

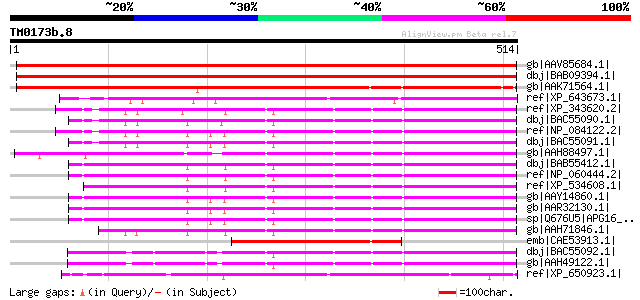
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0173b.8
(514 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAV85684.1| At5g50230 [Arabidopsis thaliana] gi|40823395|gb|A... 766 0.0
dbj|BAB09394.1| unnamed protein product [Arabidopsis thaliana] g... 766 0.0
gb|AAK71564.1| putative WD-repeat protein [Oryza sativa (japonic... 559 e-158
ref|XP_643673.1| hypothetical protein DDB0215341 [Dictyostelium ... 251 5e-65
ref|XP_343620.2| PREDICTED: similar to Apg16L alpha [Rattus norv... 207 6e-52
dbj|BAC55090.1| Apg16L alpha [Mus musculus] 205 2e-51
ref|NP_084122.2| APG16L beta isoform [Mus musculus] gi|62510494|... 202 2e-50
dbj|BAC55091.1| Apg16L beta [Mus musculus] 202 3e-50
gb|AAH88497.1| Unknown (protein for MGC:97562) [Xenopus tropical... 201 5e-50
dbj|BAB55412.1| unnamed protein product [Homo sapiens] 199 2e-49
ref|NP_060444.2| APG16 autophagy 16-like isoform 2 [Homo sapiens] 199 2e-49
ref|XP_534608.1| PREDICTED: similar to APG16L beta [Canis famili... 196 1e-48
gb|AAY14860.1| unknown [Homo sapiens] gi|31742530|ref|NP_110430.... 196 2e-48
gb|AAR32130.1| APG16L beta [Homo sapiens] 196 2e-48
sp|Q676U5|APG16_HUMAN Autophagy protein 16-like (APG16-like) 196 2e-48
gb|AAH71846.1| APG16L protein [Homo sapiens] 193 1e-47
emb|CAE53913.1| putative WD-repeat protein [Triticum aestivum] 192 2e-47
dbj|BAC55092.1| Apg16L gamma [Mus musculus] 184 4e-45
gb|AAH49122.1| Apg16l protein [Mus musculus] 184 4e-45
ref|XP_650923.1| WD repeat protein [Entamoeba histolytica HM-1:I... 176 2e-42
>gb|AAV85684.1| At5g50230 [Arabidopsis thaliana] gi|40823395|gb|AAR92280.1|
At5g50230 [Arabidopsis thaliana]
Length = 509
Score = 766 bits (1977), Expect = 0.0
Identities = 358/506 (70%), Positives = 445/506 (87%)
Query: 8 QEEIAQKAINHALKSLRKRHLLEEAAHAPAFLALSRPIVSQGSEWKEKAENLQVELQQCY 67
QEE A +AIN AL++LRKRHLLEE AHAPA ALS+P++SQGSEWKEK E L+ ELQQCY
Sbjct: 3 QEEKAMEAINDALRALRKRHLLEEGAHAPAISALSKPLISQGSEWKEKTEKLETELQQCY 62
Query: 68 KAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELTEMRDEFSQLKVDLEQKIKELE 127
KAQSRLSEQLV+EVA+SR SKA++QEK+ I DLQKELT+ R++ ++L+ +LE+K K ++
Sbjct: 63 KAQSRLSEQLVIEVAESRTSKAILQEKELLINDLQKELTQRREDCTRLQEELEEKTKTVD 122
Query: 128 VIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANALYEEMVERLR 187
V+++EN E+++QLE+MT++ +KAE ENKMLIDRWML+KM+DAERLNEAN LYEEM+ +L+
Sbjct: 123 VLIAENLEIRSQLEEMTSRVQKAETENKMLIDRWMLQKMQDAERLNEANDLYEEMLAKLK 182
Query: 188 ASGLEQLARQQVDGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGGCASILFEFNSSRLIT 247
A+GLE LARQQVDG+VRR E+G + F++S IPSTC R++AHEGGC SI+FE+NS L T
Sbjct: 183 ANGLETLARQQVDGIVRRNEDGTDHFVESTIPSTCANRIHAHEGGCGSIVFEYNSGTLFT 242
Query: 248 GGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVR 307
GGQD++VK+WDTN+G++ +L+G +G++LD+ +THDN+SVIAA+SSNNL+VWD++SGRVR
Sbjct: 243 GGQDRAVKMWDTNSGTLIKSLYGSLGNILDMAVTHDNKSVIAATSSNNLFVWDVSSGRVR 302
Query: 308 HTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMD 367
HTLTGH DKVCAVDVSK SSRHVVSAAYDRTIK+WDL KGYCTNT++F SNCNA+C S+D
Sbjct: 303 HTLTGHTDKVCAVDVSKFSSRHVVSAAYDRTIKLWDLHKGYCTNTVLFTSNCNAICLSID 362
Query: 368 GQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDV 427
G T++SGH+DGNLRLWDIQ+GKLLSEVA HS AVTS+SLSRNGN +LTSGRDNVHN+FD
Sbjct: 363 GLTVFSGHMDGNLRLWDIQTGKLLSEVAGHSSAVTSVSLSRNGNRILTSGRDNVHNVFDT 422
Query: 428 RSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTS 487
R+LE+C + R +GNR+ASNWSRSCISPD+ +VAAGSADGSVH+WS+ K IVS LKE TS
Sbjct: 423 RTLEICGTLRASGNRLASNWSRSCISPDDDYVAAGSADGSVHVWSLSKGNIVSILKEQTS 482
Query: 488 SVLCCRWSGIGKPLASADKNGIVCIW 513
+LCC WSGIGKPLASADKNG VC W
Sbjct: 483 PILCCSWSGIGKPLASADKNGYVCTW 508
>dbj|BAB09394.1| unnamed protein product [Arabidopsis thaliana]
gi|15240637|ref|NP_199834.1| transducin family protein /
WD-40 repeat family protein [Arabidopsis thaliana]
Length = 515
Score = 766 bits (1977), Expect = 0.0
Identities = 358/506 (70%), Positives = 445/506 (87%)
Query: 8 QEEIAQKAINHALKSLRKRHLLEEAAHAPAFLALSRPIVSQGSEWKEKAENLQVELQQCY 67
QEE A +AIN AL++LRKRHLLEE AHAPA ALS+P++SQGSEWKEK E L+ ELQQCY
Sbjct: 9 QEEKAMEAINDALRALRKRHLLEEGAHAPAISALSKPLISQGSEWKEKTEKLETELQQCY 68
Query: 68 KAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELTEMRDEFSQLKVDLEQKIKELE 127
KAQSRLSEQLV+EVA+SR SKA++QEK+ I DLQKELT+ R++ ++L+ +LE+K K ++
Sbjct: 69 KAQSRLSEQLVIEVAESRTSKAILQEKELLINDLQKELTQRREDCTRLQEELEEKTKTVD 128
Query: 128 VIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANALYEEMVERLR 187
V+++EN E+++QLE+MT++ +KAE ENKMLIDRWML+KM+DAERLNEAN LYEEM+ +L+
Sbjct: 129 VLIAENLEIRSQLEEMTSRVQKAETENKMLIDRWMLQKMQDAERLNEANDLYEEMLAKLK 188
Query: 188 ASGLEQLARQQVDGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGGCASILFEFNSSRLIT 247
A+GLE LARQQVDG+VRR E+G + F++S IPSTC R++AHEGGC SI+FE+NS L T
Sbjct: 189 ANGLETLARQQVDGIVRRNEDGTDHFVESTIPSTCANRIHAHEGGCGSIVFEYNSGTLFT 248
Query: 248 GGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVR 307
GGQD++VK+WDTN+G++ +L+G +G++LD+ +THDN+SVIAA+SSNNL+VWD++SGRVR
Sbjct: 249 GGQDRAVKMWDTNSGTLIKSLYGSLGNILDMAVTHDNKSVIAATSSNNLFVWDVSSGRVR 308
Query: 308 HTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMD 367
HTLTGH DKVCAVDVSK SSRHVVSAAYDRTIK+WDL KGYCTNT++F SNCNA+C S+D
Sbjct: 309 HTLTGHTDKVCAVDVSKFSSRHVVSAAYDRTIKLWDLHKGYCTNTVLFTSNCNAICLSID 368
Query: 368 GQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDV 427
G T++SGH+DGNLRLWDIQ+GKLLSEVA HS AVTS+SLSRNGN +LTSGRDNVHN+FD
Sbjct: 369 GLTVFSGHMDGNLRLWDIQTGKLLSEVAGHSSAVTSVSLSRNGNRILTSGRDNVHNVFDT 428
Query: 428 RSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTS 487
R+LE+C + R +GNR+ASNWSRSCISPD+ +VAAGSADGSVH+WS+ K IVS LKE TS
Sbjct: 429 RTLEICGTLRASGNRLASNWSRSCISPDDDYVAAGSADGSVHVWSLSKGNIVSILKEQTS 488
Query: 488 SVLCCRWSGIGKPLASADKNGIVCIW 513
+LCC WSGIGKPLASADKNG VC W
Sbjct: 489 PILCCSWSGIGKPLASADKNGYVCTW 514
>gb|AAK71564.1| putative WD-repeat protein [Oryza sativa (japonica cultivar-group)]
gi|50918651|ref|XP_469722.1| putative WD-repeat protein
[Oryza sativa (japonica cultivar-group)]
Length = 509
Score = 559 bits (1441), Expect = e-158
Identities = 280/510 (54%), Positives = 379/510 (73%), Gaps = 8/510 (1%)
Query: 8 QEEIAQKAINHALKSLRKRHLLEEAAHAPAFLALSRPIVSQGSEWKEKAENLQVELQQCY 67
+ E ++AI AL+SLR+RHL+EE AH PA AL+RP +Q EWKEKAE ++ELQQCY
Sbjct: 3 EAEAGKEAIRRALRSLRRRHLVEEGAHRPAIEALARPFAAQAVEWKEKAEKHELELQQCY 62
Query: 68 KAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELTEMRDEFSQLKVDLEQKIKELE 127
KAQSRLSEQLV E+ + +ASKAL++EK+ I +Q EL + R+E +QLK LE+K L+
Sbjct: 63 KAQSRLSEQLVTEIEEGKASKALLKEKETLITTMQTELEQTREENTQLKQSLEEKTSALD 122
Query: 128 VIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANALYEEMVERLR 187
+I+ E+ +KA+LEQ + K AE EN+ LIDRWMLEKMKDAERLNEANA+YEEMV +L+
Sbjct: 123 LIIQEHQAVKAELEQALTKQKVAEDENRNLIDRWMLEKMKDAERLNEANAMYEEMVLKLK 182
Query: 188 AS---GLEQLARQQVDGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGGCASILFEFNSSR 244
++ G++ A Q+ DG++RR E G +++ IPSTC+ + AH+GGC SI+F+ N+ +
Sbjct: 183 SAGVGGIQHNALQEADGIIRRSEAGYMDIMETPIPSTCRITIRAHDGGCGSIIFQHNTDK 242
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSG 304
LI+GGQDQ+VK+W +TG++ S L GC+GSV DL +T+DN+ VIAA SSN L+VW++N G
Sbjct: 243 LISGGQDQTVKIWSAHTGALTSTLQGCLGSVNDLAVTNDNKFVIAACSSNKLFVWEVNGG 302
Query: 305 RVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCF 364
R RHTLTGH V +VD S V S + S++ D TIK+WDL G+C +TI+ SN N+L F
Sbjct: 303 RPRHTLTGHTKNVSSVDASWVKSCVLASSSNDHTIKIWDLQSGFCKSTIMSGSNANSLAF 362
Query: 365 SMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNL 424
+DG T+ SGH DG+LRLWDI+S K S+ AH L V+S+S+SRN N +LTSG+DNVHNL
Sbjct: 363 -IDGVTLCSGHRDGHLRLWDIRSAKCTSQTFAH-LDVSSVSVSRNRNFILTSGKDNVHNL 420
Query: 425 FDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWS-VHKNEIVSTLK 483
FD R++EVC F+ GNRV S+W R CISPD + +AAG+ DGSV+IWS + K+ + + L+
Sbjct: 421 FDPRTMEVCGKFKAMGNRVVSSWGRPCISPDENSIAAGANDGSVYIWSRLKKDGVPTILQ 480
Query: 484 EHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
H+SSV+ W G+G PLA+ADK+ I IW
Sbjct: 481 GHSSSVVSSSWCGLG-PLATADKHHIY-IW 508
>ref|XP_643673.1| hypothetical protein DDB0215341 [Dictyostelium discoideum]
gi|2407788|gb|AAB70659.1| TipD [Dictyostelium
discoideum] gi|3122952|sp|O15736|TIPD_DICDI Protein tipD
gi|60471772|gb|EAL69727.1| hypothetical protein
DDB0215341 [Dictyostelium discoideum]
gi|7489899|pir||T08602 protein TipD - slime mold
(Dictyostelium discoideum)
Length = 612
Score = 251 bits (640), Expect = 5e-65
Identities = 166/499 (33%), Positives = 257/499 (51%), Gaps = 53/499 (10%)
Query: 51 EWKEKAENLQVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELTEMRD 110
E ++K LQ +L YK AD+ +S L+ +K+ DLQ EL
Sbjct: 132 EMEQKLFKLQEDLTNSYKRN-----------ADNASSILLLNDKNK---DLQNELMSKEI 177
Query: 111 EFSQLKVDLEQ---KIKELEVIVSEN-----------SELKAQLEQMTAQCKKAEAENKM 156
E +++ ++Q IK LE++V E S L+ + ++ K E EN
Sbjct: 178 EIERIRSTIQQDLDSIKRLEMVVIEKENVSQIIRDELSSLQTEFLHNESKVVKLEQENSS 237
Query: 157 LIDRWMLEKMKDAERLNEANALYEEMVER---LRASGLEQLARQQVDGVVRRCE------ 207
L++RW+ +K ++A ++NEAN Y++MVE+ A QL+ + VV+ +
Sbjct: 238 LVERWLRKKNEEASKMNEANDFYQKMVEQRDSTPAKAAVQLSESISNLVVKLPDANDVPI 297
Query: 208 ----EGAEFFLDSNIPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGS 263
E F ++ +PS K R H + F + L TGG D+ VKVWD +G
Sbjct: 298 PIVLERGVFSSEAMLPSKAKKRWTGHNSEIYCMAFNSIGNLLATGGGDKCVKVWDVISGQ 357
Query: 264 VCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVS 323
S L G S++ ++ + ++ S++ S+ N+ +W+ GR RHTLTGH KV
Sbjct: 358 QKSTLLGASQSIVSVSFSPNDESILGTSNDNSARLWNTELGRSRHTLTGHIGKVYTGKF- 416
Query: 324 KVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNAL-CFSMDGQTIYSGHVDGNLRL 382
++S VV+ ++DRTIK+WDL KGYCT TI S+CN L G + SGHVD ++R
Sbjct: 417 -INSNRVVTGSHDRTIKLWDLQKGYCTRTIFCFSSCNDLVILGGSGTHLASGHVDHSVRF 475
Query: 383 WDIQSG---KLLSEVAAHSLAVTSISLS-RNGNVVLTSGRDNVHNLFDVRSLEVCSSFRD 438
WD +G ++LS + H +TSI+ S N N +LT+ RD+ + D+R+ + +F+D
Sbjct: 476 WDSNAGEPTQVLSSI--HEGQITSITNSPTNTNQILTNSRDHTLKIIDIRTFDTIRTFKD 533
Query: 439 TGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTL-KEHT--SSVLCCRWS 495
R NW+++ SPD ++A+GS DGS+ IW + V L K H SSV CC WS
Sbjct: 534 PEYRNGLNWTKASWSPDGRYIASGSIDGSICIWDATNGKTVKVLTKVHNNGSSVCCCSWS 593
Query: 496 GIGKPLASADKNGIVCIWK 514
+ SADK+ + W+
Sbjct: 594 PLANIFISADKDKNIIQWE 612
>ref|XP_343620.2| PREDICTED: similar to Apg16L alpha [Rattus norvegicus]
Length = 603
Score = 207 bits (527), Expect = 6e-52
Identities = 145/525 (27%), Positives = 253/525 (47%), Gaps = 70/525 (13%)
Query: 47 SQGSEWKEKAENLQVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELT 106
SQ E + Q EL + +K + L+ QLV+++ + +Q+KD I + ++
Sbjct: 88 SQLQEMAQLKMKHQEELTELHKKRGELA-QLVIDLNNQ------MQQKDKEIQTNEAKIA 140
Query: 107 EMRDEFSQLK---VDLEQKIKELEV----IVSENSELKAQLEQMTAQCKKAEAENKMLID 159
E S L+ +DL K+++LEV + E L+ + + +K EN+ L+
Sbjct: 141 ECLQTISDLEADCLDLRTKLQDLEVANQTLKDEYDALQITFTALEEKLRKTTEENQELVT 200
Query: 160 RWMLEKMKDAERLN---EANALYEEMVERLRASGLE-QLARQQVDGVVRRCEEGAEFFLD 215
RWM EK ++A RLN E ++ ++ +++ R + L+ +LA + + ++ E +D
Sbjct: 201 RWMAEKAQEANRLNAENEKDSRWDIQLQQRRQARLQKELAEAAKEPLPVEQDDDIEVIVD 260
Query: 216 SN-------------------------------------------IPSTCKFRLNAHEGG 232
+P+T + +AH+G
Sbjct: 261 ETSDHTEETSPVRAISRAATRRSVSSIPVPQDVVDTHPASGKDVRVPTTASYVFDAHDGE 320
Query: 233 CASILFEFNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGCIGSVLDLTITHDNRSVIA 289
++ F S L TGG D+ VK+W+ G C +L G + + ++A
Sbjct: 321 VNAVQFSPGSRLLATGGMDRRVKLWEA-FGDKCEFKGSLSGSNAGITSIEFDSAGAYLLA 379
Query: 290 ASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYC 349
AS+ +W ++ R+RHTLTGH KV + ++R +VS ++DRT+K+WDL C
Sbjct: 380 ASNDFASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVC 438
Query: 350 TNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRN 409
T+ S+CN + + Q + SGH D +R WDI+S ++ E+ +T++ L+
Sbjct: 439 IKTVFAGSSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESVVREMELLG-KITALDLNPE 495
Query: 410 GNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVH 469
+L+ RD++ + D+R+ + SF G + S+W+R SPD S+VAAGSADGS++
Sbjct: 496 RTELLSCSRDDLLKIIDLRTNAIKQSFSAPGFKCGSDWTRVVFSPDGSYVAAGSADGSLY 555
Query: 470 IWSVHKNEIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
+WSV ++ L K+H+SS+ W+ G + S DK +W
Sbjct: 556 VWSVLTGKVEKVLSKQHSSSINAVAWAPSGLHVVSVDKGSRAVLW 600
>dbj|BAC55090.1| Apg16L alpha [Mus musculus]
Length = 588
Score = 205 bits (522), Expect = 2e-51
Identities = 142/505 (28%), Positives = 244/505 (48%), Gaps = 63/505 (12%)
Query: 60 QVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELTEMRDEFSQLK--- 116
Q EL + +K + L+ QLV+++ + +Q+KD I + +++E S L+
Sbjct: 93 QEELTELHKKRGELA-QLVIDLNNQ------MQQKDKEIQMNEAKISEYLQTISDLETNC 145
Query: 117 VDLEQKIKELEV----IVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERL 172
+DL K+++LEV + E L+ + + +K EN+ L+ RWM EK ++A RL
Sbjct: 146 LDLRTKLQDLEVANQTLKDEYDALQITFTALEEKLRKTTEENQELVTRWMAEKAQEANRL 205
Query: 173 NEANAL-----YEEMVERLRASGLEQLARQQVDGVVRRCEEGAEFF-------------- 213
N N + + L + E L +Q D + +E ++
Sbjct: 206 NAENEKDSRRRQARLQKELAEAAKEPLPVEQDDDIEVIVDETSDHTEETSPVRAVSRAAT 265
Query: 214 ---------------------LDSNIPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQ 252
D +P+T + +AH+G ++ F S L TGG D+
Sbjct: 266 RRSVSSIPVPQDIMDTHPASGKDVRVPTTASYVFDAHDGEVNAVQFSPGSRLLATGGMDR 325
Query: 253 SVKVWDTNTGSVCS---NLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHT 309
VK+W+ G C +L G + + ++AAS+ +W ++ R+RHT
Sbjct: 326 RVKLWEA-FGDKCEFKGSLSGSNAGITSIEFDSAGAYLLAASNDFASRIWTVDDYRLRHT 384
Query: 310 LTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQ 369
LTGH KV + ++R +VS ++DRT+K+WDL C T+ S+CN + + Q
Sbjct: 385 LTGHSGKVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVCIKTVFAGSSCNDIVCTE--Q 441
Query: 370 TIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRS 429
+ SGH D +R WDI+S ++ E+ +T++ L+ +L+ RD++ + D+R+
Sbjct: 442 CVMSGHFDKKIRFWDIRSESVVREMELLG-KITALDLNPERTELLSCSRDDLLKVIDLRT 500
Query: 430 LEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTL-KEHTSS 488
V +F G + S+W+R SPD S+VAAGSA+GS+++WSV ++ L K+H+SS
Sbjct: 501 NAVKQTFSAPGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYVWSVLTGKVEKVLSKQHSSS 560
Query: 489 VLCCRWSGIGKPLASADKNGIVCIW 513
+ W+ G + S DK +W
Sbjct: 561 INAVAWAPSGLHVVSVDKGSRAVLW 585
>ref|NP_084122.2| APG16L beta isoform [Mus musculus]
gi|62510494|sp|Q8C0J2|APG16_MOUSE Autophagy protein
16-like (APG16-like) gi|26326915|dbj|BAC27201.1| unnamed
protein product [Mus musculus]
Length = 607
Score = 202 bits (514), Expect = 2e-50
Identities = 148/537 (27%), Positives = 250/537 (45%), Gaps = 82/537 (15%)
Query: 47 SQGSEWKEKAENLQVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELT 106
SQ E + Q EL + +K + L+ QLV+++ + +Q+KD I + +++
Sbjct: 80 SQLQEMAQLRIKHQEELTELHKKRGELA-QLVIDLNNQ------MQQKDKEIQMNEAKIS 132
Query: 107 EMRDEFSQLK---VDLEQKIKELEV----IVSENSELKAQLEQMTAQCKKAEAENKMLID 159
E S L+ +DL K+++LEV + E L+ + + +K EN+ L+
Sbjct: 133 EYLQTISDLETNCLDLRTKLQDLEVANQTLKDEYDALQITFTALEEKLRKTTEENQELVT 192
Query: 160 RWMLEKMKDAERLNEANAL-----YEEMVERLRASGLEQLARQQVDGV------------ 202
RWM EK ++A RLN N + + L + E L +Q D +
Sbjct: 193 RWMAEKAQEANRLNAENEKDSRRRQARLQKELAEAAKEPLPVEQDDDIEVIVDETSDHTE 252
Query: 203 ------------VRRCEEGAEFFLDS------------------------------NIPS 220
+R + A LDS +P+
Sbjct: 253 ETSPVRAVSRAATKRLSQPAGGLLDSITNIFGRRSVSSIPVPQDIMDTHPASGKDVRVPT 312
Query: 221 TCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGCIGSVLD 277
T + +AH+G ++ F S L TGG D+ VK+W+ G C +L G +
Sbjct: 313 TASYVFDAHDGEVNAVQFSPGSRLLATGGMDRRVKLWEA-FGDKCEFKGSLSGSNAGITS 371
Query: 278 LTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDR 337
+ ++AAS+ +W ++ R+RHTLTGH KV + ++R +VS ++DR
Sbjct: 372 IEFDSAGAYLLAASNDFASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDR 430
Query: 338 TIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAH 397
T+K+WDL C T+ S+CN + + Q + SGH D +R WDI+S ++ E+
Sbjct: 431 TLKLWDLRSKVCIKTVFAGSSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESVVREMELL 488
Query: 398 SLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNS 457
+T++ L+ +L+ RD++ + D+R+ V +F G + S+W+R SPD S
Sbjct: 489 G-KITALDLNPERTELLSCSRDDLLKVIDLRTNAVKQTFSAPGFKCGSDWTRVVFSPDGS 547
Query: 458 HVAAGSADGSVHIWSVHKNEIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
+VAAGSA+GS+++WSV ++ L K+H+SS+ W+ G + S DK +W
Sbjct: 548 YVAAGSAEGSLYVWSVLTGKVEKVLSKQHSSSINAVAWAPSGLHVVSVDKGSRAVLW 604
>dbj|BAC55091.1| Apg16L beta [Mus musculus]
Length = 607
Score = 202 bits (513), Expect = 3e-50
Identities = 145/524 (27%), Positives = 246/524 (46%), Gaps = 82/524 (15%)
Query: 60 QVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELTEMRDEFSQLK--- 116
Q EL + +K + L+ QLV+++ + +Q+KD I + +++E S L+
Sbjct: 93 QEELTELHKKRGELA-QLVIDLNNQ------MQQKDKEIQMNEAKISEYLQTISDLETNC 145
Query: 117 VDLEQKIKELEV----IVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERL 172
+DL K+++LEV + E L+ + + +K EN+ L+ RWM EK ++A RL
Sbjct: 146 LDLRTKLQDLEVANQTLKDEYDALQITFTALEEKLRKTTEENQELVTRWMAEKAQEANRL 205
Query: 173 NEANAL-----YEEMVERLRASGLEQLARQQVDGV------------------------V 203
N N + + L + E L +Q D +
Sbjct: 206 NAENEKDSRRRQARLQKELAEAAKEPLPVEQDDDIEVIVDETSDHTEETSPVRAVSRAAT 265
Query: 204 RRCEEGAEFFLDS------------------------------NIPSTCKFRLNAHEGGC 233
+R + A LDS +P+T + +AH+G
Sbjct: 266 KRLSQPAGGLLDSITNIFGRRSVSSIPVPQDIMDTHPASGKDVRVPTTASYVFDAHDGEV 325
Query: 234 ASILFEFNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGCIGSVLDLTITHDNRSVIAA 290
++ F S L TGG D+ VK+W+ G C +L G + + ++AA
Sbjct: 326 NAVQFSPGSRLLATGGMDRRVKLWEA-FGDKCEFKGSLSGSNAGITSIEFDSAGAYLLAA 384
Query: 291 SSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCT 350
S+ +W ++ R+RHTLTGH KV + ++R +VS ++DRT+K+WDL C
Sbjct: 385 SNDFASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVCI 443
Query: 351 NTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNG 410
T+ S+CN + + Q + SGH D +R WDI+S ++ E+ +T++ L+
Sbjct: 444 KTVFAGSSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESVVREMELLG-KITALDLNPER 500
Query: 411 NVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHI 470
+L+ RD++ + D+R+ V +F G + S+W+R SPD S+VAAGSA+GS+++
Sbjct: 501 TELLSCSRDDLLKVIDLRTNAVKQTFSAPGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYV 560
Query: 471 WSVHKNEIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
WSV ++ L K+H+SS+ W+ G + S DK +W
Sbjct: 561 WSVLTGKVEKVLSKQHSSSINAVAWAPSGLHVVSVDKGSRAVLW 604
>gb|AAH88497.1| Unknown (protein for MGC:97562) [Xenopus tropicalis]
gi|62510473|sp|Q5I0B9|APG16_XENTR Autophagy protein
16-like (APG16-like)
Length = 537
Score = 201 bits (511), Expect = 5e-50
Identities = 145/525 (27%), Positives = 250/525 (47%), Gaps = 33/525 (6%)
Query: 6 KSQEEIAQKAINHALKSLRKRHL---LEEAAHAPAFLALSRPIVSQGSEWKEKAENLQVE 62
+ Q ++ ++ I+ + L K L L + A + SR S G + LQ
Sbjct: 26 REQRQVFEELISQYKRLLEKSDLQSVLADKLQAEKYEQQSRHDSSPGPDGMRNDMLLQDM 85
Query: 63 LQQCYKAQSRLSE---------QLVVEVADSRASKAL-VQEKDNAIADLQKELTEMRDEF 112
Q K Q L+E Q V+E+ + K +Q + IA + + ++ E
Sbjct: 86 AQMRIKHQEELTELHKKRGELAQTVIELNNQMQQKDKEIQANEEKIAKYLQTIQDLETEC 145
Query: 113 SQLKVDLEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERL 172
L+ L++ + + + E L+ + + +K +N+ L+ RWM EK ++A RL
Sbjct: 146 QDLRNKLQELERANQTLKDEYDALQITFTALEDKLRKTTEDNQELVSRWMAEKAQEANRL 205
Query: 173 NEANALYEEMVERLRASGLEQLARQQVDGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGG 232
N N ++ R S G R + +P+T + +AH+G
Sbjct: 206 NAENE--KDSKRRSLNSFPASPDCADTPGAGNR---------EVRVPATAVYSFDAHDGE 254
Query: 233 CASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLT-ITHDNRS--VIA 289
++ F S L TGG D+ VK+WD G+ C GS +T I D+ ++A
Sbjct: 255 VNAVRFSPGSRLLATGGMDRRVKLWDV-IGNKCEAKGSLTGSNAGITSIEFDSAGSYLLA 313
Query: 290 ASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYC 349
AS+ +W ++ R+RHTLTGH KV + ++R +VS ++DRT+K+WDL C
Sbjct: 314 ASNDFASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVC 372
Query: 350 TNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRN 409
T+ S+CN + + Q + SGH D +R WDI++ ++ E+ +T++ L+
Sbjct: 373 IKTVFAGSSCNDIVCTE--QCVMSGHFDKKIRFWDIRTECIVRELELQG-RITALDLNPE 429
Query: 410 GNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVH 469
+L+ RD++ + D+R+ V +F G + S+W+R SPD S+V+AGSA+G+++
Sbjct: 430 RTQLLSCSRDDLIKITDLRANAVQQTFSAPGFKCGSDWTRVIFSPDGSYVSAGSAEGTLY 489
Query: 470 IWSVHKNEIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
W+V ++ K+H+SS+ WS G + S DK +W
Sbjct: 490 FWNVLTGKVERMFSKQHSSSINAVAWSPSGTHVVSVDKGSRGVLW 534
>dbj|BAB55412.1| unnamed protein product [Homo sapiens]
Length = 504
Score = 199 bits (506), Expect = 2e-49
Identities = 137/499 (27%), Positives = 239/499 (47%), Gaps = 51/499 (10%)
Query: 60 QVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNA-IADLQKELTEMRDEFSQLKVD 118
Q EL + +K + L+ QLV+++ + K + + A IA+ + ++++ E L+
Sbjct: 9 QEELTELHKKRGELA-QLVIDLNNQMQRKDREMQMNEAKIAECLQTISDLETECLDLRTR 67
Query: 119 LEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANAL 178
L + + + E L+ + + +K EN+ L+ RWM EK ++A RLN N
Sbjct: 68 LCDLERANQTLKDEYDALQITFTALEGKLRKTTEENQELVTRWMAEKAQEANRLNAENEK 127
Query: 179 -----YEEMVERLRASGLEQLARQQVDGVVRRCEEGAEFFLDSN---------------- 217
+ + L + E L +Q D + +E ++ +++
Sbjct: 128 DSRRRQARLQKELAEAAKEPLPVEQDDDIEVIVDETSDHTEETSPVRAISRAATRRSVSS 187
Query: 218 -------------------IPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWD 258
+P+T +AH+G + F S L TGG D+ VK+W+
Sbjct: 188 FPVPQDNVDTHPGSGKEVRVPATALCVFDAHDGEVNAAQFSPGSRLLATGGMDRRVKLWE 247
Query: 259 TNTGSVCS---NLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKD 315
G C +L G + + ++AAS+ +W ++ R+RHTLTGH
Sbjct: 248 V-FGEKCEFKGSLSGSNAGITSIEFDSAGSYLLAASNDFASRIWTVDDYRLRHTLTGHSG 306
Query: 316 KVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGH 375
KV + ++R +VS ++DRT+K+WDL C T+ S+CN + + Q + SGH
Sbjct: 307 KVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVCIKTVFAGSSCNDIVCTE--QCVMSGH 363
Query: 376 VDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSS 435
D +R WDI+S ++ E+ +T++ L+ +L+ RD++ + D+R+ + +
Sbjct: 364 FDKKIRFWDIRSESIVREMELLG-KITALDLNPERTELLSCSRDDLLKVIDLRTNAIKQT 422
Query: 436 FRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTL-KEHTSSVLCCRW 494
F TG + S+W+R SPD S+VAAGSA+GS++IWSV ++ L K+H+SS+ W
Sbjct: 423 FSATGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYIWSVLTGKVEKVLSKQHSSSINAVAW 482
Query: 495 SGIGKPLASADKNGIVCIW 513
S G + S DK +W
Sbjct: 483 SPSGSHVVSVDKGCKAVLW 501
>ref|NP_060444.2| APG16 autophagy 16-like isoform 2 [Homo sapiens]
Length = 504
Score = 199 bits (505), Expect = 2e-49
Identities = 136/499 (27%), Positives = 239/499 (47%), Gaps = 51/499 (10%)
Query: 60 QVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNA-IADLQKELTEMRDEFSQLKVD 118
Q EL + +K + L+ QLV+++ + K + + A IA+ + ++++ E L+
Sbjct: 9 QEELTELHKKRGELA-QLVIDLNNQMQRKDREMQMNEAKIAECLQTISDLETECLDLRTK 67
Query: 119 LEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANAL 178
L + + + E L+ + + +K EN+ L+ RWM EK ++A RLN N
Sbjct: 68 LCDLERANQTLKDEYDALQITFTALEGKLRKTTEENQELVTRWMAEKAQEANRLNAENEK 127
Query: 179 -----YEEMVERLRASGLEQLARQQVDGVVRRCEEGAEFFLDSN---------------- 217
+ + L + E L +Q D + +E ++ +++
Sbjct: 128 DSRRRQARLQKELAEAAKEPLPVEQDDDIEVIVDETSDHTEETSPVRAISRAATRRSVSS 187
Query: 218 -------------------IPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWD 258
+P+T +AH+G ++ F S L TGG D+ VK+W+
Sbjct: 188 FPVPQDNVDTHPGSGKEVRVPATALCVFDAHDGEVNAVQFSPGSRLLATGGMDRRVKLWE 247
Query: 259 TNTGSVCS---NLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKD 315
G C +L G + + ++AAS+ +W ++ R+RHTLTGH
Sbjct: 248 V-FGEKCEFKGSLSGSNAGITSIEFDSAGSYLLAASNDFASRIWTVDDYRLRHTLTGHSG 306
Query: 316 KVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGH 375
KV + ++R +VS ++DRT+K+WDL C T+ S+CN + + Q + SGH
Sbjct: 307 KVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVCIKTVFAGSSCNDIVCTE--QCVMSGH 363
Query: 376 VDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSS 435
D +R WDI+S ++ E+ +T++ L+ +L+ RD++ + D+R+ + +
Sbjct: 364 FDKKIRFWDIRSESIVREMELLG-KITALDLNPERTELLSCSRDDLLKVIDLRTNAIKQT 422
Query: 436 FRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTL-KEHTSSVLCCRW 494
F G + S+W+R SPD S+VAAGSA+GS++IWSV ++ L K+H+SS+ W
Sbjct: 423 FSAPGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYIWSVLTGKVEKVLSKQHSSSINAVAW 482
Query: 495 SGIGKPLASADKNGIVCIW 513
S G + S DK +W
Sbjct: 483 SPSGSHVVSVDKGCKAVLW 501
>ref|XP_534608.1| PREDICTED: similar to APG16L beta [Canis familiaris]
Length = 616
Score = 196 bits (498), Expect = 1e-48
Identities = 131/483 (27%), Positives = 229/483 (47%), Gaps = 50/483 (10%)
Query: 76 QLVVEVADSRASKALVQEKDNA-IADLQKELTEMRDEFSQLKVDLEQKIKELEVIVSENS 134
QLV+++ + K + + A IA+ + ++++ E +L+ L+ + + + E
Sbjct: 136 QLVIDLNNQMQQKDREMQMNEAKIAECLQTISDLETECQELRSKLQDLERANQTLKDEYD 195
Query: 135 ELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANAL-----YEEMVERLRAS 189
L+ + + +K EN+ L+ RWM EK ++A RLN N + + L +
Sbjct: 196 ALQITFTALEEKLRKTTEENQELVTRWMAEKAQEANRLNAENEKDSRRRQARLQKELAEA 255
Query: 190 GLEQLARQQVDGVVRRCEEGAEFFLDSN-------------------------------- 217
E L +Q D + +E ++ +++
Sbjct: 256 AKEPLPVEQDDDIEVIVDETSDHAEETSPVRAISRAATRRSVSSFPVPQDNVDTHPGSSK 315
Query: 218 ---IPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGC 271
+P+T +AH+G ++ F S L TGG D+ VK+W+ G C +L G
Sbjct: 316 EVRVPTTAMCIFDAHDGEVNAVQFSPGSRLLATGGMDRRVKLWEV-FGDKCEFKGSLSGS 374
Query: 272 IGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVV 331
+ + ++AAS+ +W ++ R+RHTLTGH KV + ++R +V
Sbjct: 375 NAGITSIEFDSAGSYLLAASNDFASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IV 433
Query: 332 SAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLL 391
S ++DRT+K+WDL C T+ S+CN + + Q + SGH D +R WDI+S ++
Sbjct: 434 SGSHDRTLKLWDLRSKVCIKTVFAGSSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESIV 491
Query: 392 SEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSC 451
E+ +T++ L+ +L+ RD++ + D+R V +F G + S+W+R
Sbjct: 492 REMELLG-KITALDLNPERTELLSCSRDDLLKIIDLRINAVRQTFSAPGFKCGSDWTRVV 550
Query: 452 ISPDNSHVAAGSADGSVHIWSVHKNEIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIV 510
SPD +VAAGSA+GS++IWSV ++ L K+H SS+ WS G + S DK
Sbjct: 551 FSPDGGYVAAGSAEGSLYIWSVLSGKVEKVLSKQHGSSINAVAWSPSGSHVVSVDKGSKA 610
Query: 511 CIW 513
+W
Sbjct: 611 VLW 613
>gb|AAY14860.1| unknown [Homo sapiens] gi|31742530|ref|NP_110430.4| APG16 autophagy
16-like isoform 1 [Homo sapiens]
Length = 523
Score = 196 bits (497), Expect = 2e-48
Identities = 140/518 (27%), Positives = 239/518 (46%), Gaps = 70/518 (13%)
Query: 60 QVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNA-IADLQKELTEMRDEFSQLKVD 118
Q EL + +K + L+ QLV+++ + K + + A IA+ + ++++ E L+
Sbjct: 9 QEELTELHKKRGELA-QLVIDLNNQMQRKDREMQMNEAKIAECLQTISDLETECLDLRTK 67
Query: 119 LEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANAL 178
L + + + E L+ + + +K EN+ L+ RWM EK ++A RLN N
Sbjct: 68 LCDLERANQTLKDEYDALQITFTALEGKLRKTTEENQELVTRWMAEKAQEANRLNAENEK 127
Query: 179 -----YEEMVERLRASGLEQLARQQVDGV------------------------VRRCEEG 209
+ + L + E L +Q D + +R +
Sbjct: 128 DSRRRQARLQKELAEAAKEPLPVEQDDDIEVIVDETSDHTEETSPVRAISRAATKRLSQP 187
Query: 210 AEFFLDS------------------------------NIPSTCKFRLNAHEGGCASILFE 239
A LDS +P+T +AH+G ++ F
Sbjct: 188 AGGLLDSITNIFGRRSVSSFPVPQDNVDTHPGSGKEVRVPATALCVFDAHDGEVNAVQFS 247
Query: 240 FNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGCIGSVLDLTITHDNRSVIAASSSNNL 296
S L TGG D+ VK+W+ G C +L G + + ++AAS+
Sbjct: 248 PGSRLLATGGMDRRVKLWEV-FGEKCEFKGSLSGSNAGITSIEFDSAGSYLLAASNDFAS 306
Query: 297 YVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFA 356
+W ++ R+RHTLTGH KV + ++R +VS ++DRT+K+WDL C T+
Sbjct: 307 RIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVCIKTVFAG 365
Query: 357 SNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTS 416
S+CN + + Q + SGH D +R WDI+S ++ E+ +T++ L+ +L+
Sbjct: 366 SSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESIVREMELLG-KITALDLNPERTELLSC 422
Query: 417 GRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKN 476
RD++ + D+R+ + +F G + S+W+R SPD S+VAAGSA+GS++IWSV
Sbjct: 423 SRDDLLKVIDLRTNAIKQTFSAPGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYIWSVLTG 482
Query: 477 EIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++ L K+H+SS+ WS G + S DK +W
Sbjct: 483 KVEKVLSKQHSSSINAVAWSPSGSHVVSVDKGCKAVLW 520
>gb|AAR32130.1| APG16L beta [Homo sapiens]
Length = 607
Score = 196 bits (497), Expect = 2e-48
Identities = 140/518 (27%), Positives = 239/518 (46%), Gaps = 70/518 (13%)
Query: 60 QVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNA-IADLQKELTEMRDEFSQLKVD 118
Q EL + +K + L+ QLV+++ + K + + A IA+ + ++++ E L+
Sbjct: 93 QEELTELHKKRGELA-QLVIDLNNQMQRKDREMQMNEAKIAECLQTISDLETECLDLRTK 151
Query: 119 LEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANAL 178
L + + + E L+ + + +K EN+ L+ RWM EK ++A RLN N
Sbjct: 152 LCDLERANQTLKDEYDALQITFTALEGKLRKTTEENQELVTRWMAEKAQEANRLNAENEK 211
Query: 179 -----YEEMVERLRASGLEQLARQQVDGV------------------------VRRCEEG 209
+ + L + E L +Q D + +R +
Sbjct: 212 DSRRRQARLQKELAEAAKEPLPVEQDDDIEVIVDETSDHTEETSPVRAISRAATKRLSQP 271
Query: 210 AEFFLDS------------------------------NIPSTCKFRLNAHEGGCASILFE 239
A LDS +P+T +AH+G ++ F
Sbjct: 272 AGGLLDSITNIFGRRSVSSFPVPQDNVDAHPGSGKEVRVPATALCVFDAHDGEVNAVQFS 331
Query: 240 FNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGCIGSVLDLTITHDNRSVIAASSSNNL 296
S L TGG D+ VK+W+ G C +L G + + ++AAS+
Sbjct: 332 PGSRLLATGGMDRRVKLWEV-FGEKCEFKGSLSGSNAGITSIEFDSAGSYLLAASNDFAS 390
Query: 297 YVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFA 356
+W ++ R+RHTLTGH KV + ++R +VS ++DRT+K+WDL C T+
Sbjct: 391 RIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVCIKTVFAG 449
Query: 357 SNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTS 416
S+CN + + Q + SGH D +R WDI+S ++ E+ +T++ L+ +L+
Sbjct: 450 SSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESIVREMELLG-KITALDLNPERTELLSC 506
Query: 417 GRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKN 476
RD++ + D+R+ + +F G + S+W+R SPD S+VAAGSA+GS++IWSV
Sbjct: 507 SRDDLLKVIDLRTNAIKQTFSAPGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYIWSVLTG 566
Query: 477 EIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++ L K+H+SS+ WS G + S DK +W
Sbjct: 567 KVEKVLSKQHSSSINAVAWSPSGSHVVSVDKGCKAVLW 604
>sp|Q676U5|APG16_HUMAN Autophagy protein 16-like (APG16-like)
Length = 607
Score = 196 bits (497), Expect = 2e-48
Identities = 140/518 (27%), Positives = 239/518 (46%), Gaps = 70/518 (13%)
Query: 60 QVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNA-IADLQKELTEMRDEFSQLKVD 118
Q EL + +K + L+ QLV+++ + K + + A IA+ + ++++ E L+
Sbjct: 93 QEELTELHKKRGELA-QLVIDLNNQMQRKDREMQMNEAKIAECLQTISDLETECLDLRTK 151
Query: 119 LEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEANAL 178
L + + + E L+ + + +K EN+ L+ RWM EK ++A RLN N
Sbjct: 152 LCDLERANQTLKDEYDALQITFTALEGKLRKTTEENQELVTRWMAEKAQEANRLNAENEK 211
Query: 179 -----YEEMVERLRASGLEQLARQQVDGV------------------------VRRCEEG 209
+ + L + E L +Q D + +R +
Sbjct: 212 DSRRRQARLQKELAEAAKEPLPVEQDDDIEVIVDETSDHTEETSPVRAISRAATKRLSQP 271
Query: 210 AEFFLDS------------------------------NIPSTCKFRLNAHEGGCASILFE 239
A LDS +P+T +AH+G ++ F
Sbjct: 272 AGGLLDSITNIFGRRSVSSFPVPQDNVDTHPGSGKEVRVPATALCVFDAHDGEVNAVQFS 331
Query: 240 FNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGCIGSVLDLTITHDNRSVIAASSSNNL 296
S L TGG D+ VK+W+ G C +L G + + ++AAS+
Sbjct: 332 PGSRLLATGGMDRRVKLWEV-FGEKCEFKGSLSGSNAGITSIEFDSAGSYLLAASNDFAS 390
Query: 297 YVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFA 356
+W ++ R+RHTLTGH KV + ++R +VS ++DRT+K+WDL C T+
Sbjct: 391 RIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVCIKTVFAG 449
Query: 357 SNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTS 416
S+CN + + Q + SGH D +R WDI+S ++ E+ +T++ L+ +L+
Sbjct: 450 SSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESIVREMELLG-KITALDLNPERTELLSC 506
Query: 417 GRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKN 476
RD++ + D+R+ + +F G + S+W+R SPD S+VAAGSA+GS++IWSV
Sbjct: 507 SRDDLLKVIDLRTNAIKQTFSAPGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYIWSVLTG 566
Query: 477 EIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++ L K+H+SS+ WS G + S DK +W
Sbjct: 567 KVEKVLSKQHSSSINAVAWSPSGSHVVSVDKGCKAVLW 604
>gb|AAH71846.1| APG16L protein [Homo sapiens]
Length = 475
Score = 193 bits (491), Expect = 1e-47
Identities = 132/474 (27%), Positives = 225/474 (46%), Gaps = 56/474 (11%)
Query: 91 VQEKDNAIADLQKELTEMRDEFSQLK---VDLEQKIKELE----VIVSENSELKAQLEQM 143
+Q KD + + ++ E S L+ +DL K+ +LE + E L+ +
Sbjct: 4 MQRKDREMQMNEAKIAECLQTISDLETECLDLRTKLCDLERANQTLKDEYDALQITFTAL 63
Query: 144 TAQCKKAEAENKMLIDRWMLEKMKDAERLNEANAL-----YEEMVERLRASGLEQLARQQ 198
+ +K EN+ L+ RWM EK ++A RLN N + + L + E L +Q
Sbjct: 64 EGKLRKTTEENQELVTRWMAEKAQEANRLNAENEKDSRRRQARLQKELAEAAKEPLPVEQ 123
Query: 199 VDGVVRRCEEGAEFFLDSN-----------------------------------IPSTCK 223
D + +E ++ +++ +P+T
Sbjct: 124 DDDIEVIVDETSDHTEETSPVRAISRAATRRSVSSFPVPQDNVDTHPGSGKEVRVPATAL 183
Query: 224 FRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGCIGSVLDLTI 280
+AH+G ++ F S L TGG D+ VK+W+ G C +L G + +
Sbjct: 184 CVFDAHDGEVNAVQFSPGSRLLATGGMDRRVKLWEV-FGEKCEFKGSLSGSNAGITSIEF 242
Query: 281 THDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIK 340
++AAS+ +W ++ R+RHTLTGH KV + ++R +VS ++DRT+K
Sbjct: 243 DSAGSYLLAASNDFASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDRTLK 301
Query: 341 VWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLA 400
+WDL C T+ S+CN + + Q + SGH D +R WDI+S ++ E+
Sbjct: 302 LWDLRSKVCIKTVFAGSSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESIVREMELLG-K 358
Query: 401 VTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVA 460
+T++ L+ +L+ RD++ + D+R+ + +F G + S+W+R SPD S+VA
Sbjct: 359 ITALDLNPERTELLSCSRDDLLKVIDLRTNAIKQTFSAPGFKCGSDWTRVVFSPDGSYVA 418
Query: 461 AGSADGSVHIWSVHKNEIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
AGSA+GS++IWSV ++ L K+H+SS+ WS G + S DK +W
Sbjct: 419 AGSAEGSLYIWSVLTGKVEKVLSKQHSSSINAVAWSPSGSHVVSVDKGCKAVLW 472
>emb|CAE53913.1| putative WD-repeat protein [Triticum aestivum]
Length = 188
Score = 192 bits (489), Expect = 2e-47
Identities = 90/172 (52%), Positives = 123/172 (71%), Gaps = 1/172 (0%)
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
+ AH+GGC SI+F+ N+ +LI+GGQDQ+VK+W +TG++ S L GC+G+V DL +T+DN+
Sbjct: 1 IRAHDGGCGSIIFQNNTDKLISGGQDQTVKIWSAHTGALTSTLQGCLGTVNDLAVTNDNK 60
Query: 286 SVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
VIAA SSN L+VW++N GR RHTLTGH VC+VD S V S + S++ DRTIK+WDL
Sbjct: 61 FVIAACSSNKLFVWEVNGGRPRHTLTGHTKNVCSVDASWVKSLVIASSSNDRTIKIWDLQ 120
Query: 346 KGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAH 397
G+C +TI+ + F G I SGH DG+L+ DI+SGK + VAAH
Sbjct: 121 TGFCKSTIMSSKQPQCTSFH-PGDAICSGHRDGSLKFHDIRSGKCFATVAAH 171
>dbj|BAC55092.1| Apg16L gamma [Mus musculus]
Length = 623
Score = 184 bits (468), Expect = 4e-45
Identities = 132/461 (28%), Positives = 225/461 (48%), Gaps = 22/461 (4%)
Query: 59 LQVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNAI--ADLQKELTEMRDEFSQLK 116
L+ +L++ + L + + E A EKD+ A LQKEL E E ++
Sbjct: 176 LEEKLRKTTEENQELVTRWMAEKAQEANRLNAENEKDSRRRQARLQKELAEAAKEPLPVE 235
Query: 117 VDLEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEAN 176
D ++EVIV E S+ + + A +A + +L+ + + L+E+
Sbjct: 236 QD-----DDIEVIVDETSDHTEETSPVRA-VSRAATKRLSQPAGGLLDSITNIFGLSESP 289
Query: 177 ALYEEMVERLRASGLEQLARQQVDGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGGCASI 236
L + R + + Q D + G D +P+T + +AH+G ++
Sbjct: 290 LLGHHSSDAARRRSVSSIPVPQ-DIMDTHPASGK----DVRVPTTASYVFDAHDGEVNAV 344
Query: 237 LFEFNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGCIGSVLDLTITHDNRSVIAASSS 293
F S L TGG D+ VK+W+ G C +L G + + ++AAS+
Sbjct: 345 QFSPGSRLLATGGMDRRVKLWEA-FGDKCEFKGSLSGSNAGITSIEFDSAGAYLLAASND 403
Query: 294 NNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTI 353
+W ++ R+RHTLTGH KV + ++R +VS ++DRT+K+WDL C T+
Sbjct: 404 FASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVCIKTV 462
Query: 354 IFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVV 413
S+CN + + Q + SGH D +R WDI+S ++ E+ +T++ L+ +
Sbjct: 463 FAGSSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESVVREMELLG-KITALDLNPERTEL 519
Query: 414 LTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSV 473
L+ RD++ + D+R+ V +F G + S+W+R SPD S+VAAGSA+GS+++WSV
Sbjct: 520 LSCSRDDLLKVIDLRTNAVKQTFSAPGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYVWSV 579
Query: 474 HKNEIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++ L K+H+SS+ W+ G + S DK +W
Sbjct: 580 LTGKVEKVLSKQHSSSINAVAWAPSGLHVVSVDKGSRAVLW 620
>gb|AAH49122.1| Apg16l protein [Mus musculus]
Length = 623
Score = 184 bits (468), Expect = 4e-45
Identities = 132/461 (28%), Positives = 225/461 (48%), Gaps = 22/461 (4%)
Query: 59 LQVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNAI--ADLQKELTEMRDEFSQLK 116
L+ +L++ + L + + E A EKD+ A LQKEL E E ++
Sbjct: 176 LEEKLRKTTEENQELVTRWMAEKAQEANRLNAENEKDSRRRQARLQKELAEAAKEPLPVE 235
Query: 117 VDLEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERLNEAN 176
D ++EVIV E S+ + + A +A + +L+ + + L+E+
Sbjct: 236 QD-----DDIEVIVDETSDHTEETSPVRA-VSRAATKRLSQPAGGLLDSITNIFGLSESP 289
Query: 177 ALYEEMVERLRASGLEQLARQQVDGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGGCASI 236
L + R + + Q D + G D +P+T + +AH+G ++
Sbjct: 290 LLGHHSSDAARRRSVSSIPVPQ-DIMDTHPASGK----DVRVPTTASYVFDAHDGEVNAV 344
Query: 237 LFEFNSSRLITGGQDQSVKVWDTNTGSVCS---NLHGCIGSVLDLTITHDNRSVIAASSS 293
F S L TGG D+ VK+W+ G C +L G + + ++AAS+
Sbjct: 345 QFSPGSRLLATGGMDRRVKLWEA-FGDKCEFKGSLSGSNAGITSIEFDSAGAYLLAASND 403
Query: 294 NNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTI 353
+W ++ R+RHTLTGH KV + ++R +VS ++DRT+K+WDL C T+
Sbjct: 404 FASRIWTVDDYRLRHTLTGHSGKVLSAKFLLDNAR-IVSGSHDRTLKLWDLRSKVCIKTV 462
Query: 354 IFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVV 413
S+CN + + Q + SGH D +R WDI+S ++ E+ +T++ L+ +
Sbjct: 463 FAGSSCNDIVCTE--QCVMSGHFDKKIRFWDIRSESVVREMELLG-KITALDLNPERTEL 519
Query: 414 LTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSV 473
L+ RD++ + D+R+ V +F G + S+W+R SPD S+VAAGSA+GS+++WSV
Sbjct: 520 LSCSRDDLLKVIDLRTNAVKQTFSAPGFKCGSDWTRVVFSPDGSYVAAGSAEGSLYVWSV 579
Query: 474 HKNEIVSTL-KEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++ L K+H+SS+ W+ G + S DK +W
Sbjct: 580 LTGKVEKVLSKQHSSSINAVAWAPSGLHVVSVDKGSRAVLW 620
>ref|XP_650923.1| WD repeat protein [Entamoeba histolytica HM-1:IMSS]
gi|56467592|gb|EAL45537.1| WD repeat protein [Entamoeba
histolytica HM-1:IMSS]
Length = 516
Score = 176 bits (445), Expect = 2e-42
Identities = 123/474 (25%), Positives = 221/474 (45%), Gaps = 25/474 (5%)
Query: 53 KEKAENLQVELQQCYKAQSRLSEQLVVEVADSRASKALVQEKDNAIADLQKELTEMRDEF 112
K K EN+ + ++ +K +LS VE + L Q K N + Q+E++ ++ +
Sbjct: 55 KLKQENIDI-VKDYHKISVKLSN---VEQEKIAVEQELEQTK-NMVKLKQEEVSVLQTKN 109
Query: 113 SQLKVDLEQKIKELEVIVSENSELKAQLEQMTAQCKKAEAENKMLIDRWMLEKMKDAERL 172
++ D++ KI+ + VI E+S + ++ K+ EN ++D E K + L
Sbjct: 110 DEVLKDIQSKIEMINVIRQESSAHQINRIRLEDDLKRLNIENHRIMD----EYAKLEQEL 165
Query: 173 NEANALYEEMVERLRASGLEQLARQQVDGVVRRCEEGAEFFLD------SNIPSTCKFRL 226
LY ++ + Q + + F ++ S +P+ CK ++
Sbjct: 166 RVTKELYSKIQTNPSITQQPQGNNPPTQSYLSSNATTSSFQVNGMNVSASKLPTECKRQI 225
Query: 227 NAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRS 286
H + N + L + D++VK+WD TG + S++ G + S L+ +
Sbjct: 226 TGHSSDILCFRYNSNGTVLASASADKTVKLWDVATGKIKSSVGGVLQSFTHLSFSPMGDM 285
Query: 287 VIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK 346
++A S+ + VW L + R+RH+LTGH KV + + +++ ++DRT+K WD+ K
Sbjct: 286 LLATSNDSTAKVWYLANSRLRHSLTGHSGKVTCGEF--FDTDKIMTGSHDRTLKTWDVNK 343
Query: 347 GYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLS-EVAAHSLAVTSIS 405
GYC T + S+CN + G + +GH D +R WDI+S + + H+ +V+ I
Sbjct: 344 GYCLKTTVCFSSCNCMMMGGMGNLVLTGHCDNTIRFWDIRSKECIDINKNIHTSSVSDII 403
Query: 406 LSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSAD 465
+G ++ G+DN+ D +V SSF V S+ SR C SPD ++ AGS++
Sbjct: 404 NVLDGRYFVSIGKDNIIQYVDGVERKVISSFSHPDFTVTSS-SRMCCSPDGKYIIAGSSN 462
Query: 466 GSVHIWSVHKNEIVSTLKEH-----TSSVLCCRWSGIGKPLASADKNGIVCIWK 514
G V W ++ + LK + W+ + + S N IV IW+
Sbjct: 463 GEVFCWDTQSKKLETVLKPKLTQPTKAGCYAVSWNPVQSQIVSGHANKIV-IWE 515
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.129 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 809,072,088
Number of Sequences: 2540612
Number of extensions: 31873041
Number of successful extensions: 248709
Number of sequences better than 10.0: 12464
Number of HSP's better than 10.0 without gapping: 4225
Number of HSP's successfully gapped in prelim test: 8549
Number of HSP's that attempted gapping in prelim test: 156275
Number of HSP's gapped (non-prelim): 53985
length of query: 514
length of database: 863,360,394
effective HSP length: 133
effective length of query: 381
effective length of database: 525,458,998
effective search space: 200199878238
effective search space used: 200199878238
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0173b.8


