

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.13
(515 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
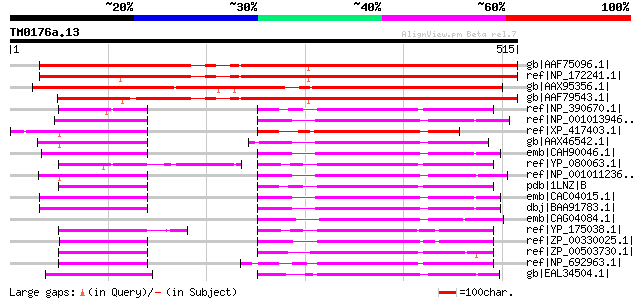
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF75096.1| It is a member of GTP1/OBG family PF|01018. [Arab... 540 e-152
ref|NP_172241.1| GTP1/OBG family protein [Arabidopsis thaliana] 530 e-149
gb|AAX95356.1| F22G5.1-related [Oryza sativa (japonica cultivar-... 491 e-137
gb|AAF79543.1| F22G5.1 [Arabidopsis thaliana] 482 e-134
ref|NP_390670.1| GTPase activity [Bacillus subtilis subsp. subti... 176 2e-42
ref|NP_001013946.1| GTP binding protein 5 [Rattus norvegicus] gi... 176 2e-42
ref|XP_417403.1| PREDICTED: similar to GTP binding protein 5; GT... 175 3e-42
gb|AAX46542.1| GTP binding protein 5 [Bos taurus] gi|62460448|re... 175 3e-42
emb|CAH90046.1| hypothetical protein [Pongo pygmaeus] 175 4e-42
ref|YP_080063.1| GTPase Obg [Bacillus licheniformis ATCC 14580] ... 174 5e-42
ref|NP_001011236.1| hypothetical protein LOC496677 [Xenopus trop... 174 6e-42
pdb|1LNZ|B Chain B, Structure Of The Obg Gtp-Binding Protein gi|... 174 8e-42
emb|CAC04015.1| GTPBP5 [Homo sapiens] gi|22477163|gb|AAH36716.1|... 173 1e-41
dbj|BAA91783.1| unnamed protein product [Homo sapiens] 173 1e-41
emb|CAG04084.1| unnamed protein product [Tetraodon nigroviridis] 170 9e-41
ref|YP_175038.1| Spo0B-associated GTP-binding protein [Bacillus ... 168 3e-40
ref|ZP_00330025.1| COG0536: Predicted GTPase [Moorella thermoace... 168 4e-40
ref|ZP_00503730.1| GTP1/OBG sub-domain [Clostridium thermocellum... 167 6e-40
ref|NP_692963.1| Spo0B-associated GTP-binding protein [Oceanobac... 166 1e-39
gb|EAL34504.1| GA12249-PA [Drosophila pseudoobscura] 165 4e-39
>gb|AAF75096.1| It is a member of GTP1/OBG family PF|01018. [Arabidopsis thaliana]
gi|25406984|pir||F86210 hypothetical protein [imported] -
Arabidopsis thaliana
Length = 1029
Score = 540 bits (1390), Expect = e-152
Identities = 286/490 (58%), Positives = 355/490 (72%), Gaps = 33/490 (6%)
Query: 31 YSDAPHKKSKLAPLQERRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGD 90
YSD KK K+APLQE RM D+F + A+ G+GGSGCSS RSR DR G+ DGGNGGRGGD
Sbjct: 568 YSDFSEKKGKVAPLQETRMRDRFTLYARGGEGGSGCSSVRRSRADRYGKPDGGNGGRGGD 627
Query: 91 VILECSPRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVIHLVNGDIPSI 150
VILEC+ VWDFSGLQ H+ GK G G+SKN+IG RG DKV+ VP+GTVIHL G+IPS
Sbjct: 628 VILECTHAVWDFSGLQPHIKGGKAGHGTSKNRIGNRGEDKVLLVPIGTVIHLQEGEIPSQ 687
Query: 151 VKTESSADTDPWEISGALVDDLPDHGNGSISNATNGEEVKTIHPTGCSSSQAAEKNVEKS 210
V+ ES D DPW++ G+LV+D N + T S++ + + E+
Sbjct: 688 VENESPKDLDPWDLPGSLVEDPASEENSDVHQET--------------MSESDQDDTEQE 733
Query: 211 VKSGHVASTDVLSQLSISNGTPKFGTEYIGEEQEEQEILYNVAELTEEGQQMIVARGGEG 270
L+ G PK ++ +++E +I YNVAELT++GQ++I+ARGGEG
Sbjct: 734 -------------SLTRQLGMPK-EADFEDDDEEIDQIRYNVAELTQQGQRVIIARGGEG 779
Query: 271 GPGNLSTSKDSRKPITTKAGACRQHISNLEDS---DSVCSSLNAGMPGSENVLILELKSI 327
G GN+S ++ R K+ + ++ ++ED D SS+ AG+ GSE VLILELKSI
Sbjct: 780 GLGNVSATRYVRGSKFAKSTIRQTNLRSMEDDAEEDDERSSIKAGLLGSEAVLILELKSI 839
Query: 328 ADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKG 387
ADV VGMPNAGKSTLLGA+SRAKP VGHYAFTTLRPNLGN+N+DDFS+TVADIPGLIKG
Sbjct: 840 ADVGLVGMPNAGKSTLLGALSRAKPRVGHYAFTTLRPNLGNVNYDDFSMTVADIPGLIKG 899
Query: 388 AHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQLKDLILELEYHQDGLSNR 447
AHQNRGLGH FLRHIERTKVLAYVVDLA+ L G +G+ PW+QL+DL++ELE+H++GLS+R
Sbjct: 900 AHQNRGLGHNFLRHIERTKVLAYVVDLASGLDGCEGLTPWQQLRDLVMELEFHEEGLSDR 959
Query: 448 PSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLEEGIPELKAGLRMLV--NGEMSY 505
SLIVANKIDEEGA+E EL+RRV GV IF VCAVLEEG+ ELK GL+MLV NGE S
Sbjct: 960 SSLIVANKIDEEGAEERLKELKRRVKGVKIFPVCAVLEEGVAELKDGLKMLVDGNGEPSE 1019
Query: 506 KLCLDQILID 515
+L L+ I +D
Sbjct: 1020 RLKLENICVD 1029
>ref|NP_172241.1| GTP1/OBG family protein [Arabidopsis thaliana]
Length = 1016
Score = 530 bits (1364), Expect = e-149
Identities = 286/512 (55%), Positives = 357/512 (68%), Gaps = 55/512 (10%)
Query: 31 YSDAPHKKSKLAPLQERRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGD 90
YSD KK K+APLQE RM D+F + A+ G+GGSGCSS RSR DR G+ DGGNGGRGGD
Sbjct: 533 YSDFSEKKGKVAPLQETRMRDRFTLYARGGEGGSGCSSVRRSRADRYGKPDGGNGGRGGD 592
Query: 91 VILECSPRVWDFSGLQHHLIA----------------------GKGGPGSSKNKIGTRGA 128
VILEC+ VWDFSGLQ H+++ GK G G+SKN+IG RG
Sbjct: 593 VILECTHAVWDFSGLQPHIVSVSVTLFVFSGNMSHENFSELKGGKAGHGTSKNRIGNRGE 652
Query: 129 DKVVHVPVGTVIHLVNGDIPSIVKTESSADTDPWEISGALVDDLPDHGNGSISNATNGEE 188
DKV+ VP+GTVIHL G+IPS V+ ES D DPW++ G+LV+D N + T
Sbjct: 653 DKVLLVPIGTVIHLQEGEIPSQVENESPKDLDPWDLPGSLVEDPASEENSDVHQET---- 708
Query: 189 VKTIHPTGCSSSQAAEKNVEKSVKSGHVASTDVLSQLSISNGTPKFGTEYIGEEQEEQEI 248
S++ + + E+ L+ G PK ++ +++E +I
Sbjct: 709 ----------MSESDQDDTEQE-------------SLTRQLGMPK-EADFEDDDEEIDQI 744
Query: 249 LYNVAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDS---DSV 305
YNVAELT++GQ++I+ARGGEGG GN+S ++ R K+ + ++ ++ED D
Sbjct: 745 RYNVAELTQQGQRVIIARGGEGGLGNVSATRYVRGSKFAKSTIRQTNLRSMEDDAEEDDE 804
Query: 306 CSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPN 365
SS+ AG+ GSE VLILELKSIADV VGMPNAGKSTLLGA+SRAKP VGHYAFTTLRPN
Sbjct: 805 RSSIKAGLLGSEAVLILELKSIADVGLVGMPNAGKSTLLGALSRAKPRVGHYAFTTLRPN 864
Query: 366 LGNLNFDDFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIP 425
LGN+N+DDFS+TVADIPGLIKGAHQNRGLGH FLRHIERTKVLAYVVDLA+ L G +G+
Sbjct: 865 LGNVNYDDFSMTVADIPGLIKGAHQNRGLGHNFLRHIERTKVLAYVVDLASGLDGCEGLT 924
Query: 426 PWEQLKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLE 485
PW+QL+DL++ELE+H++GLS+R SLIVANKIDEEGA+E EL+RRV GV IF VCAVLE
Sbjct: 925 PWQQLRDLVMELEFHEEGLSDRSSLIVANKIDEEGAEERLKELKRRVKGVKIFPVCAVLE 984
Query: 486 EGIPELKAGLRMLV--NGEMSYKLCLDQILID 515
EG+ ELK GL+MLV NGE S +L L+ I +D
Sbjct: 985 EGVAELKDGLKMLVDGNGEPSERLKLENICVD 1016
>gb|AAX95356.1| F22G5.1-related [Oryza sativa (japonica cultivar-group)]
Length = 664
Score = 491 bits (1265), Expect = e-137
Identities = 264/510 (51%), Positives = 343/510 (66%), Gaps = 48/510 (9%)
Query: 24 SATVYSCY--SDAPHKKSKLAPLQERRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRAD 81
+AT CY ++ +K K APLQ R M+D+F++ AK GDGG+GC S RSR DRQG+ D
Sbjct: 19 AATGIGCYYATEPEGRKPKTAPLQSRGMVDRFRLRAKGGDGGNGCISLRRSRSDRQGKPD 78
Query: 82 GGNGGRGGDVILECSPRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVIH 141
GGNGGRGGDVILECS VWDFSGLQHH+ A +G G SKN+IGTRG+DK+ VPVGTVIH
Sbjct: 79 GGNGGRGGDVILECSRSVWDFSGLQHHMKASRGANGVSKNQIGTRGSDKIAQVPVGTVIH 138
Query: 142 LVNGDIPSIVKTESSADTDPWEISGALVDDLPDHGNGSISNATNGEEVKTIHPTGCSSSQ 201
LV G+ PS+ + + DPW+I A V+ P + S G + S
Sbjct: 139 LVEGEQPSLSVNKPTRALDPWDIPDA-VEHSPFSSSRIGSKMMKGLDSSRSSQHISSKKN 197
Query: 202 AAEKNVEKS---------------VKSGHVASTDVLSQLSI--------------SNGTP 232
AE + E+ V++ T Q+ + +
Sbjct: 198 TAENDCERGNRNHRGKEPYYMTEFVRTEDYDGTSYPHQVGVDENDQFDDEDDEFWEDDEE 257
Query: 233 KFGTEYIGEEQEEQE-ILYNVAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGA 291
+ E + EE+EE+E + Y+VAE+T+ GQ++I+ARGGEGG GN K+
Sbjct: 258 ELDMEEVTEEKEEEEDVRYSVAEMTKPGQRLIIARGGEGGLGNACILKE----------- 306
Query: 292 CRQHISNLEDSDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAK 351
+S + + +SL+ G PG+E LILELKSIADV VGMPNAGKSTLL A+SRA+
Sbjct: 307 --MWLSKAHKEEEM-ASLSTGHPGTETYLILELKSIADVGLVGMPNAGKSTLLSALSRAR 363
Query: 352 PAVGHYAFTTLRPNLGNLNFDD-FSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAY 410
P + YAFTTLRPN+G+L ++D FS+ VADIPGLIKGAH+NRGLGHAFLRHIERTKVLAY
Sbjct: 364 PEIADYAFTTLRPNIGSLTYEDYFSVKVADIPGLIKGAHENRGLGHAFLRHIERTKVLAY 423
Query: 411 VVDLAAALHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQR 470
V+DLAA L+GRKG+PPWEQL+DL++ELE++Q+GL+ RPSLIVANKIDEEGADE+Y EL++
Sbjct: 424 VLDLAATLNGRKGVPPWEQLRDLVVELEHYQEGLTKRPSLIVANKIDEEGADEMYEELKK 483
Query: 471 RVHGVPIFRVCAVLEEGIPELKAGLRMLVN 500
RV GVP+F +CA+L+EG+P+L+ GLR L++
Sbjct: 484 RVQGVPMFPICAILQEGVPDLRVGLRDLMD 513
>gb|AAF79543.1| F22G5.1 [Arabidopsis thaliana]
Length = 445
Score = 482 bits (1241), Expect = e-134
Identities = 264/478 (55%), Positives = 335/478 (69%), Gaps = 44/478 (9%)
Query: 49 MIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQHH 108
M D+F + A+ G+GGSGCSS RSR DR G+ DGGNGGRGGDVILEC+ VWDFSGLQ H
Sbjct: 1 MRDRFTLYARGGEGGSGCSSVRRSRADRYGKPDGGNGGRGGDVILECTHAVWDFSGLQPH 60
Query: 109 LIAGK------GGPGSSKNKIGTRGADKVVHVPVGTVIHLVNGDIPSIVKTESSADTDPW 162
+++ G S +N ++ V+ VP+GTVIHL G+IPS V+ ES D DPW
Sbjct: 61 IVSVSVTLFVFSGNMSHENF-----SELVLLVPIGTVIHLQEGEIPSQVENESPKDLDPW 115
Query: 163 EISGALVDDLPDHGNGSISNATNGEEVKTIHPTGCSSSQAAEKNVEKSVKSGHVASTDVL 222
++ G+LV+D N + T S++ + + E+
Sbjct: 116 DLPGSLVEDPASEENSDVHQET--------------MSESDQDDTEQE------------ 149
Query: 223 SQLSISNGTPKFGTEYIGEEQEEQEILYNVAELTEEGQQMIVARGGEGGPGNLSTSKDSR 282
L+ G PK ++ +++E +I YNVAELT++GQ++I+ARGGEGG GN+S ++ R
Sbjct: 150 -SLTRQLGMPK-EADFEDDDEEIDQIRYNVAELTQQGQRVIIARGGEGGLGNVSATRYVR 207
Query: 283 KPITTKAGACRQHISNLEDS---DSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAG 339
K+ + ++ ++ED D SS+ AG+ GSE VLILELKSIADV VGMPNAG
Sbjct: 208 GSKFAKSTIRQTNLRSMEDDAEEDDERSSIKAGLLGSEAVLILELKSIADVGLVGMPNAG 267
Query: 340 KSTLLGAISRAKPAVGHYAFTTLRPNLGNLNFDDFSITVADIPGLIKGAHQNRGLGHAFL 399
KSTLLGA+SRAKP VGHYAFTTLRPNLGN+N+DDFS+TVADIPGLIKGAHQNRGLGH FL
Sbjct: 268 KSTLLGALSRAKPRVGHYAFTTLRPNLGNVNYDDFSMTVADIPGLIKGAHQNRGLGHNFL 327
Query: 400 RHIERTKVLAYVVDLAAALHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANKIDEE 459
RHIERTKVLAYVVDLA+ L G +G+ PW+QL+DL++ELE+H++GLS+R SLIVANKIDEE
Sbjct: 328 RHIERTKVLAYVVDLASGLDGCEGLTPWQQLRDLVMELEFHEEGLSDRSSLIVANKIDEE 387
Query: 460 GADEVYLELQRRVHGVPIFRVCAVLEEGIPELKAGLRMLV--NGEMSYKLCLDQILID 515
GA+E EL+RRV GV IF VCAVLEEG+ ELK GL+MLV NGE S +L L+ I +D
Sbjct: 388 GAEERLKELKRRVKGVKIFPVCAVLEEGVAELKDGLKMLVDGNGEPSERLKLENICVD 445
>ref|NP_390670.1| GTPase activity [Bacillus subtilis subsp. subtilis str. 168]
gi|2635257|emb|CAB14752.1| GTPase activity [Bacillus
subtilis subsp. subtilis str. 168]
gi|129021|sp|P20964|OBG_BACSU Spo0B-associated
GTP-binding protein gi|508979|gb|AAA22505.1| GTP-binding
protein
Length = 428
Score = 176 bits (446), Expect = 2e-42
Identities = 103/242 (42%), Positives = 139/242 (56%), Gaps = 27/242 (11%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNA 311
+A+LTE GQ+ ++ARGG GG GN T A Q N
Sbjct: 104 IADLTEHGQRAVIARGGRGGRGN--------SRFATPANPAPQLSEN------------- 142
Query: 312 GMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNF 371
G PG E ++LELK +ADV VG P+ GKSTLL +S AKP + Y FTTL PNLG +
Sbjct: 143 GEPGKERYIVLELKVLADVGLVGFPSVGKSTLLSVVSSAKPKIADYHFTTLVPNLGMVET 202
Query: 372 DD-FSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
DD S +AD+PGLI+GAHQ GLGH FLRHIERT+V+ +V+D++ G +G P++
Sbjct: 203 DDGRSFVMADLPGLIEGAHQGVGLGHQFLRHIERTRVIVHVIDMS----GLEGRDPYDDY 258
Query: 431 KDLILELEYHQDGLSNRPSLIVANKID-EEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
+ EL + L+ RP +IVANK+D E A+ + ++ P+F + AV EG+
Sbjct: 259 LTINQELSEYNLRLTERPQIIVANKMDMPEAAENLEAFKEKLTDDYPVFPISAVTREGLR 318
Query: 490 EL 491
EL
Sbjct: 319 EL 320
Score = 69.3 bits (168), Expect = 3e-10
Identities = 37/94 (39%), Positives = 54/94 (57%), Gaps = 4/94 (4%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSP---RVWDFSGLQ 106
+D+ K+ K GDGG+G +F R ++ +G GG+GG+GGDV+ E + DF +
Sbjct: 3 VDQVKVYVKGGDGGNGMVAFRREKYVPKGGPAGGDGGKGGDVVFEVDEGLRTLMDFR-YK 61
Query: 107 HHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
H A +G G SKN+ G D V+ VP GTV+
Sbjct: 62 KHFKAIRGEHGMSKNQHGRNADDMVIKVPPGTVV 95
>ref|NP_001013946.1| GTP binding protein 5 [Rattus norvegicus]
gi|53734242|gb|AAH83707.1| GTP binding protein 5 [Rattus
norvegicus]
Length = 406
Score = 176 bits (446), Expect = 2e-42
Identities = 107/258 (41%), Positives = 148/258 (56%), Gaps = 32/258 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGN-LSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLN 310
VA+L+ G + + A GG GG GN + D+R P+T
Sbjct: 170 VADLSHLGDEYVAALGGAGGKGNRFFLANDNRAPVTC----------------------T 207
Query: 311 AGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLN 370
G PG E VL LELK++A VG PNAGKS+LL AIS AKPAV Y FTTL P++G ++
Sbjct: 208 PGQPGQERVLYLELKTMAHAGMVGFPNAGKSSLLRAISNAKPAVASYPFTTLNPHVGIVH 267
Query: 371 FDDF-SITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQ 429
++ + VADIPG+I+GAHQN+GLG +FLRHIER + YVVDL+ PW Q
Sbjct: 268 YEGHQQVAVADIPGIIRGAHQNKGLGLSFLRHIERCRFFLYVVDLSLP-------EPWTQ 320
Query: 430 LKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
+ DL ELE +++G S R +I+ANKID A L+LQ R+ G + + A+ E +
Sbjct: 321 VDDLKYELEKYEEGPSERSHVIIANKIDLPQARAHLLQLQDRL-GQEVIALSALTGENLE 379
Query: 490 ELKAGLRMLVNGEMSYKL 507
+L L++L + + +L
Sbjct: 380 QLLLHLKVLHDAHIEAEL 397
Score = 69.7 bits (169), Expect = 2e-10
Identities = 35/95 (36%), Positives = 48/95 (49%)
Query: 46 ERRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGL 105
+R +D +++ + G GGSG S FH G DGG+GG GG +IL +V S +
Sbjct: 69 KRHFVDHRRVLFRGGSGGSGMSCFHSEPRKEFGGPDGGDGGNGGHIILRVDQQVKSLSSV 128
Query: 106 QHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
G G SKN G GA + VPVGT++
Sbjct: 129 LSQYQGFSGEDGGSKNCFGRGGATLYIQVPVGTLV 163
>ref|XP_417403.1| PREDICTED: similar to GTP binding protein 5; GTP-binding protein 5
(putative) [Gallus gallus]
Length = 387
Score = 175 bits (444), Expect = 3e-42
Identities = 100/207 (48%), Positives = 129/207 (62%), Gaps = 29/207 (14%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNA 311
VA+LT+ G++ + A GG GG GN R +SN + + ++
Sbjct: 151 VADLTQHGEEYVAAYGGAGGKGN------------------RFFLSNEKRAPTL---FTP 189
Query: 312 GMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNF 371
G PG E VL LELK+ A VG PNAGKS+LL A+S AKPAV Y FTTL P++G + +
Sbjct: 190 GEPGQERVLHLELKTTAHAGLVGFPNAGKSSLLRALSNAKPAVAAYPFTTLNPHVGIVRY 249
Query: 372 DDF-SITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
+D+ + VADIPGLI+GAHQNRGLG AFLRHIER L YVVDL+ + PW QL
Sbjct: 250 EDYEQVAVADIPGLIRGAHQNRGLGMAFLRHIERCCFLLYVVDLSVS-------QPWIQL 302
Query: 431 KDLILELEYHQDGLSNRPSLIVANKID 457
+DL ELE ++ GLS RP +++ NKID
Sbjct: 303 QDLKYELEQYKKGLSTRPCVVIGNKID 329
Score = 60.1 bits (144), Expect = 2e-07
Identities = 45/145 (31%), Positives = 69/145 (47%), Gaps = 6/145 (4%)
Query: 1 MLVFQARSLRRIEAFTQTCINRRSATVYSCYSDAPHKKSKLAP-LQERRM----IDKFKI 55
ML+ A SLR + + + + R A SC + + +++ + + E+++ +D ++
Sbjct: 1 MLLRCALSLRGLSGW-RPVLGREPAFSTSCAACSKNRRLRQKRVISEKKLTRYFVDHRRV 59
Query: 56 IAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQHHLIAGKGG 115
G GG G SF G DGGNGG GG VIL+ ++ S + G
Sbjct: 60 RVVGGQGGGGSQSFCSEPRKIFGGPDGGNGGDGGHVILKADQQMKSLSSVLPFYQGFHGE 119
Query: 116 PGSSKNKIGTRGADKVVHVPVGTVI 140
G SKN G GA V VPVGT++
Sbjct: 120 RGGSKNCYGANGACVYVKVPVGTLV 144
>gb|AAX46542.1| GTP binding protein 5 [Bos taurus] gi|62460448|ref|NP_001014874.1|
GTP binding protein 5 [Bos taurus]
Length = 454
Score = 175 bits (444), Expect = 3e-42
Identities = 110/246 (44%), Positives = 138/246 (55%), Gaps = 34/246 (13%)
Query: 243 QEEQEILYNVAELTEEGQQMIVARGGEGGPGN-LSTSKDSRKPITTKAGACRQHISNLED 301
+E E+L A+L+ G + I A GG GG GN + D+R P T
Sbjct: 164 KEGNEVL---ADLSRPGDEFIAAVGGTGGKGNRFFLANDNRAPTTC-------------- 206
Query: 302 SDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTT 361
G PG E VL LELK++A VG PNAGKS+LL AIS A+PAV Y FTT
Sbjct: 207 --------TPGQPGQERVLFLELKTVAHAGLVGFPNAGKSSLLRAISNARPAVAAYPFTT 258
Query: 362 LRPNLGNLNFDDF-SITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHG 420
L P++G ++++D I VADIPG+I+GAHQNRGLG AFLRHIER L ++VDL+
Sbjct: 259 LNPHVGIVHYEDHQQIAVADIPGIIQGAHQNRGLGLAFLRHIERCPFLLFLVDLSVP--- 315
Query: 421 RKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRV 480
PW QL DL ELE + +GLS RP +VANKID A +LQ + I
Sbjct: 316 ----EPWTQLDDLKYELEQYDEGLSKRPYTVVANKIDLPQARARLPQLQAHLGQEAIAPA 371
Query: 481 CAVLEE 486
AV+ E
Sbjct: 372 AAVVAE 377
Score = 68.2 bits (165), Expect = 6e-10
Identities = 38/117 (32%), Positives = 57/117 (48%), Gaps = 5/117 (4%)
Query: 29 SCYSDAPHKKS-KLAPLQERRM----IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGG 83
SC H++ + PL E+++ +D +++ + G GG+G S FH G DGG
Sbjct: 47 SCADCTKHQEPPRKQPLSEKKLKQHFVDHRRVLVRGGHGGNGVSCFHSEPRKEFGGPDGG 106
Query: 84 NGGRGGDVILECSPRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
+GG GG V+L +V S + G G KN G GA + VPVGT++
Sbjct: 107 DGGNGGHVVLRVDQQVKSLSSVLSRYQGFDGEDGGRKNCFGRNGAVLYIRVPVGTLV 163
>emb|CAH90046.1| hypothetical protein [Pongo pygmaeus]
Length = 406
Score = 175 bits (443), Expect = 4e-42
Identities = 106/249 (42%), Positives = 144/249 (57%), Gaps = 32/249 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGN-LSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLN 310
VA+L+ G + I A GG GG GN + ++R P+T
Sbjct: 170 VADLSRVGDEYIAALGGAGGKGNRFFLANNNRAPVTC----------------------T 207
Query: 311 AGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLN 370
G PG + VL LELK++A VG PNAGKS+LL AIS A+PAV Y FTTL+P++G ++
Sbjct: 208 PGQPGQQRVLHLELKTVAHAGMVGFPNAGKSSLLRAISNARPAVASYPFTTLKPHVGIVH 267
Query: 371 FD-DFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQ 429
++ I VADIPG+I+GAHQNRGLG AFLRHIER + L +VVDL+ PW Q
Sbjct: 268 YEGHLQIAVADIPGIIRGAHQNRGLGSAFLRHIERCRFLLFVVDLSQP-------EPWTQ 320
Query: 430 LKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
+ DL ELE ++ GLS RP I+ANKID A +L+ + G + + A+ E +
Sbjct: 321 VDDLKYELEMYEKGLSERPHAIIANKIDLPEAQANLSQLRDHL-GQEVIVLSALTGENLE 379
Query: 490 ELKAGLRML 498
+L L++L
Sbjct: 380 QLLLHLKVL 388
Score = 68.2 bits (165), Expect = 6e-10
Identities = 37/108 (34%), Positives = 52/108 (47%)
Query: 33 DAPHKKSKLAPLQERRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVI 92
+ P KK +R +D +++ G+GG+G S FH G DGG+GG GG VI
Sbjct: 56 ELPGKKPLSEKKLKRYFVDYRRVLVCGGNGGAGASCFHSEPRKEFGGPDGGDGGNGGHVI 115
Query: 93 LECSPRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
L +V S + G G SKN G GA + VP+GT++
Sbjct: 116 LRADQQVKSLSSVLSRYQGFSGEDGGSKNCFGRSGAVLYIRVPMGTLV 163
>ref|YP_080063.1| GTPase Obg [Bacillus licheniformis ATCC 14580]
gi|52004483|gb|AAU24425.1| GTPase Obg [Bacillus
licheniformis ATCC 14580] gi|52786651|ref|YP_092480.1|
Obg [Bacillus licheniformis ATCC 14580]
gi|52349153|gb|AAU41787.1| Obg [Bacillus licheniformis
DSM 13]
Length = 428
Score = 174 bits (442), Expect = 5e-42
Identities = 100/242 (41%), Positives = 140/242 (57%), Gaps = 27/242 (11%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNA 311
+A+LTE GQ+ ++A+GG GG GN T A Q N
Sbjct: 104 IADLTEHGQEAVIAKGGRGGRGNTR--------FATPANPAPQLSEN------------- 142
Query: 312 GMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNF 371
G PG E ++LELK +ADV VG P+ GKSTLL +S AKP + Y FTTL PNLG +
Sbjct: 143 GEPGKERYIVLELKVLADVGLVGFPSVGKSTLLSVVSSAKPKIADYHFTTLNPNLGMVET 202
Query: 372 DD-FSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
+D S +AD+PGLI+GAH+ GLGH FLRHIERT+V+ +V+D++ G +G P+E
Sbjct: 203 EDGRSFVMADLPGLIEGAHEGVGLGHQFLRHIERTRVIVHVIDMS----GLEGRDPYEDY 258
Query: 431 KDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRV-HGVPIFRVCAVLEEGIP 489
+ ELE + L+ RP +IVANK+D A+E + ++ P+F + AV +G+
Sbjct: 259 VTINKELEQYNLRLTERPQIIVANKMDMPDAEENLKAFKEKLTDDYPVFPISAVTRQGLR 318
Query: 490 EL 491
+L
Sbjct: 319 DL 320
Score = 73.2 bits (178), Expect = 2e-11
Identities = 58/190 (30%), Positives = 85/190 (44%), Gaps = 26/190 (13%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVIL---ECSPRVWDFSGLQ 106
+D+ KI K GDGG+G +F R ++ +G GG+GG+GGDV+ E + DF Q
Sbjct: 3 VDQVKIYVKGGDGGNGMVAFRREKYVPKGGPAGGDGGKGGDVVFKVDEGLSTLMDFR-YQ 61
Query: 107 HHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVIHLVNGDIPSIVKTESSADTDPWEISG 166
H A +G G SKN+ G D VV VP GTV+ D D ++
Sbjct: 62 RHFKAARGEHGMSKNQHGRNAEDMVVKVPPGTVV----------------IDDDTKQV-- 103
Query: 167 ALVDDLPDHGNGS-ISNATNGEEVKTIHPTGCSSSQAAEKNVEKSVKSGHVASTDVLSQL 225
+ DL +HG + I+ G T T + + +N E + V VL+ +
Sbjct: 104 --IADLTEHGQEAVIAKGGRGGRGNTRFATPANPAPQLSENGEPGKERYIVLELKVLADV 161
Query: 226 SISNGTPKFG 235
+ G P G
Sbjct: 162 GLV-GFPSVG 170
>ref|NP_001011236.1| hypothetical protein LOC496677 [Xenopus tropicalis]
gi|56556285|gb|AAH87807.1| Hypothetical LOC496677
[Xenopus tropicalis]
Length = 405
Score = 174 bits (441), Expect = 6e-42
Identities = 107/256 (41%), Positives = 141/256 (54%), Gaps = 32/256 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGN-LSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLN 310
+A+L++ G + + ARGG GG GN S ++R P+T
Sbjct: 172 LADLSKAGDEFLAARGGVGGKGNRFFLSNENRAPMTA----------------------T 209
Query: 311 AGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLN 370
G PG E VL LELK++A V VG PNAGKS+LL +S A+PAV Y FTTL P++G +
Sbjct: 210 PGQPGEERVLHLELKTMAHVGMVGFPNAGKSSLLRLLSNARPAVAAYPFTTLNPHVGIIK 269
Query: 371 FDDF-SITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQ 429
+ D+ I VAD PG+I GAHQNRGLG AFLRHIER ++L +V+DL+ PW Q
Sbjct: 270 YRDYVQIAVADTPGIIDGAHQNRGLGFAFLRHIERCRILLFVLDLSHK-------EPWAQ 322
Query: 430 LKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
L L ELE + + L RP +IVANK+D A E L+ + V A+ E
Sbjct: 323 LASLRYELEQYNEDLVQRPHVIVANKLDLPAARETLRRLRHETDS-NVIGVSALTGENAE 381
Query: 490 ELKAGLRMLVNGEMSY 505
EL LR L +G +
Sbjct: 382 ELILHLRELYDGHRQH 397
Score = 65.9 bits (159), Expect = 3e-09
Identities = 37/117 (31%), Positives = 54/117 (45%), Gaps = 6/117 (5%)
Query: 30 CYSDAPHKK--SKLAPLQERRM----IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGG 83
C A H+ K L E+++ +D ++ G GG+G FH G DGG
Sbjct: 49 CPRRAKHRSLTGKKPDLSEKKLTRYFVDHRRVRVVGGAGGNGACCFHSEPRKEYGGPDGG 108
Query: 84 NGGRGGDVILECSPRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
+GG GG ++L+ PRV S + G PG S N G G + VP+GT++
Sbjct: 109 DGGNGGQIVLKADPRVKSLSTVTPVYKGNNGEPGRSANCFGRNGESIYIKVPLGTLV 165
>pdb|1LNZ|B Chain B, Structure Of The Obg Gtp-Binding Protein
gi|24158881|pdb|1LNZ|A Chain A, Structure Of The Obg
Gtp-Binding Protein
Length = 342
Score = 174 bits (440), Expect = 8e-42
Identities = 103/242 (42%), Positives = 136/242 (55%), Gaps = 27/242 (11%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNA 311
+A+LTE GQ+ ++ARGG GG GN T A Q N
Sbjct: 104 IADLTEHGQRAVIARGGRGGRGN--------SRFATPANPAPQLSEN------------- 142
Query: 312 GMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNF 371
G PG E ++LELK +ADV VG P+ GKSTLL +S AKP + Y FTTL PNLG +
Sbjct: 143 GEPGKERYIVLELKVLADVGLVGFPSVGKSTLLSVVSSAKPKIADYHFTTLVPNLGXVET 202
Query: 372 DD-FSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
DD S AD+PGLI+GAHQ GLGH FLRHIERT+V+ +V+D + G +G P++
Sbjct: 203 DDGRSFVXADLPGLIEGAHQGVGLGHQFLRHIERTRVIVHVIDXS----GLEGRDPYDDY 258
Query: 431 KDLILELEYHQDGLSNRPSLIVANKID-EEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
+ EL + L+ RP +IVANK D E A+ + ++ P+F + AV EG+
Sbjct: 259 LTINQELSEYNLRLTERPQIIVANKXDXPEAAENLEAFKEKLTDDYPVFPISAVTREGLR 318
Query: 490 EL 491
EL
Sbjct: 319 EL 320
Score = 69.7 bits (169), Expect = 2e-10
Identities = 35/93 (37%), Positives = 53/93 (56%), Gaps = 2/93 (2%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH-- 107
+D+ K+ K GDGG+G +F R ++ +G GG+GG+GGDV+ E + ++
Sbjct: 3 VDQVKVYVKGGDGGNGXVAFRREKYVPKGGPAGGDGGKGGDVVFEVDEGLRTLXDFRYKK 62
Query: 108 HLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
H A +G G SKN+ G D V+ VP GTV+
Sbjct: 63 HFKAIRGEHGXSKNQHGRNADDXVIKVPPGTVV 95
>emb|CAC04015.1| GTPBP5 [Homo sapiens] gi|22477163|gb|AAH36716.1| GTP binding
protein 5 [Homo sapiens] gi|24308117|ref|NP_056481.1|
GTP binding protein 5 [Homo sapiens]
gi|32469779|sp|Q9H4K7|GTPB5_HUMAN Putative GTP-binding
protein 5
Length = 406
Score = 173 bits (438), Expect = 1e-41
Identities = 107/249 (42%), Positives = 144/249 (56%), Gaps = 32/249 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGN-LSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLN 310
VA+L+ G + I A GG GG GN + ++R P+T
Sbjct: 170 VADLSCVGDEYIAALGGAGGKGNRFFLANNNRAPVTC----------------------T 207
Query: 311 AGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLN 370
G PG + VL LELK++A VG PNAGKS+LL AIS A+PAV Y FTTL+P++G ++
Sbjct: 208 PGQPGQQRVLHLELKTVAHAGMVGFPNAGKSSLLRAISNARPAVASYPFTTLKPHVGIVH 267
Query: 371 FD-DFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQ 429
++ I VADIPG+I+GAHQNRGLG AFLRHIER + L +VVDL+ PW Q
Sbjct: 268 YEGHLQIAVADIPGIIRGAHQNRGLGSAFLRHIERCRFLLFVVDLSQP-------EPWTQ 320
Query: 430 LKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
+ DL ELE ++ GLS RP IVANKID A +L+ + G + + A+ E +
Sbjct: 321 VDDLKYELEMYEKGLSARPHAIVANKIDLPEAQANLSQLRDHL-GQEVIVLSALTGENLE 379
Query: 490 ELKAGLRML 498
+L L++L
Sbjct: 380 QLLLHLKVL 388
Score = 69.3 bits (168), Expect = 3e-10
Identities = 38/110 (34%), Positives = 53/110 (47%)
Query: 31 YSDAPHKKSKLAPLQERRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGD 90
+ + P KK +R +D +++ G+GG+G S FH G DGG+GG GG
Sbjct: 54 HQELPGKKLLSEKKLKRYFVDYRRVLVCGGNGGAGASCFHSEPRKEFGGPDGGDGGNGGH 113
Query: 91 VILECSPRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
VIL +V S + G G SKN G GA + VPVGT++
Sbjct: 114 VILRVDQQVKSLSSVLSRYQGFSGEDGGSKNCFGRSGAVLYIRVPVGTLV 163
>dbj|BAA91783.1| unnamed protein product [Homo sapiens]
Length = 401
Score = 173 bits (438), Expect = 1e-41
Identities = 107/249 (42%), Positives = 144/249 (56%), Gaps = 32/249 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGN-LSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLN 310
VA+L+ G + I A GG GG GN + ++R P+T
Sbjct: 170 VADLSCVGDEYIAALGGAGGKGNRFFLANNNRAPVTC----------------------T 207
Query: 311 AGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLN 370
G PG + VL LELK++A VG PNAGKS+LL AIS A+PAV Y FTTL+P++G ++
Sbjct: 208 PGQPGQQRVLHLELKTVAHAGMVGFPNAGKSSLLRAISNARPAVASYPFTTLKPHVGIVH 267
Query: 371 FD-DFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQ 429
++ I VADIPG+I+GAHQNRGLG AFLRHIER + L +VVDL+ PW Q
Sbjct: 268 YEGHLQIAVADIPGIIRGAHQNRGLGSAFLRHIERCRFLLFVVDLSQP-------EPWTQ 320
Query: 430 LKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
+ DL ELE ++ GLS RP IVANKID A +L+ + G + + A+ E +
Sbjct: 321 VDDLKYELEMYEKGLSARPHAIVANKIDLPEAQANLSQLRDHL-GQEVIVLSALTGENLE 379
Query: 490 ELKAGLRML 498
+L L++L
Sbjct: 380 QLLLHLKVL 388
Score = 69.3 bits (168), Expect = 3e-10
Identities = 38/110 (34%), Positives = 53/110 (47%)
Query: 31 YSDAPHKKSKLAPLQERRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGD 90
+ + P KK +R +D +++ G+GG+G S FH G DGG+GG GG
Sbjct: 54 HQELPGKKLLSEKKLKRYFVDYRRVLVCGGNGGAGASCFHSEPRKEFGGPDGGDGGNGGH 113
Query: 91 VILECSPRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
VIL +V S + G G SKN G GA + VPVGT++
Sbjct: 114 VILRVDQQVKSLSSVLSRYQGFSGEDGGSKNCFGRSGAVLYIRVPVGTLV 163
>emb|CAG04084.1| unnamed protein product [Tetraodon nigroviridis]
Length = 381
Score = 170 bits (431), Expect = 9e-41
Identities = 105/252 (41%), Positives = 146/252 (57%), Gaps = 32/252 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGN-LSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLN 310
V +L++ GQ+ + A GG GG GN + ++R P+T+ GA
Sbjct: 154 VTDLSQHGQEFMAASGGAGGKGNRFFLTNENRAPMTSTPGA------------------- 194
Query: 311 AGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLN 370
G E +L LEL+++A VG PNAGKS+LL AIS AKPAV Y FTTL+P++G +N
Sbjct: 195 ---QGQERLLHLELRTMAHAGLVGFPNAGKSSLLRAISNAKPAVAAYPFTTLKPHVGIVN 251
Query: 371 F-DDFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQ 429
+ D + VADIPG+I+GAH NRGLG +FLRHIER + L +V+D+++ PW Q
Sbjct: 252 YRDHVQVAVADIPGIIRGAHLNRGLGLSFLRHIERCRFLLFVLDMSSP-------DPWAQ 304
Query: 430 LKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLEEGIP 489
L+DL EL++++ LS RP I+ANK+D A LE R + + V AV +
Sbjct: 305 LQDLHHELDHYEPDLSQRPQAIIANKMDLPDA-RSKLEALRSLVTRRVIPVSAVTGQNTE 363
Query: 490 ELKAGLRMLVNG 501
EL LR L +G
Sbjct: 364 ELILHLRELYDG 375
Score = 45.8 bits (107), Expect = 0.003
Identities = 36/123 (29%), Positives = 46/123 (37%), Gaps = 30/123 (24%)
Query: 47 RRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVIL----ECS-PRVWD 101
R +D+ + AG GG G SSFH G DGGNGG GG +I+ +C+ +W
Sbjct: 1 RYFVDQRCVKLVAGSGGKGVSSFHSEPRKEWGGPDGGNGGDGGSIIIKGRSQCTCTELWL 60
Query: 102 FSGLQHHLI-----------------------AGKGGPGSSKNKIGTRGADKVVHVPVGT 138
S Q + G G SKN G G HV V
Sbjct: 61 VSMFQRENVVFFCPLTADRFVKSLAQVYPVYKGEDGQSGGSKNCFGKNG--NTTHVHVSR 118
Query: 139 VIH 141
+H
Sbjct: 119 CVH 121
>ref|YP_175038.1| Spo0B-associated GTP-binding protein [Bacillus clausii KSM-K16]
gi|56909550|dbj|BAD64077.1| Spo0B-associated GTP-binding
protein [Bacillus clausii KSM-K16]
Length = 428
Score = 168 bits (426), Expect = 3e-40
Identities = 99/242 (40%), Positives = 140/242 (56%), Gaps = 27/242 (11%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNA 311
+A+LT GQ+ +VA+GG GG GN+ T A H N
Sbjct: 104 LADLTTHGQRAVVAKGGRGGRGNVR--------FVTPANPAPDHAEN------------- 142
Query: 312 GMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNF 371
G PG E L+LELK +ADV VG P+ GKSTLL +S AKP + Y FTT+ PNLG ++
Sbjct: 143 GEPGEERNLLLELKVLADVGLVGFPSVGKSTLLSIVSAAKPKIAEYHFTTITPNLGVVDT 202
Query: 372 DD-FSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
D S +AD+PGLI+GAH+ GLGH FLRHIERT+V+ ++VD+ +A+ GR + + ++
Sbjct: 203 QDGRSFVMADLPGLIEGAHEGVGLGHQFLRHIERTRVIVHIVDM-SAMEGRDPVEDYHKI 261
Query: 431 KDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRV-HGVPIFRVCAVLEEGIP 489
+ EL + L+ RP L+VANK+D A+E + + IF V A+ ++G+
Sbjct: 262 NE---ELSQYNYRLTERPQLVVANKMDMPEANENLKRFKEALGEETKIFPVSAITKDGVR 318
Query: 490 EL 491
EL
Sbjct: 319 EL 320
Score = 71.2 bits (173), Expect = 7e-11
Identities = 46/133 (34%), Positives = 65/133 (48%), Gaps = 22/133 (16%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH-- 107
+DK K+ AK GDGG+G ++ R ++ G GG+GGRG VILE + ++
Sbjct: 3 VDKVKVYAKGGDGGNGMVAYRREKYVPDGGPAGGDGGRGASVILEVDEGLRTLMDFRYNK 62
Query: 108 HLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVIHLVNGDIPSIVKTESSADTDPWEISGA 167
H +G G SKN G + D VV VP GT++ TD E +GA
Sbjct: 63 HFKGKRGEHGMSKNMHGKKAEDLVVKVPPGTMV------------------TD--EETGA 102
Query: 168 LVDDLPDHGNGSI 180
L+ DL HG ++
Sbjct: 103 LLADLTTHGQRAV 115
>ref|ZP_00330025.1| COG0536: Predicted GTPase [Moorella thermoacetica ATCC 39073]
Length = 423
Score = 168 bits (425), Expect = 4e-40
Identities = 104/244 (42%), Positives = 135/244 (54%), Gaps = 32/244 (13%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLS--TSKDSRKPITTKAGACRQHISNLEDSDSVCSSL 309
+A+L E+GQ+++VA GG GG GN TS D K
Sbjct: 104 LADLVEDGQRVVVAAGGRGGRGNARFVTSTDRAPTFAEK--------------------- 142
Query: 310 NAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNL 369
G PG E L+LELK +ADV +G+PNAGKSTLL IS A+P + Y FTTL PNLG +
Sbjct: 143 --GEPGEERWLVLELKLLADVGLIGLPNAGKSTLLARISAARPKIADYPFTTLTPNLGVV 200
Query: 370 NFDD-FSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWE 428
+D S VADIPGLI GAHQ GLG FLRHIERT+VL +V+D + G + W
Sbjct: 201 RLEDGDSFVVADIPGLIAGAHQGAGLGLKFLRHIERTRVLVHVLDTSQP--GEDVLAGWR 258
Query: 429 QLKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRV-HGVPIFRVCAVLEEG 487
+ D EL ++ L+ RP ++ ANK+D G +E L+ R+ IF + A EG
Sbjct: 259 TVND---ELAHYNPELARRPQVVAANKMDIPGGEEKVAFLRERLGDSYRIFPISAATGEG 315
Query: 488 IPEL 491
+ EL
Sbjct: 316 VQEL 319
Score = 70.1 bits (170), Expect = 2e-10
Identities = 37/92 (40%), Positives = 52/92 (56%), Gaps = 2/92 (2%)
Query: 51 DKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH--H 108
D+ KI K GDGG+G +F R ++ +G +GG+GGRGG VILE + ++ H
Sbjct: 4 DEAKIYVKGGDGGNGIVAFRREKYVPRGGPNGGDGGRGGSVILEADAGLRTLVDFRYRAH 63
Query: 109 LIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
A +G G KNK G D ++ VPVG V+
Sbjct: 64 YRAERGQHGQGKNKHGRSAPDLILRVPVGVVV 95
>ref|ZP_00503730.1| GTP1/OBG sub-domain [Clostridium thermocellum ATCC 27405]
gi|67851847|gb|EAM47410.1| GTP1/OBG sub-domain
[Clostridium thermocellum ATCC 27405]
Length = 424
Score = 167 bits (424), Expect = 6e-40
Identities = 100/244 (40%), Positives = 145/244 (58%), Gaps = 30/244 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNA 311
+A+L + G+++++A+GG+GG GN + +R+ V S
Sbjct: 104 LADLVKPGKKVVIAKGGKGGAGNQHFATPTRQ---------------------VPSFAKP 142
Query: 312 GMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNF 371
G PG E +ILELK +ADV +G PN GKST+L ++ A+P + +Y FTT+ PNLG +N
Sbjct: 143 GEPGEELWVILELKLLADVGLIGFPNVGKSTILSMVTAAQPKIANYHFTTINPNLGVVNI 202
Query: 372 D-DFSITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
D + + +ADIPGLI+GAHQ GLGH FL+HIERTK+L +VVD++ G +G P +
Sbjct: 203 DAENAFVMADIPGLIEGAHQGVGLGHEFLKHIERTKLLIHVVDIS----GSEGRDPVQDF 258
Query: 431 KDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRV---HGVPIFRVCAVLEEG 487
+ + EL+ + L RP +I ANK+D GA+E LE R+V G IF V A +G
Sbjct: 259 EVINEELKKYNPVLCERPQIIAANKMDVTGAEE-NLEKFRKVIEPRGYKIFPVSAASNKG 317
Query: 488 IPEL 491
+ EL
Sbjct: 318 LKEL 321
Score = 69.3 bits (168), Expect = 3e-10
Identities = 36/93 (38%), Positives = 51/93 (54%), Gaps = 2/93 (2%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH-- 107
+D+ +I KAGDGG G SFHR ++ +G DGG+GG+GGDVI + ++
Sbjct: 3 VDRARIYIKAGDGGDGAISFHREKYISKGGPDGGDGGKGGDVIFVVDEGLRTLQDFRYKT 62
Query: 108 HLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
A G G S N G G D ++ VP GT++
Sbjct: 63 RYRAEDGQNGGSSNCSGRSGEDLIIKVPPGTLV 95
>ref|NP_692963.1| Spo0B-associated GTP-binding protein [Oceanobacillus iheyensis
HTE831] gi|22777726|dbj|BAC13998.1| Spo0B-associated
GTP-binding protein [Oceanobacillus iheyensis HTE831]
Length = 426
Score = 166 bits (421), Expect = 1e-39
Identities = 98/259 (37%), Positives = 147/259 (55%), Gaps = 32/259 (12%)
Query: 235 GTEYIGEEQEEQEILYNVAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQ 294
GT I E+ E +A+LT+ Q+ ++ +GG GG GN T+ R
Sbjct: 92 GTTVIDEDTGEV-----IADLTKHEQEAVIVKGGRGGRGN------------TRFATPRN 134
Query: 295 HISNLEDSDSVCSSLNAGMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAV 354
++ ++ G PG E + +ELK IADV VG P+ GKSTLL +S AKP +
Sbjct: 135 PAPDMAEN---------GEPGQERNIKVELKLIADVGLVGFPSVGKSTLLSVVSAAKPKI 185
Query: 355 GHYAFTTLRPNLGNLNFDDF-SITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVD 413
Y FTTL PNLG ++ D S +AD+PGLI+GA Q GLGH FLRHIERT+V+ +V+D
Sbjct: 186 ADYHFTTLSPNLGVVDTQDSRSFVLADLPGLIEGASQGIGLGHQFLRHIERTRVILHVID 245
Query: 414 LAAALHGRKGIPPWEQLKDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVH 473
+A G +G P+E K + EL + + L +RP +I ANK+D A E ++ + +
Sbjct: 246 MA----GTEGRDPYEDYKKINQELSDYDEKLMDRPQIIAANKMDMPNAQENLIQFKNELE 301
Query: 474 -GVPIFRVCAVLEEGIPEL 491
+P++ + A+ ++G+ +L
Sbjct: 302 DDIPVYEISALTKDGLRDL 320
Score = 52.4 bits (124), Expect = 3e-05
Identities = 27/93 (29%), Positives = 47/93 (50%), Gaps = 2/93 (2%)
Query: 50 IDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECSPRVWDFSGLQH-- 107
+D+ + KAGDGG+G ++ R ++ +G GG+GG G +V+ + + ++
Sbjct: 3 VDQVSVYVKAGDGGNGLVAYRREKYVPKGGPAGGDGGNGSNVVFKVDEGLNTLMEFRYNR 62
Query: 108 HLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVI 140
H +G G SK + G V+ VP GT +
Sbjct: 63 HFKGKRGENGMSKTQHGRNADPLVIPVPPGTTV 95
>gb|EAL34504.1| GA12249-PA [Drosophila pseudoobscura]
Length = 382
Score = 165 bits (417), Expect = 4e-39
Identities = 98/247 (39%), Positives = 137/247 (54%), Gaps = 30/247 (12%)
Query: 252 VAELTEEGQQMIVARGGEGGPGNLSTSKDSRKPITTKAGACRQHISNLEDSDSVCSSLNA 311
V +L + ARGG GG GN + D E S VC
Sbjct: 152 VGDLGHADLMFVAARGGAGGKGNRFFTTDK------------------ETSPKVCEY--- 190
Query: 312 GMPGSENVLILELKSIADVSFVGMPNAGKSTLLGAISRAKPAVGHYAFTTLRPNLGNLNF 371
G G + +EL+S+ADV +G PNAGKSTLL A++RAKP V YAFTTLRP+LG + +
Sbjct: 191 GPTGEDISYTIELRSMADVGLIGYPNAGKSTLLNALTRAKPKVAPYAFTTLRPHLGTVQY 250
Query: 372 DDF-SITVADIPGLIKGAHQNRGLGHAFLRHIERTKVLAYVVDLAAALHGRKGIPPWEQL 430
DD IT+AD+PGL+ AH N+GLG FL+H ER +L +V+D +A PW
Sbjct: 251 DDLVQITIADLPGLVPDAHLNKGLGIQFLKHAERCTLLLFVLDASAP-------EPWTHY 303
Query: 431 KDLILELEYHQDGLSNRPSLIVANKIDEEGADEVYLELQRRVHGVPIFRVCAVLEEGIPE 490
+ L+ EL L++RP L+VANKID E + + ELQRR+ P+ + A + + + +
Sbjct: 304 EQLMHELRQFGGSLASRPQLVVANKIDMEDSAANFEELQRRLQN-PVLGISAKMGQNLGQ 362
Query: 491 LKAGLRM 497
L + +R+
Sbjct: 363 LLSSIRL 369
Score = 62.8 bits (151), Expect = 3e-08
Identities = 36/109 (33%), Positives = 56/109 (51%)
Query: 37 KKSKLAPLQERRMIDKFKIIAKAGDGGSGCSSFHRSRHDRQGRADGGNGGRGGDVILECS 96
KK K + + D ++ A G GG GC SF + + + DGG+GG GG V+ + S
Sbjct: 41 KKPKSTRKEAQYFSDAKRVRAVGGKGGDGCVSFLQLWCNERAGPDGGDGGNGGHVVFQAS 100
Query: 97 PRVWDFSGLQHHLIAGKGGPGSSKNKIGTRGADKVVHVPVGTVIHLVNG 145
V +F+ + L A +G G++K+ G V+ VP+GT+I G
Sbjct: 101 NDVRNFNHVGSVLRAEEGERGNAKDCHGKNAKHAVIKVPIGTIIRNAQG 149
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 913,843,827
Number of Sequences: 2540612
Number of extensions: 42638277
Number of successful extensions: 107185
Number of sequences better than 10.0: 1924
Number of HSP's better than 10.0 without gapping: 1122
Number of HSP's successfully gapped in prelim test: 804
Number of HSP's that attempted gapping in prelim test: 103494
Number of HSP's gapped (non-prelim): 2796
length of query: 515
length of database: 863,360,394
effective HSP length: 133
effective length of query: 382
effective length of database: 525,458,998
effective search space: 200725337236
effective search space used: 200725337236
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0176a.13


