1| putative leucoanthocyanidin
dioxygenase [Oryza sativa (japonica cultivar-group)]
Length = 368
Score = 426 bits (1094), Expect = e-118
Identities = 202/355 (56%), Positives = 260/355 (72%), Gaps = 12/355 (3%)
Query: 11 WPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPD 70
WPEP+VRVQ+LSE+ +IP+RY++P T+RPS +S +++ NIP++D+
Sbjct: 10 WPEPVVRVQALSESGAATIPDRYVRPETERPSSSSEANAVANINIPVVDMSS-------- 61
Query: 71 HPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPK 130
+ + ++EAC+EWGFFQ VNHGV L+ AR WR FF PME+KQ+Y NSP
Sbjct: 62 ---SPGTAAAAVAEACREWGFFQAVNHGVPAALLRRARGVWRGFFQQPMEVKQRYGNSPA 118
Query: 131 TYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLC 190
TYEGYGSRLGV+KGAILDW DYYFLH P L +KWP LPP RE EY E+ +LC
Sbjct: 119 TYEGYGSRLGVDKGAILDWGDYYFLHVRPPHLLSPHKWPHLPPDLRETTTEYSEEVRRLC 178
Query: 191 GRLMKVLSLNLELEEDFLQNGFGG-EDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTL 249
RLM V+++ L +EE LQ FGG E G C+RVN+YP+CP+P+LTLGLSSHSDPGGMT+
Sbjct: 179 ERLMAVMAVGLGVEEGRLQEAFGGREGAGVCVRVNYYPRCPQPDLTLGLSSHSDPGGMTV 238
Query: 250 LLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERV 309
LL DD+V GLQVR W+TV PVP AFI+N+GDQIQV++NA+Y+SVEHRV+VN+ +ER+
Sbjct: 239 LLVDDRVKGLQVRHAGAWVTVDPVPDAFIINVGDQIQVVTNALYRSVEHRVVVNAAEERL 298
Query: 310 SLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESL 364
S+A FYNPR D+P+ P ELV ++P LY+ MTFD+YR +IR GP GKS V+ L
Sbjct: 299 SIATFYNPRSDLPVAPLPELVSPERPPLYSPMTFDDYRLYIRRNGPRGKSQVDRL 353
>ref|NP_915344.1| leucoanthocyanidin dioxygenase-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 385
Score = 417 bits (1071), Expect = e-115
Identities = 210/377 (55%), Positives = 273/377 (71%), Gaps = 30/377 (7%)
Query: 11 WPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPD 70
WPEP+VRVQ+L+++ +++IP Y+KP DRP+ + +SS A+ IP++DL +G D
Sbjct: 8 WPEPVVRVQALADSGLEAIPRCYVKPPCDRPAPEADDASSGAS-IPVVDLG----NGGDD 62
Query: 71 HPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPK 130
A ++ AC+ WGFFQ+VNHGV P+LM ARE W FF LP++ KQ+YANSP+
Sbjct: 63 EGGQLAEA---VAAACRGWGFFQVVNHGVRPELMRAAREAWHGFFRLPLQEKQKYANSPR 119
Query: 131 TYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNK-WPALPPSCREVFDEYGRELVKL 189
TYEGYGSRLGVEKGAILDW DYYFL P + K K WPA P C+EV +EYGRE++KL
Sbjct: 120 TYEGYGSRLGVEKGAILDWGDYYFLVLSPDAAKSPAKYWPANPGICKEVSEEYGREVIKL 179
Query: 190 CGRLMKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTL 249
C RLM++LS +L L+E Q FGG D GA LR N+YP+CP+P+LTLGLS+HSDPG +T+
Sbjct: 180 CERLMRLLSASLGLDETRFQEAFGGADCGAGLRANYYPRCPQPDLTLGLSAHSDPGILTV 239
Query: 250 LLPDDQVTGLQVRKGD-NWITVKPVPHAFIVNIGDQI-------------------QVLS 289
LL DD V GLQVR+ D +W+TV+P+P AFIVN+GDQI Q+LS
Sbjct: 240 LLADDHVRGLQVRRRDGHWVTVQPLPDAFIVNVGDQIEELISSCTVHGWNSWTDDVQILS 299
Query: 290 NAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPAKELVKEDKPAL-YTAMTFDEYRR 348
N++YKSVEHRVIVN+ +ER+SLA FYNPRGD+P+ PA ELV ++P+L Y MTFDEYR
Sbjct: 300 NSMYKSVEHRVIVNAEEERISLALFYNPRGDVPVAPAPELVTPERPSLYYRPMTFDEYRV 359
Query: 349 FIRMKGPCGKSHVESLK 365
++R GP GK+ +E+LK
Sbjct: 360 YVRKNGPKGKAQLEALK 376
>gb|AAX95314.1| leucoanthocyanidin dioxygenase-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 391
Score = 389 bits (1000), Expect = e-107
Identities = 189/362 (52%), Positives = 252/362 (69%), Gaps = 6/362 (1%)
Query: 8 PLNWPEPIVRVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHG 67
P WPEP+V VQSLSE + ++P +YIKP DRP + + S ++P +D+ L
Sbjct: 10 PDAWPEPVVPVQSLSEAGVSAVPPQYIKPPQDRPVLPAPSL-----DVPTVDVAAFLDLD 64
Query: 68 DPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYAN 127
A A L+ ++EAC + GFFQ+VNHGV ++ R WR+FF L ME K+ +N
Sbjct: 65 GAAAACAAAEQLKNLAEACSKHGFFQVVNHGVQASTVERMRGAWRRFFALEMEEKKACSN 124
Query: 128 SPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELV 187
SP EGYGSR GVEKGA+LDW DYYFL+ LP +K NKWP P RE+ ++YGR+L+
Sbjct: 125 SPSAPEGYGSRAGVEKGALLDWGDYYFLNILPREIKRRNKWPKSPHDLREITEDYGRDLM 184
Query: 188 KLCGRLMKVLSLNLELEEDFLQNGFGGED-IGACLRVNFYPKCPKPELTLGLSSHSDPGG 246
LC L+K +SL+L L E+ L FG +D I AC+RVN+YPKCP+PELTLG+SSHSD GG
Sbjct: 185 NLCEVLLKAMSLSLGLGENQLHAAFGSDDGISACMRVNYYPKCPQPELTLGISSHSDAGG 244
Query: 247 MTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNK 306
+ +LL DD+V G QV KGD W TV+P+P+AF+VN+GDQIQ++SN YKSVEHR + +S+
Sbjct: 245 IAVLLADDRVKGTQVLKGDTWYTVQPIPNAFLVNVGDQIQIISNDKYKSVEHRAVASSDD 304
Query: 307 ERVSLAFFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKS 366
R ++AFF NP G++PI PA +LV PALYT + FDEYRRF R +G GKS +E++K+
Sbjct: 305 ARFTVAFFCNPSGNLPIGPAAQLVSSQSPALYTPIVFDEYRRFSRRRGLKGKSQLEAMKN 364
Query: 367 PR 368
+
Sbjct: 365 SK 366
>gb|AAS21058.1| flavonol synthase [Ginkgo biloba]
Length = 340
Score = 259 bits (662), Expect = 9e-68
Identities = 135/335 (40%), Positives = 207/335 (61%), Gaps = 12/335 (3%)
Query: 17 RVQSLSETCIDSIPERYIKPLTDRPSINSSSSSSDA--NNIPIIDLRGLLYHGDPDHPEA 74
RVQ ++E+ +IP +++P+ +RP IN++ + IP+ID+ L + PE
Sbjct: 5 RVQYVAESRPQTIPLEFVRPVEERP-INTTFNDDIGLGRQIPVIDMCSL------EAPEL 57
Query: 75 RASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPK--TY 132
R T ++I+ A KEWG FQ++NH +SP L + ++FF LP E K+ YA + + +
Sbjct: 58 RVKTFKEIARASKEWGIFQVINHAISPLLFESLETVGKQFFQLPQEEKEAYACTGEDGSS 117
Query: 133 EGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGR 192
GYG++L WSD++F P SL+D++KWP P S EV +EY + ++ + +
Sbjct: 118 TGYGTKLACTTDGRQGWSDFFFHMLWPPSLRDFSKWPQKPSSYIEVTEEYSKGILGVLNK 177
Query: 193 LMKVLSLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLP 252
L+ LS++LEL+E L++ GGE++ L++N+YP CP+PE+ G+ H+D +T+L P
Sbjct: 178 LLSALSISLELQESALKDALGGENLEMELKINYYPTCPQPEVAFGVVPHTDMSALTILKP 237
Query: 253 DDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLA 312
+D V GLQV K + WIT VP+A I++IGDQIQ+LSN +KSV HR +VN K R+S
Sbjct: 238 ND-VPGLQVWKDEKWITAHYVPNALIIHIGDQIQILSNGKFKSVLHRSLVNKEKVRMSWP 296
Query: 313 FFYNPRGDIPIEPAKELVKEDKPALYTAMTFDEYR 347
F +P D I P KEL+ + P LY A T+ EY+
Sbjct: 297 VFCSPPLDTVIGPLKELIDDSNPPLYNARTYREYK 331
>gb|AAP54811.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|21717150|gb|AAM76343.1| unknown protein [Oryza sativa
(japonica cultivar-group)] gi|37536444|ref|NP_922524.1|
unknown protein [Oryza sativa (japonica cultivar-group)]
gi|18057095|gb|AAL58118.1| putative flavanone
3-hydroxylase [Oryza sativa (japonica cultivar-group)]
Length = 342
Score = 243 bits (621), Expect = 5e-63
Identities = 131/329 (39%), Positives = 195/329 (58%), Gaps = 17/329 (5%)
Query: 19 QSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAST 78
Q LS D++P +Y++P + RP ++ S + IP++DL PD RA+
Sbjct: 10 QLLSTAVHDTMPGKYVRPESQRPRLDLVVSDA---RIPVVDL------ASPD----RAAV 56
Query: 79 LRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQ-YANSPKTYEGYGS 137
+ + +AC+ GFFQ+VNHG+ L+ E R+FF LP E K + Y++ P +
Sbjct: 57 VSAVGDACRTHGFFQVVNHGIDAALIASVMEVGREFFRLPAEEKAKLYSDDPAKKIRLST 116
Query: 138 RLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVL 197
V K + +W DY LH PL + WP+ PPS +E+ Y E+ +L RL + +
Sbjct: 117 SFNVRKETVHNWRDYLRLHCYPLH-QFVPDWPSNPPSFKEIIGTYCTEVRELGFRLYEAI 175
Query: 198 SLNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVT 257
S +L LE +++ G ++ + VN+YP+CP+PELT GL +H+DP +T+LL DDQV
Sbjct: 176 SESLGLEGGYMRETLGEQE--QHMAVNYYPQCPEPELTYGLPAHTDPNALTILLMDDQVA 233
Query: 258 GLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNP 317
GLQV WI V P P A ++NIGDQ+Q LSN Y+SV HR +VNS++ER+S+A F P
Sbjct: 234 GLQVLNDGKWIAVNPQPGALVINIGDQLQALSNGKYRSVWHRAVVNSDRERMSVASFLCP 293
Query: 318 RGDIPIEPAKELVKEDKPALYTAMTFDEY 346
+ + PAK+L+ +D PA+Y T+DEY
Sbjct: 294 CNSVELGPAKKLITDDSPAVYRNYTYDEY 322
>emb|CAD41169.2| OSJNBa0064M23.14 [Oryza sativa (japonica cultivar-group)]
gi|50928227|ref|XP_473641.1| OSJNBa0064M23.14 [Oryza
sativa (japonica cultivar-group)]
Length = 340
Score = 242 bits (617), Expect = 1e-62
Identities = 130/322 (40%), Positives = 195/322 (60%), Gaps = 18/322 (5%)
Query: 27 DSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARASTLRQISEAC 86
+++PE Y +P +DRP + ++ S NIP+IDL PD P A +I++AC
Sbjct: 14 ETLPEGYARPESDRPRLAEVATDS---NIPLIDL------ASPDKPRVIA----EIAQAC 60
Query: 87 KEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQ-YANSPKTYEGYGSRLGVEKGA 145
+ +GFFQ+ NHG++ +L++ +FF LP E K++ Y++ P + V K
Sbjct: 61 RTYGFFQVTNHGIAEELLEKVMAVALEFFRLPPEEKEKLYSDEPSKKIRLSTSFNVRKET 120
Query: 146 ILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLELEE 205
+ +W DY LH PL + +WP+ P +E+ Y RE+ +L RL+ +S++L LEE
Sbjct: 121 VHNWRDYLRLHCHPLE-EFVPEWPSNPAQFKEIMSTYCREVRQLGLRLLGAISVSLGLEE 179
Query: 206 DFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQV-RKG 264
D+++ G ++ + VN+YP+CP+P+LT GL H+DP +T+LLPD V GLQV R G
Sbjct: 180 DYIEKVLGEQE--QHMAVNYYPRCPEPDLTYGLPKHTDPNALTILLPDPHVAGLQVLRDG 237
Query: 265 DNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIE 324
D WI V P P+A +VN+GDQIQ LSN YKSV HR +VN +ER+S+A F P I
Sbjct: 238 DQWIVVNPRPNALVVNLGDQIQALSNDAYKSVWHRAVVNPVQERMSVASFMCPCNSAVIS 297
Query: 325 PAKELVKEDKPALYTAMTFDEY 346
PA++LV + +Y + T+DEY
Sbjct: 298 PARKLVADGDAPVYRSFTYDEY 319
>gb|AAQ65160.1| At4g10500 [Arabidopsis thaliana] gi|7267747|emb|CAB78173.1|
putative Fe(II)/ascorbate oxidase [Arabidopsis thaliana]
gi|4539410|emb|CAB40043.1| putative Fe(II)/ascorbate
oxidase [Arabidopsis thaliana] gi|4115914|gb|AAD03425.1|
contains similarity to Iron/Ascorbate family of
oxidoreductases (Pfam: PF00671, Score=297.8, E=1.3e-85,
N=1) [Arabidopsis thaliana] gi|15235126|ref|NP_192788.1|
oxidoreductase, 2OG-Fe(II) oxygenase family protein
[Arabidopsis thaliana] gi|51972019|dbj|BAD44674.1|
putative Fe(II)/ascorbate oxidase [Arabidopsis thaliana]
gi|51971553|dbj|BAD44441.1| putative Fe(II)/ascorbate
oxidase [Arabidopsis thaliana] gi|7433239|pir||T04185
hypothetical protein F7L13.80 - Arabidopsis thaliana
Length = 349
Score = 238 bits (607), Expect = 2e-61
Identities = 140/342 (40%), Positives = 196/342 (56%), Gaps = 17/342 (4%)
Query: 29 IPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARASTLRQISEACKE 88
IP Y++P++DRP+++ SS D+ IP+IDLR L H RA ++Q++ AC
Sbjct: 20 IPSNYVRPISDRPNLSEVESSGDS--IPLIDLRDL-------HGPNRAVIVQQLASACST 70
Query: 89 WGFFQIVNHGVSPDLMDMARETWRKFFHLP-MELKQQYANSPKTYEGYGSRLGVEKGAIL 147
+GFFQI NHGV ++ + R+FFH P E + Y+ P + V +L
Sbjct: 71 YGFFQIKNHGVPDTTVNKMQTVAREFFHQPESERVKHYSADPTKTTRLSTSFNVGADKVL 130
Query: 148 DWSDYYFLHYLPLSLKDY-NKWPALPPSCREVFDEYGRELVKLCGRLMKVLSLNLELEED 206
+W D+ LH P+ +D+ +WP+ P S REV EY + L RL++ +S +L LE D
Sbjct: 131 NWRDFLRLHCFPI--EDFIEEWPSSPISFREVTAEYATSVRALVLRLLEAISESLGLESD 188
Query: 207 FLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTGLQVRKGDN 266
+ N G + N+YP CP+PELT GL H DP +T+LL DQV+GLQV K D
Sbjct: 189 HISNILGKH--AQHMAFNYYPPCPEPELTYGLPGHKDPTVITVLL-QDQVSGLQVFKDDK 245
Query: 267 WITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPRGDIPIEPA 326
W+ V P+P+ FIVNIGDQ+QV+SN YKSV HR +VN+ ER+S+ FY P D I PA
Sbjct: 246 WVAVSPIPNTFIVNIGDQMQVISNDKYKSVLHRAVVNTENERLSIPTFYFPSTDAVIGPA 305
Query: 327 KELVKE-DKPALYTAMTFDEYRRFIRMKGPCGKSHVESLKSP 367
ELV E D A+Y F EY + S +++ K+P
Sbjct: 306 HELVNEQDSLAIYRTYPFVEYWDKFWNRSLATASCLDAFKAP 347
>gb|AAD50032.1| SRG1 Protein [Arabidopsis thaliana] gi|479047|emb|CAA55654.1| SRG1
[Arabidopsis thaliana] gi|22655018|gb|AAM98100.1|
At1g17020/F6I1.30 [Arabidopsis thaliana]
gi|15219988|ref|NP_173145.1| oxidoreductase, 2OG-Fe(II)
oxygenase family protein [Arabidopsis thaliana]
gi|15081819|gb|AAK82564.1| F6I1.30/F6I1.30 [Arabidopsis
thaliana] gi|629561|pir||S44261 SRG1 protein -
Arabidopsis thaliana
Length = 358
Score = 237 bits (605), Expect = 4e-61
Identities = 121/347 (34%), Positives = 202/347 (57%), Gaps = 8/347 (2%)
Query: 19 QSLSETCIDSIPERYIKPLTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPEARAST 78
+ + E I ++P RY++ D+ ++ IPIID++ L D S
Sbjct: 19 EMVKEKTITTVPPRYVRSDQDKTEVDDDFDVKI--EIPIIDMKRLCSSTTMD------SE 70
Query: 79 LRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKTYEGYGSR 138
+ ++ ACKEWGFFQ+VNHG+ +D + + FF+LPME K+++ P EG+G
Sbjct: 71 VEKLDFACKEWGFFQLVNHGIDSSFLDKVKSEIQDFFNLPMEEKKKFWQRPDEIEGFGQA 130
Query: 139 LGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCGRLMKVLS 198
V + LDW+D +F P+ L+ + +P LP R+ + Y E+ + L+ ++
Sbjct: 131 FVVSEDQKLDWADLFFHTVQPVELRKPHLFPKLPLPFRDTLEMYSSEVQSVAKILIAKMA 190
Query: 199 LNLELEEDFLQNGFGGEDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLLLPDDQVTG 258
LE++ + L+ F D +R+N+YP CP+P+ +GL+ HSD G+T+L+ + V G
Sbjct: 191 RALEIKPEELEKLFDDVDSVQSMRMNYYPPCPQPDQVIGLTPHSDSVGLTVLMQVNDVEG 250
Query: 259 LQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVSLAFFYNPR 318
LQ++K W+ VKP+P+AFIVNIGD +++++N Y+S+EHR +VNS KER+S+A F+N
Sbjct: 251 LQIKKDGKWVPVKPLPNAFIVNIGDVLEIITNGTYRSIEHRGVVNSEKERLSIATFHNVG 310
Query: 319 GDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGPCGKSHVESLK 365
+ PAK LV+ K A + +T EY + + GK+++++L+
Sbjct: 311 MYKEVGPAKSLVERQKVARFKRLTMKEYNDGLFSRTLDGKAYLDALR 357
>emb|CAA50498.1| anthocyanidin hydroxylase [Malus sp.] gi|4588783|gb|AAD26205.1|
anthocyanidin synthase [Malus x domestica]
gi|1730107|sp|P51091|LDOX_MALDO Leucoanthocyanidin
dioxygenase (LDOX) (Leucocyanidin oxygenase)
(Leucoanthocyanidin hydroxylase) (Anthocyanidin
synthase)
Length = 357
Score = 236 bits (601), Expect = 1e-60
Identities = 125/337 (37%), Positives = 202/337 (59%), Gaps = 12/337 (3%)
Query: 17 RVQSLSETCIDSIPERYIKP---LTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPE 73
RV++L+ + I +IP+ YI+P L + I +++ +P IDL+ + + D+ +
Sbjct: 10 RVETLAGSGISTIPKEYIRPKDELVNIGDIFEQEKNNEGPQVPTIDLKEI----ESDNEK 65
Query: 74 ARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKT-- 131
RA ++ +A +WG +VNHG+S +LMD R+ + FF LP+E K++YAN +
Sbjct: 66 VRAKCREKLKKAAVDWGVMHLVNHGISDELMDKVRKAGKAFFDLPIEQKEKYANDQASGK 125
Query: 132 YEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCG 191
+GYGS+L L+W DY+F P +D + WP P E EY ++L +L
Sbjct: 126 IQGYGSKLANNASGQLEWEDYFFHCVYPEDKRDLSIWPQTPADYIEATAEYAKQLRELAT 185
Query: 192 RLMKVLSLNLELEEDFLQNGFGG-EDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLL 250
+++KVLSL L L+E L+ GG E++ +++N+YPKCP+PEL LG+ +H+D +T +
Sbjct: 186 KVLKVLSLGLGLDEGRLEKEVGGLEELLLQMKINYYPKCPQPELALGVEAHTDVSALTFI 245
Query: 251 LPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVS 310
L + V GLQ+ W+T K VP++ +++IGD +++LSN YKS+ HR +VN K R+S
Sbjct: 246 L-HNMVPGLQLFYEGKWVTAKCVPNSIVMHIGDTLEILSNGKYKSILHRGMVNKEKVRIS 304
Query: 311 LAFFYN-PRGDIPIEPAKELVKEDKPALYTAMTFDEY 346
A F P+ I ++P E V ED+PA++ TF E+
Sbjct: 305 WAVFCEPPKEKIILKPLPETVSEDEPAMFPPRTFAEH 341
>dbj|BAB92998.1| anthocyanidin synthase [Malus x domestica]
Length = 357
Score = 235 bits (600), Expect = 1e-60
Identities = 125/337 (37%), Positives = 202/337 (59%), Gaps = 12/337 (3%)
Query: 17 RVQSLSETCIDSIPERYIKP---LTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPE 73
RV++L+ + I +IP+ YI+P L + I +++ +P IDL+ + + D+ +
Sbjct: 10 RVETLAGSGISTIPKEYIRPKDELVNIGDIFEQEKNNEGPQVPTIDLKEI----ESDNEK 65
Query: 74 ARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKT-- 131
RA ++ +A +WG +VNHG+S +LMD R+ + FF LP+E K++YAN +
Sbjct: 66 VRAKCREKLKKATVDWGVMHLVNHGISDELMDKVRKAGKAFFDLPIEQKEKYANDQASGK 125
Query: 132 YEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCG 191
+GYGS+L L+W DY+F P +D + WP P E EY ++L +L
Sbjct: 126 IQGYGSKLANNASGQLEWEDYFFHCVYPEDKRDLSIWPQTPADYIEATAEYAKQLRELAT 185
Query: 192 RLMKVLSLNLELEEDFLQNGFGG-EDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLL 250
+++KVLSL L L+E L+ GG E++ +++N+YPKCP+PEL LG+ +H+D +T +
Sbjct: 186 KVLKVLSLGLGLDEGRLEKEVGGLEELLLQMKINYYPKCPQPELALGVEAHTDVSALTFI 245
Query: 251 LPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVS 310
L + V GLQ+ W+T K VP++ +++IGD +++LSN YKS+ HR +VN K R+S
Sbjct: 246 L-HNMVPGLQLFYEGKWVTAKCVPNSIVMHIGDTLEILSNGKYKSILHRGMVNKEKVRIS 304
Query: 311 LAFFYN-PRGDIPIEPAKELVKEDKPALYTAMTFDEY 346
A F P+ I ++P E V ED+PA++ TF E+
Sbjct: 305 WAVFCEPPKEKIILKPLPETVSEDEPAMFPPRTFAEH 341
>gb|AAS99853.1| anthocyanidin synthase [Allium cepa]
Length = 352
Score = 234 bits (598), Expect = 2e-60
Identities = 133/352 (37%), Positives = 202/352 (56%), Gaps = 25/352 (7%)
Query: 14 PIVRVQSLSETCIDSIPERYIKPLTDRPSINSS----SSSSDANNIPIIDLRGLLYHGDP 69
P RV++LS++ + SIP YI+P +R + + +S+ IPIIDL
Sbjct: 11 PAPRVETLSKSNLHSIPLEYIRPEHERACLGDALEQLHNSNSGPQIPIIDLDS------- 63
Query: 70 DHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYAN-- 127
+ ++++A KEWG IVNHG+S +LM+ R FF+LP+E K++YAN
Sbjct: 64 ------RDCIEKVTKAAKEWGVMHIVNHGISSELMEKVRAAGMAFFNLPLEAKEEYANDQ 117
Query: 128 SPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELV 187
S +GYGS+L L+W DY+F P D + WP P E+ E+G +L
Sbjct: 118 SKGKIQGYGSKLANNASGQLEWEDYFFHLIFPDDKVDLSVWPKQPSDYIEIMQEFGSQLR 177
Query: 188 KLCGRLMKVLSLNLELE-EDFLQNGFGG-EDIGACLRVNFYPKCPKPELTLGLSSHSDPG 245
L +++ +LSL L+L +D L+ G ED+ L++N+YPKCP+P L LG+ +H+D
Sbjct: 178 ILASKMLSILSLGLQLPTKDRLEQELKGPEDLLLQLKINYYPKCPQPHLALGVEAHTDVS 237
Query: 246 GMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSN 305
++ +L ++ V GLQV W+T K VP + IV++GD +++LSN IYKSV HR +VN
Sbjct: 238 ALSFILHNN-VPGLQVFYEGEWVTAKLVPDSLIVHVGDSLEILSNGIYKSVLHRGLVNKE 296
Query: 306 KERVSLAFFYNPRGD-IPIEPAKELVKEDKPALYTAMTFDEY--RRFIRMKG 354
K R+S A F P D + ++P E+V +D PA YT TF ++ R+ + KG
Sbjct: 297 KVRISWAVFCEPPKDAVVLKPLDEVVTDDAPARYTPRTFAQHLERKLFKKKG 348
>dbj|BAB21477.1| anthocyanidin synthase [Torenia fournieri]
Length = 375
Score = 234 bits (598), Expect = 2e-60
Identities = 135/367 (36%), Positives = 209/367 (56%), Gaps = 17/367 (4%)
Query: 6 ISPLNWPEPIVRVQSLSETCIDSIPERYIKP------LTDRPSINSSSSSSDANNIPIID 59
+SP + P P RV+ LS + I +IP+ Y++ +TD S + + D+ ++PIID
Sbjct: 2 VSPAS-PSP-ARVELLSNSGIKAIPKEYVRTEEELRSITDIFSKEDGAKNIDSPDLPIID 59
Query: 60 LRGLLYHGDPDHPEARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPM 119
L + D E R ++ EA EWG ++NHG+S +L++ ++ +FF LP+
Sbjct: 60 LSKI----DSSDEETRKKGHEELKEAAIEWGVMHLINHGISDELINRVKKAGGEFFDLPV 115
Query: 120 ELKQQYAN--SPKTYEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCRE 177
E K++YAN S +GYGS+L G IL+W DY+F P +D WP P
Sbjct: 116 EEKEKYANDQSSGNVQGYGSKLANNAGGILEWEDYFFHCVYPEEKRDMAIWPKDPQDYIP 175
Query: 178 VFDEYGRELVKLCGRLMKVLSLNLELEEDFLQNGFGG-EDIGACLRVNFYPKCPKPELTL 236
EY +E+ L +++ VLSL L L++D L+ GG +D+ +++N+YPKCP+PEL L
Sbjct: 176 ATTEYAKEIRSLTTKILSVLSLGLGLDQDRLEKEVGGKDDLTLQMKINYYPKCPQPELAL 235
Query: 237 GLSSHSDPGGMTLLLPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSV 296
G+ +H+D +T +L + V GLQV W+T VP + I+++GD I++LSN +YKSV
Sbjct: 236 GVEAHTDVSALTFIL-HNMVPGLQVLYKGEWVTASCVPDSIILHVGDTIEILSNGMYKSV 294
Query: 297 EHRVIVNSNKERVSLAFFYN-PRGDIPIEPAKELVKEDKPALYTAMTFDEYRRFIRMKGP 355
HR +VN + RVS A F P+ I ++P E V ED+PAL+ TF ++ + K
Sbjct: 295 LHRGLVNRERVRVSWAVFCEPPKEKIVLKPLPETVGEDRPALFPPRTFAQHMKHKLFKKS 354
Query: 356 CGKSHVE 362
+ H E
Sbjct: 355 DDEVHDE 361
>gb|AAU12368.1| anthocyanidin synthase [Fragaria x ananassa]
Length = 383
Score = 234 bits (597), Expect = 3e-60
Identities = 124/337 (36%), Positives = 197/337 (57%), Gaps = 12/337 (3%)
Query: 17 RVQSLSETCIDSIPERYIKP---LTDRPSINSSSSSSDANNIPIIDLRGLLYHGDPDHPE 73
RV+SL+ + I +IP+ Y++P L + I S++ +P IDL+ + D D +
Sbjct: 10 RVESLASSGISTIPKEYVRPEEELVNIGDIFEDEKSTEGPQVPTIDLKEI----DSDDIK 65
Query: 74 ARASTLRQISEACKEWGFFQIVNHGVSPDLMDMARETWRKFFHLPMELKQQYANSPKT-- 131
R + EA EWG ++NHG+S +LM+ ++ + FF LP+E K++YAN +
Sbjct: 66 VREKCREDLKEAAVEWGVMHLINHGISDELMERVKKAGKAFFDLPIEQKEKYANDQASGK 125
Query: 132 YEGYGSRLGVEKGAILDWSDYYFLHYLPLSLKDYNKWPALPPSCREVFDEYGRELVKLCG 191
+GYGS+L L+W DY+F P +D + WP P EY +EL L
Sbjct: 126 IQGYGSKLANNASGQLEWEDYFFHCVYPEDKRDLSIWPQTPSDYIVATSEYAKELRGLAT 185
Query: 192 RLMKVLSLNLELEEDFLQNGFGG-EDIGACLRVNFYPKCPKPELTLGLSSHSDPGGMTLL 250
+++ +LSL L LEE L+ GG E++ +++N+YPKCP+PEL LG+ +H+D +T +
Sbjct: 186 KILSILSLGLGLEEGRLEKEVGGLEELLLQMKINYYPKCPQPELALGVEAHTDISALTFI 245
Query: 251 LPDDQVTGLQVRKGDNWITVKPVPHAFIVNIGDQIQVLSNAIYKSVEHRVIVNSNKERVS 310
L + V GLQ+ G W+T K VP++ +++IGD +++LSN YKS+ HR +VN K R+S
Sbjct: 246 L-HNMVPGLQLFYGGKWVTAKCVPNSIVMHIGDTLEILSNGKYKSILHRGLVNKEKVRIS 304
Query: 311 LAFFYN-PRGDIPIEPAKELVKEDKPALYTAMTFDEY 346
A F P+ I ++P E V E++PA++ TF E+
Sbjct: 305 WAVFCEPPKEKIILKPLPETVSEEEPAIFPPRTFFEH 341
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 690,726,631
Number of Sequences: 2540612
Number of extensions: 31057358
Number of successful extensions: 70141
Number of sequences better than 10.0: 1090
Number of HSP's better than 10.0 without gapping: 991
Number of HSP's successfully gapped in prelim test: 99
Number of HSP's that attempted gapping in prelim test: 66748
Number of HSP's gapped (non-prelim): 1196
length of query: 368
length of database: 863,360,394
effective HSP length: 129
effective length of query: 239
effective length of database: 535,621,446
effective search space: 128013525594
effective search space used: 128013525594
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0204.4


