

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0185.2
(122 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
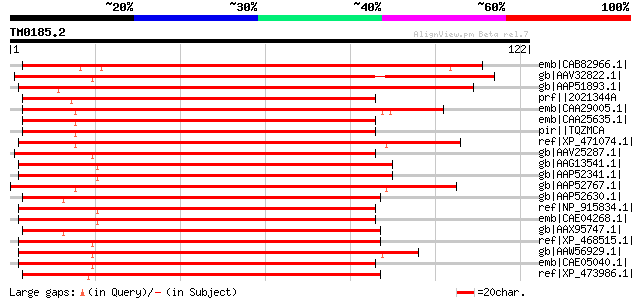
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB82966.1| putative protein [Arabidopsis thaliana] gi|11357... 110 6e-24
gb|AAV32822.1| transposase [Zea mays] 110 6e-24
gb|AAP51893.1| putative Tam3-like transposon protein [Oryza sati... 109 1e-23
prf||2021344A activator-like transposable element 107 7e-23
emb|CAA29005.1| ORFa [Zea mays] gi|136125|sp|P08770|TRA1_MAIZE P... 106 1e-22
emb|CAA25635.1| unnamed protein product [Zea mays] gi|7489762|pi... 105 3e-22
pir||TQZMCA probable transposase - maize transposon Ac9 gi|13614... 105 3e-22
ref|XP_471074.1| OSJNBa0020P07.19 [Oryza sativa (japonica cultiv... 103 1e-21
gb|AAV25287.1| hypothetical protein [Oryza sativa (japonica cult... 102 2e-21
gb|AAG13541.1| putative Tam3-transposase [Oryza sativa (japonica... 100 6e-21
gb|AAP52341.1| putative transposable element [Oryza sativa (japo... 100 6e-21
gb|AAP52767.1| putative activator-like transposable element [Ory... 100 8e-21
gb|AAP52630.1| putative transposase [Oryza sativa (japonica cult... 99 2e-20
ref|NP_915834.1| P0003D09.25 [Oryza sativa (japonica cultivar-gr... 99 2e-20
emb|CAE04268.1| OSJNBb0103I08.10 [Oryza sativa (japonica cultiva... 99 2e-20
gb|AAX95747.1| hAT family dimerisation domain, putative [Oryza s... 99 2e-20
ref|XP_468515.1| hypothetical protein [Oryza sativa (japonica cu... 99 2e-20
gb|AAW56929.1| unknown protein [Oryza sativa (japonica cultivar-... 99 2e-20
emb|CAE05040.1| OSJNBa0049H08.1 [Oryza sativa (japonica cultivar... 98 5e-20
ref|XP_473986.1| OSJNBa0089N06.8 [Oryza sativa (japonica cultiva... 95 4e-19
>emb|CAB82966.1| putative protein [Arabidopsis thaliana] gi|11357980|pir||T48044
hypothetical protein T12C14.220 - Arabidopsis thaliana
Length = 705
Score = 110 bits (276), Expect = 6e-24
Identities = 56/119 (47%), Positives = 81/119 (68%), Gaps = 11/119 (9%)
Query: 4 NELTKYLEDGLE--ERGSL----DILNWWKLNASRYPILASIARELLAIPISTVASESTF 57
NEL KYL+D L SL D+L+WWK N+S+YPI++ +ARE+LAIP+S+VASES F
Sbjct: 574 NELVKYLKDELHVTTENSLGLPFDLLDWWKTNSSKYPIMSLMAREVLAIPVSSVASESAF 633
Query: 58 SAGGRVVDPYRSSLTPKTLEALICTQDWIKGKYSKSLLSNEEVME-----LEKYEQGII 111
S GGR++D YRS LTP +EAL+ TQDW++ + + + +E ++ E+GI+
Sbjct: 634 STGGRILDQYRSCLTPDMVEALVLTQDWLRASLRSEAMKSLDKLEEENKFMDSLEEGIL 692
>gb|AAV32822.1| transposase [Zea mays]
Length = 674
Score = 110 bits (276), Expect = 6e-24
Identities = 55/114 (48%), Positives = 81/114 (70%), Gaps = 3/114 (2%)
Query: 2 SKNELTKYLEDGLEERG-SLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAG 60
+K+EL KYL + E+ LD+L WWK + R+PIL+ +AR++LAIPIS+VASES FS G
Sbjct: 534 NKSELDKYLAEETEDTEMKLDLLVWWKASEQRFPILSRLARDVLAIPISSVASESAFSTG 593
Query: 61 GRVVDPYRSSLTPKTLEALICTQDWIKGKYSKSLLSNEEVMELEKYEQGIITNY 114
GR++D +RSSLTP LE+L+CTQDW+ +++ + E + EL K E+ + +
Sbjct: 594 GRILDDFRSSLTPFMLESLVCTQDWL--RWTIPIDITENIEELTKLEEELFEEF 645
>gb|AAP51893.1| putative Tam3-like transposon protein [Oryza sativa (japonica
cultivar-group)] gi|37530608|ref|NP_919606.1| putative
Tam3-like transposon protein [Oryza sativa (japonica
cultivar-group)] gi|16924038|gb|AAL31650.1| Putative
Tam3-like transposon protein [Oryza sativa]
Length = 737
Score = 109 bits (273), Expect = 1e-23
Identities = 54/108 (50%), Positives = 76/108 (70%), Gaps = 1/108 (0%)
Query: 3 KNELTKYL-EDGLEERGSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGG 61
K+E+ KYL ED + DIL WWK N++R+PIL+ +A +LLAIPI++VASES FSAGG
Sbjct: 616 KSEVDKYLSEDNEPDTPKFDILKWWKANSTRFPILSHLACDLLAIPITSVASESAFSAGG 675
Query: 62 RVVDPYRSSLTPKTLEALICTQDWIKGKYSKSLLSNEEVMELEKYEQG 109
R +D +R+SLTP+ +E L+C DW++G S+ + E M L + E G
Sbjct: 676 RTLDDFRTSLTPRMVERLVCANDWLRGGNYVSVEEDSEQMALLEEELG 723
>prf||2021344A activator-like transposable element
Length = 804
Score = 107 bits (267), Expect = 7e-23
Identities = 52/84 (61%), Positives = 63/84 (74%), Gaps = 1/84 (1%)
Query: 4 NELTKYLEDG-LEERGSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGGR 62
NEL KY+ D L G DIL WWK + YPILA IAR+LLA+ +STVASES FSAGGR
Sbjct: 667 NELDKYMADPPLRFSGQFDILAWWKNQSDEYPILAKIARDLLAVQVSTVASESAFSAGGR 726
Query: 63 VVDPYRSSLTPKTLEALICTQDWI 86
VVDP+RS L P+ ++ALIC +DW+
Sbjct: 727 VVDPFRSRLDPEMVQALICMKDWV 750
>emb|CAA29005.1| ORFa [Zea mays] gi|136125|sp|P08770|TRA1_MAIZE Putative AC
transposase (ORFA)
Length = 807
Score = 106 bits (265), Expect = 1e-22
Identities = 55/105 (52%), Positives = 75/105 (71%), Gaps = 6/105 (5%)
Query: 4 NELTKYLEDGL-EERGSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGGR 62
NEL KY+ + L + G DIL+WW+ + YPIL IAR++LAI +STVASES FSAGGR
Sbjct: 668 NELDKYMSEPLLKHSGQFDILSWWRGRVAEYPILTQIARDVLAIQVSTVASESAFSAGGR 727
Query: 63 VVDPYRSSLTPKTLEALICTQDWI----KG-KYSKSLLSNEEVME 102
VVDPYR+ L + +EALICT+DW+ KG Y +++ + EV++
Sbjct: 728 VVDPYRNRLGSEIVEALICTKDWVAASRKGATYFPTMIGDLEVLD 772
>emb|CAA25635.1| unnamed protein product [Zea mays] gi|7489762|pir||T03954
hypothetical protein - maize transposable element Ac
Length = 221
Score = 105 bits (262), Expect = 3e-22
Identities = 50/84 (59%), Positives = 64/84 (75%), Gaps = 1/84 (1%)
Query: 4 NELTKYLEDGL-EERGSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGGR 62
NEL KY+ + L + G DIL+WW+ + YPIL IAR++LAI +STVASES FSAGGR
Sbjct: 111 NELDKYMSEPLLKHSGQFDILSWWRGRVAEYPILTQIARDVLAIQVSTVASESAFSAGGR 170
Query: 63 VVDPYRSSLTPKTLEALICTQDWI 86
VVDPYR+ L + +EALICT+DW+
Sbjct: 171 VVDPYRNRLGSEIVEALICTKDWV 194
>pir||TQZMCA probable transposase - maize transposon Ac9
gi|136140|sp|P03010|TRA9_MAIZE PUTATIVE AC9 TRANSPOSASE
Length = 839
Score = 105 bits (262), Expect = 3e-22
Identities = 50/84 (59%), Positives = 64/84 (75%), Gaps = 1/84 (1%)
Query: 4 NELTKYLEDGL-EERGSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGGR 62
NEL KY+ + L + G DIL+WW+ + YPIL IAR++LAI +STVASES FSAGGR
Sbjct: 637 NELDKYMSEPLLKHSGQFDILSWWRGRVAEYPILTQIARDVLAIQVSTVASESAFSAGGR 696
Query: 63 VVDPYRSSLTPKTLEALICTQDWI 86
VVDPYR+ L + +EALICT+DW+
Sbjct: 697 VVDPYRNRLGSEIVEALICTKDWV 720
>ref|XP_471074.1| OSJNBa0020P07.19 [Oryza sativa (japonica cultivar-group)]
gi|38344876|emb|CAE01302.2| OSJNBa0020P07.19 [Oryza
sativa (japonica cultivar-group)]
Length = 741
Score = 103 bits (257), Expect = 1e-21
Identities = 49/111 (44%), Positives = 76/111 (68%), Gaps = 7/111 (6%)
Query: 3 KNELTKYLEDGL---EERGSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSA 59
K EL Y+E L + + DIL+WWKL + +PIL +AR+ L I +STVASES FSA
Sbjct: 603 KTELQIYMEQPLLLWTSKDTFDILSWWKLKQAEFPILCKLARDFLCIQVSTVASESAFSA 662
Query: 60 GGRVVDPYRSSLTPKTLEALICTQDWIK----GKYSKSLLSNEEVMELEKY 106
GG VVDP+R+ L P+ ++AL+CT+DWIK G ++++++ ++ +E++
Sbjct: 663 GGHVVDPFRTRLDPEAMQALVCTKDWIKAANNGYKTQAIINELDIESVERH 713
>gb|AAV25287.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|57863883|gb|AAW56923.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 676
Score = 102 bits (255), Expect = 2e-21
Identities = 47/86 (54%), Positives = 67/86 (77%), Gaps = 1/86 (1%)
Query: 2 SKNELTKYLEDGLEERG-SLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAG 60
+K+E YLE+ +EE S D+L+WWK +A ++P+L+S+AR+ LAIP+STV+SE FS G
Sbjct: 572 TKSEFAIYLEEDVEEDSESFDVLDWWKRHAQKFPVLSSMARDFLAIPLSTVSSELAFSCG 631
Query: 61 GRVVDPYRSSLTPKTLEALICTQDWI 86
GR++ RSSLTP+ LEALIC +DW+
Sbjct: 632 GRILGDTRSSLTPEMLEALICAKDWL 657
>gb|AAG13541.1| putative Tam3-transposase [Oryza sativa (japonica cultivar-group)]
gi|31432800|gb|AAP54387.1| putative Tam3-transposase
[Oryza sativa (japonica cultivar-group)]
gi|37535596|ref|NP_922100.1| putative Tam3-transposase
[Oryza sativa (japonica cultivar-group)]
Length = 568
Score = 100 bits (250), Expect = 6e-21
Identities = 47/90 (52%), Positives = 65/90 (72%), Gaps = 2/90 (2%)
Query: 3 KNELTKYLEDGLEERGS--LDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAG 60
K+EL +YLED L + IL+WWK+ + +P L +AR++ AIP+STVASES FS
Sbjct: 467 KSELDRYLEDELVSINTENFKILDWWKVAGTSFPTLRKVARDIFAIPVSTVASESAFSTS 526
Query: 61 GRVVDPYRSSLTPKTLEALICTQDWIKGKY 90
GRV+ +RS LTP+ LEAL+C+QDW++ KY
Sbjct: 527 GRVLSEHRSRLTPELLEALMCSQDWLRNKY 556
>gb|AAP52341.1| putative transposable element [Oryza sativa (japonica
cultivar-group)] gi|37531504|ref|NP_920054.1| putative
transposable element [Oryza sativa (japonica
cultivar-group)] gi|21671885|gb|AAM74247.1| Putative
transposable element [Oryza sativa (japonica
cultivar-group)]
Length = 772
Score = 100 bits (250), Expect = 6e-21
Identities = 47/90 (52%), Positives = 65/90 (72%), Gaps = 2/90 (2%)
Query: 3 KNELTKYLEDGLEERGS--LDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAG 60
K+EL +YLED L + IL+WWK+ + +P L +AR++ AIP+STVASES FS
Sbjct: 656 KSELDRYLEDELVSINTENFKILDWWKVAGTSFPTLRKVARDIFAIPVSTVASESAFSTS 715
Query: 61 GRVVDPYRSSLTPKTLEALICTQDWIKGKY 90
GRV+ +RS LTP+ LEAL+C+QDW++ KY
Sbjct: 716 GRVLSEHRSRLTPELLEALMCSQDWLRNKY 745
>gb|AAP52767.1| putative activator-like transposable element [Oryza sativa
(japonica cultivar-group)] gi|37532356|ref|NP_920480.1|
putative activator-like transposable element [Oryza
sativa (japonica cultivar-group)]
gi|20270084|gb|AAM18172.1| Putative activator-like
transposable element [Oryza sativa (japonica
cultivar-group)]
Length = 737
Score = 100 bits (249), Expect = 8e-21
Identities = 51/115 (44%), Positives = 76/115 (65%), Gaps = 10/115 (8%)
Query: 1 GSKNELTKYLEDGL-----EERGSLDILNWWKLNASRYPILASIARELLAIPISTVASES 55
G+K+EL Y++ L +++ +IL+WW L PIL+ +AR++LAI +STVASES
Sbjct: 597 GTKSELEVYMDQPLLEWDIKDKSPFNILHWWSLKQHELPILSRLARDVLAIQVSTVASES 656
Query: 56 TFSAGGRVVDPYRSSLTPKTLEALICTQDWIK-----GKYSKSLLSNEEVMELEK 105
FSAGGRV+DP+RS L P+ ++ALICT+DW G S+L+ ++ LE+
Sbjct: 657 AFSAGGRVIDPFRSCLDPEIVQALICTKDWTAASRKGGNVVGSILTEMDMENLER 711
>gb|AAP52630.1| putative transposase [Oryza sativa (japonica cultivar-group)]
gi|37532082|ref|NP_920343.1| putative transposase [Oryza
sativa (japonica cultivar-group)]
Length = 997
Score = 99.4 bits (246), Expect = 2e-20
Identities = 49/91 (53%), Positives = 65/91 (70%), Gaps = 7/91 (7%)
Query: 4 NELTKYLE-------DGLEERGSLDILNWWKLNASRYPILASIARELLAIPISTVASEST 56
NEL Y++ D E DIL WWK N +PIL+++AR+++A+ ISTVASES
Sbjct: 865 NELDVYMKEKPIRWVDPTGEGVEFDILAWWKNNQMTFPILSTLARDVMAVQISTVASESA 924
Query: 57 FSAGGRVVDPYRSSLTPKTLEALICTQDWIK 87
FSAGGRVV P+RSSL P+ +EAL+CT+DWI+
Sbjct: 925 FSAGGRVVGPFRSSLHPEMIEALVCTKDWIR 955
>ref|NP_915834.1| P0003D09.25 [Oryza sativa (japonica cultivar-group)]
Length = 639
Score = 99.4 bits (246), Expect = 2e-20
Identities = 45/85 (52%), Positives = 64/85 (74%), Gaps = 1/85 (1%)
Query: 3 KNELTKYLEDGLEERGS-LDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGG 61
K+E+ YLE+ E+ D+L WWK N+ ++P+LA +AR+ LAIP+STV SES FS+GG
Sbjct: 547 KSEIDTYLEEVCEDDSEDFDVLAWWKKNSKKFPVLAIMARDFLAIPLSTVPSESAFSSGG 606
Query: 62 RVVDPYRSSLTPKTLEALICTQDWI 86
R++ RSSLTP+ LEAL+C +DW+
Sbjct: 607 RILGDTRSSLTPEMLEALVCAKDWL 631
>emb|CAE04268.1| OSJNBb0103I08.10 [Oryza sativa (japonica cultivar-group)]
gi|50927186|ref|XP_473369.1| OSJNBb0103I08.10 [Oryza
sativa (japonica cultivar-group)]
Length = 349
Score = 99.4 bits (246), Expect = 2e-20
Identities = 45/85 (52%), Positives = 64/85 (74%), Gaps = 1/85 (1%)
Query: 3 KNELTKYLEDGLEERGS-LDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGG 61
K+E+ YLE+ E+ D+L WWK N+ ++P+LA +AR+ LAIP+STV SES FS+GG
Sbjct: 257 KSEIDTYLEEVCEDDSEDFDVLAWWKKNSKKFPVLAIMARDFLAIPLSTVPSESAFSSGG 316
Query: 62 RVVDPYRSSLTPKTLEALICTQDWI 86
R++ RSSLTP+ LEAL+C +DW+
Sbjct: 317 RILGDTRSSLTPEMLEALVCAKDWL 341
>gb|AAX95747.1| hAT family dimerisation domain, putative [Oryza sativa (japonica
cultivar-group)]
Length = 989
Score = 99.4 bits (246), Expect = 2e-20
Identities = 49/91 (53%), Positives = 65/91 (70%), Gaps = 7/91 (7%)
Query: 4 NELTKYLE-------DGLEERGSLDILNWWKLNASRYPILASIARELLAIPISTVASEST 56
NEL Y++ D E DIL WWK N +PIL+++AR+++A+ ISTVASES
Sbjct: 857 NELDVYMKEKPIRWVDPTGEGVEFDILAWWKNNQMTFPILSTLARDVMAVQISTVASESA 916
Query: 57 FSAGGRVVDPYRSSLTPKTLEALICTQDWIK 87
FSAGGRVV P+RSSL P+ +EAL+CT+DWI+
Sbjct: 917 FSAGGRVVGPFRSSLHPEMIEALVCTKDWIR 947
>ref|XP_468515.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|48716460|dbj|BAD23067.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 561
Score = 99.0 bits (245), Expect = 2e-20
Identities = 46/89 (51%), Positives = 63/89 (70%), Gaps = 4/89 (4%)
Query: 3 KNELTKYLEDGLEERG----SLDILNWWKLNASRYPILASIARELLAIPISTVASESTFS 58
K ELT+YLED +E DIL WW++N+ +YPIL+ +A +LLA+P S+VASES FS
Sbjct: 283 KTELTRYLEDIPQENDFPDDDFDILQWWRVNSCKYPILSRMALDLLAVPASSVASESAFS 342
Query: 59 AGGRVVDPYRSSLTPKTLEALICTQDWIK 87
G R++ YRS L T+EAL+C QDW++
Sbjct: 343 TGSRIISDYRSRLASGTVEALVCLQDWMR 371
>gb|AAW56929.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 542
Score = 99.0 bits (245), Expect = 2e-20
Identities = 49/99 (49%), Positives = 67/99 (67%), Gaps = 5/99 (5%)
Query: 3 KNELTKYLEDGLEERG-SLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGG 61
K+EL KYL++ + R DI++WWK+ R+P L +AR++LA+P+S V SES FS G
Sbjct: 430 KSELDKYLDEPVIPRAVDFDIIDWWKIMGKRFPTLQMLARDILAVPVSRVVSESAFSIAG 489
Query: 62 RVVDPYRSSLTPKTLEALICTQDWI----KGKYSKSLLS 96
RV+ P RS L P TLEAL+C+QDW+ +G S LLS
Sbjct: 490 RVLSPQRSKLLPDTLEALMCSQDWLHAENRGAGSSGLLS 528
>emb|CAE05040.1| OSJNBa0049H08.1 [Oryza sativa (japonica cultivar-group)]
gi|38346748|emb|CAD40758.2| OSJNBa0081G05.11 [Oryza
sativa (japonica cultivar-group)]
gi|50923507|ref|XP_472114.1| OSJNBa0081G05.11 [Oryza
sativa (japonica cultivar-group)]
Length = 557
Score = 97.8 bits (242), Expect = 5e-20
Identities = 44/85 (51%), Positives = 65/85 (75%), Gaps = 1/85 (1%)
Query: 3 KNELTKYLEDGLEERG-SLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGG 61
K+E YLE+ +EE S D+L+WWK A ++P+L+++A++ LAIP+STV+SES FS G
Sbjct: 454 KSEFAIYLEEDVEEDSESFDVLDWWKRQAQKFPVLSAMAKDFLAIPLSTVSSESAFSCRG 513
Query: 62 RVVDPYRSSLTPKTLEALICTQDWI 86
R++ RSSLTP+ L+ALIC +DW+
Sbjct: 514 RILGDTRSSLTPEMLDALICAKDWL 538
>ref|XP_473986.1| OSJNBa0089N06.8 [Oryza sativa (japonica cultivar-group)]
gi|39546238|emb|CAE04247.3| OSJNBa0089N06.8 [Oryza
sativa (japonica cultivar-group)]
Length = 802
Score = 94.7 bits (234), Expect = 4e-19
Identities = 43/85 (50%), Positives = 64/85 (74%), Gaps = 1/85 (1%)
Query: 4 NELTKYLEDGLEER-GSLDILNWWKLNASRYPILASIARELLAIPISTVASESTFSAGGR 62
+EL YLEDGL R DILNWW ++++YP LA+IAR++LA+P S V SE+ FS+ G
Sbjct: 718 SELDDYLEDGLVPRKDDFDILNWWMTHSTKYPTLATIARDILAMPASAVQSEAAFSSSGP 777
Query: 63 VVDPYRSSLTPKTLEALICTQDWIK 87
V+ ++S+L +T+EAL+CT+DW++
Sbjct: 778 VIPKHQSTLNIRTIEALVCTRDWMR 802
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.139 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 193,537,253
Number of Sequences: 2540612
Number of extensions: 6638176
Number of successful extensions: 17240
Number of sequences better than 10.0: 227
Number of HSP's better than 10.0 without gapping: 206
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 17020
Number of HSP's gapped (non-prelim): 230
length of query: 122
length of database: 863,360,394
effective HSP length: 98
effective length of query: 24
effective length of database: 614,380,418
effective search space: 14745130032
effective search space used: 14745130032
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0185.2


