

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0195b.3
(495 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
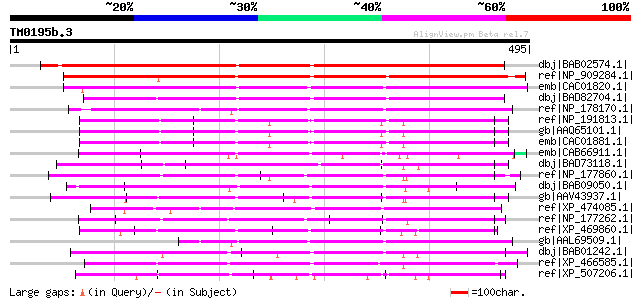
dbj|BAB02574.1| unnamed protein product [Arabidopsis thaliana] g... 526 e-148
ref|NP_909284.1| hypothetical protein [Oryza sativa (japonica cu... 463 e-129
emb|CAC01820.1| putative protein [Arabidopsis thaliana] gi|15242... 271 3e-71
dbj|BAD82704.1| pentatricopeptide (PPR) repeat-containing protei... 250 6e-65
ref|NP_178170.1| pentatricopeptide (PPR) repeat-containing prote... 225 3e-57
ref|NP_191813.1| pentatricopeptide (PPR) repeat-containing prote... 218 3e-55
gb|AAQ65101.1| At3g62470 [Arabidopsis thaliana] gi|7340718|emb|C... 218 3e-55
emb|CAC01881.1| putative protein [Arabidopsis thaliana] gi|15241... 218 3e-55
emb|CAB66911.1| putative protein [Arabidopsis thaliana] gi|15229... 207 8e-52
dbj|BAD73118.1| pentatricopeptide (PPR) repeat-containing protei... 198 4e-49
ref|NP_177860.1| pentatricopeptide (PPR) repeat-containing prote... 193 1e-47
dbj|BAB09050.1| unnamed protein product [Arabidopsis thaliana] g... 193 1e-47
gb|AAV43937.1| unknown protein [Oryza sativa (japonica cultivar-... 191 3e-47
ref|XP_474085.1| OSJNBa0033G05.8 [Oryza sativa (japonica cultiva... 185 3e-45
ref|NP_177262.1| pentatricopeptide (PPR) repeat-containing prote... 181 4e-44
ref|XP_469860.1| unknown protein [Oryza sativa (japonica cultiva... 181 6e-44
gb|AAL69509.1| unknown protein [Arabidopsis thaliana] 177 5e-43
dbj|BAB01242.1| unnamed protein product [Arabidopsis thaliana] g... 177 7e-43
ref|XP_466585.1| putative pentatricopeptide (PPR) repeat-contain... 174 6e-42
ref|XP_507206.1| PREDICTED OSJNBa0091C18.36 gene product [Oryza ... 172 3e-41
>dbj|BAB02574.1| unnamed protein product [Arabidopsis thaliana]
gi|15232517|ref|NP_188138.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
Length = 523
Score = 526 bits (1354), Expect = e-148
Identities = 258/443 (58%), Positives = 329/443 (74%), Gaps = 7/443 (1%)
Query: 30 PPLFTRRFLHSQPEHAAGAGAFVQNLLKFRRDKPTDQVERALDLCGFDLNDDLVLDVLRR 89
P F RF + + +A V N++K R ++++R LD CG DL ++LVL+V+ R
Sbjct: 62 PDKFPNRFNDDKDKQSA---LDVHNIIKHHRGSSPEKIKRILDKCGIDLTEELVLEVVNR 118
Query: 90 HRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNE 149
+RSDWKPA + K + +S + NEILD+LGKM RFEE HQVFDEMS R+G VNE
Sbjct: 119 NRSDWKPAYILSQLVVKQSVHLSSSMLYNEILDVLGKMRRFEEFHQVFDEMSKRDGFVNE 178
Query: 150 DTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHS 209
T+ LL R+AAAHKV+EA+ +F R++FG+D DL AF LLMWLCRYKHVE AETLF S
Sbjct: 179 KTYEVLLNRYAAAHKVDEAVGVFERRKEFGIDDDLVAFHGLLMWLCRYKHVEFAETLFCS 238
Query: 210 KAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKG 269
+ REF DIK N+ILNGWCVLGN HEAKR WKDI+ASKCRPD+ +Y T I ALTKKG
Sbjct: 239 RRREFGC--DIKAMNMILNGWCVLGNVHEAKRFWKDIIASKCRPDVVSYGTMINALTKKG 296
Query: 270 KLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVAT 329
KLG A++L+R MW+ N PDV ICN +IDALCFKKR+PEALEVF+++ E+G +PNV T
Sbjct: 297 KLGKAMELYRAMWDTRRN--PDVKICNNVIDALCFKKRIPEALEVFREISEKGPDPNVVT 354
Query: 330 YNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMER 389
YNSL+KHLCKIRR EKV+ELVE+ME K GSC PN VT+S LL + ++V VLERM +
Sbjct: 355 YNSLLKHLCKIRRTEKVWELVEEMELKGGSCSPNDVTFSYLLKYSQRSKDVDIVLERMAK 414
Query: 390 NGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKD 449
N C ++ D+YNL+ RLY++WD ++ +R+ W EMER+G GPD+R+YTI IHG + GK+ +
Sbjct: 415 NKCEMTSDLYNLMFRLYVQWDKEEKVREIWSEMERSGLGPDQRTYTIRIHGLHTKGKIGE 474
Query: 450 AMRYFREMTSKGMVAEPRTEKLV 472
A+ YF+EM SKGMV EPRTE L+
Sbjct: 475 ALSYFQEMMSKGMVPEPRTEMLL 497
>ref|NP_909284.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|13872936|dbj|BAB44041.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
gi|9663979|dbj|BAB03620.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 534
Score = 463 bits (1192), Expect = e-129
Identities = 231/444 (52%), Positives = 312/444 (70%), Gaps = 15/444 (3%)
Query: 52 VQNLLKFRRDKPTD-QVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSY 110
V ++K + + D ++ L+ ++++D+VL VL++ RS+WK AL FF WA+ Y
Sbjct: 83 VSEIVKILKSRDGDSELAEVLNQFADEMDEDVVLKVLQKQRSNWKVALSFFKWAAGLPQY 142
Query: 111 APTSRVCNEILDILGKMSRFEELHQVFDEM--SHREGLVNEDTFSTLLRRFAAAHKVEEA 168
SR E+LDILG+M + + Q+FDE+ R+ +V F+ LL R+A AHKV+EA
Sbjct: 143 NHGSRAYTEMLDILGRMKKVRLMRQLFDEIPVESRQSVVTNRMFAVLLNRYAGAHKVQEA 202
Query: 169 ISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILN 228
I MFY R+ +G +LDL F+ LLM LCRYKHVE+AE LF K EF IK+WN+ILN
Sbjct: 203 IDMFYKRKDYGFELDLVGFQILLMSLCRYKHVEEAEALFLQKKDEFP--PVIKSWNIILN 260
Query: 229 GWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNC 288
GWCV G+ +AKRVW +I+ASK +PDLFTY TFI +LTK GKL TA+KLF MW +G N
Sbjct: 261 GWCVKGSLADAKRVWNEIIASKLKPDLFTYGTFINSLTKSGKLSTAVKLFTSMWEKGIN- 319
Query: 289 KPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYE 348
PDV ICNCIID LCFKKR+PEALE+F +M +RGC+ +VATYN+LIKH CKI RMEKVYE
Sbjct: 320 -PDVAICNCIIDQLCFKKRIPEALEIFGEMNDRGCQADVATYNTLIKHFCKINRMEKVYE 378
Query: 349 LVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMK 408
L++DME K S PN +TYS +L + + P++V +++RME++GC L D YNL+L LY+
Sbjct: 379 LLDDMEVKGVS--PNNMTYSYILKTTEKPKDVISLMQRMEKSGCRLDSDTYNLILNLYVS 436
Query: 409 WDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRT 468
WD + G++ WDEMERNG GPD+RS+TIM+HG + + K+ +A+ Y+R M S+GM EPRT
Sbjct: 437 WDYEKGVQLVWDEMERNGSGPDQRSFTIMVHGLHSHDKLDEALHYYRTMESRGMTPEPRT 496
Query: 469 EKLVISMNSPLKERTEKQEGKEEE 492
+ LV ++ R +K E EE
Sbjct: 497 KLLVKAI------RMKKDEPATEE 514
>emb|CAC01820.1| putative protein [Arabidopsis thaliana]
gi|15242190|ref|NP_197005.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
gi|11357721|pir||T51446 hypothetical protein F2G14_130 -
Arabidopsis thaliana
Length = 532
Score = 271 bits (693), Expect = 3e-71
Identities = 157/448 (35%), Positives = 250/448 (55%), Gaps = 11/448 (2%)
Query: 52 VQNLLKFRRDKPTDQVE--RALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADS 109
V + K +D +D+ E L+ C +++LV+++L R R+DW+ A FF WA K
Sbjct: 57 VGKISKLVKDCGSDRKELRNKLEECDVKPSNELVVEILSRVRNDWETAFTFFVWAGKQQG 116
Query: 110 YAPTSRVCNEILDILGKMSRFEELHQVFDEM-SHREGLVNEDTFSTLLRRFAAAHKVEEA 168
Y + R + ++ ILGKM +F+ + DEM LVN T ++R++ A H V +A
Sbjct: 117 YVRSVREYHSMISILGKMRKFDTAWTLIDEMRKFSPSLVNSQTLLIMIRKYCAVHDVGKA 176
Query: 169 ISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILN 228
I+ F+ ++F L++ +D F++LL LCRYK+V DA L ++ D K++N++LN
Sbjct: 177 INTFHAYKRFKLEMGIDDFQSLLSALCRYKNVSDAGHLIFCNKDKYPF--DAKSFNIVLN 234
Query: 229 GWC-VLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCN 287
GWC V+G+ EA+RVW ++ + D+ +Y++ I +K G L LKLF M E C
Sbjct: 235 GWCNVIGSPREAERVWMEMGNVGVKHDVVSYSSMISCYSKGGSLNKVLKLFDRMKKE-C- 292
Query: 288 CKPDVVICNCIIDALCFKKRVPEALEVFQDMKE-RGCEPNVATYNSLIKHLCKIRRMEKV 346
+PD + N ++ AL V EA + + M+E +G EPNV TYNSLIK LCK R+ E+
Sbjct: 293 IEPDRKVYNAVVHALAKASFVSEARNLMKTMEEEKGIEPNVVTYNSLIKPLCKARKTEEA 352
Query: 347 YELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLY 406
++ ++M K P TY + L+ EEV +L +M + GC + + Y +++R
Sbjct: 353 KQVFDEMLEK--GLFPTIRTYHAFMRILRTGEEVFELLAKMRKMGCEPTVETYIMLIRKL 410
Query: 407 MKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEP 466
+W + D + WDEM+ GPD SY +MIHG + NGK+++A Y++EM KGM
Sbjct: 411 CRWRDFDNVLLLWDEMKEKTVGPDLSSYIVMIHGLFLNGKIEEAYGYYKEMKDKGMRPNE 470
Query: 467 RTEKLVISMNSPLKERTEKQEGKEEETN 494
E ++ S S + ++ + E N
Sbjct: 471 NVEDMIQSWFSGKQYAEQRITDSKGEVN 498
>dbj|BAD82704.1| pentatricopeptide (PPR) repeat-containing protein-like [Oryza
sativa (japonica cultivar-group)]
Length = 530
Score = 250 bits (639), Expect = 6e-65
Identities = 139/403 (34%), Positives = 224/403 (55%), Gaps = 8/403 (1%)
Query: 71 LDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGKMSRF 130
L+ CG ++ +V+ VL R R+ A F WAS YAP C+ +L IL K RF
Sbjct: 104 LERCGATASEPVVVAVLDRLRNSCAAAHAAFRWASAQPGYAPGRHACHSMLAILAKHRRF 163
Query: 131 EELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTL 190
++ + D+M R L + L+RR+ AA V A++ F G + F L
Sbjct: 164 DDARALLDQM-RRSSLASPAAVMLLIRRYCAARDVAGAVAAFRALPNLGFRPGVAEFHGL 222
Query: 191 LMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWC-VLGNAHEAKRVWKDIMAS 249
L LCRYK+V+DAE L S +EF + K++NV+LNGWC ++ + EAKR W +
Sbjct: 223 LTALCRYKNVQDAEHLLLSSEKEFPF--ETKSFNVVLNGWCNMVRSVREAKRFWNAMEIK 280
Query: 250 KCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVP 309
+ D+ +Y + I +K G L T +KLF M G PD I N ++ AL + V
Sbjct: 281 GIKRDVVSYGSMISCFSKAGSLDTVMKLFNRMKEAGVI--PDRKIYNAVVYALAKGQCVN 338
Query: 310 EALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSC 369
EA + + M+E+G P+ AT+NSLI+ LCK R++++ ++++DM + S P+ T+
Sbjct: 339 EAKALVRSMEEKGVAPDTATFNSLIRPLCKARQVQEARKMLDDMLGRGLS--PSVRTFHA 396
Query: 370 LLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGP 429
LL+ + P EV +L++M+ C D + +++R + +W D + K W M NG P
Sbjct: 397 LLDVARSPIEVFDLLDKMKELQCDPEMDTFIMLIRKFCRWRQHDSVEKLWSAMPANGLSP 456
Query: 430 DRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLV 472
DR +Y ++IHG + NG+++++ +Y+ EM +KG E +TE+++
Sbjct: 457 DRSAYIVLIHGLFLNGRLEESAKYYEEMKAKGFPPEKKTEEMI 499
>ref|NP_178170.1| pentatricopeptide (PPR) repeat-containing protein [Arabidopsis
thaliana] gi|25406672|pir||E96837 unknown protein
T21F11.12 [imported] - Arabidopsis thaliana
gi|6730729|gb|AAF27119.1| unknown protein; 31926-33272
[Arabidopsis thaliana]
Length = 448
Score = 225 bits (573), Expect = 3e-57
Identities = 137/427 (32%), Positives = 218/427 (50%), Gaps = 19/427 (4%)
Query: 57 KFRRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRV 116
KFR + DQ + V + L + +DW+ AL FFNW + + T+
Sbjct: 33 KFRSQEEEDQSS---------YDQKTVCEALTCYSNDWQKALEFFNWVERESGFRHTTET 83
Query: 117 CNEILDILGKMSRFEELHQVFDEM-SHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTR 175
N ++DILGK FE + + M + E + N TF + +R+ AH V+EAI +
Sbjct: 84 FNRVIDILGKYFEFEISWALINRMIGNTESVPNHVTFRIVFKRYVTAHLVQEAIDAYDKL 143
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSK---AREFQLHRDIKTWNVILNGWCV 232
+ F L D +F L+ LC +KHV +AE L K F + + K N+IL GW
Sbjct: 144 DDFNLR-DETSFYNLVDALCEHKHVVEAEELCFGKNVIGNGFSVS-NTKIHNLILRGWSK 201
Query: 233 LGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDV 292
LG + K WK + DLF+Y+ ++ + K GK A+KL++ M + K DV
Sbjct: 202 LGWWGKCKEYWKKMDTEGVTKDLFSYSIYMDIMCKSGKPWKAVKLYKEMKSR--RMKLDV 259
Query: 293 VICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVED 352
V N +I A+ + V + VF++M+ERGCEPNVAT+N++IK LC+ RM Y ++++
Sbjct: 260 VAYNTVIRAIGASQGVEFGIRVFREMRERGCEPNVATHNTIIKLLCEDGRMRDAYRMLDE 319
Query: 353 MERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQ 412
M ++ C P+++TY CL + L+ P E+ + RM R+G D Y +++R + +W
Sbjct: 320 MPKR--GCQPDSITYMCLFSRLEKPSEILSLFGRMIRSGVRPKMDTYVMLMRKFERWGFL 377
Query: 413 DGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLV 472
+ W M+ +G PD +Y +I + G + A Y EM +G+ R E +
Sbjct: 378 QPVLYVWKTMKESGDTPDSAAYNAVIDALIQKGMLDMAREYEEEMIERGLSPRRRPELVE 437
Query: 473 ISMNSPL 479
S++ L
Sbjct: 438 KSLDETL 444
>ref|NP_191813.1| pentatricopeptide (PPR) repeat-containing protein [Arabidopsis
thaliana]
Length = 599
Score = 218 bits (556), Expect = 3e-55
Identities = 130/399 (32%), Positives = 214/399 (53%), Gaps = 10/399 (2%)
Query: 67 VERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGK 126
+E LD DL+ DL+++VL R R KPA FF WA++ +A SR N ++ IL K
Sbjct: 148 MEAVLDEMKLDLSHDLIVEVLERFRHARKPAFRFFCWAAERQGFAHASRTYNSMMSILAK 207
Query: 127 MSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDA 186
+FE + V +EM + GL+ +TF+ ++ FAAA + ++A+ +F +++ + ++
Sbjct: 208 TRQFETMVSVLEEMGTK-GLLTMETFTIAMKAFAAAKERKKAVGIFELMKKYKFKIGVET 266
Query: 187 FRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDI 246
LL L R K ++A+ LF F ++ T+ V+LNGWC + N EA R+W D+
Sbjct: 267 INCLLDSLGRAKLGKEAQVLFDKLKERFT--PNMMTYTVLLNGWCRVRNLIEAARIWNDM 324
Query: 247 MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKK 306
+ +PD+ + ++ L + K A+KLF M ++G C P+V +I C +
Sbjct: 325 IDHGLKPDIVAHNVMLEGLLRSMKKSDAIKLFHVMKSKG-PC-PNVRSYTIMIRDFCKQS 382
Query: 307 RVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVT 366
+ A+E F DM + G +P+ A Y LI ++++ VYEL+++M+ K P+ T
Sbjct: 383 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHP--PDGKT 440
Query: 367 YSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEME 423
Y+ L+ + K PE + +M +N S +N++++ Y N + R WDEM
Sbjct: 441 YNALIKLMANQKMPEHGTRIYNKMIQNEIEPSIHTFNMIMKSYFVARNYEMGRAVWDEMI 500
Query: 424 RNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
+ G PD SYT++I G GK ++A RY EM KGM
Sbjct: 501 KKGICPDDNSYTVLIRGLISEGKSREACRYLEEMLDKGM 539
Score = 65.9 bits (159), Expect = 3e-09
Identities = 71/305 (23%), Positives = 131/305 (42%), Gaps = 15/305 (4%)
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQ-LHRDIKTWNVILNGWCVLG 234
++ LDL D +L R++H F A E Q +T+N +++ +L
Sbjct: 153 DEMKLDLSHDLIVEVLE---RFRHARKPAFRFFCWAAERQGFAHASRTYNSMMS---ILA 206
Query: 235 NAHEAKRVWKDI--MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDV 292
+ + + + M +K + T+ +KA + A+ +F M + K V
Sbjct: 207 KTRQFETMVSVLEEMGTKGLLTMETFTIAMKAFAAAKERKKAVGIFELM--KKYKFKIGV 264
Query: 293 VICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVED 352
NC++D+L K EA +F +KER PN+ TY L+ C++R + + + D
Sbjct: 265 ETINCLLDSLGRAKLGKEAQVLFDKLKER-FTPNMMTYTVLLNGWCRVRNLIEAARIWND 323
Query: 353 M--ERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWD 410
M K + + V LL S+K + + + M+ G + Y +++R + K
Sbjct: 324 MIDHGLKPDIVAHNVMLEGLLRSMKKSDAIK-LFHVMKSKGPCPNVRSYTIMIRDFCKQS 382
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+ + + +D+M +G PD YT +I G K+ +EM KG + +T
Sbjct: 383 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYN 442
Query: 471 LVISM 475
+I +
Sbjct: 443 ALIKL 447
>gb|AAQ65101.1| At3g62470 [Arabidopsis thaliana] gi|7340718|emb|CAB82961.1|
putative protein [Arabidopsis thaliana]
gi|15228767|ref|NP_191806.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
gi|11357975|pir||T48039 hypothetical protein T12C14.170
- Arabidopsis thaliana
Length = 599
Score = 218 bits (555), Expect = 3e-55
Identities = 129/399 (32%), Positives = 214/399 (53%), Gaps = 10/399 (2%)
Query: 67 VERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGK 126
+E LD DL+ DL+++VL R R KPA FF WA++ +A SR N ++ IL K
Sbjct: 148 MEAVLDEMKLDLSHDLIVEVLERFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMSILAK 207
Query: 127 MSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDA 186
+FE + V +EM + GL+ +TF+ ++ FAAA + ++A+ +F +++ + ++
Sbjct: 208 TRQFETMVSVLEEMGTK-GLLTMETFTIAMKAFAAAKERKKAVGIFELMKKYKFKIGVET 266
Query: 187 FRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDI 246
LL L R K ++A+ LF F ++ T+ V+LNGWC + N EA R+W D+
Sbjct: 267 INCLLDSLGRAKLGKEAQVLFDKLKERFT--PNMMTYTVLLNGWCRVRNLIEAARIWNDM 324
Query: 247 MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKK 306
+ +PD+ + ++ L + K A+KLF M ++G C P+V +I C +
Sbjct: 325 IDQGLKPDIVAHNVMLEGLLRSRKKSDAIKLFHVMKSKG-PC-PNVRSYTIMIRDFCKQS 382
Query: 307 RVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVT 366
+ A+E F DM + G +P+ A Y LI ++++ VYEL+++M+ K P+ T
Sbjct: 383 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHP--PDGKT 440
Query: 367 YSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEME 423
Y+ L+ + K PE + +M +N S +N++++ Y N + R W+EM
Sbjct: 441 YNALIKLMANQKMPEHATRIYNKMIQNEIEPSIHTFNMIMKSYFMARNYEMGRAVWEEMI 500
Query: 424 RNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
+ G PD SYT++I G GK ++A RY EM KGM
Sbjct: 501 KKGICPDDNSYTVLIRGLIGEGKSREACRYLEEMLDKGM 539
Score = 68.2 bits (165), Expect = 6e-10
Identities = 72/305 (23%), Positives = 132/305 (42%), Gaps = 15/305 (4%)
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQ-LHRDIKTWNVILNGWCVLG 234
++ LDL D +L R++H F A E Q D +T+N +++ +L
Sbjct: 153 DEMKLDLSHDLIVEVLE---RFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMS---ILA 206
Query: 235 NAHEAKRVWKDI--MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDV 292
+ + + + M +K + T+ +KA + A+ +F M + K V
Sbjct: 207 KTRQFETMVSVLEEMGTKGLLTMETFTIAMKAFAAAKERKKAVGIFELM--KKYKFKIGV 264
Query: 293 VICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVED 352
NC++D+L K EA +F +KER PN+ TY L+ C++R + + + D
Sbjct: 265 ETINCLLDSLGRAKLGKEAQVLFDKLKER-FTPNMMTYTVLLNGWCRVRNLIEAARIWND 323
Query: 353 M--ERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWD 410
M + K + + V LL S K + + + M+ G + Y +++R + K
Sbjct: 324 MIDQGLKPDIVAHNVMLEGLLRSRKKSDAIK-LFHVMKSKGPCPNVRSYTIMIRDFCKQS 382
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+ + + +D+M +G PD YT +I G K+ +EM KG + +T
Sbjct: 383 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYN 442
Query: 471 LVISM 475
+I +
Sbjct: 443 ALIKL 447
>emb|CAC01881.1| putative protein [Arabidopsis thaliana]
gi|15241508|ref|NP_196986.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
gi|11358457|pir||T51427 hypothetical protein T9L3_120 -
Arabidopsis thaliana
Length = 598
Score = 218 bits (555), Expect = 3e-55
Identities = 130/399 (32%), Positives = 214/399 (53%), Gaps = 10/399 (2%)
Query: 67 VERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGK 126
+E LD DL+ DL+++VL R R KPA FF WA++ +A SR N ++ IL K
Sbjct: 147 MEAVLDEMKLDLSHDLIVEVLERFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMSILAK 206
Query: 127 MSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDA 186
+FE + V +EM + GL+ +TF+ ++ FAAA + ++A+ +F +++ + ++
Sbjct: 207 TRQFETMVSVLEEMGTK-GLLTMETFTIAMKAFAAAKERKKAVGIFELMKKYKFKIGVET 265
Query: 187 FRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDI 246
LL L R K ++A+ LF F ++ T+ V+LNGWC + N EA R+W D+
Sbjct: 266 INCLLDSLGRAKLGKEAQVLFDKLKERFT--PNMMTYTVLLNGWCRVRNLIEAARIWNDM 323
Query: 247 MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKK 306
+ +PD+ + ++ L + K A+KLF M ++G C P+V +I C +
Sbjct: 324 IDHGLKPDIVAHNVMLEGLLRSMKKSDAIKLFHVMKSKG-PC-PNVRSYTIMIRDFCKQS 381
Query: 307 RVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVT 366
+ A+E F DM + G +P+ A Y LI ++++ VYEL+++M+ K P+ T
Sbjct: 382 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHP--PDGKT 439
Query: 367 YSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEME 423
Y+ L+ + K PE + +M +N S +N++++ Y N + R WDEM
Sbjct: 440 YNALIKLMANQKMPEHGTRIYNKMIQNEIEPSIHTFNMIMKSYFVARNYEMGRAVWDEMI 499
Query: 424 RNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
+ G PD SYT++I G GK ++A RY EM KGM
Sbjct: 500 KKGICPDDNSYTVLIRGLISEGKSREACRYLEEMLDKGM 538
Score = 68.9 bits (167), Expect = 3e-10
Identities = 72/305 (23%), Positives = 132/305 (42%), Gaps = 15/305 (4%)
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQ-LHRDIKTWNVILNGWCVLG 234
++ LDL D +L R++H F A E Q D +T+N +++ +L
Sbjct: 152 DEMKLDLSHDLIVEVLE---RFRHARKPAFRFFCWAAERQGFAHDSRTYNSMMS---ILA 205
Query: 235 NAHEAKRVWKDI--MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDV 292
+ + + + M +K + T+ +KA + A+ +F M + K V
Sbjct: 206 KTRQFETMVSVLEEMGTKGLLTMETFTIAMKAFAAAKERKKAVGIFELM--KKYKFKIGV 263
Query: 293 VICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVED 352
NC++D+L K EA +F +KER PN+ TY L+ C++R + + + D
Sbjct: 264 ETINCLLDSLGRAKLGKEAQVLFDKLKER-FTPNMMTYTVLLNGWCRVRNLIEAARIWND 322
Query: 353 M--ERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWD 410
M K + + V LL S+K + + + M+ G + Y +++R + K
Sbjct: 323 MIDHGLKPDIVAHNVMLEGLLRSMKKSDAIK-LFHVMKSKGPCPNVRSYTIMIRDFCKQS 381
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+ + + +D+M +G PD YT +I G K+ +EM KG + +T
Sbjct: 382 SMETAIEYFDDMVDSGLQPDAAVYTCLITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYN 441
Query: 471 LVISM 475
+I +
Sbjct: 442 ALIKL 446
>emb|CAB66911.1| putative protein [Arabidopsis thaliana]
gi|15229604|ref|NP_190542.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
gi|11358047|pir||T46039 hypothetical protein T16K5.80 -
Arabidopsis thaliana
Length = 1184
Score = 207 bits (526), Expect = 8e-52
Identities = 134/421 (31%), Positives = 220/421 (51%), Gaps = 11/421 (2%)
Query: 66 QVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILG 125
++E AL+ G DL L++ VL R FF WA+K Y + VC ++ IL
Sbjct: 83 KLELALNESGIDLRPGLIIRVLSRCGDAGNLGYRFFLWATKQPGYFHSYEVCKSMVMILS 142
Query: 126 KMSRFEELHQVFDEMSHREG-LVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDL 184
KM +F + + +EM L+ + F L+RRFA+A+ V++A+ + ++GL+ D
Sbjct: 143 KMRQFGAVWGLIEEMRKTNPELIEPELFVVLMRRFASANMVKKAVEVLDEMPKYGLEPDE 202
Query: 185 DAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWK 244
F LL LC+ V++A +F +F +++ + +L GWC G EAK V
Sbjct: 203 YVFGCLLDALCKNGSVKEASKVFEDMREKFP--PNLRYFTSLLYGWCREGKLMEAKEVLV 260
Query: 245 DIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALC- 303
+ + PD+ + + GK+ A L M G +P+V +I ALC
Sbjct: 261 QMKEAGLEPDIVVFTNLLSGYAHAGKMADAYDLMNDMRKRGF--EPNVNCYTVLIQALCR 318
Query: 304 FKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPN 363
+KR+ EA+ VF +M+ GCE ++ TY +LI CK ++K Y +++DM RKKG MP+
Sbjct: 319 TEKRMDEAMRVFVEMERYGCEADIVTYTALISGFCKWGMIDKGYSVLDDM-RKKG-VMPS 376
Query: 364 AVTYSCLLNSLKGPE---EVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWD 420
VTY ++ + + E E ++E+M+R GC IYN+V+RL K + W+
Sbjct: 377 QVTYMQIMVAHEKKEQFEECLELIEKMKRRGCHPDLLIYNVVIRLACKLGEVKEAVRLWN 436
Query: 421 EMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVISMNSPLK 480
EME NG P ++ IMI+G G + +A +F+EM S+G+ + P+ L +N+ ++
Sbjct: 437 EMEANGLSPGVDTFVIMINGFTSQGFLIEACNHFKEMVSRGIFSAPQYGTLKSLLNNLVR 496
Query: 481 E 481
+
Sbjct: 497 D 497
Score = 70.1 bits (170), Expect = 2e-10
Identities = 86/413 (20%), Positives = 159/413 (37%), Gaps = 64/413 (15%)
Query: 125 GKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHK-VEEAISMFYTREQFGLDLD 183
GKM+ + + + ++M R N + ++ L++ K ++EA+ +F E++G + D
Sbjct: 285 GKMA---DAYDLMNDMRKRGFEPNVNCYTVLIQALCRTEKRMDEAMRVFVEMERYGCEAD 341
Query: 184 LDAFRTLLMWLCRYKHVEDAETLF---------------------HSKAREFQ------- 215
+ + L+ C++ ++ ++ H K +F+
Sbjct: 342 IVTYTALISGFCKWGMIDKGYSVLDDMRKKGVMPSQVTYMQIMVAHEKKEQFEECLELIE 401
Query: 216 ------LHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKG 269
H D+ +NV++ C LG EA R+W ++ A+ P + T+ I T +G
Sbjct: 402 KMKRRGCHPDLLIYNVVIRLACKLGEVKEAVRLWNEMEANGLSPGVDTFVIMINGFTSQG 461
Query: 270 KLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQ--DMKERGCEPNV 327
L A F+ M + G P +++ L ++ A +V+ K CE NV
Sbjct: 462 FLIEACNHFKEMVSRGIFSAPQYGTLKSLLNNLVRDDKLEMAKDVWSCISNKTSSCELNV 521
Query: 328 ATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCL---LNSLKGPEEVPGVL 384
+ + I L +++ DM + MP TY+ L LN L +
Sbjct: 522 SAWTIWIHALYAKGHVKEACSYCLDM--MEMDLMPQPNTYAKLMKGLNKLYNRTIAAEIT 579
Query: 385 ERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNG--CGPDRRSYTIMIHGHY 442
E++ + + ++Y K +D + K + + G G D + Y
Sbjct: 580 EKVVKMASE-----REMSFKMYKKKGEEDLIEKAKPKGNKEGKKKGTDHQRY-------- 626
Query: 443 ENGKMKDAMRYFREMTSKGMVAEPRTEKLVISMNSPL--KERTEKQEGKEEET 493
G++ A R T +A R S +SP K T K EEE+
Sbjct: 627 -KGRVSLARNRLRSETPSSFLARDRLRSKTPS-SSPFSSKRHTPKTSEIEEES 677
>dbj|BAD73118.1| pentatricopeptide (PPR) repeat-containing protein-like [Oryza
sativa (japonica cultivar-group)]
Length = 540
Score = 198 bits (503), Expect = 4e-49
Identities = 121/421 (28%), Positives = 212/421 (49%), Gaps = 8/421 (1%)
Query: 45 AAGAGAFVQNLLKFRRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWA 104
A G + + + +P ++ LD G ++ LV +VL+ + AL FF WA
Sbjct: 85 APGVSEAAERVCRVVSAQPEHRIAPVLDALGVTVSPQLVAEVLKNLSNAGILALAFFRWA 144
Query: 105 SKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHK 164
+ + ++ + +++ LGK+ +F + + + M R L ++DTF ++RR+A A K
Sbjct: 145 ERQQGFRYSAEGFHNLIEALGKIKQFRLVWSLVEAMRCRSCL-SKDTFKIIVRRYARARK 203
Query: 165 VEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWN 224
V+EA+ F FGL DL + L+ L + K V+ A +F R+ + D+KT+
Sbjct: 204 VKEAVETFEKMSSFGLKTDLSDYNWLIDILSKSKQVKKAHAIFKEMKRKGRFIPDLKTYT 263
Query: 225 VILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNE 284
V++ GW + K V+++++ + +PD+ Y I A K GK A+K+F M
Sbjct: 264 VLMEGWGHEKDLLMLKAVYQEMLDAGIKPDVVAYGMLISAFCKSGKCDEAIKVFHEMEES 323
Query: 285 GCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRME 344
GC P V +I+ L +R+ EAL+ FQ KE G V T N++I C+
Sbjct: 324 GCMPSPHVYC--MLINGLGSMERLDEALKYFQLSKESGFPMEVPTCNAVIGAYCRALEFH 381
Query: 345 KVYELVEDMERKKGSCMPNAVTYSCLLNSL---KGPEEVPGVLERMERNGCSLSDDIYNL 401
+++V++M +K PN TY +LN L + EE + +RMER+GC + Y +
Sbjct: 382 HAFKMVDEM--RKSGIGPNTRTYDIILNHLIKSEKIEEAYNLFQRMERDGCEPELNTYTM 439
Query: 402 VLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKG 461
++ ++ + D K W +M+ G P ++ +I+G ++++A YF+EM KG
Sbjct: 440 MVGMFCSNERVDMALKVWKQMKEKGVLPCMHMFSALINGLCFENRLEEACVYFQEMLDKG 499
Query: 462 M 462
+
Sbjct: 500 I 500
Score = 89.4 bits (220), Expect = 2e-16
Identities = 72/312 (23%), Positives = 139/312 (44%), Gaps = 9/312 (2%)
Query: 168 AISMF-YTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVI 226
A++ F + Q G + F L+ L + K +L + L +D T+ +I
Sbjct: 137 ALAFFRWAERQQGFRYSAEGFHNLIEALGKIKQFRLVWSLVEAMRCRSCLSKD--TFKII 194
Query: 227 LNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGC 286
+ + EA ++ + + + DL Y I L+K ++ A +F+ M +G
Sbjct: 195 VRRYARARKVKEAVETFEKMSSFGLKTDLSDYNWLIDILSKSKQVKKAHAIFKEMKRKG- 253
Query: 287 NCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKV 346
PD+ +++ +K + V+Q+M + G +P+V Y LI CK + ++
Sbjct: 254 RFIPDLKTYTVLMEGWGHEKDLLMLKAVYQEMLDAGIKPDVVAYGMLISAFCKSGKCDEA 313
Query: 347 YELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERME---RNGCSLSDDIYNLVL 403
++ +ME + CMP+ Y L+N L E + L+ + +G + N V+
Sbjct: 314 IKVFHEME--ESGCMPSPHVYCMLINGLGSMERLDEALKYFQLSKESGFPMEVPTCNAVI 371
Query: 404 RLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMV 463
Y + K DEM ++G GP+ R+Y I+++ ++ K+++A F+ M G
Sbjct: 372 GAYCRALEFHHAFKMVDEMRKSGIGPNTRTYDIILNHLIKSEKIEEAYNLFQRMERDGCE 431
Query: 464 AEPRTEKLVISM 475
E T +++ M
Sbjct: 432 PELNTYTMMVGM 443
Score = 74.3 bits (181), Expect = 8e-12
Identities = 49/229 (21%), Positives = 108/229 (46%), Gaps = 3/229 (1%)
Query: 126 KMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLD 185
K + +E +VF EM + + + L+ + +++EA+ F ++ G +++
Sbjct: 306 KSGKCDEAIKVFHEMEESGCMPSPHVYCMLINGLGSMERLDEALKYFQLSKESGFPMEVP 365
Query: 186 AFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKD 245
++ CR A + + R+ + + +T+++ILN EA +++
Sbjct: 366 TCNAVIGAYCRALEFHHAFKMV-DEMRKSGIGPNTRTYDIILNHLIKSEKIEEAYNLFQR 424
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFK 305
+ C P+L TY + ++ ALK+++ M +G P + + + +I+ LCF+
Sbjct: 425 MERDGCEPELNTYTMMVGMFCSNERVDMALKVWKQMKEKGV--LPCMHMFSALINGLCFE 482
Query: 306 KRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDME 354
R+ EA FQ+M ++G P +++L + L + R+ E+ + +E
Sbjct: 483 NRLEEACVYFQEMLDKGIRPPGQLFSNLKEALVEGGRITLAQEVTQRLE 531
>ref|NP_177860.1| pentatricopeptide (PPR) repeat-containing protein [Arabidopsis
thaliana] gi|25406495|pir||F96802 hypothetical protein
F2P24.7 [imported] - Arabidopsis thaliana
gi|11079489|gb|AAG29201.1| hypothetical protein
[Arabidopsis thaliana]
Length = 481
Score = 193 bits (490), Expect = 1e-47
Identities = 117/427 (27%), Positives = 219/427 (50%), Gaps = 9/427 (2%)
Query: 38 LHSQPEHAAGAGAFVQNLLKFRRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPA 97
L+S E +N+ K P ++ ALD G ++ ++V DVL R R+
Sbjct: 22 LYSSSEQVRDVADVAKNISKVLMSSPQLVLDSALDQSGLRVSQEVVEDVLNRFRNAGLLT 81
Query: 98 LVFFNWASKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLR 157
FF W+ K Y + R + +++ K+ +++ + + + M ++ ++N +TF ++R
Sbjct: 82 YRFFQWSEKQRHYEHSVRAYHMMIESTAKIRQYKLMWDLINAMRKKK-MLNVETFCIVMR 140
Query: 158 RFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLH 217
++A A KV+EAI F E++ L +L AF LL LC+ K+V A+ +F + F
Sbjct: 141 KYARAQKVDEAIYAFNVMEKYDLPPNLVAFNGLLSALCKSKNVRKAQEVFENMRDRFT-- 198
Query: 218 RDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKL 277
D KT++++L GW N +A+ V+++++ + C PD+ TY+ + L K G++ AL +
Sbjct: 199 PDSKTYSILLEGWGKEPNLPKAREVFREMIDAGCHPDIVTYSIMVDILCKAGRVDEALGI 258
Query: 278 FRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHL 337
R M + CKP I + ++ + R+ EA++ F +M+ G + +VA +NSLI
Sbjct: 259 VRSM--DPSICKPTTFIYSVLVHTYGTENRLEEAVDTFLEMERSGMKADVAVFNSLIGAF 316
Query: 338 CKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL--KGPEEVPGVLERMERNGCSLS 395
CK RM+ VY ++++M+ K PN+ + + +L L +G ++ + R C
Sbjct: 317 CKANRMKNVYRVLKEMKSK--GVTPNSKSCNIILRHLIERGEKDEAFDVFRKMIKVCEPD 374
Query: 396 DDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFR 455
D Y +V++++ + + K W M + G P +++++I+G E + A
Sbjct: 375 ADTYTMVIKMFCEKKEMETADKVWKYMRKKGVFPSMHTFSVLINGLCEERTTQKACVLLE 434
Query: 456 EMTSKGM 462
EM G+
Sbjct: 435 EMIEMGI 441
Score = 84.7 bits (208), Expect = 6e-15
Identities = 69/253 (27%), Positives = 125/253 (49%), Gaps = 13/253 (5%)
Query: 240 KRVWKDI--MASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNC 297
K +W I M K ++ T+ ++ + K+ A+ F M E + P++V N
Sbjct: 115 KLMWDLINAMRKKKMLNVETFCIVMRKYARAQKVDEAIYAFNVM--EKYDLPPNLVAFNG 172
Query: 298 IIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKK 357
++ ALC K V +A EVF++M++R P+ TY+ L++ K + K E+ +M
Sbjct: 173 LLSALCKSKNVRKAQEVFENMRDR-FTPDSKTYSILLEGWGKEPNLPKAREVFREMI--D 229
Query: 358 GSCMPNAVTYSCLLNSLKGP---EEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDG 414
C P+ VTYS +++ L +E G++ M+ + C + IY++++ Y + +
Sbjct: 230 AGCHPDIVTYSIMVDILCKAGRVDEALGIVRSMDPSICKPTTFIYSVLVHTYGTENRLEE 289
Query: 415 LRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVIS 474
T+ EMER+G D + +I + +MK+ R +EM SKG+ ++ +++
Sbjct: 290 AVDTFLEMERSGMKADVAVFNSLIGAFCKANRMKNVYRVLKEMKSKGVTPNSKSCNIIL- 348
Query: 475 MNSPLKERTEKQE 487
L ER EK E
Sbjct: 349 --RHLIERGEKDE 359
Score = 43.1 bits (100), Expect = 0.020
Identities = 31/153 (20%), Positives = 70/153 (45%), Gaps = 2/153 (1%)
Query: 116 VCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTR 175
V N ++ K +R + +++V EM + N + + +LR + +EA +F
Sbjct: 308 VFNSLIGAFCKANRMKNVYRVLKEMKSKGVTPNSKSCNIILRHLIERGEKDEAFDVFRKM 367
Query: 176 EQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGN 235
+ + D D + ++ C K +E A+ ++ R+ + + T++V++NG C
Sbjct: 368 IKV-CEPDADTYTMVIKMFCEKKEMETADKVW-KYMRKKGVFPSMHTFSVLINGLCEERT 425
Query: 236 AHEAKRVWKDIMASKCRPDLFTYATFIKALTKK 268
+A + ++++ RP T+ + L K+
Sbjct: 426 TQKACVLLEEMIEMGIRPSGVTFGRLRQLLIKE 458
>dbj|BAB09050.1| unnamed protein product [Arabidopsis thaliana]
gi|15239161|ref|NP_201383.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
Length = 637
Score = 193 bits (490), Expect = 1e-47
Identities = 132/432 (30%), Positives = 216/432 (49%), Gaps = 12/432 (2%)
Query: 55 LLKFRRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTS 114
L KF P ++E AL+ G +L L+ VL R FF WA+K Y +
Sbjct: 90 LRKFHSRVP--KLELALNESGVELRPGLIERVLNRCGDAGNLGYRFFVWAAKQPRYCHSI 147
Query: 115 RVCNEILDILGKMSRFEELHQVFDEM-SHREGLVNEDTFSTLLRRFAAAHKVEEAISMFY 173
V ++ IL KM +F + + +EM L+ + F L++RFA+A V++AI +
Sbjct: 148 EVYKSMVKILSKMRQFGAVWGLIEEMRKENPQLIEPELFVVLVQRFASADMVKKAIEVLD 207
Query: 174 TREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVL 233
+FG + D F LL LC++ V+DA LF F ++ ++ + +L GWC +
Sbjct: 208 EMPKFGFEPDEYVFGCLLDALCKHGSVKDAAKLFEDMRMRFPVN--LRYFTSLLYGWCRV 265
Query: 234 GNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVV 293
G EAK V + + PD+ Y + GK+ A L R M G +P+
Sbjct: 266 GKMMEAKYVLVQMNEAGFEPDIVDYTNLLSGYANAGKMADAYDLLRDMRRRGF--EPNAN 323
Query: 294 ICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDM 353
+I ALC R+ EA++VF +M+ CE +V TY +L+ CK +++K Y +++DM
Sbjct: 324 CYTVLIQALCKVDRMEEAMKVFVEMERYECEADVVTYTALVSGFCKWGKIDKCYIVLDDM 383
Query: 354 ERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMER-NGCSLSDDI--YNLVLRLYMKWD 410
+K MP+ +TY ++ + + E LE ME+ DI YN+V+RL K
Sbjct: 384 IKK--GLMPSELTYMHIMVAHEKKESFEECLELMEKMRQIEYHPDIGIYNVVIRLACKLG 441
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+ W+EME NG P ++ IMI+G G + +A +F+EM ++G+ + +
Sbjct: 442 EVKEAVRLWNEMEENGLSPGVDTFVIMINGLASQGCLLEASDHFKEMVTRGLFSVSQYGT 501
Query: 471 LVISMNSPLKER 482
L + +N+ LK++
Sbjct: 502 LKLLLNTVLKDK 513
Score = 70.5 bits (171), Expect = 1e-10
Identities = 74/323 (22%), Positives = 134/323 (40%), Gaps = 16/323 (4%)
Query: 110 YAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAI 169
+ P + ++ L K+ R EE +VF EM E + T++ L+ F K+++
Sbjct: 318 FEPNANCYTVLIQALCKVDRMEEAMKVFVEMERYECEADVVTYTALVSGFCKWGKIDKCY 377
Query: 170 SMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFH--SKAREFQLHRDIKTWNVIL 227
+ + GL + + T + + ++ E E K R+ + H DI +NV++
Sbjct: 378 IVLDDMIKKGL---MPSELTYMHIMVAHEKKESFEECLELMEKMRQIEYHPDIGIYNVVI 434
Query: 228 NGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCN 287
C LG EA R+W ++ + P + T+ I L +G L A F+ M G
Sbjct: 435 RLACKLGEVKEAVRLWNEMEENGLSPGVDTFVIMINGLASQGCLLEASDHFKEMVTRGLF 494
Query: 288 CKPDVVICNCIIDALCFKKRVPEALEVFQDMKERG-CEPNVATYNSLIKHL-CKIRRMEK 345
+++ + K++ A +V+ + +G CE NV ++ I L K E
Sbjct: 495 SVSQYGTLKLLLNTVLKDKKLEMAKDVWSCITSKGACELNVLSWTIWIHALFSKGYEKEA 554
Query: 346 VYELVEDMERKKGSCMPNAVTYSCLLNSLKG--PEEVPGVLERMERNGCSLSDDIYNLVL 403
+E +E MP T++ L+ LK E G + RN + + +
Sbjct: 555 CSYCIEMIEM---DFMPQPDTFAKLMKGLKKLYNREFAGEITEKVRNMAAERE----MSF 607
Query: 404 RLYMKWDNQDGLRKTWDEMERNG 426
++Y + QD K + +R G
Sbjct: 608 KMYKRRGVQDLTEKAKSKQDREG 630
>gb|AAV43937.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|55168028|gb|AAV43896.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 632
Score = 191 bits (486), Expect = 3e-47
Identities = 123/432 (28%), Positives = 222/432 (50%), Gaps = 16/432 (3%)
Query: 40 SQPEHAAGAGAFVQNLLKFRRDKPTD-QVERALDLCGFDLNDDLVLDVLRRHRSDWKPAL 98
S PEH A + +++ D +E AL L++ LVL VL R + +P+
Sbjct: 142 SHPEHVGRVCAAIADVVAAGAGAGADASLEAALTALSPPLSETLVLAVLDRFKHAHRPSH 201
Query: 99 VFFNWASKADS----YAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFST 154
FF WA+ A + +A T+ +++ ILGK +F+ + + EM +EG + D F
Sbjct: 202 RFFRWAAAAAAASGGFAHTTITYCKMVHILGKARQFQSMVALIQEMG-KEGALCMDAFKI 260
Query: 155 LLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREF 214
++ FAAA +++ A+ +F G D +++F LL+ L + +A +F +
Sbjct: 261 AIKSFAAAGEIKNAVGVFEMMRTHGFDDGVESFNCLLVALAQEGLGREANQVFDRMRDRY 320
Query: 215 QLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTA 274
D++++ ++ WC N EA RVW +++ + +PD+ + T I+ L + + A
Sbjct: 321 A--PDLRSYTALMLAWCNARNLVEAGRVWNEMLENGLKPDVVVHNTMIEGLLRGQRRPEA 378
Query: 275 LKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLI 334
+K+F M +G P+V +I C + ++ A+ F++M++ GC+P+VATY L+
Sbjct: 379 VKMFELMKAKGP--APNVWTYTMLIRDHCKRGKMDMAMRCFEEMQDVGCQPDVATYTCLL 436
Query: 335 KHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKG---PEEVPGVLERMERNG 391
+RM++V L+E+M +K C P+ TY+ L+ L P++ + ++M + G
Sbjct: 437 VGYGNAKRMDRVTALLEEMTQK--GCPPDGRTYNALIKLLTNRNMPDDAARIYKKMIKKG 494
Query: 392 CSLSDDIYNLVLRLYMKWDNQDGLR-KTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDA 450
+ YN++++ Y + W+EM R G PD SYT+ I+GH +G+ ++A
Sbjct: 495 LEPTIHTYNMMMKSYFLGGRNYMMGCAVWEEMHRKGICPDVNSYTVFINGHIRHGRPEEA 554
Query: 451 MRYFREMTSKGM 462
+Y EM KGM
Sbjct: 555 CKYIEEMIQKGM 566
Score = 69.3 bits (168), Expect = 3e-10
Identities = 55/218 (25%), Positives = 103/218 (47%), Gaps = 10/218 (4%)
Query: 262 IKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKER 321
IK+ G++ A+ +F M G + V NC++ AL + EA +VF M++R
Sbjct: 262 IKSFAAAGEIKNAVGVFEMMRTHGFD--DGVESFNCLLVALAQEGLGREANQVFDRMRDR 319
Query: 322 GCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL----KGP 377
P++ +Y +L+ C R + + + +M + P+ V ++ ++ L + P
Sbjct: 320 YA-PDLRSYTALMLAWCNARNLVEAGRVWNEM--LENGLKPDVVVHNTMIEGLLRGQRRP 376
Query: 378 EEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIM 437
E V + E M+ G + + Y +++R + K D + ++EM+ GC PD +YT +
Sbjct: 377 EAVK-MFELMKAKGPAPNVWTYTMLIRDHCKRGKMDMAMRCFEEMQDVGCQPDVATYTCL 435
Query: 438 IHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVISM 475
+ G+ +M EMT KG + RT +I +
Sbjct: 436 LVGYGNAKRMDRVTALLEEMTQKGCPPDGRTYNALIKL 473
Score = 48.9 bits (115), Expect = 4e-04
Identities = 52/246 (21%), Positives = 104/246 (42%), Gaps = 8/246 (3%)
Query: 112 PTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISM 171
P V N +++ L + R E ++F+ M + N T++ L+R K++ A+
Sbjct: 357 PDVVVHNTMIEGLLRGQRRPEAVKMFELMKAKGPAPNVWTYTMLIRDHCKRGKMDMAMRC 416
Query: 172 FYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWC 231
F + G D+ + LL+ K ++ L ++ D +T+N ++
Sbjct: 417 FEEMQDVGCQPDVATYTCLLVGYGNAKRMDRVTALLEEMTQK-GCPPDGRTYNALIKLLT 475
Query: 232 VLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGK---LGTALKLFRGMWNEGCNC 288
+A R++K ++ P + TY +K+ G+ +G A+ + M +G C
Sbjct: 476 NRNMPDDAARIYKKMIKKGLEPTIHTYNMMMKSYFLGGRNYMMGCAV--WEEMHRKGI-C 532
Query: 289 KPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYE 348
PDV I+ R EA + ++M ++G + YN K + + +YE
Sbjct: 533 -PDVNSYTVFINGHIRHGRPEEACKYIEEMIQKGMKTPQIDYNKFAADFSKAGKPDILYE 591
Query: 349 LVEDME 354
L + ++
Sbjct: 592 LAQKVK 597
>ref|XP_474085.1| OSJNBa0033G05.8 [Oryza sativa (japonica cultivar-group)]
gi|38344884|emb|CAD41907.2| OSJNBa0033G05.8 [Oryza
sativa (japonica cultivar-group)]
Length = 442
Score = 185 bits (469), Expect = 3e-45
Identities = 123/400 (30%), Positives = 201/400 (49%), Gaps = 16/400 (4%)
Query: 78 LNDDLVLDVLRRHRSDWKPALVFFNWASKAD--SYAPTSRVCNEILDILGKMSRFEELHQ 135
L+ VL+ L + +DW+ AL FF+W++ D + PT +D+LGK F
Sbjct: 31 LDATAVLETLSLYANDWRRALEFFHWSASPDGANVPPTPATVARAVDVLGKHFEFPLATS 90
Query: 136 VFDEMSHREGLVNEDTF-----STLLRRFAAAHKVEEAISMF-YTREQFGLDLDLDAFRT 189
+ +SH + + +F +LL R AAA+ +++AI F T GL D +F
Sbjct: 91 LL--VSHHDPGRADPSFLRPALRSLLNRLAAANLIDDAIRAFDSTAGSIGLR-DEASFHA 147
Query: 190 LLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMAS 249
L+ LC ++ V++A L K KT+N++L GW +++W D+ +
Sbjct: 148 LVDALCDHRRVDEAHHLCFGKDPP-PFPPVTKTYNLLLRGWAKTRAWARLRQLWFDMDSR 206
Query: 250 KCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVP 309
DL +Y+ ++ AL K GK A K+F+ M +G DVV N I ++ + V
Sbjct: 207 GVAKDLHSYSIYMDALAKSGKPWKAFKVFKEMKQKGMAI--DVVAYNTAIHSVGLAQGVD 264
Query: 310 EALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSC 369
A+ +++ M + GC+PN +T+N+++K LCK R ++ Y V+ M K PN +TY C
Sbjct: 265 FAIRLYRQMVDAGCKPNASTFNTIVKLLCKEGRFKEGYAFVQQMH--KFGIEPNVLTYHC 322
Query: 370 LLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGP 429
L P+EV G+ E+M GC D Y ++++ + +W + W ME+ G P
Sbjct: 323 FFQYLSRPQEVLGLFEKMLERGCRPRMDTYVMLIKRFGRWGFLRPVFIVWKAMEKQGLSP 382
Query: 430 DRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTE 469
D +Y +I + G + A +Y EM SKG+ +PR E
Sbjct: 383 DAFAYNSLIDALLQKGMVDLARKYDEEMLSKGLSPKPRKE 422
>ref|NP_177262.1| pentatricopeptide (PPR) repeat-containing protein [Arabidopsis
thaliana] gi|12323433|gb|AAG51696.1| hypothetical
protein; 31939-30407 [Arabidopsis thaliana]
gi|25406098|pir||A96735 hypothetical protein F23N20.5
[imported] - Arabidopsis thaliana
Length = 510
Score = 181 bits (460), Expect = 4e-44
Identities = 116/397 (29%), Positives = 199/397 (49%), Gaps = 6/397 (1%)
Query: 66 QVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILG 125
+VE L+ L+ L+ +VL++ + AL F WA + T+ N +++ LG
Sbjct: 80 KVETLLNEASVKLSPALIEEVLKKLSNAGVLALSVFKWAENQKGFKHTTSNYNALIESLG 139
Query: 126 KMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLD 185
K+ +F+ + + D+M ++ L++++TF+ + RR+A A KV+EAI F+ E+FG ++
Sbjct: 140 KIKQFKLIWSLVDDMKAKK-LLSKETFALISRRYARARKVKEAIGAFHKMEEFGFKMESS 198
Query: 186 AFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKD 245
F +L L + ++V DA+ +F K ++ + DIK++ ++L GW N V ++
Sbjct: 199 DFNRMLDTLSKSRNVGDAQKVF-DKMKKKRFEPDIKSYTILLEGWGQELNLLRVDEVNRE 257
Query: 246 IMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFK 305
+ PD+ Y I A K K A++ F M E NCKP I +I+ L +
Sbjct: 258 MKDEGFEPDVVAYGIIINAHCKAKKYEEAIRFFNEM--EQRNCKPSPHIFCSLINGLGSE 315
Query: 306 KRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAV 365
K++ +ALE F+ K G TYN+L+ C +RME Y+ V++M K PNA
Sbjct: 316 KKLNDALEFFERSKSSGFPLEAPTYNALVGAYCWSQRMEDAYKTVDEMRLK--GVGPNAR 373
Query: 366 TYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERN 425
TY +L+ L + E + C + Y +++R++ + D K WDEM+
Sbjct: 374 TYDIILHHLIRMQRSKEAYEVYQTMSCEPTVSTYEIMVRMFCNKERLDMAIKIWDEMKGK 433
Query: 426 GCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
G P ++ +I K+ +A YF EM G+
Sbjct: 434 GVLPGMHMFSSLITALCHENKLDEACEYFNEMLDVGI 470
Score = 81.6 bits (200), Expect = 5e-14
Identities = 61/254 (24%), Positives = 118/254 (46%), Gaps = 6/254 (2%)
Query: 102 NWASKADSYAPTSRVCNEILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAA 161
N K + + P I++ K ++EE + F+EM R + F +L+ +
Sbjct: 255 NREMKDEGFEPDVVAYGIIINAHCKAKKYEEAIRFFNEMEQRNCKPSPHIFCSLINGLGS 314
Query: 162 AHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIK 221
K+ +A+ F + G L+ + L+ C + +EDA + R + + +
Sbjct: 315 EKKLNDALEFFERSKSSGFPLEAPTYNALVGAYCWSQRMEDAYKTV-DEMRLKGVGPNAR 373
Query: 222 TWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGM 281
T+++IL+ + + EA V++ + C P + TY ++ K +L A+K++ M
Sbjct: 374 TYDIILHHLIRMQRSKEAYEVYQTM---SCEPTVSTYEIMVRMFCNKERLDMAIKIWDEM 430
Query: 282 WNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIR 341
+G P + + + +I ALC + ++ EA E F +M + G P ++ L + L
Sbjct: 431 KGKGV--LPGMHMFSSLITALCHENKLDEACEYFNEMLDVGIRPPGHMFSRLKQTLLDEG 488
Query: 342 RMEKVYELVEDMER 355
R +KV +LV M+R
Sbjct: 489 RKDKVTDLVVKMDR 502
Score = 56.2 bits (134), Expect = 2e-06
Identities = 40/172 (23%), Positives = 77/172 (44%), Gaps = 7/172 (4%)
Query: 306 KRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAV 365
++V EA+ F M+E G + + +N ++ L K R + ++ + M++K+ P+
Sbjct: 176 RKVKEAIGAFHKMEEFGFKMESSDFNRMLDTLSKSRNVGDAQKVFDKMKKKRFE--PDIK 233
Query: 366 TYSCLLNSLKGPE----EVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDE 421
+Y+ LL G E V V M+ G Y +++ + K + + ++E
Sbjct: 234 SYTILLEGW-GQELNLLRVDEVNREMKDEGFEPDVVAYGIIINAHCKAKKYEEAIRFFNE 292
Query: 422 MERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVI 473
ME+ C P + +I+G K+ DA+ +F S G E T ++
Sbjct: 293 MEQRNCKPSPHIFCSLINGLGSEKKLNDALEFFERSKSSGFPLEAPTYNALV 344
>ref|XP_469860.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|14488357|gb|AAK63924.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 484
Score = 181 bits (458), Expect = 6e-44
Identities = 113/401 (28%), Positives = 205/401 (50%), Gaps = 13/401 (3%)
Query: 67 VERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWA--SKADSYAPTSRVCNEILDIL 124
VE LD G + D+ VL R + A FF WA K A T R + ++ L
Sbjct: 52 VEHELDHSGVRVTPDVAERVLERLDNAGMLAYRFFEWARRQKRGGCAHTVRSYHTVVASL 111
Query: 125 GKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDL 184
K+ +++ + V M REG VN +TF ++R++A A KV+EA+ F E++G+ +L
Sbjct: 112 AKIRQYQLMWDVVAVM-RREGAVNVETFGIIMRKYARAQKVDEAVYTFNVMEKYGVVPNL 170
Query: 185 DAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWK 244
AF +LL LC+ K+V A+ +F F D KT++++L GW N + + V+
Sbjct: 171 AAFNSLLGALCKSKNVRKAQEIFDKMNSRFS--PDAKTYSILLEGWGRAPNLPKMREVYS 228
Query: 245 DIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCF 304
+++ + C PD+ TY + +L K G++ A+++ + M + G C+P I + ++
Sbjct: 229 EMLDAGCEPDIVTYGIMVDSLCKTGRVEEAVRVVQDMTSRG--CQPTTYIYSVLVHTYGV 286
Query: 305 KKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNA 364
+ R+ +A+ F DM++ G P++ YN+L+ CK ++ E + ++ DME N+
Sbjct: 287 EMRIEDAVATFLDMEKDGIVPDIVVYNALVSAFCKAKKFENAFRVLNDMEGH--GITTNS 344
Query: 365 VTYSCLLN---SLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDE 421
T++ +LN SL +E V RM + C D Y ++++++ + D + K W
Sbjct: 345 RTWNIILNHLISLGRDDEAYKVFRRMIK-CCQPDCDTYTMMIKMFCENDKVEMALKVWKY 403
Query: 422 MERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGM 462
M P +++++I+G + ++ A +M KG+
Sbjct: 404 MRLKQFLPSMHTFSVLINGLCDKREVSQACVLLEDMIEKGI 444
Score = 95.1 bits (235), Expect = 4e-18
Identities = 67/234 (28%), Positives = 117/234 (49%), Gaps = 4/234 (1%)
Query: 120 ILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFG 179
++D L K R EE +V +M+ R +S L+ + ++E+A++ F E+ G
Sbjct: 245 MVDSLCKTGRVEEAVRVVQDMTSRGCQPTTYIYSVLVHTYGVEMRIEDAVATFLDMEKDG 304
Query: 180 LDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEA 239
+ D+ + L+ C+ K E+A + + + + +TWN+ILN LG EA
Sbjct: 305 IVPDIVVYNALVSAFCKAKKFENAFRVLNDMEGH-GITTNSRTWNIILNHLISLGRDDEA 363
Query: 240 KRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCII 299
+V++ M C+PD TY IK + K+ ALK+++ M + P + + +I
Sbjct: 364 YKVFRR-MIKCCQPDCDTYTMMIKMFCENDKVEMALKVWKYMRLK--QFLPSMHTFSVLI 420
Query: 300 DALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDM 353
+ LC K+ V +A + +DM E+G P +T+ L + L K R + + LVE M
Sbjct: 421 NGLCDKREVSQACVLLEDMIEKGIRPPGSTFGKLRQLLLKEGRKDVLDFLVEKM 474
Score = 70.5 bits (171), Expect = 1e-10
Identities = 54/218 (24%), Positives = 96/218 (43%), Gaps = 9/218 (4%)
Query: 251 CRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPE 310
C + +Y T + +L K + + M EG +V I+ ++V E
Sbjct: 97 CAHTVRSYHTVVASLAKIRQYQLMWDVVAVMRREGA---VNVETFGIIMRKYARAQKVDE 153
Query: 311 ALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCL 370
A+ F M++ G PN+A +NSL+ LCK + + K E+ + M + P+A TYS L
Sbjct: 154 AVYTFNVMEKYGVVPNLAAFNSLLGALCKSKNVRKAQEIFDKMNSRFS---PDAKTYSIL 210
Query: 371 LNSLKGPEEVPGVLE---RMERNGCSLSDDIYNLVLRLYMKWDNQDGLRKTWDEMERNGC 427
L +P + E M GC Y +++ K + + +M GC
Sbjct: 211 LEGWGRAPNLPKMREVYSEMLDAGCEPDIVTYGIMVDSLCKTGRVEEAVRVVQDMTSRGC 270
Query: 428 GPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAE 465
P Y++++H + +++DA+ F +M G+V +
Sbjct: 271 QPTTYIYSVLVHTYGVEMRIEDAVATFLDMEKDGIVPD 308
>gb|AAL69509.1| unknown protein [Arabidopsis thaliana]
Length = 320
Score = 177 bits (450), Expect = 5e-43
Identities = 107/321 (33%), Positives = 170/321 (52%), Gaps = 9/321 (2%)
Query: 162 AHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSK---AREFQLHR 218
AH V+EAI + + F L D +F L+ LC +KHV +AE L K F +
Sbjct: 2 AHLVQEAIDAYDKLDDFNLR-DETSFYNLVDALCEHKHVVEAEELCFGKNVIGNGFSVS- 59
Query: 219 DIKTWNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLF 278
+ K N+IL GW LG + K WK + DLF+Y+ ++ + K GK A+KL+
Sbjct: 60 NTKIHNLILRGWSKLGWWGKCKEYWKKMDTEGVTKDLFSYSIYMDIMCKSGKPWKAVKLY 119
Query: 279 RGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLC 338
+ M + K DVV N +I A+ + V + VF++M+ERGCEPNVAT+N++IK LC
Sbjct: 120 KEMKSR--RMKLDVVAYNTVIRAIGASQGVEFGIRVFREMRERGCEPNVATHNTIIKLLC 177
Query: 339 KIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDI 398
+ RM Y ++++M ++ C P+++TY CL + L+ P E+ + RM R+G D
Sbjct: 178 EDGRMRDAYRMLDEMPKR--GCQPDSITYMCLFSRLEKPSEILSLFGRMIRSGVRPKMDT 235
Query: 399 YNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMT 458
Y +++R + +W + W M+ +G PD +Y +I + G + A Y EM
Sbjct: 236 YVMLMRKFERWGFLQPVLYVWKTMKESGDTPDSAAYNAVIDALIQKGMLDMAREYEEEMI 295
Query: 459 SKGMVAEPRTEKLVISMNSPL 479
+G+ R E + S++ L
Sbjct: 296 ERGLSPRRRPELVEKSLDETL 316
>dbj|BAB01242.1| unnamed protein product [Arabidopsis thaliana]
gi|15228825|ref|NP_188906.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
Length = 562
Score = 177 bits (449), Expect = 7e-43
Identities = 121/425 (28%), Positives = 212/425 (49%), Gaps = 17/425 (4%)
Query: 59 RRDKPTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCN 118
++D + V + L C + + LVL VLRR + W A FF WA+ Y + N
Sbjct: 111 KKDTSHEDVVKELSKCDVVVTESLVLQVLRRFSNGWNQAYGFFIWANSQTGYVHSGHTYN 170
Query: 119 EILDILGKMSRFEELHQVFDEMSHRE--GLVNEDTFSTLLRRFAAAHKVEEAISMFYTRE 176
++D+LGK F+ + ++ +EM+ E LV DT S ++RR A + K +A+ F E
Sbjct: 171 AMVDVLGKCRNFDLMWELVNEMNKNEESKLVTLDTMSKVMRRLAKSGKYNKAVDAFLEME 230
Query: 177 Q-FGLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQ-LHRDIKTWNVILNGWCVLG 234
+ +G+ D A +L+ L + +E A +F + F + D +T+N++++G+C
Sbjct: 231 KSYGVKTDTIAMNSLMDALVKENSIEHAHEVF---LKLFDTIKPDARTFNILIHGFCKAR 287
Query: 235 NAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVI 294
+A+ + + ++ PD+ TY +F++A K+G ++ M GCN P+VV
Sbjct: 288 KFDDARAMMDLMKVTEFTPDVVTYTSFVEAYCKEGDFRRVNEMLEEMRENGCN--PNVVT 345
Query: 295 CNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDME 354
++ +L K+V EAL V++ MKE GC P+ Y+SLI L K R + E+ EDM
Sbjct: 346 YTIVMHSLGKSKQVAEALGVYEKMKEDGCVPDAKFYSSLIHILSKTGRFKDAAEIFEDMT 405
Query: 355 RKKGSCMPNAVTYSCLLNSL---KGPEEVPGVLERM---ERNGCSLSDDIYNLVLRLYMK 408
+ + + Y+ ++++ E +L+RM E CS + + Y +L++
Sbjct: 406 NQ--GVRRDVLVYNTMISAALHHSRDEMALRLLKRMEDEEGESCSPNVETYAPLLKMCCH 463
Query: 409 WDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRT 468
L M +N D +Y ++I G +GK+++A +F E KGMV T
Sbjct: 464 KKKMKLLGILLHHMVKNDVSIDVSTYILLIRGLCMSGKVEEACLFFEEAVRKGMVPRDST 523
Query: 469 EKLVI 473
K+++
Sbjct: 524 CKMLV 528
Score = 64.3 bits (155), Expect = 8e-09
Identities = 61/278 (21%), Positives = 115/278 (40%), Gaps = 44/278 (15%)
Query: 222 TWNVILNGWCVLGNAHEAKRVWKDIMASKCRPD-----LFTYATFIKALTKKGKLGTALK 276
T+N +++ VLG +W+ + + L T + ++ L K GK A+
Sbjct: 168 TYNAMVD---VLGKCRNFDLMWELVNEMNKNEESKLVTLDTMSKVMRRLAKSGKYNKAVD 224
Query: 277 LFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKH 336
F M + K D + N ++DAL + + A EVF + + +P+ T+N LI
Sbjct: 225 AFLEM-EKSYGVKTDTIAMNSLMDALVKENSIEHAHEVFLKLFDT-IKPDARTFNILIHG 282
Query: 337 LCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSD 396
CK R+ + +++ M K P+ VTY+ + +
Sbjct: 283 FCKARKFDDARAMMDLM--KVTEFTPDVVTYTSFVEA----------------------- 317
Query: 397 DIYNLVLRLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFRE 456
Y K + + + +EM NGC P+ +YTI++H ++ ++ +A+ + +
Sbjct: 318 ---------YCKEGDFRRVNEMLEEMRENGCNPNVVTYTIVMHSLGKSKQVAEALGVYEK 368
Query: 457 MTSKGMVAEPRTEKLVISMNSPLKERTEKQEGKEEETN 494
M G V + + +I + S + E E+ TN
Sbjct: 369 MKEDGCVPDAKFYSSLIHILSKTGRFKDAAEIFEDMTN 406
>ref|XP_466585.1| putative pentatricopeptide (PPR) repeat-containing protein [Oryza
sativa (japonica cultivar-group)]
gi|47848296|dbj|BAD22160.1| putative pentatricopeptide
(PPR) repeat-containing protein [Oryza sativa (japonica
cultivar-group)]
Length = 454
Score = 174 bits (441), Expect = 6e-42
Identities = 117/418 (27%), Positives = 202/418 (47%), Gaps = 13/418 (3%)
Query: 72 DLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNEILDILGKMSRFE 131
D G L ++V +L + DWK AL FF WA D Y T+ CN ++D+LGKM + +
Sbjct: 28 DCSGIRLTGNIVDTLLFKFGDDWKSALGFFQWAQSRDDYRHTAYACNRMVDLLGKMRQID 87
Query: 132 ELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLL 191
++ ++ +M H GLV +T + +RR A A + ++ + +F E GL+ + + LL
Sbjct: 88 QMWELLSDM-HGRGLVTVETVAKSIRRLAGARRWKDVVLLFDKLEDMGLERNTETMNVLL 146
Query: 192 MWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMASKC 251
LC+ + +E A +F + D T+N+ ++GWC + EA +++
Sbjct: 147 DVLCKERKIEVAREVF--AVLSPHIPPDAYTFNIFVHGWCSIRRIDEAMWTIEEMKRRGF 204
Query: 252 RPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEA 311
P + TY T ++A K+ ++ M ++GC+ P+V+ I+ +L +R EA
Sbjct: 205 PPSVITYTTVLEAYCKQRNFRRVYEVLDSMGSQGCH--PNVITYTMIMTSLAKCERFEEA 262
Query: 312 LEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELVEDMERKKGSCMPNAVTYSCLL 371
L V MK GC+P+ YNSLI L K + + ++ +E N TY+ ++
Sbjct: 263 LSVSHRMKSSGCKPDTLFYNSLINLLGKSGHLFEASQVFR-VEMPMNGVSHNLATYNTMI 321
Query: 372 NSL---KGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWDNQ-DGLRKTWDEM-ERNG 426
+ ++ VL+ ME C Y +LRL++ Q D +R E+ ++
Sbjct: 322 SIFCYYGRDDDALNVLKEMEAQSCKPDIQSYRPLLRLFLSRRGQADTVRHLLSELTSKHN 381
Query: 427 CGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEKLVISMNSPLKERTE 484
G D +YT++IHG G A + F EM S + PR++ V+ ++ + E
Sbjct: 382 LGLDLDTYTLLIHGLCRVGDTVWAYQLFDEMVSSEIA--PRSKTCVMLLDEAQRTNME 437
>ref|XP_507206.1| PREDICTED OSJNBa0091C18.36 gene product [Oryza sativa (japonica
cultivar-group)]
Length = 511
Score = 172 bits (435), Expect = 3e-41
Identities = 121/425 (28%), Positives = 211/425 (49%), Gaps = 26/425 (6%)
Query: 63 PTDQVERALDLCGFDLNDDLVLDVLRRHRSDWKPALVFFNWASKADSYAPTSRVCNE--- 119
P + + L G L DL++ +L R R K AL + A A +SR +
Sbjct: 49 PAESPLQLLSGGGVSLTGDLLVQLLLRLRGASKLALSLLHAARLHPPPAASSRAADAYDA 108
Query: 120 ILDILGKMSRFEELHQVFDEMSHREGLVNEDTFSTLLRRFAAAHKVEEAISMFYTREQF- 178
++D LG+ +F+ ++ E + +G TF+ L RR+ AA +A+ F E F
Sbjct: 109 VVDALGRARQFDAAWRLVVEAA-ADGAATPRTFAVLARRYVAAGMTRQAVRAFDDMEAFV 167
Query: 179 GLDLDLDAFRTLLMWLCRYKHVEDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHE 238
G + D F TLL LC+YK+ + A +F+ R+++ + K + V++ GWC +
Sbjct: 168 GREPDAAEFTTLLDTLCKYKYPKVAAEVFNK--RKYKYEPNEKMYTVLIYGWCKVNRNDM 225
Query: 239 AKRVWKDIMASKCRPDLFTYATFIKALTKKGKLG----------TALKLFRGMWNEGCNC 288
A++ KD++ P++ TY + + + L A L + M G
Sbjct: 226 AQKFLKDMIYHGIEPNIVTYNILLNGICRHASLHPDYRFDRTVRAAEDLLKEMHQRGI-- 283
Query: 289 KPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYE 348
+P+V + I+ + L +F+ MKERG P VATY S+IK L R++
Sbjct: 284 EPNVTSYSVILHVYSRAHKPELCLCMFRSMKERGICPTVATYTSVIKCLASCGRLDDAES 343
Query: 349 LVEDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLE---RMERNGCSLSDDI--YNLVL 403
L+++M +G C P+ TY+C +G ++V G L+ +M+ G + DI YN++L
Sbjct: 344 LLDEMA-SEGVC-PSPATYNCFFKEYRGRKDVNGALQLYNKMKAPGSPATPDIHTYNILL 401
Query: 404 RLYMKWDNQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMV 463
+++K + D + + W++M + GPD SYT+++HG +N K ++A ++F EM +G +
Sbjct: 402 GMFIKLNQHDTVMQVWNDMCESTVGPDLNSYTLLVHGLCDNKKWREACQFFMEMIERGFL 461
Query: 464 AEPRT 468
+ T
Sbjct: 462 PQKIT 466
Score = 64.7 bits (156), Expect = 6e-09
Identities = 53/225 (23%), Positives = 105/225 (46%), Gaps = 13/225 (5%)
Query: 142 HREGLV-NEDTFSTLLRRFAAAHKVEEAISMFYTREQFGLDLDLDAFRTLLMWLCRYKHV 200
H+ G+ N ++S +L ++ AHK E + MF + ++ G+ + + +++ L +
Sbjct: 279 HQRGIEPNVTSYSVILHVYSRAHKPELCLCMFRSMKERGICPTVATYTSVIKCLASCGRL 338
Query: 201 EDAETLFHSKAREFQLHRDIKTWNVILNGWCVLGNAHEAKRVWKDIMA--SKCRPDLFTY 258
+DAE+L A E + T+N + + + A +++ + A S PD+ TY
Sbjct: 339 DDAESLLDEMASE-GVCPSPATYNCFFKEYRGRKDVNGALQLYNKMKAPGSPATPDIHTY 397
Query: 259 ATFIKALTKKGKLGTALKLFRGMWNEGCNCK--PDVVICNCIIDALCFKKRVPEALEVFQ 316
+ K + T +++ WN+ C PD+ ++ LC K+ EA + F
Sbjct: 398 NILLGMFIKLNQHDTVMQV----WNDMCESTVGPDLNSYTLLVHGLCDNKKWREACQFFM 453
Query: 317 DMKERGCEPNVATYNSLIKHLCK---IRRMEKVYELVEDMERKKG 358
+M ERG P T+ +L + L + +R ++ + V++ K G
Sbjct: 454 EMIERGFLPQKITFETLYRGLIQADMLRTWRRLKKRVDEEAAKFG 498
Score = 55.8 bits (133), Expect = 3e-06
Identities = 61/243 (25%), Positives = 98/243 (40%), Gaps = 28/243 (11%)
Query: 233 LGNAHEAKRVWKDIM--ASKCRPDLFTYATFIKALTKKGKLGTALKLFRGMWNEGCNCKP 290
LG A + W+ ++ A+ T+A + G A++ F M +P
Sbjct: 113 LGRARQFDAAWRLVVEAAADGAATPRTFAVLARRYVAAGMTRQAVRAFDDM-EAFVGREP 171
Query: 291 DVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLCKIRRMEKVYELV 350
D ++D LC K A EVF K + EPN Y LI CK+ R + + +
Sbjct: 172 DAAEFTTLLDTLCKYKYPKVAAEVFNKRKYK-YEPNEKMYTVLIYGWCKVNRNDMAQKFL 230
Query: 351 EDMERKKGSCMPNAVTYSCLLNSLKGPEEVPGVLERMERNGCSLSDDIYNLVLRLYMKWD 410
+DM PN VTY+ LLN + R+ D ++ +R
Sbjct: 231 KDMIYH--GIEPNIVTYNILLNGIC-------------RHASLHPDYRFDRTVRAA---- 271
Query: 411 NQDGLRKTWDEMERNGCGPDRRSYTIMIHGHYENGKMKDAMRYFREMTSKGMVAEPRTEK 470
+D L+ EM + G P+ SY++++H + K + + FR M +G+ T
Sbjct: 272 -EDLLK----EMHQRGIEPNVTSYSVILHVYSRAHKPELCLCMFRSMKERGICPTVATYT 326
Query: 471 LVI 473
VI
Sbjct: 327 SVI 329
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 870,760,030
Number of Sequences: 2540612
Number of extensions: 37458887
Number of successful extensions: 108826
Number of sequences better than 10.0: 1285
Number of HSP's better than 10.0 without gapping: 736
Number of HSP's successfully gapped in prelim test: 558
Number of HSP's that attempted gapping in prelim test: 88491
Number of HSP's gapped (non-prelim): 6914
length of query: 495
length of database: 863,360,394
effective HSP length: 132
effective length of query: 363
effective length of database: 527,999,610
effective search space: 191663858430
effective search space used: 191663858430
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0195b.3


