

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0178.14
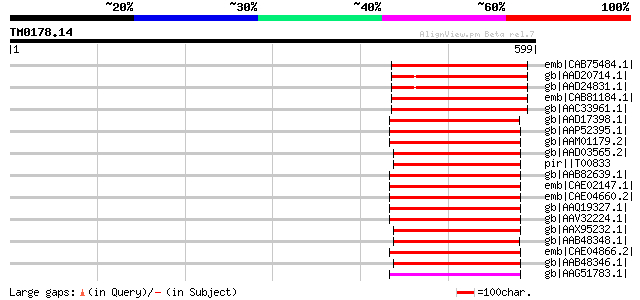
(599 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB75484.1| putative protein [Arabidopsis thaliana] gi|11357... 150 9e-35
gb|AAD20714.1| putative non-LTR retroelement reverse transcripta... 149 3e-34
gb|AAD24831.1| putative non-LTR retroelement reverse transcripta... 149 3e-34
emb|CAB81184.1| putative protein [Arabidopsis thaliana] gi|45393... 149 3e-34
gb|AAC33961.1| contains similarity to reverse trancriptase (Pfam... 149 3e-34
gb|AAD17398.1| putative non-LTR retroelement reverse transcripta... 147 1e-33
gb|AAP52395.1| putative retroelement [Oryza sativa (japonica cul... 147 1e-33
gb|AAM01179.2| Putative retroelement [Oryza sativa (japonica cul... 147 1e-33
gb|AAD03565.2| putative non-LTR retroelement reverse transcripta... 145 3e-33
pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabi... 145 3e-33
gb|AAB82639.1| putative non-LTR retroelement reverse transcripta... 144 6e-33
emb|CAE02147.1| OSJNBa0081G05.2 [Oryza sativa (japonica cultivar... 142 4e-32
emb|CAE04660.2| OSJNBa0061G20.16 [Oryza sativa (japonica cultiva... 142 4e-32
gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultiva... 140 1e-31
gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cult... 139 3e-31
gb|AAX95232.1| retrotransposon protein, putative, unclassified [... 138 4e-31
gb|AAB48348.1| reverse transcriptase 137 1e-30
emb|CAE04866.2| OSJNBa0086O06.14 [Oryza sativa (japonica cultiva... 135 5e-30
gb|AAB48346.1| reverse transcriptase 134 8e-30
gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Ara... 134 1e-29
>emb|CAB75484.1| putative protein [Arabidopsis thaliana] gi|11357921|pir||T47495
hypothetical protein F9K21.130 - Arabidopsis thaliana
Length = 851
Score = 150 bits (380), Expect = 9e-35
Identities = 70/155 (45%), Positives = 102/155 (65%)
Query: 436 KFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITH 495
K GPDGL FYK WD V + + FF + H+ + +N T + +IPK+ NP++++
Sbjct: 65 KAPGPDGLTARFYKQCWDIVGNDVIKEVKLFFESSHMKTSVNHTNICMIPKIQNPQTLSD 124
Query: 496 YRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRK 555
YRPI+ CN YKVIS+ +V RLK LN +++ +Q+AFI ++I DN+MIA E H++ +
Sbjct: 125 YRPIALCNVLYKVISKCMVNRLKAHLNSIVSDSQAAFIPGRIINDNVMIAHEIMHSLKVR 184
Query: 556 GRASKEHVALKLDMSKAYERVEWSFLEKNPPCFRF 590
R SK ++A+K D+SKAY+RVEW FLE F F
Sbjct: 185 KRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGF 219
>gb|AAD20714.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25412331|pir||G84649 hypothetical protein
At2g25550 [imported] - Arabidopsis thaliana
Length = 1750
Score = 149 bits (376), Expect = 3e-34
Identities = 74/156 (47%), Positives = 103/156 (65%), Gaps = 2/156 (1%)
Query: 436 KFEGPDGLNGLFYKNHWDTV-HEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESIT 494
K GPDGL FYKN WD V ++V E + FF T + IN T + +IPK+ NP +++
Sbjct: 826 KAPGPDGLTARFYKNCWDIVGYDVILEV-KKFFETSFMKPSINHTNICMIPKITNPTTLS 884
Query: 495 HYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILR 554
YRPI+ CN YKVIS+ +V RLK LN +++ +Q+AFI ++I DN+MIA E H++
Sbjct: 885 DYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKV 944
Query: 555 KGRASKEHVALKLDMSKAYERVEWSFLEKNPPCFRF 590
+ R SK ++A+K D+SKAY+RVEW FLE F F
Sbjct: 945 RKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGF 980
>gb|AAD24831.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25408166|pir||G84721 hypothetical protein
At2g31520 [imported] - Arabidopsis thaliana
Length = 1524
Score = 149 bits (376), Expect = 3e-34
Identities = 74/156 (47%), Positives = 103/156 (65%), Gaps = 2/156 (1%)
Query: 436 KFEGPDGLNGLFYKNHWDTV-HEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESIT 494
K GPDGL FYKN WD V ++V E + FF T + IN T + +IPK+ NP +++
Sbjct: 600 KAPGPDGLTARFYKNCWDIVGYDVILEV-KKFFETSFMKPSINHTNICMIPKITNPTTLS 658
Query: 495 HYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILR 554
YRPI+ CN YKVIS+ +V RLK LN +++ +Q+AFI ++I DN+MIA E H++
Sbjct: 659 DYRPIALCNVLYKVISKCLVNRLKSHLNSIVSDSQAAFIPGRIINDNVMIAHEVMHSLKV 718
Query: 555 KGRASKEHVALKLDMSKAYERVEWSFLEKNPPCFRF 590
+ R SK ++A+K D+SKAY+RVEW FLE F F
Sbjct: 719 RKRVSKTYMAVKTDVSKAYDRVEWDFLETTMRLFGF 754
>emb|CAB81184.1| putative protein [Arabidopsis thaliana] gi|4539357|emb|CAB40051.1|
putative protein [Arabidopsis thaliana]
gi|7486147|pir||T04278 hypothetical protein F25I24.40 -
Arabidopsis thaliana
Length = 1294
Score = 149 bits (375), Expect = 3e-34
Identities = 68/155 (43%), Positives = 104/155 (66%)
Query: 436 KFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITH 495
K GPDGL FYK+ W+ V + + FF T ++ IN T + +IPK+ NPE+++
Sbjct: 806 KAPGPDGLTARFYKSCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSD 865
Query: 496 YRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRK 555
YRPI+ CN YK+IS+ +V RLKG L+ +++ +Q+AFI +++ DN+MIA E H++ +
Sbjct: 866 YRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTR 925
Query: 556 GRASKEHVALKLDMSKAYERVEWSFLEKNPPCFRF 590
R S+ ++A+K D+SKAY+RVEW+FLE F F
Sbjct: 926 KRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGF 960
>gb|AAC33961.1| contains similarity to reverse trancriptase (Pfam: rvt.hmm, score:
42.57) [Arabidopsis thaliana] gi|7486711|pir||T01893
hypothetical protein F8M12.22 - Arabidopsis thaliana
Length = 1662
Score = 149 bits (375), Expect = 3e-34
Identities = 68/155 (43%), Positives = 104/155 (66%)
Query: 436 KFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITH 495
K GPDGL FYK+ W+ V + + FF T ++ IN T + +IPK+ NPE+++
Sbjct: 826 KAPGPDGLTARFYKSCWEIVGPDVIKEVKIFFRTSYMKQSINHTNICMIPKITNPETLSD 885
Query: 496 YRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRK 555
YRPI+ CN YK+IS+ +V RLKG L+ +++ +Q+AFI +++ DN+MIA E H++ +
Sbjct: 886 YRPIALCNVLYKIISKCLVERLKGHLDAIVSDSQAAFIPGRLVNDNVMIAHEMMHSLKTR 945
Query: 556 GRASKEHVALKLDMSKAYERVEWSFLEKNPPCFRF 590
R S+ ++A+K D+SKAY+RVEW+FLE F F
Sbjct: 946 KRVSQSYMAVKTDVSKAYDRVEWNFLETTMRLFGF 980
>gb|AAD17398.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411644|pir||C84530 hypothetical protein
At2g15540 [imported] - Arabidopsis thaliana
Length = 1225
Score = 147 bits (371), Expect = 1e-33
Identities = 69/148 (46%), Positives = 98/148 (65%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDG+ FY+++WD + ++F STG ++NET + LIPK P +
Sbjct: 389 PDKAPGPDGMTAFFYQHYWDLTGPDLIKLVQNFHSTGFFDERLNETNICLIPKTERPRKM 448
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
+RPIS CN +YKVIS+++ +RLK L +LI+ QSAF+ ++I DNI+IAQE FH +
Sbjct: 449 AEFRPISLCNVSYKVISKVLSSRLKRLLPELISETQSAFVAERLITDNILIAQENFHALR 508
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFL 581
K+++A+K DMSKAY+RVEWSFL
Sbjct: 509 TNPACKKKYMAIKTDMSKAYDRVEWSFL 536
>gb|AAP52395.1| putative retroelement [Oryza sativa (japonica cultivar-group)]
gi|37531612|ref|NP_920108.1| putative retroelement [Oryza
sativa (japonica cultivar-group)]
Length = 1694
Score = 147 bits (370), Expect = 1e-33
Identities = 68/150 (45%), Positives = 97/150 (64%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDG G FY+ +W + + A + FFS G +PS +NET + LIPK P+ +
Sbjct: 1056 PLKAPGPDGFPGRFYQRNWAILKDDIVRAVQEFFSLGTMPSGVNETAIVLIPKTEQPQEL 1115
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
+RPIS CN YK++S+ +V RL+ L+ L++ NQSAF+ ++I DN +IA E FH+I
Sbjct: 1116 KDFRPISLCNVVYKIVSKCLVNRLRPILDDLVSQNQSAFVPGRLITDNALIAFEYFHHIQ 1175
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLEK 583
R + A KLD+SKAY+RV+W FLE+
Sbjct: 1176 RNKNPENAYSAYKLDLSKAYDRVDWEFLEQ 1205
>gb|AAM01179.2| Putative retroelement [Oryza sativa (japonica cultivar-group)]
Length = 1888
Score = 147 bits (370), Expect = 1e-33
Identities = 68/150 (45%), Positives = 97/150 (64%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDG G FY+ +W + + A + FFS G +PS +NET + LIPK P+ +
Sbjct: 1013 PLKAPGPDGFPGRFYQRNWAILKDDIVRAVQEFFSLGTMPSGVNETAIVLIPKTEQPQEL 1072
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
+RPIS CN YK++S+ +V RL+ L+ L++ NQSAF+ ++I DN +IA E FH+I
Sbjct: 1073 KDFRPISLCNVVYKIVSKCLVNRLRPILDDLVSQNQSAFVPGRLITDNALIAFEYFHHIQ 1132
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLEK 583
R + A KLD+SKAY+RV+W FLE+
Sbjct: 1133 RNKNPENAYSAYKLDLSKAYDRVDWEFLEQ 1162
>gb|AAD03565.2| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411819|pir||H84557 hypothetical protein
At2g17910 [imported] - Arabidopsis thaliana
Length = 1344
Score = 145 bits (367), Expect = 3e-33
Identities = 68/144 (47%), Positives = 92/144 (63%)
Query: 439 GPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITHYRP 498
GPDG LF++ HWD V FF TG LP N T + LIPK+ +P+ ++ RP
Sbjct: 425 GPDGFTALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRP 484
Query: 499 ISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRKGRA 558
IS C+ YK+IS+I+ RLK L +++ QSAF+ ++I DNI++A E H++ R
Sbjct: 485 ISLCSVLYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRI 544
Query: 559 SKEHVALKLDMSKAYERVEWSFLE 582
SKEH+A K DMSKAY+RVEW FLE
Sbjct: 545 SKEHMAFKTDMSKAYDRVEWPFLE 568
>pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabidopsis thaliana
(fragment)
Length = 1365
Score = 145 bits (367), Expect = 3e-33
Identities = 68/144 (47%), Positives = 92/144 (63%)
Query: 439 GPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITHYRP 498
GPDG LF++ HWD V FF TG LP N T + LIPK+ +P+ ++ RP
Sbjct: 446 GPDGFTALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRP 505
Query: 499 ISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRKGRA 558
IS C+ YK+IS+I+ RLK L +++ QSAF+ ++I DNI++A E H++ R
Sbjct: 506 ISLCSVLYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRI 565
Query: 559 SKEHVALKLDMSKAYERVEWSFLE 582
SKEH+A K DMSKAY+RVEW FLE
Sbjct: 566 SKEHMAFKTDMSKAYDRVEWPFLE 589
>gb|AAB82639.1| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25408936|pir||A84888 hypothetical protein
At2g45230 [imported] - Arabidopsis thaliana
Length = 1374
Score = 144 bits (364), Expect = 6e-33
Identities = 67/150 (44%), Positives = 98/150 (64%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDG+NG Y+ W+T+ + E ++FF +G + +N+T + LIPK+ E +
Sbjct: 442 PHKCPGPDGMNGFLYQQFWETMGDQITEMVQAFFRSGSIEEGMNKTNICLIPKILKAEKM 501
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
T +RPIS CN YKVI +++ RLK L LI+ Q+AF+ ++I DNI+IA E H +
Sbjct: 502 TDFRPISLCNVIYKVIGKLMANRLKKILPSLISETQAAFVKGRLISDNILIAHELLHALS 561
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLEK 583
+ S+E +A+K D+SKAY+RVEW FLEK
Sbjct: 562 SNNKCSEEFIAIKTDISKAYDRVEWPFLEK 591
>emb|CAE02147.1| OSJNBa0081G05.2 [Oryza sativa (japonica cultivar-group)]
gi|50923489|ref|XP_472105.1| OSJNBa0081G05.2 [Oryza
sativa (japonica cultivar-group)]
Length = 1711
Score = 142 bits (357), Expect = 4e-32
Identities = 68/149 (45%), Positives = 95/149 (63%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDG FY+ +W T+ A R+FF +G +P +N+T + LIPK P +
Sbjct: 994 PLKSPGPDGFPARFYQRNWGTLKSDIILAVRNFFQSGLMPEGVNDTAIVLIPKKDQPIDL 1053
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
YRPIS CN YKV+S+ +V RL+ L+ L++ QSAFI +MI DN ++A E FH+I
Sbjct: 1054 KDYRPISLCNVVYKVVSKCLVNRLRPILDDLVSKEQSAFIQGRMITDNALLAFECFHSIQ 1113
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLE 582
+ +A+ A KLD+SKAY+RV+W FLE
Sbjct: 1114 KNKKANSAACAYKLDLSKAYDRVDWRFLE 1142
>emb|CAE04660.2| OSJNBa0061G20.16 [Oryza sativa (japonica cultivar-group)]
Length = 1285
Score = 142 bits (357), Expect = 4e-32
Identities = 68/149 (45%), Positives = 95/149 (63%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDG FY+ +W T+ A R+FF +G +P +N+T + LIPK P +
Sbjct: 994 PLKSPGPDGFPARFYQRNWGTLKSDIILAVRNFFQSGLMPEGVNDTAIVLIPKKDQPIDL 1053
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
YRPIS CN YKV+S+ +V RL+ L+ L++ QSAFI +MI DN ++A E FH+I
Sbjct: 1054 KDYRPISLCNVVYKVVSKCLVNRLRPILDDLVSKEQSAFIQGRMITDNALLAFECFHSIQ 1113
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLE 582
+ +A+ A KLD+SKAY+RV+W FLE
Sbjct: 1114 KNKKANSAACAYKLDLSKAYDRVDWRFLE 1142
>gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultivar-group)]
Length = 2367
Score = 140 bits (353), Expect = 1e-31
Identities = 66/149 (44%), Positives = 94/149 (62%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDG FY+ +W T+ A R FF TG +P +N+T + LIPK P +
Sbjct: 1176 PLKSPGPDGFPARFYQRNWGTIKADIIGAVRRFFQTGLMPEGVNDTAIVLIPKKEQPVDL 1235
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
+RPIS CN YKV+S+ +V RL+ L+ L+++ QSAF+ +MI DN ++A E FH +
Sbjct: 1236 RDFRPISLCNVVYKVVSKCLVNRLRPILDDLVSVEQSAFVQGRMITDNALLAFECFHAMQ 1295
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLE 582
+ +A+ A KLD+SKAY+RV+W FLE
Sbjct: 1296 KNKKANHAACAYKLDLSKAYDRVDWRFLE 1324
>gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|45267888|gb|AAS55787.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 1936
Score = 139 bits (349), Expect = 3e-31
Identities = 67/149 (44%), Positives = 94/149 (62%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K PDG FY+ +W T+ A R+FF +G +P +N+T + LIPK P +
Sbjct: 1082 PLKSPRPDGFPARFYQRNWGTLKSDIILAVRNFFQSGLMPKGVNDTAIVLIPKKDQPIDL 1141
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
YRPIS CN YKV+S+ +V RL+ L+ L++ QSAFI +MI DN ++A E FH+I
Sbjct: 1142 KDYRPISLCNVVYKVVSKCLVNRLRPILDDLVSKEQSAFIQGRMITDNALLAFECFHSIQ 1201
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLE 582
+ +A+ A KLD+SKAY+RV+W FLE
Sbjct: 1202 KNKKANSAACAYKLDLSKAYDRVDWRFLE 1230
>gb|AAX95232.1| retrotransposon protein, putative, unclassified [Oryza sativa
(japonica cultivar-group)] gi|62733010|gb|AAX95129.1|
retrotransposon protein, putative, unclassified [Oryza
sativa (japonica cultivar-group)]
Length = 1324
Score = 138 bits (348), Expect = 4e-31
Identities = 69/145 (47%), Positives = 90/145 (61%)
Query: 439 GPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITHYRP 498
G DG+ LFYK W+ V + + F G +P N+TVV LIPKVPNPE I RP
Sbjct: 904 GSDGMPALFYKQFWECVGDDIVHEVKDFLGGGEMPDSWNDTVVVLIPKVPNPERIKDLRP 963
Query: 499 ISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRKGRA 558
IS CN YK+ S+++ RLK L+++I+ Q AF+ +MI DNI++A E H + K R
Sbjct: 964 ISLCNVVYKIASKVLANRLKPLLSEIISPIQIAFVPQRMITDNILLAYELTHFLKTKRRG 1023
Query: 559 SKEHVALKLDMSKAYERVEWSFLEK 583
S A+K MSKAY+RVEW FLEK
Sbjct: 1024 SVGFAAVKQGMSKAYDRVEWGFLEK 1048
>gb|AAB48348.1| reverse transcriptase
Length = 199
Score = 137 bits (345), Expect = 1e-30
Identities = 65/143 (45%), Positives = 92/143 (63%)
Query: 439 GPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITHYRP 498
GPDG+ LFY+ WD + ++F S+ ++NET + LIPK+ P + +RP
Sbjct: 3 GPDGMTALFYQRFWDLTGPDLVKVVQNFHSSASFDERLNETNICLIPKIDRPRRMAEFRP 62
Query: 499 ISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRKGRA 558
IS CN +YKVIS+++ +RLK L LI+ QSAF+ ++I DNI++AQE FH +
Sbjct: 63 ISLCNVSYKVISKVLSSRLKKILPDLISETQSAFVAGRLITDNILVAQENFHALRTNPAC 122
Query: 559 SKEHVALKLDMSKAYERVEWSFL 581
K+ +A+K DMSKAY RVEWSFL
Sbjct: 123 RKKFMAIKTDMSKAYGRVEWSFL 145
>emb|CAE04866.2| OSJNBa0086O06.14 [Oryza sativa (japonica cultivar-group)]
gi|50928373|ref|XP_473714.1| OSJNBa0086O06.14 [Oryza
sativa (japonica cultivar-group)]
Length = 1205
Score = 135 bits (339), Expect = 5e-30
Identities = 63/149 (42%), Positives = 94/149 (62%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDG F++ +W + E R FF TG +N+TV+ +IPK P +
Sbjct: 430 PLKAPGPDGFPARFFQRNWGVLKRDVIEGVREFFETGEWKEGMNDTVIVMIPKTNAPVEM 489
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
+RP+S CN YKV+++ +V RL+ L ++I+ QSAF+ +MI DN ++A E FH+I
Sbjct: 490 KDFRPVSLCNVIYKVVAKCLVNRLRPLLQEIISETQSAFVPGRMITDNALVAFECFHSIH 549
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLE 582
+ R S++ ALKLD+SKAY+RV+W FL+
Sbjct: 550 KCTRESQDFCALKLDLSKAYDRVDWGFLD 578
>gb|AAB48346.1| reverse transcriptase
Length = 199
Score = 134 bits (337), Expect = 8e-30
Identities = 61/145 (42%), Positives = 96/145 (66%)
Query: 439 GPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESITHYRP 498
GPDG FY + WD + C R FF + + ++IN+T + LIPK+ +P+ ++ YRP
Sbjct: 3 GPDGXXAAFYHHLWDLIGNDVCLMVRHFFESDVMDNQINQTQICLIPKIIDPKHMSDYRP 62
Query: 499 ISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNILRKGRA 558
IS C +YK+IS+I++ RLK L +I+ +Q+AF+ + I DN+++A E H++ +
Sbjct: 63 ISLCTASYKIISKILIKRLKQCLGDVISDSQAAFVPGRNISDNVLVAHELLHSLKSRREC 122
Query: 559 SKEHVALKLDMSKAYERVEWSFLEK 583
+VA+K D+SKAY+RVEW+FLEK
Sbjct: 123 QSGYVAVKTDISKAYDRVEWNFLEK 147
>gb|AAG51783.1| reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
gi|25405244|pir||E96519 probable reverse transcriptase,
16838-20266 [imported] - Arabidopsis thaliana
Length = 1142
Score = 134 bits (336), Expect = 1e-29
Identities = 65/149 (43%), Positives = 88/149 (58%)
Query: 434 PRKFEGPDGLNGLFYKNHWDTVHEVFCEAARSFFSTGHLPSKINETVVTLIPKVPNPESI 493
P K GPDG+ LF++ W + SF G ++N T + LIPK P +
Sbjct: 226 PEKAPGPDGMTALFFQKSWAIIKSDLLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRM 285
Query: 494 THYRPISCCNFTYKVISRIIVTRLKGGLNQLITLNQSAFIGVKMIQDNIMIAQEAFHNIL 553
T RPIS CN YKVIS+I+ RLK L LI+ QSAF+ ++I DNI+IAQE FH +
Sbjct: 286 TELRPISLCNVGYKVISKILCQRLKTVLPNLISETQSAFVDGRLISDNILIAQEMFHGLR 345
Query: 554 RKGRASKEHVALKLDMSKAYERVEWSFLE 582
+ +A+K DMSKAY++VEW+F+E
Sbjct: 346 TNSSCKDKFMAIKTDMSKAYDQVEWNFIE 374
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.350 0.155 0.548
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 888,989,483
Number of Sequences: 2540612
Number of extensions: 32593485
Number of successful extensions: 113263
Number of sequences better than 10.0: 644
Number of HSP's better than 10.0 without gapping: 423
Number of HSP's successfully gapped in prelim test: 221
Number of HSP's that attempted gapping in prelim test: 112251
Number of HSP's gapped (non-prelim): 764
length of query: 599
length of database: 863,360,394
effective HSP length: 134
effective length of query: 465
effective length of database: 522,918,386
effective search space: 243157049490
effective search space used: 243157049490
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0178.14


