

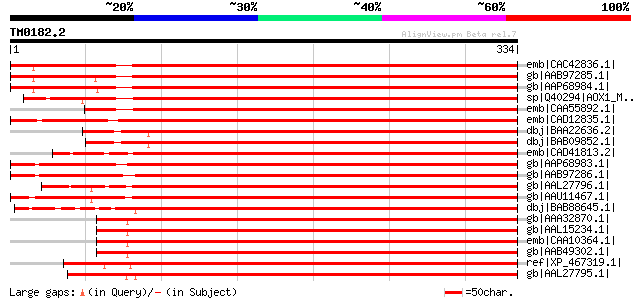
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0182.2
(334 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC42836.1| putative alternative oxidase [Vigna unguiculata] 537 e-151
gb|AAB97285.1| alternative oxidase 2a [Glycine max] gi|3913067|s... 531 e-149
gb|AAP68984.1| alternative oxidase 2a [Glycine max] 531 e-149
sp|Q40294|AOX1_MANIN Alternative oxidase, mitochondrial precursor 495 e-139
emb|CAA55892.1| alternative oxidase [Mangifera indica] 487 e-136
emb|CAD12835.1| putative alternative oxidase [Vigna unguiculata] 459 e-128
dbj|BAA22636.2| alternative oxidase [Arabidopsis thaliana] gi|67... 457 e-127
dbj|BAB09852.1| alternative oxidase 2 [Arabidopsis thaliana] 456 e-127
emb|CAD41813.2| OSJNBa0083N12.11 [Oryza sativa (japonica cultiva... 447 e-124
gb|AAP68983.1| alternative oxidase 2b [Glycine max] 447 e-124
gb|AAB97286.1| alternative oxidase 2b [Glycine max] gi|3023305|s... 446 e-124
gb|AAL27796.1| alternative oxidase AOX2 precursor [Zea mays] gi|... 446 e-124
gb|AAU11467.1| mitochondrial alternative oxidase 1 [Saccharum of... 445 e-123
dbj|BAB88645.1| alternative oxidase [Triticum aestivum] 444 e-123
gb|AAA32870.1| oxidase 442 e-123
gb|AAL15234.1| putative alternative oxidase 1a precursor [Arabid... 442 e-123
emb|CAA10364.1| alternative oxidase [Arabidopsis thaliana] gi|11... 442 e-123
gb|AAB49302.1| alternative oxidase [Arabidopsis thaliana] 442 e-123
ref|XP_467319.1| alternative oxidase 1c [Oryza sativa (japonica ... 437 e-121
gb|AAL27795.1| alternative oxidase AOX1 precursor [Zea mays] 436 e-121
>emb|CAC42836.1| putative alternative oxidase [Vigna unguiculata]
Length = 329
Score = 537 bits (1384), Expect = e-151
Identities = 272/339 (80%), Positives = 285/339 (83%), Gaps = 15/339 (4%)
Query: 1 MKHLALSYALRRAL----NCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWNRMMSSQA- 55
MK +ALS +RRAL NCN G TAV A E R NG W R M+SQA
Sbjct: 1 MKFIALSCTVRRALLNGRNCNGLGSTAVMAYAAPETRFLCAGAANGGLFYWRRSMASQAE 60
Query: 56 APEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYR 115
A EK +EKAE E SVV SSYWGISRP+I REDGTEWPWNCFMPWETY
Sbjct: 61 AKLPEKDKEKAEAEK----------SVVESSYWGISRPRIMREDGTEWPWNCFMPWETYH 110
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGM 175
+LSIDLTKHHVPKNFLDKVAYRTVKLLRIPTD+FFQRRYGCRAMMLETVAAVPGMVGGM
Sbjct: 111 SNLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDLFFQRRYGCRAMMLETVAAVPGMVGGM 170
Query: 176 LLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFF 235
LLHLRSLRKFQQSGGWIKAL+EEAENERMHLMTMVELV+PKWYER LV+ VQGVFFNAFF
Sbjct: 171 LLHLRSLRKFQQSGGWIKALMEEAENERMHLMTMVELVKPKWYERLLVIAVQGVFFNAFF 230
Query: 236 VLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKD 295
VLY+LSPKVAHR+VGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDA LKD
Sbjct: 231 VLYILSPKVAHRIVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDAKLKD 290
Query: 296 VITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
VITVIRADEAHHRDVNHFASDIHF GKELREAPAP+GYH
Sbjct: 291 VITVIRADEAHHRDVNHFASDIHFQGKELREAPAPIGYH 329
>gb|AAB97285.1| alternative oxidase 2a [Glycine max]
gi|3913067|sp|Q41266|AOX2_SOYBN Alternative oxidase 2,
mitochondrial precursor
Length = 333
Score = 531 bits (1368), Expect = e-149
Identities = 268/343 (78%), Positives = 286/343 (83%), Gaps = 19/343 (5%)
Query: 1 MKHLALSYALRRAL------NCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWNRMMSSQ 54
MK AL+ +RRAL N NR G A+ A E R G NG F W R M S
Sbjct: 1 MKLTALNSTVRRALLNGRNQNGNRLGSAALMPYAAAETRLLCAGGANGWFFYWKRTMVSP 60
Query: 55 A---APEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPW 111
A PE+EK++EKA+ E SVV SSYWGISRPK+ REDGTEWPWNCFMPW
Sbjct: 61 AEAKVPEKEKEKEKAKAEK----------SVVESSYWGISRPKVVREDGTEWPWNCFMPW 110
Query: 112 ETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGM 171
E+YR ++SIDLTKHHVPKN LDKVAYRTVKLLRIPTD+FF+RRYGCRAMMLETVAAVPGM
Sbjct: 111 ESYRSNVSIDLTKHHVPKNVLDKVAYRTVKLLRIPTDLFFKRRYGCRAMMLETVAAVPGM 170
Query: 172 VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFF 231
VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELV+PKWYER LVL VQGVFF
Sbjct: 171 VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVKPKWYERLLVLAVQGVFF 230
Query: 232 NAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDA 291
NAFFVLY+LSPKVAHR+VGYLEEEAIHSYTEYLKD+ESGAIENVPAPAIAIDYWRLPKDA
Sbjct: 231 NAFFVLYILSPKVAHRIVGYLEEEAIHSYTEYLKDLESGAIENVPAPAIAIDYWRLPKDA 290
Query: 292 TLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
LKDVITVIRADEAHHRDVNHFASDIHF GKELREAPAP+GYH
Sbjct: 291 RLKDVITVIRADEAHHRDVNHFASDIHFQGKELREAPAPIGYH 333
>gb|AAP68984.1| alternative oxidase 2a [Glycine max]
Length = 333
Score = 531 bits (1367), Expect = e-149
Identities = 268/343 (78%), Positives = 286/343 (83%), Gaps = 19/343 (5%)
Query: 1 MKHLALSYALRRAL------NCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWNRMMSSQ 54
MK AL+ +RRAL N NR G A+ A E R G NG F W R M S
Sbjct: 1 MKLTALNSTVRRALLNGRNQNGNRLGSAALMPYAAAETRLLCAGGANGWFFYWKRTMVSP 60
Query: 55 AA---PEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPW 111
A PE+EK++EKA+ E SVV SSYWGISRPK+ REDGTEWPWNCFMPW
Sbjct: 61 AEAKLPEKEKEKEKAKAEK----------SVVESSYWGISRPKVVREDGTEWPWNCFMPW 110
Query: 112 ETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGM 171
E+YR ++SIDLTKHHVPKN LDKVAYRTVKLLRIPTD+FF+RRYGCRAMMLETVAAVPGM
Sbjct: 111 ESYRSNVSIDLTKHHVPKNVLDKVAYRTVKLLRIPTDLFFKRRYGCRAMMLETVAAVPGM 170
Query: 172 VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFF 231
VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELV+PKWYER LVL VQGVFF
Sbjct: 171 VGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVKPKWYERLLVLAVQGVFF 230
Query: 232 NAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDA 291
NAFFVLY+LSPKVAHR+VGYLEEEAIHSYTEYLKD+ESGAIENVPAPAIAIDYWRLPKDA
Sbjct: 231 NAFFVLYILSPKVAHRIVGYLEEEAIHSYTEYLKDLESGAIENVPAPAIAIDYWRLPKDA 290
Query: 292 TLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
LKDVITVIRADEAHHRDVNHFASDIHF GKELREAPAP+GYH
Sbjct: 291 RLKDVITVIRADEAHHRDVNHFASDIHFQGKELREAPAPIGYH 333
>sp|Q40294|AOX1_MANIN Alternative oxidase, mitochondrial precursor
Length = 318
Score = 495 bits (1275), Expect = e-139
Identities = 242/328 (73%), Positives = 273/328 (82%), Gaps = 16/328 (4%)
Query: 10 LRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVFSC---WNRMMSSQAAPEEEKKEEKA 66
+R LN R+G + A +R V NG+ S W RM+S+ E + KE+K
Sbjct: 4 MRGLLNGGRYGNRYI--WTAISLRHPEVMEGNGLESAVMQWRRMLSNAGGAEAQVKEQKE 61
Query: 67 EKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTKHH 126
EK+ +VS+YWGISRPKITREDG+EWPWNCFMPWETYR DLSIDL KHH
Sbjct: 62 EKKD-----------AMVSNYWGISRPKITREDGSEWPWNCFMPWETYRSDLSIDLKKHH 110
Query: 127 VPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQ 186
VP+ F+DK AYRTVK+LR+PTD+FFQRRYGCRAMMLETVAAVPGMVGGMLLHL+SLRK +
Sbjct: 111 VPRTFMDKFAYRTVKILRVPTDIFFQRRYGCRAMMLETVAAVPGMVGGMLLHLKSLRKLE 170
Query: 187 QSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAH 246
QSGGWIKALLEEAENERMHLMTMVELVQPKWYER LVL VQGVFFN+FFVLY+LSPK+AH
Sbjct: 171 QSGGWIKALLEEAENERMHLMTMVELVQPKWYERLLVLAVQGVFFNSFFVLYVLSPKLAH 230
Query: 247 RVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITVIRADEAH 306
R+VGYLEEEAIHSYTEYLKDI+SGAI+N+PAPAIAIDYWRLPKDATLKDVITV+RADEAH
Sbjct: 231 RIVGYLEEEAIHSYTEYLKDIDSGAIKNIPAPAIAIDYWRLPKDATLKDVITVVRADEAH 290
Query: 307 HRDVNHFASDIHFHGKELREAPAPLGYH 334
HRDVNHFASD+ GKELR+APAP+GYH
Sbjct: 291 HRDVNHFASDVQVQGKELRDAPAPVGYH 318
>emb|CAA55892.1| alternative oxidase [Mangifera indica]
Length = 274
Score = 487 bits (1253), Expect = e-136
Identities = 229/285 (80%), Positives = 255/285 (89%), Gaps = 11/285 (3%)
Query: 50 MMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFM 109
M+S+ E + KE+K EK+ +VS+YWGISRPKITREDG+EWPWNCFM
Sbjct: 1 MLSNAGGAEAQVKEQKEEKKD-----------AMVSNYWGISRPKITREDGSEWPWNCFM 49
Query: 110 PWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVP 169
PWETYR DLSIDL KHHVP+ F+DK AYRTVK+LR+PTD+FFQRRYGCRAMMLETVAAVP
Sbjct: 50 PWETYRSDLSIDLKKHHVPRTFMDKFAYRTVKILRVPTDIFFQRRYGCRAMMLETVAAVP 109
Query: 170 GMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGV 229
GMVGGMLLHL+SLRK +QSGGWIKALLEEAENERMHLMTMVELVQPKWYER LVL VQGV
Sbjct: 110 GMVGGMLLHLKSLRKLEQSGGWIKALLEEAENERMHLMTMVELVQPKWYERLLVLAVQGV 169
Query: 230 FFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPK 289
FFN+FFVLY+LSPK+AHR+VGYLEEEAIHSYTEYLKDI+SGAI+N+PAPAIAIDYWRLPK
Sbjct: 170 FFNSFFVLYVLSPKLAHRIVGYLEEEAIHSYTEYLKDIDSGAIKNIPAPAIAIDYWRLPK 229
Query: 290 DATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
DATLKDVITV+RADEAHHRDVNHFASD+ GKELR+APAP+GYH
Sbjct: 230 DATLKDVITVVRADEAHHRDVNHFASDVQVQGKELRDAPAPVGYH 274
>emb|CAD12835.1| putative alternative oxidase [Vigna unguiculata]
Length = 326
Score = 459 bits (1180), Expect = e-128
Identities = 225/335 (67%), Positives = 261/335 (77%), Gaps = 10/335 (2%)
Query: 1 MKHLALSYALRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVF-SCWNRMMSSQAAPEE 59
MKH + A R L R + R P + G F S + R MS+ ++
Sbjct: 1 MKHTLVRSAARALLGGGR---SYYRHAPTAAIVEPTRQHGGGAFGSFYLRRMSTLPDIKD 57
Query: 60 EKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLS 119
EEK + E K ++ + V+SSYWGISRPK+ REDGTEWPWNCFMPW+TY D+S
Sbjct: 58 HNSEEK------KNEVKDDNTNAVISSYWGISRPKVRREDGTEWPWNCFMPWDTYHSDVS 111
Query: 120 IDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHL 179
ID+TKHH PK+ DKVA+R+VK LR+ +D++F+ RYGC AMMLET+AAVPGMVGGMLLHL
Sbjct: 112 IDVTKHHTPKSLTDKVAFRSVKFLRVLSDLYFKERYGCHAMMLETIAAVPGMVGGMLLHL 171
Query: 180 RSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYL 239
+SLRKFQ SGGWIKALLEEAENERMHLMTMVELVQPKW+ER L+ T QGVFFNAFFV YL
Sbjct: 172 KSLRKFQHSGGWIKALLEEAENERMHLMTMVELVQPKWHERLLIFTAQGVFFNAFFVFYL 231
Query: 240 LSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITV 299
LSPK AHR VGYLEEEA+ SYT++L+ IESG +ENVPAPAIAIDYWRLPKDATLKDV+TV
Sbjct: 232 LSPKAAHRFVGYLEEEAVISYTQHLEAIESGKVENVPAPAIAIDYWRLPKDATLKDVVTV 291
Query: 300 IRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
IRADEAHHRDVNHFASDIH GKELR+APAP+GYH
Sbjct: 292 IRADEAHHRDVNHFASDIHHQGKELRDAPAPIGYH 326
>dbj|BAA22636.2| alternative oxidase [Arabidopsis thaliana]
gi|67633916|gb|AAY78882.1| mitochondrial alternative
oxidase 2 [Arabidopsis thaliana]
gi|30697967|ref|NP_201226.2| alternative oxidase 2,
mitochondrial (AOX2) [Arabidopsis thaliana]
gi|21264382|sp|O22049|AOX2_ARATH Alternative oxidase 2,
mitochondrial precursor
Length = 353
Score = 457 bits (1176), Expect = e-127
Identities = 219/288 (76%), Positives = 249/288 (86%), Gaps = 6/288 (2%)
Query: 49 RMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGI--SRPKITREDGTEWPWN 106
R M +A EKK+E + + + GSV V SYWGI ++ KITR+DG++WPWN
Sbjct: 70 RWMGMSSASAMEKKDENLTVK----KGQNGGGSVAVPSYWGIETAKMKITRKDGSDWPWN 125
Query: 107 CFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVA 166
CFMPWETY+ +LSIDL KHHVPKN DKVAYR VKLLRIPTD+FFQRRYGCRAMMLETVA
Sbjct: 126 CFMPWETYQANLSIDLKKHHVPKNIADKVAYRIVKLLRIPTDIFFQRRYGCRAMMLETVA 185
Query: 167 AVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTV 226
AVPGMVGGMLLHL+S+RKF+ SGGWIKALLEEAENERMHLMTM+ELV+PKWYER LV+ V
Sbjct: 186 AVPGMVGGMLLHLKSIRKFEHSGGWIKALLEEAENERMHLMTMMELVKPKWYERLLVMLV 245
Query: 227 QGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWR 286
QG+FFN+FFV Y++SP++AHRVVGYLEEEAIHSYTE+LKDI++G IENV APAIAIDYWR
Sbjct: 246 QGIFFNSFFVCYVISPRLAHRVVGYLEEEAIHSYTEFLKDIDNGKIENVAAPAIAIDYWR 305
Query: 287 LPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
LPKDATLKDV+TVIRADEAHHRDVNHFASDI GKELREA AP+GYH
Sbjct: 306 LPKDATLKDVVTVIRADEAHHRDVNHFASDIRNQGKELREAAAPIGYH 353
>dbj|BAB09852.1| alternative oxidase 2 [Arabidopsis thaliana]
Length = 282
Score = 456 bits (1172), Expect = e-127
Identities = 218/286 (76%), Positives = 248/286 (86%), Gaps = 6/286 (2%)
Query: 51 MSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSYWGI--SRPKITREDGTEWPWNCF 108
M +A EKK+E + + + GSV V SYWGI ++ KITR+DG++WPWNCF
Sbjct: 1 MGMSSASAMEKKDENLTVK----KGQNGGGSVAVPSYWGIETAKMKITRKDGSDWPWNCF 56
Query: 109 MPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAV 168
MPWETY+ +LSIDL KHHVPKN DKVAYR VKLLRIPTD+FFQRRYGCRAMMLETVAAV
Sbjct: 57 MPWETYQANLSIDLKKHHVPKNIADKVAYRIVKLLRIPTDIFFQRRYGCRAMMLETVAAV 116
Query: 169 PGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQG 228
PGMVGGMLLHL+S+RKF+ SGGWIKALLEEAENERMHLMTM+ELV+PKWYER LV+ VQG
Sbjct: 117 PGMVGGMLLHLKSIRKFEHSGGWIKALLEEAENERMHLMTMMELVKPKWYERLLVMLVQG 176
Query: 229 VFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLP 288
+FFN+FFV Y++SP++AHRVVGYLEEEAIHSYTE+LKDI++G IENV APAIAIDYWRLP
Sbjct: 177 IFFNSFFVCYVISPRLAHRVVGYLEEEAIHSYTEFLKDIDNGKIENVAAPAIAIDYWRLP 236
Query: 289 KDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
KDATLKDV+TVIRADEAHHRDVNHFASDI GKELREA AP+GYH
Sbjct: 237 KDATLKDVVTVIRADEAHHRDVNHFASDIRNQGKELREAAAPIGYH 282
>emb|CAD41813.2| OSJNBa0083N12.11 [Oryza sativa (japonica cultivar-group)]
gi|50928459|ref|XP_473757.1| OSJNBa0083N12.11 [Oryza
sativa (japonica cultivar-group)]
gi|3218544|dbj|BAA28772.1| alternative oxidase [Oryza
sativa (japonica cultivar-group)]
gi|6467126|dbj|BAA86963.1| alternative oxidase [Oryza
sativa] gi|3218546|dbj|BAA28773.1| alternative oxidase
[Oryza sativa (japonica cultivar-group)]
Length = 332
Score = 447 bits (1151), Expect = e-124
Identities = 219/308 (71%), Positives = 252/308 (81%), Gaps = 9/308 (2%)
Query: 29 ATEVRRFLVSGENGVFSCWN-RMMSSQAAPEEEKKEEKAEKESLRTEAKKNDGSVVVSSY 87
A R FL GE V W R+MS+ + E A+ E+ + +A+K VVV+SY
Sbjct: 32 AAAARPFLAGGE-AVPGVWGLRLMSTSSVASTEAA---AKAEAKKADAEKE---VVVNSY 84
Query: 88 WGISRPK-ITREDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIP 146
WGI + K + REDGTEW W+CF PWETY D SIDLTKHHVPK LDK+AY TVK LR P
Sbjct: 85 WGIEQSKKLVREDGTEWKWSCFRPWETYTADTSIDLTKHHVPKTLLDKIAYWTVKSLRFP 144
Query: 147 TDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHL 206
TD+FFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLR+F+QSGGWI+ LLEEAENERMHL
Sbjct: 145 TDIFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRRFEQSGGWIRTLLEEAENERMHL 204
Query: 207 MTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKD 266
MT +E+ PKWYER LV+TVQGVFFNA+F+ YLLSPK AHRVVGYLEEEAIHSYTE+LKD
Sbjct: 205 MTFMEVANPKWYERALVITVQGVFFNAYFLGYLLSPKFAHRVVGYLEEEAIHSYTEFLKD 264
Query: 267 IESGAIENVPAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELRE 326
+E+G I+NVPAPAIAIDYWRLP +ATLKDV+TV+RADEAHHRDVNHFASDIH+ G EL++
Sbjct: 265 LEAGKIDNVPAPAIAIDYWRLPANATLKDVVTVVRADEAHHRDVNHFASDIHYQGMELKQ 324
Query: 327 APAPLGYH 334
PAP+GYH
Sbjct: 325 TPAPIGYH 332
>gb|AAP68983.1| alternative oxidase 2b [Glycine max]
Length = 326
Score = 447 bits (1149), Expect = e-124
Identities = 218/335 (65%), Positives = 259/335 (77%), Gaps = 10/335 (2%)
Query: 1 MKHLALSYALRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWN-RMMSSQAAPEE 59
MK++ + A R L G + RQL + G F ++ R MS+ ++
Sbjct: 1 MKNVLVRSAARALLGGG--GRSYYRQLSTAAIVEQRHQHGGGAFGSFHLRRMSTLPEVKD 58
Query: 60 EKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLS 119
+ EEK + + + + VV+SYWGI+RPK+ REDGTEWPWNCFMPW++Y D+S
Sbjct: 59 QHSEEKKNEVN-------DTSNAVVTSYWGITRPKVRREDGTEWPWNCFMPWDSYHSDVS 111
Query: 120 IDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHL 179
ID+TKHH PK+ DKVA+R VK LR+ +D++F+ RYGC AMMLET+AAVPGMVGGMLLHL
Sbjct: 112 IDVTKHHTPKSLTDKVAFRAVKFLRVLSDIYFKERYGCHAMMLETIAAVPGMVGGMLLHL 171
Query: 180 RSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYL 239
+SLRKFQ SGGWIKALLEEAENERMHLMTMVELV+P W+ER L+ T QGVFFNAFFV YL
Sbjct: 172 KSLRKFQHSGGWIKALLEEAENERMHLMTMVELVKPSWHERLLIFTAQGVFFNAFFVFYL 231
Query: 240 LSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITV 299
LSPK AHR VGYLEEEA+ SYT++L IESG +ENVPAPAIAIDYWRLPKDATLKDV+TV
Sbjct: 232 LSPKAAHRFVGYLEEEAVISYTQHLNAIESGKVENVPAPAIAIDYWRLPKDATLKDVVTV 291
Query: 300 IRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
IRADEAHHRDVNHFASDIH GKEL+EAPAP+GYH
Sbjct: 292 IRADEAHHRDVNHFASDIHHQGKELKEAPAPIGYH 326
>gb|AAB97286.1| alternative oxidase 2b [Glycine max]
gi|3023305|sp|O03376|AOX3_SOYBN Alternative oxidase 3,
mitochondrial precursor
Length = 326
Score = 446 bits (1148), Expect = e-124
Identities = 219/335 (65%), Positives = 259/335 (76%), Gaps = 10/335 (2%)
Query: 1 MKHLALSYALRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWN-RMMSSQAAPEE 59
MK++ + A R L G + RQL + G F ++ R MS+ ++
Sbjct: 1 MKNVLVRSAARALLGGG--GRSYYRQLSTAAIVEQRHQHGGGAFGSFHLRRMSTLPEVKD 58
Query: 60 EKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLS 119
+ EEK + + + A VV+SYWGI+RPK+ REDGTEWPWNCFMPW++Y D+S
Sbjct: 59 QHSEEKKNEVNGTSNA-------VVTSYWGITRPKVRREDGTEWPWNCFMPWDSYHSDVS 111
Query: 120 IDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHL 179
ID+TKHH PK+ DKVA+R VK LR+ +D++F+ RYGC AMMLET+AAVPGMVGGMLLHL
Sbjct: 112 IDVTKHHTPKSLTDKVAFRAVKFLRVLSDIYFKERYGCHAMMLETIAAVPGMVGGMLLHL 171
Query: 180 RSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYL 239
+SLRKFQ SGGWIKALLEEAENERMHLMTMVELV+P W+ER L+ T QGVFFNAFFV YL
Sbjct: 172 KSLRKFQHSGGWIKALLEEAENERMHLMTMVELVKPSWHERLLIFTAQGVFFNAFFVFYL 231
Query: 240 LSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITV 299
LSPK AHR VGYLEEEA+ SYT++L IESG +ENVPAPAIAIDYWRLPKDATLKDV+TV
Sbjct: 232 LSPKAAHRFVGYLEEEAVISYTQHLNAIESGKVENVPAPAIAIDYWRLPKDATLKDVVTV 291
Query: 300 IRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
IRADEAHHRDVNHFASDIH GKEL+EAPAP+GYH
Sbjct: 292 IRADEAHHRDVNHFASDIHHQGKELKEAPAPIGYH 326
>gb|AAL27796.1| alternative oxidase AOX2 precursor [Zea mays]
gi|39984776|gb|AAR36136.1| alternative oxidase 1a [Zea
mays]
Length = 329
Score = 446 bits (1148), Expect = e-124
Identities = 225/316 (71%), Positives = 256/316 (80%), Gaps = 9/316 (2%)
Query: 22 TAVRQLPATEVRRFLVSGENGVFSCWNRMMS--SQAAPEEEKKEEKAEKESLRTEAKKND 79
TA PA R L G NGV + R+MS S AAP E K E A K S KK
Sbjct: 20 TAAAISPAAASRPLLAGG-NGVPAVMLRLMSTSSPAAPTEAKDE--AAKASKVGGDKK-- 74
Query: 80 GSVVVSSYWGISRP-KITREDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFLDKVAYR 138
+VV++SYWGI + K+ R+DGTEW W CF PWETY D SIDLT+HH PK +DKVAY
Sbjct: 75 -AVVINSYWGIEQNNKLARDDGTEWKWTCFRPWETYTADTSIDLTRHHEPKTLMDKVAYW 133
Query: 139 TVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEE 198
TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLR+F+QSGGWI+ALLEE
Sbjct: 134 TVKSLRFPTDIFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRRFEQSGGWIRALLEE 193
Query: 199 AENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIH 258
AENERMHLMT +E+ +P+WYER LV+TVQGVFFNA+F+ YLLSPK AHRVVGYLEEEAIH
Sbjct: 194 AENERMHLMTFMEVAKPRWYERALVITVQGVFFNAYFLGYLLSPKFAHRVVGYLEEEAIH 253
Query: 259 SYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIH 318
SYTEYLKD+E+G IENVPAPAIAIDYWRLP +ATLKDV+TV+RADEAHHRDVNHFASDIH
Sbjct: 254 SYTEYLKDLEAGKIENVPAPAIAIDYWRLPANATLKDVVTVVRADEAHHRDVNHFASDIH 313
Query: 319 FHGKELREAPAPLGYH 334
G +L+++PAP+GYH
Sbjct: 314 CQGMQLKQSPAPIGYH 329
>gb|AAU11467.1| mitochondrial alternative oxidase 1 [Saccharum officinarum]
Length = 331
Score = 445 bits (1144), Expect = e-123
Identities = 225/337 (66%), Positives = 260/337 (76%), Gaps = 10/337 (2%)
Query: 1 MKHLALSYALRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWNRMMS--SQAAPE 58
M A S LRRA TA PA R L G N V + R+MS S AA
Sbjct: 2 MSSRAGSVLLRRA---GSRLFTAAAVSPAAASRPLLAGGNNVVQAVMVRLMSTSSPAAVS 58
Query: 59 EEKKEEKAEKESLRTEAKKNDGSVVVSSYWGISRP-KITREDGTEWPWNCFMPWETYRPD 117
E K+E A+ + K +VV++SYWGI + K+ R+DGTEW W CF PWETY D
Sbjct: 59 EATKDEAAKASKEGGDKK----AVVINSYWGIEQNNKLVRDDGTEWKWTCFRPWETYTAD 114
Query: 118 LSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLL 177
SIDLT+HH K +DK+AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGMLL
Sbjct: 115 TSIDLTRHHELKTLMDKIAYWTVKSLRFPTDIFFQRRYGCRAMMLETVAAVPGMVGGMLL 174
Query: 178 HLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVL 237
HLRSLR+F+QSGGWI+ALLEEAENERMHLMT +E+ +P+WYER LV+TVQGVFFNA+F+
Sbjct: 175 HLRSLRRFEQSGGWIRALLEEAENERMHLMTFMEVAKPRWYERALVITVQGVFFNAYFLG 234
Query: 238 YLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVI 297
YLLSPK AHRVVGYLEEEAIHSYTEYLKD+E+G IENVPAPAIAIDYWRLP +ATLKDV+
Sbjct: 235 YLLSPKFAHRVVGYLEEEAIHSYTEYLKDVEAGKIENVPAPAIAIDYWRLPANATLKDVV 294
Query: 298 TVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
TV+RADEAHHRDVNHFASDIH G +L+++PAP+GYH
Sbjct: 295 TVVRADEAHHRDVNHFASDIHCQGMQLKQSPAPIGYH 331
>dbj|BAB88645.1| alternative oxidase [Triticum aestivum]
Length = 328
Score = 444 bits (1141), Expect = e-123
Identities = 223/335 (66%), Positives = 260/335 (77%), Gaps = 15/335 (4%)
Query: 4 LALSYALRRALNCNRHGLTAVRQLPATEVRRFLVSGENGVFSCWNRMMSSQAAPEEEKKE 63
+A S LRRA L A PA R + G G W RMMS+ AA + K+
Sbjct: 5 MAGSVLLRRA-GAGAGRLFATTASPAA---RTALGGGGGA---WVRMMSTSAA--SQVKD 55
Query: 64 EKAEKESLRTEAKKNDGS---VVVSSYWGISRPK-ITREDGTEWPWNCFMPWETYRPDLS 119
E A+ ++ EA K DG V +SSYWGI + K + REDGTEW W+CF PWETY D S
Sbjct: 56 EAAK--GVKAEAAKGDGEKKEVAISSYWGIDQSKKLVREDGTEWKWSCFRPWETYTADTS 113
Query: 120 IDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHL 179
IDLTKHHVP LDK+AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGMLLHL
Sbjct: 114 IDLTKHHVPNTMLDKIAYYTVKSLRFPTDIFFQRRYGCRAMMLETVAAVPGMVGGMLLHL 173
Query: 180 RSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYL 239
RSLR+F+QSGGWI+ALLEEAENERMHLMT +E+ QP+WYER LV+ VQGVFFNA+F YL
Sbjct: 174 RSLRRFEQSGGWIRALLEEAENERMHLMTFMEVAQPRWYERALVIAVQGVFFNAYFFGYL 233
Query: 240 LSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITV 299
+SPK AHRVVGYLEEEA+HSYTE+LKD++ G I+NVPAPAIAIDYWRLP +ATLKDV+TV
Sbjct: 234 ISPKFAHRVVGYLEEEAVHSYTEFLKDLDDGKIDNVPAPAIAIDYWRLPANATLKDVVTV 293
Query: 300 IRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
+RADEAHHRDVNHFASD+++ G +L+ PAP+GYH
Sbjct: 294 VRADEAHHRDVNHFASDVYYQGMQLKATPAPIGYH 328
>gb|AAA32870.1| oxidase
Length = 305
Score = 442 bits (1138), Expect = e-123
Identities = 204/279 (73%), Positives = 241/279 (86%), Gaps = 2/279 (0%)
Query: 58 EEEKKEEKAEKESLRTEAK--KNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYR 115
EE+ ++K E ES +A N G ++SYWG+ KIT+EDG+EW WNCF PWETY+
Sbjct: 27 EEDANQKKTENESTGGDAAGGNNKGDKGIASYWGVEPNKITKEDGSEWKWNCFRPWETYK 86
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGM 175
D++IDL KHHVP FLD++AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGM
Sbjct: 87 ADITIDLKKHHVPTTFLDRIAYWTVKSLRWPTDLFFQRRYGCRAMMLETVAAVPGMVGGM 146
Query: 176 LLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFF 235
LLH +SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +PKWYER LV+TVQGVFFNA+F
Sbjct: 147 LLHCKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPKWYERALVITVQGVFFNAYF 206
Query: 236 VLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKD 295
+ YL+SPK AHR+VGYLEEEAIHSYTE+LK+++ G IENVPAPAIAIDYWRLP DATL+D
Sbjct: 207 LGYLISPKFAHRMVGYLEEEAIHSYTEFLKELDKGNIENVPAPAIAIDYWRLPADATLRD 266
Query: 296 VITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+ V+RADEAHHRDVNHFASDIH+ G+EL+EAPAP+GYH
Sbjct: 267 VVMVVRADEAHHRDVNHFASDIHYQGRELKEAPAPIGYH 305
>gb|AAL15234.1| putative alternative oxidase 1a precursor [Arabidopsis thaliana]
gi|13877807|gb|AAK43981.1| putative alternative oxidase
1a precursor [Arabidopsis thaliana]
gi|2506083|dbj|BAA22625.1| alternative oxidase
[Arabidopsis thaliana] gi|9293872|dbj|BAB01775.1|
alternative oxidase 1a precursor [Arabidopsis thaliana]
gi|3915639|sp|Q39219|AOX1A_ARATH Alternative oxidase 1a,
mitochondrial precursor gi|15228734|ref|NP_188876.1|
alternative oxidase 1a, mitochondrial (AOX1A)
[Arabidopsis thaliana]
Length = 354
Score = 442 bits (1138), Expect = e-123
Identities = 204/279 (73%), Positives = 241/279 (86%), Gaps = 2/279 (0%)
Query: 58 EEEKKEEKAEKESLRTEAK--KNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYR 115
EE+ ++K E ES +A N G ++SYWG+ KIT+EDG+EW WNCF PWETY+
Sbjct: 76 EEDANQKKTENESTGGDAAGGNNKGDKGIASYWGVEPNKITKEDGSEWKWNCFRPWETYK 135
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGM 175
D++IDL KHHVP FLD++AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGM
Sbjct: 136 ADITIDLKKHHVPTTFLDRIAYWTVKSLRWPTDLFFQRRYGCRAMMLETVAAVPGMVGGM 195
Query: 176 LLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFF 235
LLH +SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +PKWYER LV+TVQGVFFNA+F
Sbjct: 196 LLHCKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPKWYERALVITVQGVFFNAYF 255
Query: 236 VLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKD 295
+ YL+SPK AHR+VGYLEEEAIHSYTE+LK+++ G IENVPAPAIAIDYWRLP DATL+D
Sbjct: 256 LGYLISPKFAHRMVGYLEEEAIHSYTEFLKELDKGNIENVPAPAIAIDYWRLPADATLRD 315
Query: 296 VITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+ V+RADEAHHRDVNHFASDIH+ G+EL+EAPAP+GYH
Sbjct: 316 VVMVVRADEAHHRDVNHFASDIHYQGRELKEAPAPIGYH 354
>emb|CAA10364.1| alternative oxidase [Arabidopsis thaliana] gi|11278947|pir||T51615
alternative respiratory pathway oxidase (EC 1.1.3.-)
[similarity] - Arabidopsis thaliana
Length = 353
Score = 442 bits (1138), Expect = e-123
Identities = 204/279 (73%), Positives = 241/279 (86%), Gaps = 2/279 (0%)
Query: 58 EEEKKEEKAEKESLRTEAK--KNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYR 115
EE+ ++K E ES +A N G ++SYWG+ KIT+EDG+EW WNCF PWETY+
Sbjct: 75 EEDANQKKTENESTGGDAAGGNNKGDKGIASYWGVEPNKITKEDGSEWKWNCFRPWETYK 134
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGM 175
D++IDL KHHVP FLD++AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGM
Sbjct: 135 ADITIDLKKHHVPTTFLDRIAYWTVKSLRWPTDLFFQRRYGCRAMMLETVAAVPGMVGGM 194
Query: 176 LLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFF 235
LLH +SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +PKWYER LV+TVQGVFFNA+F
Sbjct: 195 LLHCKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPKWYERALVITVQGVFFNAYF 254
Query: 236 VLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKD 295
+ YL+SPK AHR+VGYLEEEAIHSYTE+LK+++ G IENVPAPAIAIDYWRLP DATL+D
Sbjct: 255 LGYLISPKFAHRMVGYLEEEAIHSYTEFLKELDKGNIENVPAPAIAIDYWRLPADATLRD 314
Query: 296 VITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+ V+RADEAHHRDVNHFASDIH+ G+EL+EAPAP+GYH
Sbjct: 315 VVMVVRADEAHHRDVNHFASDIHYQGRELKEAPAPIGYH 353
>gb|AAB49302.1| alternative oxidase [Arabidopsis thaliana]
Length = 287
Score = 442 bits (1138), Expect = e-123
Identities = 204/279 (73%), Positives = 241/279 (86%), Gaps = 2/279 (0%)
Query: 58 EEEKKEEKAEKESLRTEAK--KNDGSVVVSSYWGISRPKITREDGTEWPWNCFMPWETYR 115
EE+ ++K E ES +A N G ++SYWG+ KIT+EDG+EW WNCF PWETY+
Sbjct: 9 EEDANQKKTENESTGGDAAGGNNKGDKGIASYWGVEPNKITKEDGSEWKWNCFRPWETYK 68
Query: 116 PDLSIDLTKHHVPKNFLDKVAYRTVKLLRIPTDVFFQRRYGCRAMMLETVAAVPGMVGGM 175
D++IDL KHHVP FLD++AY TVK LR PTD+FFQRRYGCRAMMLETVAAVPGMVGGM
Sbjct: 69 ADITIDLKKHHVPTTFLDRIAYWTVKSLRWPTDLFFQRRYGCRAMMLETVAAVPGMVGGM 128
Query: 176 LLHLRSLRKFQQSGGWIKALLEEAENERMHLMTMVELVQPKWYERFLVLTVQGVFFNAFF 235
LLH +SLR+F+QSGGWIKALLEEAENERMHLMT +E+ +PKWYER LV+TVQGVFFNA+F
Sbjct: 129 LLHCKSLRRFEQSGGWIKALLEEAENERMHLMTFMEVAKPKWYERALVITVQGVFFNAYF 188
Query: 236 VLYLLSPKVAHRVVGYLEEEAIHSYTEYLKDIESGAIENVPAPAIAIDYWRLPKDATLKD 295
+ YL+SPK AHR+VGYLEEEAIHSYTE+LK+++ G IENVPAPAIAIDYWRLP DATL+D
Sbjct: 189 LGYLISPKFAHRMVGYLEEEAIHSYTEFLKELDKGNIENVPAPAIAIDYWRLPADATLRD 248
Query: 296 VITVIRADEAHHRDVNHFASDIHFHGKELREAPAPLGYH 334
V+ V+RADEAHHRDVNHFASDIH+ G+EL+EAPAP+GYH
Sbjct: 249 VVMVVRADEAHHRDVNHFASDIHYQGRELKEAPAPIGYH 287
>ref|XP_467319.1| alternative oxidase 1c [Oryza sativa (japonica cultivar-group)]
gi|41052978|dbj|BAD07888.1| alternative oxidase 1c
[Oryza sativa (japonica cultivar-group)]
gi|41052670|dbj|BAD07517.1| alternative oxidase 1c
[Oryza sativa (japonica cultivar-group)]
gi|16902308|dbj|BAB71945.1| alternative oxidase 1c
[Oryza sativa (japonica cultivar-group)]
gi|16902306|dbj|BAB71944.1| alternative oxidase 1c
[Oryza sativa (japonica cultivar-group)]
Length = 345
Score = 437 bits (1123), Expect = e-121
Identities = 211/311 (67%), Positives = 242/311 (76%), Gaps = 12/311 (3%)
Query: 36 LVSGENGVFSCWNRMMSSQAAPEEEK----KEEKAEKESLRTEAKKN--------DGSVV 83
L G + W R++S+ AA +E+ KE + + EA K S V
Sbjct: 35 LAGAGGGGVALWARLLSTSAAAAKEETAASKENTGSTAAAKAEATKAAKEGPASATASPV 94
Query: 84 VSSYWGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLL 143
SSYWGI K+ +DG EW W+CF PWETY PD +IDL KHH PK LDKVAY TVK L
Sbjct: 95 ASSYWGIEASKLASKDGVEWKWSCFRPWETYSPDTTIDLKKHHEPKVLLDKVAYWTVKAL 154
Query: 144 RIPTDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENER 203
R+PTD+FFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLR+F+ SGGWI+ALLEEAENER
Sbjct: 155 RVPTDIFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRRFEHSGGWIRALLEEAENER 214
Query: 204 MHLMTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEY 263
MHLMT +E+ +P+WYER LVL VQGVFFNA+F+ YLLSPK+AHRVVGYLEEEAIHSYTEY
Sbjct: 215 MHLMTFMEVAKPRWYERALVLAVQGVFFNAYFLGYLLSPKLAHRVVGYLEEEAIHSYTEY 274
Query: 264 LKDIESGAIENVPAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKE 323
LKDIE+G IENVPAP IAIDYWRLP ATLKDV+ V+RADEAHHRDVNHFASD+HF G +
Sbjct: 275 LKDIEAGKIENVPAPPIAIDYWRLPAGATLKDVVVVVRADEAHHRDVNHFASDVHFQGMD 334
Query: 324 LREAPAPLGYH 334
L++ PAPL YH
Sbjct: 335 LKDIPAPLDYH 345
>gb|AAL27795.1| alternative oxidase AOX1 precursor [Zea mays]
Length = 347
Score = 436 bits (1121), Expect = e-121
Identities = 209/308 (67%), Positives = 244/308 (78%), Gaps = 12/308 (3%)
Query: 39 GENGVFSCWNRMMSSQAAPEEEKKEEKAEKESLRTEA----------KKNDGS--VVVSS 86
GE G W R+ + E K+E A K + + A K+ DG VVSS
Sbjct: 40 GERGGALVWVRVRLLSTSAAEAKEEVAASKGNSGSTAAAKAEAVEAAKEGDGKRDKVVSS 99
Query: 87 YWGISRPKITREDGTEWPWNCFMPWETYRPDLSIDLTKHHVPKNFLDKVAYRTVKLLRIP 146
YWG++ K+ +DG EW W+CF PWE Y+PD +IDL +HH PK LDK+AY TVKLLR+P
Sbjct: 100 YWGVAPSKLMNKDGAEWRWSCFRPWEAYKPDTTIDLNRHHEPKVLLDKIAYWTVKLLRVP 159
Query: 147 TDVFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRKFQQSGGWIKALLEEAENERMHL 206
TD+FFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLR+F+ SGGWI+ALLEEAENERMHL
Sbjct: 160 TDIFFQRRYGCRAMMLETVAAVPGMVGGMLLHLRSLRRFEHSGGWIRALLEEAENERMHL 219
Query: 207 MTMVELVQPKWYERFLVLTVQGVFFNAFFVLYLLSPKVAHRVVGYLEEEAIHSYTEYLKD 266
MT +E+ +PKWYER LVL VQGVFFNA+F+ YL+SPK AHRVVGYLEEEAIHSYTEYLKD
Sbjct: 220 MTFMEVAKPKWYERALVLAVQGVFFNAYFLGYLISPKFAHRVVGYLEEEAIHSYTEYLKD 279
Query: 267 IESGAIENVPAPAIAIDYWRLPKDATLKDVITVIRADEAHHRDVNHFASDIHFHGKELRE 326
+E+G IENVPAPAIAIDYW+LP DATLKDV+ V+R+DEAHHRDVNHFASDIHF G +L+E
Sbjct: 280 LEAGKIENVPAPAIAIDYWQLPADATLKDVVVVVRSDEAHHRDVNHFASDIHFQGMQLKE 339
Query: 327 APAPLGYH 334
PAP+ YH
Sbjct: 340 TPAPIEYH 347
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.136 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 575,113,144
Number of Sequences: 2540612
Number of extensions: 24121302
Number of successful extensions: 86373
Number of sequences better than 10.0: 160
Number of HSP's better than 10.0 without gapping: 146
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 86064
Number of HSP's gapped (non-prelim): 164
length of query: 334
length of database: 863,360,394
effective HSP length: 128
effective length of query: 206
effective length of database: 538,162,058
effective search space: 110861383948
effective search space used: 110861383948
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0182.2


