

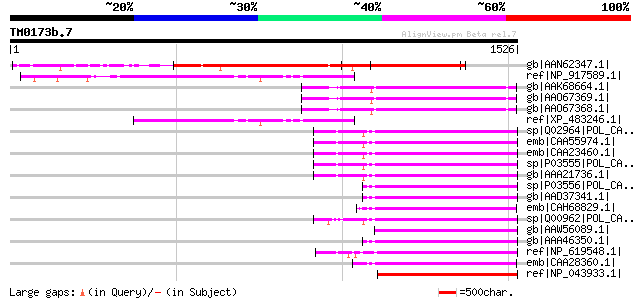
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0173b.7
(1526 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN62347.1| CTV.20 [Poncirus trifoliata] 800 0.0
ref|NP_917589.1| sumilar to Oryza sativa chromosome 10, OSJNBb00... 360 2e-97
gb|AAK68664.1| ORF I polyprotein [petunia vein clearing virus] g... 350 2e-94
gb|AAO67369.1| polyprotein 1 [Petunia vein clearing virus] 348 1e-93
gb|AAO67368.1| polyprotein 1 [Petunia vein clearing virus] 348 1e-93
ref|XP_483246.1| hypothetical protein [Oryza sativa (japonica cu... 341 1e-91
sp|Q02964|POL_CAMVE Enzymatic polyprotein [Contains: Aspartic pr... 336 4e-90
emb|CAA55974.1| unnamed protein product [Cauliflower mosaic virus] 336 4e-90
emb|CAA23460.1| unnamed protein product [Cauliflower mosaic viru... 335 5e-90
sp|P03555|POL_CAMVC Enzymatic polyprotein [Contains: Aspartic pr... 334 2e-89
gb|AAA21736.1| reverse transcriptase 332 5e-89
sp|P03556|POL_CAMVD Enzymatic polyprotein [Contains: Aspartic pr... 331 1e-88
gb|AAD37341.1| reverse transcriptase [Cauliflower mosaic virus] 331 1e-88
emb|CAH68829.1| polyprotein [Carnation etched ring virus] 330 2e-88
sp|Q00962|POL_CAMVN Enzymatic polyprotein [Contains: Aspartic pr... 328 9e-88
gb|AAW56089.1| polyprotein [Horseradish latent virus] 328 9e-88
gb|AAA46350.1| ORF5; putative 326 4e-87
ref|NP_619548.1| Enzymatic polyprotein [Contains: Aspartic prote... 325 6e-87
emb|CAA28360.1| unnamed protein product [Carnation etched ring v... 320 2e-85
ref|NP_043933.1| ORF V [Strawberry vein banding virus] gi|136061... 309 5e-82
>gb|AAN62347.1| CTV.20 [Poncirus trifoliata]
Length = 3148
Score = 800 bits (2066), Expect = 0.0
Identities = 429/910 (47%), Positives = 585/910 (64%), Gaps = 48/910 (5%)
Query: 493 NSLIFTIAQHFVGDPSLIKDRSGDLLSNLKCKSLGDFRWYKDTFLTRVYTREDSQHAFWK 552
N+ + + Q+ GDPS ++D++ +LL NL+C+ L DF+ YK TF TR++ R+D+ H WK
Sbjct: 1951 NTELQNVTQYSDGDPSHLRDKNAELLHNLRCRKLSDFQLYKTTFFTRLFLRDDANHTTWK 2010
Query: 553 EKFLAGLPKSFGDKVREKLRSQNPGGEIPYQTLSYGQLIAIIQRVALKICQDDKIQQQLT 612
EKFLA LP G+KVR +++ IPY L+YG+L++ + + LKICQD K+Q++L
Sbjct: 2011 EKFLACLPTLLGEKVRNFIKALYDN-RIPYDELTYGELVSFVNKEGLKICQDLKLQKRLK 2069
Query: 613 KEKSQNRRDLGTFCEQFGI--------QGCPKKPKPRKQDPPPKQQWRRKSSQNHDHRKP 664
+E Q++R+LG+FC+QF + C K R K + RK H
Sbjct: 2070 QEIRQSKRELGSFCKQFNYDPFKASTSKDCNGKCSSRPYKKHYKSKSHRKLFLGHRENFY 2129
Query: 665 KPRSKPHSTQAA------KTPPENRPQGKDVTCYNCGKPGHISRYCRLKRRISELHLEPE 718
K ++P+ K P+ K+ C+ CG GH ++YC++ R++ EL L+ +
Sbjct: 2130 KKPTRPYKKSRFPRKKDFKATPKTPFNFKEAICHRCGIKGHTAKYCKMNRKLHELGLDDD 2189
Query: 719 IEDKINNLLIQTSDEEESVPSDS------EVSEDLNQIQNDDDQSSSS----INVLTNEQ 768
I KI L+I++S+ E S+ DS E+ + + N D S S INVLT +Q
Sbjct: 2190 ILSKIAPLMIESSNSESSMSGDSDSLQIDELIDSDTSVSNSSDSESESYLKKINVLTKDQ 2249
Query: 769 DLIFRAIDSIPDPDEKKVYLERLKLTLE-DRPPKSPITTNKFNLKDTFKRLEKSTIKPVT 827
+ + I DP+ +K YLE+L T++ ++ S + K N D + L+K K
Sbjct: 2250 ETFLELVKHISDPNLQKEYLEKLLKTMDFNKAETSKVPIVKKNSYDLTEILDKKKTKKSV 2309
Query: 828 --IQDLQSEVHILKAEVKSLKQIQTSQ----QLILEKLTEENSNGGSSSSSSSTSTSNRA 881
IQDLQ E+ +K+E+K LK+ Q S QL+L+K +++S+ S+ S + +
Sbjct: 2310 PNIQDLQKEIKDIKSEIKDLKEKQKSDSETIQLLLQKHLQDDSDNESTHSENHIEQNVDN 2369
Query: 882 PNNNVGDFLEIINYVTIQKFYINITIIIG-DFILETPALFDTGADSSCISEGLIPTRYFE 940
+ DFL ++ VT +K+ I T+I DF ++ ALFDTGAD +CI + ++P R+ E
Sbjct: 2370 IESVPHDFLFVLKQVTTRKYLIKATLIFSNDFAIDAIALFDTGADLNCIRQDIVPKRFHE 2429
Query: 941 KTTEKLSGAEGSKLIIKYKIPSAIIKNDSLEIETSFLLVRNLTHKVIIETPFIKKLFPYN 1000
KT E+LS A SKL + K+ A I N+ E +TSF+L ++ H VI+ TPFI + PY
Sbjct: 2430 KTKERLSAANNSKLKVDSKV-EASIHNNGFEFKTSFILTNDIHHAVILGTPFINLITPYT 2488
Query: 1001 TDEKGITVQHHGQPIVFKFSKPPFVKTLNI--------------ISYKEKQINFLKEEIS 1046
+ I+ + + I+F F + P + LNI I+ K+ + FL++++S
Sbjct: 2489 VNYDSISFKVKNKKIIFPFIEKPKTRNLNIVKACSIYQNRINNLINSKQNDLTFLQKDLS 2548
Query: 1047 YKNIEVQLQQPSVKSRIENILENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTK 1106
+ IE LQ+ +K +I + I+ IC DLP+AFW RK HMV+LPYE F +KQIPTK
Sbjct: 2549 LQRIENDLQRDFIKRKIFDFKTLIEKEICADLPSAFWNRKQHMVDLPYENSFDEKQIPTK 2608
Query: 1107 ARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKP 1166
ARPIQMN +L + + EINDL++K LI +S+SPWSCAAFYVNK +EIERGTPRLVINYKP
Sbjct: 2609 ARPIQMNMDLERHCKEEINDLVKKGLIVKSRSPWSCAAFYVNKNSEIERGTPRLVINYKP 2668
Query: 1167 LNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYE 1226
LN+AL WIRYPIPNKKDLL +LH A IFSKFDMKSGFWQIQ+ KDRYKTAFTVPFGQYE
Sbjct: 2669 LNKALKWIRYPIPNKKDLLQKLHSAFIFSKFDMKSGFWQIQIHPKDRYKTAFTVPFGQYE 2728
Query: 1227 WNVMPFGLKNAPSEFQRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKK 1286
W VMPFGLKNAPSEFQRIMN+I+NP S F IVYIDDVLIFS SIDQHFKHL TF +K
Sbjct: 2729 WTVMPFGLKNAPSEFQRIMNDIYNPYSDFCIVYIDDVLIFSNSIDQHFKHLKTFYFATRK 2788
Query: 1287 NGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLN 1346
G+A+S +KVSLFQTKIRFLGH+I +GTI PI R++ F DKFPD+I+DKTQLQRFLG LN
Sbjct: 2789 AGLAISNSKVSLFQTKIRFLGHHISKGTITPIERSLAFADKFPDKILDKTQLQRFLGSLN 2848
Query: 1347 YVADFCPQLS 1356
YV DFCP +S
Sbjct: 2849 YVLDFCPNIS 2858
Score = 449 bits (1154), Expect = e-124
Identities = 215/285 (75%), Positives = 243/285 (84%)
Query: 1086 KSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAF 1145
K HMV+LPYE F +KQIPTKARPIQMN +L + + EINDL++K LI +S+SPWSCAAF
Sbjct: 1123 KQHMVDLPYENSFDEKQIPTKARPIQMNMDLERHCKEEINDLVKKGLIVKSRSPWSCAAF 1182
Query: 1146 YVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQ 1205
YVNK +EIERGTPRLVINYKPLN+AL WIRYPIPNKKDLL +LH A IFSKFDMKS FWQ
Sbjct: 1183 YVNKNSEIERGTPRLVINYKPLNKALKWIRYPIPNKKDLLQKLHSAFIFSKFDMKSVFWQ 1242
Query: 1206 IQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP*SKFAIVYIDDVLI 1265
IQ+ K+RYKTAFTVPFGQYEW VMPFGLKNAPSEFQRIMN+I+NP S F IVYIDDVLI
Sbjct: 1243 IQIHPKERYKTAFTVPFGQYEWTVMPFGLKNAPSEFQRIMNDIYNPYSDFCIVYIDDVLI 1302
Query: 1266 FSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFT 1325
FS SIDQHFKHL TF +K G+A+S +KVSLFQTKIRFLGH+I +GTI PI R++ F
Sbjct: 1303 FSNSIDQHFKHLKTFYFATRKAGLAISNSKVSLFQTKIRFLGHHISKGTITPIERSLAFA 1362
Query: 1326 DKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDP 1370
DKFPD+I+DKTQLQRFLG LNYV DFCP +S + KPLHDRLKK P
Sbjct: 1363 DKFPDKILDKTQLQRFLGSLNYVLDFCPNISRLSKPLHDRLKKKP 1407
Score = 323 bits (829), Expect = 2e-86
Identities = 268/1008 (26%), Positives = 449/1008 (43%), Gaps = 214/1008 (21%)
Query: 9 SRRNQPESSARPSISHDGSTIHLAGYVNSTP----VKTTYDKGDEDHQSTL--SPTYSSL 62
S ++QP S S S+IH Y + P + + + E+ + +L SPT+S++
Sbjct: 311 STKSQPRFSTSDIPS---SSIHSVDYTTNVPHPIYTSSQHVQSQEEKEPSLPTSPTFSAV 367
Query: 63 ETTGETSDFIGAINAEFVIDKEGIRADFYSDRWTKQREWFFANYQGKRRKKVQDRFYGFL 122
T + I I EF +DK + DFYSD +++ WFF + +R K++Q +Y F+
Sbjct: 368 -----TENVINIIEKEFELDKTLLHNDFYSDLNKEKKIWFFKQFLNQR-KEIQQIYYEFV 421
Query: 123 ELVQQNIFFFDWFHAYTIKKKIPYPYQVD--------VISWQLKDGKETL-SNLPPKTPF 173
+ +I FFDWF Y+ + I YP++ + W+L D +T+ S PP
Sbjct: 422 NFHKVHILFFDWFEIYSSENNITYPFKESNPITIRKKIPEWKLSDSDKTIESEHPPLRSL 481
Query: 174 FLKGAQSAL-VLASPFKTREGEGDVTSKDIKDLMEQANYTNKYLQTLGESQTNDSSVKGK 232
+ + + + ASP+K + + ++ +++Q N+ N L T+G+ T + K
Sbjct: 482 TIDHGEPPIQIRASPYKIPKPND--SDSNLSSIIQQNNFCNTNLNTIGKQLTRIENQFQK 539
Query: 233 GKIKIESSSSSAPPDIPKGCQLEKPLFKPFQISSRSRHSAQSLGAKNESDNELLQKVVEK 292
I + SS+S P +L++ +FKPFQ+S S+ Q ES ++ + + E+
Sbjct: 540 STITV-SSTSPIPSKSDYDKKLKEAIFKPFQVSKTSQKLVQ------ESKSDFAKAIREQ 592
Query: 293 LKLLNQVVPESPATTPVAVSAPAPPTAAPEPVIRRSSTRNASASSLNNIEEGESDVQSVK 352
L +++ S ++ V + AP P ++ V+ + + ++S L ++
Sbjct: 593 L---DRIESASSSSNKVQI-APNTPQSSKIGVLEQDDQTSIASSDLEAFKDEA------- 641
Query: 353 SIPAVNPVIHNSKNQWRKETKLYYPRATAP-DLLLEEKDSSNFKSFSANNVYEWNIDGEN 411
P +K W +L P +P DL ++ +
Sbjct: 642 ------PTPRANKIHW----ELAPPTVKSPPDLAIDNRP--------------------- 670
Query: 412 EYGITKILQNMTMVATAYETSNNCPESLIVEILVAGFCGQLKGWWDNYLTEDEKNQILTA 471
++LQ MTM A AY+T + + I EIL+AGF GQLK
Sbjct: 671 -----RVLQQMTMAANAYKTQSGTSDMAIAEILIAGFTGQLK------------------ 707
Query: 472 VKSDEEGNPIIEDGKFIFDAVNSLIFTIAQHFVGDPSLIKDRSGDLLSNLKCKSLGDFRW 531
GDPS ++D++ +LL NL+C+ L DF+
Sbjct: 708 ---------------------------------GDPSHLRDKNAELLHNLRCRKLSDFQL 734
Query: 532 YKDTFLTRVYTREDSQHAFWKEKFLAGLPKSFGDKVREKLRSQNPGGEIPYQTLSYGQLI 591
YK TF TR++ R+D+ H WKEKFLAGLP G+KVR +++ IPY L+YGQL+
Sbjct: 735 YKTTFFTRLFLRDDANHTTWKEKFLAGLPTLLGEKVRNSIKALY-DNRIPYDELTYGQLV 793
Query: 592 AIIQRVALKICQDDKIQQQLTKEKSQNRRDLGTFCEQFGIQGCPKKPKPRKQDPPPKQQW 651
+ + + LKICQD K+Q++L +E Q++R+LG+FC+QF P K
Sbjct: 794 SFVNKEGLKICQDLKLQKRLKQELRQSKRELGSFCKQFNYD-------------PFKAST 840
Query: 652 RRKSSQNHDHRKPKPRSKPHSTQAAKTPPENRPQGKDVTCYNCGKPGHISRYCRLKRRIS 711
+ + + D + ++ + + E+ K + + + L + IS
Sbjct: 841 SKDCNGDSDSLQIDELIDSDTSVSNSSDSESESYLKKINILTKDQ----ETFLELVKHIS 896
Query: 712 ELHLEPEIEDKINNLLIQTSDEEESVPSDSEVSEDLNQIQNDDDQSSSSINVLTNEQDLI 771
+ +L+ E +K+ + E VP + S DL +I + S N+ QDL
Sbjct: 897 DPNLQKEYLEKLLKTMDFNKAETSKVPIVKKNSYDLTEILDKKKTKKSVPNI----QDL- 951
Query: 772 FRAIDSIPDPDEKKVYLERLKLTLEDRPPKSPITTNKFNLKDTFKRLEKSTIKPVTIQDL 831
+ I K +KD K +KS + +
Sbjct: 952 -----------------------------QKEIKDIKSEIKD-LKEKQKSDSETI----- 976
Query: 832 QSEVHILKAEVKSLKQIQTSQQLILEKLTEENSNGGSSSSSSSTSTSNRAPNNNVGDFLE 891
QL+L+K +++S+ S+ S + + + DFL
Sbjct: 977 ---------------------QLLLQKHLQDDSDNESTHSENHIEQNVDNIESVPHDFLF 1015
Query: 892 IINYVTIQKFYINITIII-GDFILETPALFDTGADSSCISEGLIPTRYFEKTTEKLSGAE 950
++ VT +K+ I T+I DF ++ ALFDTGAD +CI + ++P R+ EKT E+LS A
Sbjct: 1016 VLKQVTTRKYLIKATLIFSNDFAIDAIALFDTGADLNCIRQDIVPKRFHEKTKERLSAAN 1075
Query: 951 GSKLIIKYKIPSAIIKNDSLEIETSFLLVRNLTHKVIIETPFIKKLFP 998
SKL + K+ A I N+ E +TSF+L ++ H VI+ TPFI + P
Sbjct: 1076 NSKLKVDSKV-EASIHNNGFEFKTSFILTNDIHHAVILGTPFINLITP 1122
>ref|NP_917589.1| sumilar to Oryza sativa chromosome 10, OSJNBb0036B06.4 [Oryza sativa
(japonica cultivar-group)] gi|20804792|dbj|BAB92476.1|
hypothetical protein [Oryza sativa (japonica
cultivar-group)] gi|18677105|dbj|BAB84832.1| hypothetical
protein [Oryza sativa (japonica cultivar-group)]
Length = 1634
Score = 360 bits (924), Expect = 2e-97
Identities = 305/1074 (28%), Positives = 476/1074 (43%), Gaps = 151/1074 (14%)
Query: 34 YVNSTPVKTTYDKGDEDHQSTLSPTYSSLETTGETSDF--------IGAINA----EFVI 81
++ S P + + Q+ + TY SDF +G + A EF I
Sbjct: 83 FLQSRPSVSLDSRPRTKPQNVVYATYEDNSDEPSISDFDINVIELDVGFVIALEEEEFEI 142
Query: 82 DKEGIRADFYSDRWTKQREWFFANYQGKRRKKVQDRFYGFLELVQQNIFFFDWFHAYTI- 140
DKE +R + + + + + R K+++ ++ + ++NIFFFDW+ I
Sbjct: 143 DKELLRREIRLPKNRAKTKRYLEEVDESFRMKIREVWHNEMREQRRNIFFFDWYENSQII 202
Query: 141 -------------KKKIPYPYQVDVIS-----WQLKDGKETLSNLPPKTPFFLK-GAQSA 181
K++ + VI W+ G + S PP L A
Sbjct: 203 HFEEFFKRKNMIKKEQKSEAEDLTVIKKVSTEWETASGNKVDSVHPPFESIQLSHNGGKA 262
Query: 182 LVLASPFKTREGEGDVTSKDIKDLMEQANYTNKYLQTLGESQTNDSSVKGKG-------- 233
L S K GE + I L+EQ NY N L++LG+ ++ +G
Sbjct: 263 CPLKSISKNTYGE-TAKVEHIGHLVEQQNYANISLRSLGQQTDRIETILMEGYKTGRPEV 321
Query: 234 KIKIESSSSSAPPDIPKGCQLEKPLFKPFQISSRSRHSAQSLGAKNESDNELLQKVVEKL 293
KI I S+S S+ Q P+F
Sbjct: 322 KINIPSNSQSS------SSQSVSPMF---------------------------------- 341
Query: 294 KLLNQVVPESPATTPVAVSAPAPPTAAPEPVIRRSSTRNASASSLNNIEEGES--DVQSV 351
VP P + E V N A LNN++ ++ ++
Sbjct: 342 ------VPTIDPNIKFGKQKAFGPAISEELV-------NELALKLNNLKVNKNINEISDN 388
Query: 352 KSIPAVNPVIHNSKNQWRKETKLYYPRATAPDLLLEEKDS-SNFKSFSANNVYEWNIDGE 410
+ VN + S T+ YYPR T DL EE N ++ + EWN+DG
Sbjct: 389 EKYDIVNKIFKPST--LTSTTRNYYPRPTYADLQFEEMPQIQNMTYYNGKEIVEWNLDGF 446
Query: 411 NEYGITKILQNMTMVATAYETSNNCPESLIVEILVAGFCGQLKGWWDNYLTEDEKNQILT 470
EY I + M M A A + N E ++V GF GQLKGWW+NYL E ++ +IL
Sbjct: 447 TEYQIFTLCHQMIMYANACIANGN-KEREAANMIVIGFSGQLKGWWNNYLNETQRQEILC 505
Query: 471 AVKSDEEGNPIIE-DGKF-----IFDAVNSLIFTIAQHFVGDPSLIKDRSGDLLSNLKCK 524
AVK D++G P+ + DG I DA+ +LI+ I HF G+ I +++ + L NLKCK
Sbjct: 506 AVKRDDQGRPLPDRDGNGNPTGNISDALATLIYNIIYHFAGNYHDIYEKNREQLINLKCK 565
Query: 525 SLGDFRWYKDTFLTRVYTREDSQHAFWKEKFLAGLPKSFGDKVREKLRSQNPGGEIPYQT 584
++ DFRWYKDTFL+++YT + FWKEK+++GLP F +KVR LR + GG I Y
Sbjct: 566 TMSDFRWYKDTFLSKLYTLPEPNQDFWKEKYISGLPPLFAEKVRNSLRKEG-GGSINYHY 624
Query: 585 LSYGQLIAIIQRVALKICQDDKIQQQLTKEKSQNRRDLGTFCEQFGIQGCPKKPKPRKQD 644
L G++ IQ V ++C D KI+ QL K++ +R++G FC QFG Q P + RK
Sbjct: 625 LDIGKITQKIQLVGAELCNDLKIKDQLKKQRILGKREMGDFCYQFGFQD-PYVYRKRKTH 683
Query: 645 PPPKQQWRRKSSQNHDHRKPKPR---SKPHSTQAAKTPPENRPQGKDVTCYNCGKPGHIS 701
P + KS + K KP+ +K TQ + K+ CY CG GHI+
Sbjct: 684 SKPMTKPNDKSKMSFQATKRKPKRIYNKNIRTQDT--------ESKETICYKCGLKGHIA 735
Query: 702 RYC---RLKRRISELHLEPEIEDKINNLLIQTSDEEESVPSDSEVSEDLNQIQ------- 751
C ++K+ I L L+ E ED L ++ + + SD E + ++N Q
Sbjct: 736 NRCFKSKVKKEIQAL-LDSESEDVKEKLEAILNNIDNDLSSDEEKNAEINCCQDSGCSCC 794
Query: 752 ---NDDDQSSSSINVLTNEQDLIFRAIDSIPDPDEKKVYLERL--KLTLEDRPPKSPITT 806
N +++S +I VLT+ ++ + ++I DP+EK+ LE+ ++ + K I
Sbjct: 795 EPDNSEEESDENILVLTSLEEFVLDTFETIQDPEEKRRVLEKFLSRVKTDKDKLKKDIQK 854
Query: 807 NKF-NLKDTFKRLEKSTIKPVTIQDLQSEVHILKAEVKSLKQIQTSQQLILEKLTEENSN 865
+KF ++ + FKRL++ K + Q ++ L + K +KQ + I ++L S
Sbjct: 855 SKFLSIDEVFKRLDEQKKK-----NEQPDLISLFEDQKIMKQ---DLEEIRKRLYMLESK 906
Query: 866 GGSSSSSSSTSTSNRAPNNNVGDFLEIINYVTIQKFYINITI-IIGDFILETPALFDTGA 924
G P + I QK+Y + I + L D+GA
Sbjct: 907 EGFHMEEKD------EPIQEDDHVVGTIQKYMKQKWYTEVMYRFIDGSYFQHITLIDSGA 960
Query: 925 DSSCISEGLIPTRYFEKTTEKLSGAEGSKLIIKYKIPSAIIKNDSLEIETSFLLVRNLTH 984
D SCI EG+IP +YF K ++ GA+G L ++Y+IP I + I+TSFLLV+NL
Sbjct: 961 DVSCIREGIIPHKYFYKAAHRIRGADGRLLTVEYQIPEIYICISEVCIKTSFLLVKNLKQ 1020
Query: 985 KVIIETPFIKKLFPYNTDEKGITVQHHGQPIVFKFSKPPFVKTLNIISYKEKQI 1038
VI+ TPF+ + P+ + I + + + +FS +I K +Q+
Sbjct: 1021 DVILGTPFLSLIRPFLVTNEDIQFKIMDKQVSLRFSSNTDEILDQLIQTKREQV 1074
>gb|AAK68664.1| ORF I polyprotein [petunia vein clearing virus]
gi|14575753|ref|NP_127504.1| ORF I polyprotein [petunia
vein clearing virus]
Length = 2179
Score = 350 bits (899), Expect = 2e-94
Identities = 225/657 (34%), Positives = 343/657 (51%), Gaps = 42/657 (6%)
Query: 877 TSNRAPNNNVGDFLEIINYVTIQKFYINITIIIGDFILETPALFDTGADSSCISEGLIPT 936
TS AP +I V I + + I D + A +DTGA S + ++P+
Sbjct: 1182 TSYEAPKTENPPLPKIHTIVEIPQTEVKIYTSKWDKPISVIAFYDTGAAYSIMDPAILPS 1241
Query: 937 RYFEKTTEKLSGAEGSKLIIKYKIPSAIIKNDSLEIETSFLLVRNLTHKVIIETPFIKKL 996
Y+ A+ L K H + IE
Sbjct: 1242 EYWIPHFRHFGTADDGILTTTVKTK----------------------HPITIE---FFPG 1276
Query: 997 FPYNTDEKGITVQHHGQPIVFKFSKPPFVKTLNIISYKEKQINFLKEEISYKNIEVQLQQ 1056
F Y T G + G+ ++ F + + N + I + + Y I +L Q
Sbjct: 1277 FKYTTKLLGSDIP--GKDLLIGFDI--YRQLNNKLRIGADGIRWKNQFKRYTEIP-RLFQ 1331
Query: 1057 PSVKSRIENILENIQSSICFDLPNAFWERKSH--------MVELPYEKDFSDKQIPTKAR 1108
+ + ++ + + I++ +C D F + SH ++LP++K+ + PTKA
Sbjct: 1332 LTTSNELQQLEDVIKNQLCADSHVDFLSKCSHPLWLNQDFFIQLPFKKN--ENINPTKAS 1389
Query: 1109 PIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLN 1168
MN E LQ +E ++L Q LI S S W+C AFYVNK++E RG RLVINY+PLN
Sbjct: 1390 HSGMNPEHLQLAIKECDELQQFDLIEPSDSQWACEAFYVNKRSEQVRGKLRLVINYQPLN 1449
Query: 1169 QALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWN 1228
L ++PIPNK L + L AK+FSKFD+KSGFWQ+ + +R KT F +P ++W
Sbjct: 1450 HFLQDDKFPIPNKLTLFSHLSKAKLFSKFDLKSGFWQLGIHPNERPKTGFCIPDRHFQWK 1509
Query: 1229 VMPFGLKNAPSEFQRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNG 1288
VMPFGLK APS FQ+ M +IF P A+VYIDD+L+FS++++ H K LN FIS++KK G
Sbjct: 1510 VMPFGLKTAPSLFQKAMIKIFQPILFSALVYIDDILLFSETLEDHIKLLNQFISLVKKFG 1569
Query: 1289 MAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYV 1348
+ +S K+ L Q KI+FLG + GT P KFPD + Q+Q+FLG +NY+
Sbjct: 1570 VMLSAKKMILAQNKIQFLGMDFADGTFSPAGHISLELQKFPDTNLSVKQIQQFLGIVNYI 1629
Query: 1349 ADFCPQLSTIIKPLHDRLKKDPPPWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETD 1408
DF P+++ I PL D LKK PP W N VKQ+K + + L++P+ + KI++TD
Sbjct: 1630 RDFIPEVTEHISPLSDMLKKKPPAWGKCQDNAVKQLKQLAQQVKSLHIPS-EGKKILQTD 1688
Query: 1409 ASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQK 1468
ASD + +L ++ K +I F S + ++Q+Y + KE+LA+ I F LI+
Sbjct: 1689 ASDQYWSAVLLEEHNGKRKICGFASGKFKVSEQHYHSTFKEILAVKNGIKKFNFFLIHTN 1748
Query: 1469 FLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTR 1525
FLV +D ++ +++ + K + + + RW S + FE++++KG N L DFL+R
Sbjct: 1749 FLVEMDMRAFPKMIRLNPKIVPNSQLL-RWAQWFSPYQFEVKHLKGKDNILADFLSR 1804
Score = 47.8 bits (112), Expect = 0.003
Identities = 92/440 (20%), Positives = 168/440 (37%), Gaps = 74/440 (16%)
Query: 324 VIRRSSTR--NASASSLNNIEEGESDVQSVKSIPAVNPVIHNSKNQWRKETKLYYPRATA 381
V+ ST+ S+SS ++ E E+ + + P I +++ T+ P
Sbjct: 781 VVSEESTQLSQLSSSSNDSPENNENTLPQTFMVRPTEPEISEVEDEVDGMTEEPIPERR- 839
Query: 382 PDLLLEEKDSSNFKSFSANNVYEWNIDGENEYGITKILQNMTMVATAYETSNNCPES--L 439
P++ + + F +FS +++ ITK + + T T L
Sbjct: 840 PEITPPKMVGTGFHTFSLDDI-----------SITKWPERIQDFHTWMLTKQLVEREPFL 888
Query: 440 IVEILVAGFCGQLKGWWDNYLTEDEKNQILTAVKSDEEGNPIIEDGKFIFDAVNSLIFTI 499
I+ A G L+ WW N + D+KN+ LT+ D N I +
Sbjct: 889 ILSEFTARLSGTLREWW-NSVGPDDKNRFLTS--QDFTWN----------------IRIL 929
Query: 500 AQHFVGDPSLIKDRSGDLLSNLKCKSLGDFRWYKDTFLTRVYTR------EDSQHAFWKE 553
+F GD S K+ + +KC S Y + R + R K+
Sbjct: 930 YSYFCGDQSQNKEELRRQIFEMKCLS------YDRKKIDRHFQRMIKLFYHIGGDISLKQ 983
Query: 554 KFLAGLPKSFGDKVREKLRSQNPGGEIPYQTLSYGQL-IAIIQRVALKICQDDKIQQQLT 612
F++ LP +++ ++ + S Q+ + I++ A + D +++
Sbjct: 984 AFISSLPPILSERISALIKERGT---------SVTQMHVGDIRQTAFYVLDDLCSKRKFF 1034
Query: 613 KEKSQNRRDLGTFCEQF-----GIQGCPKKPKPRKQDPPPKQQWRRKSSQNHDHRKPKPR 667
+ + RDL C + G +GC P ++ ++++ S + D R+ + R
Sbjct: 1035 NQMKKMSRDLEKACTKSDLIIKGDKGCSGYCNPSRRRK--YKRFKLPSFKERDGRQYRKR 1092
Query: 668 SKPHSTQAAKTPPENRPQGKDVTCYNCGKPGHISRYCRLKRRISELHLEPEIEDKINNLL 727
+ +P+ +C+ CGK GH SR C ++ + L EI+
Sbjct: 1093 RRFFRRSKTSKAMRQKPR----SCFTCGKIGHFSRNCPQNKK--SIKLISEIQKYTG--- 1143
Query: 728 IQTSDEEESVPS-DSEVSED 746
I D+ ESV S + E SED
Sbjct: 1144 IDIEDDLESVFSIEDEPSED 1163
>gb|AAO67369.1| polyprotein 1 [Petunia vein clearing virus]
Length = 1886
Score = 348 bits (892), Expect = 1e-93
Identities = 222/657 (33%), Positives = 343/657 (51%), Gaps = 42/657 (6%)
Query: 877 TSNRAPNNNVGDFLEIINYVTIQKFYINITIIIGDFILETPALFDTGADSSCISEGLIPT 936
TS AP +I V I + + + D + A +DTGA S + ++P+
Sbjct: 1183 TSYEAPKTENPPLPKIHTIVEIPQTEVKVYTSKWDKPISVIAFYDTGAAYSIMDPAILPS 1242
Query: 937 RYFEKTTEKLSGAEGSKLIIKYKIPSAIIKNDSLEIETSFLLVRNLTHKVIIETPFIKKL 996
Y+ A+ L K H + IE
Sbjct: 1243 EYWIPHFRHFGTADDGILTTTVKTK----------------------HPITIE---FFPG 1277
Query: 997 FPYNTDEKGITVQHHGQPIVFKFSKPPFVKTLNIISYKEKQINFLKEEISYKNIEVQLQQ 1056
F Y T G + G+ ++ F + + N + I + + Y I +L Q
Sbjct: 1278 FKYTTKLLGSDIP--GKDLLIGFDI--YRQLNNKLRIGADGIRWKNQFKRYTEIP-RLFQ 1332
Query: 1057 PSVKSRIENILENIQSSICFDLPNAFWERKSH--------MVELPYEKDFSDKQIPTKAR 1108
+ + ++ + + I++ +C + F + SH ++LP++++ + PTKA
Sbjct: 1333 LTTSNELQQLEDVIKNQLCAESHVDFLSKCSHPLWLNQDFFIQLPFKRN--ENINPTKAS 1390
Query: 1109 PIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLN 1168
MN E LQ +E ++L Q LI S S W+C AFYVNK++E RG RLVINY+PLN
Sbjct: 1391 HSGMNPEHLQLALKECDELQQFDLIEPSDSQWACEAFYVNKRSEQVRGKLRLVINYQPLN 1450
Query: 1169 QALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWN 1228
L ++PIPNK L + L AK+FSKFD+KSGFWQ+ + +R KT F +P ++W
Sbjct: 1451 HFLQDDKFPIPNKLTLFSHLSKAKLFSKFDLKSGFWQLGIHPNERPKTGFCIPDRHFQWK 1510
Query: 1229 VMPFGLKNAPSEFQRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNG 1288
VMPFGLK APS FQ+ M +IF P A+VYIDD+L+FS++++ H K LN FIS++KK G
Sbjct: 1511 VMPFGLKTAPSLFQKAMIKIFQPILFSALVYIDDILLFSETLEDHIKLLNQFISLVKKFG 1570
Query: 1289 MAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYV 1348
+ +S K+ L Q KI+FLG + GT P KFPD + Q+Q+FLG +NY+
Sbjct: 1571 VMLSAKKMILAQNKIQFLGMDFADGTFSPAGHISLELQKFPDTNLSVKQIQQFLGIVNYI 1630
Query: 1349 ADFCPQLSTIIKPLHDRLKKDPPPWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETD 1408
DF P+++ I PL D LKK PP W N VKQ+K + + L++P+ + KI++TD
Sbjct: 1631 RDFIPEVTEHISPLSDMLKKKPPAWGKCQDNAVKQLKQLAQQVKSLHIPS-EGKKILQTD 1689
Query: 1409 ASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQK 1468
ASD + +L ++ K +I F S + ++Q+Y + KE+LA+ I F LI+
Sbjct: 1690 ASDQYWSAVLLEEHNGKRKICGFASGKFKVSEQHYHSTFKEILAVKNGIKKFNFFLIHTN 1749
Query: 1469 FLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTR 1525
FLV +D ++ +++ + K + + + RW S + FE++++KG N L DFL+R
Sbjct: 1750 FLVEMDMRAFPKMIRLNPKIVPNSQLL-RWAQWFSPYQFEVKHLKGKDNILADFLSR 1805
Score = 47.8 bits (112), Expect = 0.003
Identities = 92/440 (20%), Positives = 168/440 (37%), Gaps = 74/440 (16%)
Query: 324 VIRRSSTR--NASASSLNNIEEGESDVQSVKSIPAVNPVIHNSKNQWRKETKLYYPRATA 381
V+ ST+ S+SS ++ E E+ + + P I +++ T+ P
Sbjct: 782 VVSEESTQLSQLSSSSNDSPENNENTLPQTFMVRPTEPEISEVEDEVDGMTEEPIPERR- 840
Query: 382 PDLLLEEKDSSNFKSFSANNVYEWNIDGENEYGITKILQNMTMVATAYETSNNCPES--L 439
P++ + + F +FS +++ ITK + + T T L
Sbjct: 841 PEITPPKMVGTGFHTFSLDDI-----------SITKWPERIQDFHTWMLTKQLVEREPFL 889
Query: 440 IVEILVAGFCGQLKGWWDNYLTEDEKNQILTAVKSDEEGNPIIEDGKFIFDAVNSLIFTI 499
I+ A G L+ WW N + D+KN+ LT+ D N I +
Sbjct: 890 ILSEFTARLSGTLREWW-NSVGPDDKNRFLTS--QDFTWN----------------IRIL 930
Query: 500 AQHFVGDPSLIKDRSGDLLSNLKCKSLGDFRWYKDTFLTRVYTR------EDSQHAFWKE 553
+F GD S K+ + +KC S Y + R + R K+
Sbjct: 931 YSYFCGDQSQNKEELRRQIFEMKCLS------YDRKKIDRHFQRMIKLFYHIGGDISLKQ 984
Query: 554 KFLAGLPKSFGDKVREKLRSQNPGGEIPYQTLSYGQL-IAIIQRVALKICQDDKIQQQLT 612
F++ LP +++ ++ + S Q+ + I++ A + D +++
Sbjct: 985 AFISSLPPILSERISALIKERGT---------SVTQMHVGDIRQTAFYVLDDLCSKRKFF 1035
Query: 613 KEKSQNRRDLGTFCEQF-----GIQGCPKKPKPRKQDPPPKQQWRRKSSQNHDHRKPKPR 667
+ + RDL C + G +GC P ++ ++++ S + D R+ + R
Sbjct: 1036 NQMKKMSRDLEKACTKSDLIIKGDKGCSGYCNPSRRRK--YKRFKLPSFKERDGRQYRKR 1093
Query: 668 SKPHSTQAAKTPPENRPQGKDVTCYNCGKPGHISRYCRLKRRISELHLEPEIEDKINNLL 727
+ +P+ +C+ CGK GH SR C ++ + L EI+
Sbjct: 1094 RRFFRKSKTSKAMRQKPR----SCFTCGKIGHFSRNCPQNKK--SIKLISEIQKYTG--- 1144
Query: 728 IQTSDEEESVPS-DSEVSED 746
I D+ ESV S + E SED
Sbjct: 1145 IDIEDDLESVFSIEDEPSED 1164
>gb|AAO67368.1| polyprotein 1 [Petunia vein clearing virus]
Length = 2180
Score = 348 bits (892), Expect = 1e-93
Identities = 222/657 (33%), Positives = 343/657 (51%), Gaps = 42/657 (6%)
Query: 877 TSNRAPNNNVGDFLEIINYVTIQKFYINITIIIGDFILETPALFDTGADSSCISEGLIPT 936
TS AP +I V I + + + D + A +DTGA S + ++P+
Sbjct: 1183 TSYEAPKTENPPLPKIHTIVEIPQTEVKVYTSKWDKPISVIAFYDTGAAYSIMDPAILPS 1242
Query: 937 RYFEKTTEKLSGAEGSKLIIKYKIPSAIIKNDSLEIETSFLLVRNLTHKVIIETPFIKKL 996
Y+ A+ L K H + IE
Sbjct: 1243 EYWIPHFRHFGTADDGILTTTVKTK----------------------HPITIE---FFPG 1277
Query: 997 FPYNTDEKGITVQHHGQPIVFKFSKPPFVKTLNIISYKEKQINFLKEEISYKNIEVQLQQ 1056
F Y T G + G+ ++ F + + N + I + + Y I +L Q
Sbjct: 1278 FKYTTKLLGSDIP--GKDLLIGFDI--YRQLNNKLRIGADGIRWKNQFKRYTEIP-RLFQ 1332
Query: 1057 PSVKSRIENILENIQSSICFDLPNAFWERKSH--------MVELPYEKDFSDKQIPTKAR 1108
+ + ++ + + I++ +C + F + SH ++LP++++ + PTKA
Sbjct: 1333 LTTSNELQQLEDVIKNQLCAESHVDFLSKCSHPLWLNQDFFIQLPFKRN--ENINPTKAS 1390
Query: 1109 PIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLN 1168
MN E LQ +E ++L Q LI S S W+C AFYVNK++E RG RLVINY+PLN
Sbjct: 1391 HSGMNPEHLQLALKECDELQQFDLIEPSDSQWACEAFYVNKRSEQVRGKLRLVINYQPLN 1450
Query: 1169 QALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWN 1228
L ++PIPNK L + L AK+FSKFD+KSGFWQ+ + +R KT F +P ++W
Sbjct: 1451 HFLQDDKFPIPNKLTLFSHLSKAKLFSKFDLKSGFWQLGIHPNERPKTGFCIPDRHFQWK 1510
Query: 1229 VMPFGLKNAPSEFQRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNG 1288
VMPFGLK APS FQ+ M +IF P A+VYIDD+L+FS++++ H K LN FIS++KK G
Sbjct: 1511 VMPFGLKTAPSLFQKAMIKIFQPILFSALVYIDDILLFSETLEDHIKLLNQFISLVKKFG 1570
Query: 1289 MAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYV 1348
+ +S K+ L Q KI+FLG + GT P KFPD + Q+Q+FLG +NY+
Sbjct: 1571 VMLSAKKMILAQNKIQFLGMDFADGTFSPAGHISLELQKFPDTNLSVKQIQQFLGIVNYI 1630
Query: 1349 ADFCPQLSTIIKPLHDRLKKDPPPWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETD 1408
DF P+++ I PL D LKK PP W N VKQ+K + + L++P+ + KI++TD
Sbjct: 1631 RDFIPEVTEHISPLSDMLKKKPPAWGKCQDNAVKQLKQLAQQVKSLHIPS-EGKKILQTD 1689
Query: 1409 ASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQK 1468
ASD + +L ++ K +I F S + ++Q+Y + KE+LA+ I F LI+
Sbjct: 1690 ASDQYWSAVLLEEHNGKRKICGFASGKFKVSEQHYHSTFKEILAVKNGIKKFNFFLIHTN 1749
Query: 1469 FLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTR 1525
FLV +D ++ +++ + K + + + RW S + FE++++KG N L DFL+R
Sbjct: 1750 FLVEMDMRAFPKMIRLNPKIVPNSQLL-RWAQWFSPYQFEVKHLKGKDNILADFLSR 1805
Score = 48.9 bits (115), Expect = 0.001
Identities = 90/439 (20%), Positives = 166/439 (37%), Gaps = 72/439 (16%)
Query: 324 VIRRSSTR--NASASSLNNIEEGESDVQSVKSIPAVNPVIHNSKNQWRKETKLYYPRATA 381
V+ ST+ S+SS ++ E E+ + + P I +++ T+ P
Sbjct: 782 VVSEESTQLSQLSSSSNDSPENNENTLPQTFMVRPTEPEISEVEDEVDGMTEEPIPERR- 840
Query: 382 PDLLLEEKDSSNFKSFSANNVYEWNIDGENEYGITKILQNMTMVATAYETSNNCPES--L 439
P++ + + F +FS +++ ITK + + T T L
Sbjct: 841 PEITPPKMVGTGFHTFSLDDI-----------SITKWPERIQDFHTWMLTKQLVEREPFL 889
Query: 440 IVEILVAGFCGQLKGWWDNYLTEDEKNQILTAVKSDEEGNPIIEDGKFIFDAVNSLIFTI 499
I+ A G L+ WW N + D+KN+ LT+ D N I +
Sbjct: 890 ILSEFTARLSGTLREWW-NSVGPDDKNRFLTS--QDFTWN----------------IRIL 930
Query: 500 AQHFVGDPSLIKDRSGDLLSNLKCKSLGDFRWYKDTFLTRVYTR------EDSQHAFWKE 553
+F GD S K+ + +KC S Y + R + R K+
Sbjct: 931 YSYFCGDQSQNKEELRRQIFEMKCLS------YDRKKIDRHFQRMIKLFYHIGGDISLKQ 984
Query: 554 KFLAGLPKSFGDKVREKLRSQNPGGEIPYQTLSYGQLIAIIQRVALKICQDDKIQQQLTK 613
F++ LP +++ ++ + G + + I++ + D +++
Sbjct: 985 AFISSLPPILSERISALIKERGTSGTQMH--------VGDIRQTGFYVLDDLCSKRKFFN 1036
Query: 614 EKSQNRRDLGTFCEQF-----GIQGCPKKPKPRKQDPPPKQQWRRKSSQNHDHRKPKPRS 668
+ + RDL C + G +GC P ++ ++++ S + D R+ + R
Sbjct: 1037 QMKKMSRDLEKACTKSDLIIKGDKGCSGYCNPSRRRK--YKRFKLPSFKERDGRQYRKRR 1094
Query: 669 KPHSTQAAKTPPENRPQGKDVTCYNCGKPGHISRYCRLKRRISELHLEPEIEDKINNLLI 728
+ +P+ +C+ CGK GH SR C ++ + L EI+ I
Sbjct: 1095 RFFRKSKTSKAMRQKPR----SCFTCGKIGHFSRNCPQNKK--SIKLISEIQKYTG---I 1145
Query: 729 QTSDEEESVPS-DSEVSED 746
D+ ESV S + E SED
Sbjct: 1146 DIEDDLESVFSIEDEPSED 1164
>ref|XP_483246.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|42408922|dbj|BAD10179.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 1275
Score = 341 bits (875), Expect = 1e-91
Identities = 229/694 (32%), Positives = 353/694 (49%), Gaps = 53/694 (7%)
Query: 372 TKLYYPRATAPDLLLEEKDS-SNFKSFSANNVYEWNIDGENEYGITKILQNMTMVATAYE 430
T+ YYPR T DL EE N ++ + EWN+DG EY I + M M A A
Sbjct: 487 TRNYYPRPTYADLQFEEMPQIQNMIYYNGKEIVEWNLDGFTEYQIFTLCHQMIMYANACI 546
Query: 431 TSNNCPESLIVEILVAGFCGQLKGWWDNYLTEDEKNQILTAVKSDEEGNPIIE-DGKF-- 487
+ N E ++V GF GQLKGWW+NYL E ++ +IL AVK D++G P+ + DG
Sbjct: 547 ANGN-KEREAANMIVIGFSGQLKGWWNNYLNETQRQEILCAVKRDDQGRPLPDRDGNGNP 605
Query: 488 ---IFDAVNSLIFTIAQHFVGDPSLIKDRSGDLLSNLKCKSLGDFRWYKDTFLTRVYTRE 544
I DA+ +LI+ I HF G+ I +++ + L NLKCK++ DFRWYKDTFL+++YT
Sbjct: 606 TGNISDALATLIYNIIYHFAGNYHDIYEKNREQLINLKCKTMSDFRWYKDTFLSKLYTLP 665
Query: 545 DSQHAFWKEKFLAGLPKSFGDKVREKLRSQNPGGEIPYQTLSYGQLIAIIQRVALKICQD 604
+ FWKEK+++GLP F +KVR LR + GG I Y L G++ IQ V ++C D
Sbjct: 666 EPNQDFWKEKYISGLPPLFAEKVRNSLRKEG-GGSINYHYLDIGKITQKIQLVGAELCND 724
Query: 605 DKIQQQLTKEKSQNRRDLGTFCEQFGIQGCPKKPKPRKQDPPPKQQWRRKSSQNHDHRKP 664
KI+ QL K++ +R++G FC QFG Q P + RK P + KS + K
Sbjct: 725 LKIKDQLKKQRILGKREMGDFCYQFGFQD-PYVHRKRKTHSKPMTKPNDKSKMSFQATKR 783
Query: 665 KPR---SKPHSTQAAKTPPENRPQGKDVTCYNCGKPGHISRYC---RLKRRISELHLEPE 718
KP+ +K TQ + K+ CY CG GHI+ C ++K+ I L L+ E
Sbjct: 784 KPKRIYNKNIRTQDT--------ESKETICYKCGLKGHIANRCFKSKVKKEIQAL-LDSE 834
Query: 719 IEDKINNLLIQTSDEEESVPSDSEVSEDLNQIQ----------NDDDQSSSSINVLTNEQ 768
ED L ++ + SD E + ++N Q N +++S +I VLT+ +
Sbjct: 835 SEDVKEKLEAILNNIDNDSSSDEEKNAEINCCQDSGCSCCEPDNSEEESDENILVLTSLE 894
Query: 769 DLIFRAIDSIPDPDEKKVYLERL--KLTLEDRPPKSPITTNKF-NLKDTFKRLEKSTIKP 825
+ + ++I DP+EK+ LE+ ++ + K I +KF ++ + FKRL++ K
Sbjct: 895 EFVLDTFETIQDPEEKRRVLEKFLSRVKTDKDKLKKDIQKSKFLSIDEVFKRLDEQKKK- 953
Query: 826 VTIQDLQSEVHILKAEVKSLKQIQTSQQLILEKLTEENSNGGSSSSSSSTSTSNRAPNNN 885
+ Q ++ L + K +KQ + I ++L S G P
Sbjct: 954 ----NEQPDLISLFEDQKIMKQ---DLEEIRKRLYMLESKEGFHMEEKD------EPIQE 1000
Query: 886 VGDFLEIINYVTIQKFYINITI-IIGDFILETPALFDTGADSSCISEGLIPTRYFEKTTE 944
+ I QK+Y + I + L D+GAD +CI EG+IP +YF K
Sbjct: 1001 DDHVVGTIQKYMKQKWYTEVMYRFIDGSYFQHITLIDSGADVNCIREGIIPHKYFYKAAH 1060
Query: 945 KLSGAEGSKLIIKYKIPSAIIKNDSLEIETSFLLVRNLTHKVIIETPFIKKLFPYNTDEK 1004
++ GA+G L ++Y+IP I + I+TSFLLV+NL VI+ TPF+ + P+ +
Sbjct: 1061 RIRGADGGLLTVEYQIPEIYICITEVCIKTSFLLVKNLKQDVILGTPFLSLIRPFLVTNE 1120
Query: 1005 GITVQHHGQPIVFKFSKPPFVKTLNIISYKEKQI 1038
I + G+ + +FS ++ K +Q+
Sbjct: 1121 DIQFKIMGKQVSLRFSSNTDEILDQLVQTKREQV 1154
Score = 45.4 bits (106), Expect = 0.015
Identities = 59/269 (21%), Positives = 107/269 (38%), Gaps = 43/269 (15%)
Query: 34 YVNSTPVKTTYDKGDEDHQSTLSPTYSSLETTGETSDF--------IGAINA----EFVI 81
++ S P + + Q+ + TY SDF +G + A EF I
Sbjct: 232 FLQSRPSVSLDSRPRTKPQNVVYATYEDNSDEPSISDFDINVIELDVGFVIAIEEDEFEI 291
Query: 82 DKEGIRADFYSDRWTKQREWFFANYQGKRRKKVQDRFYGFLELVQQNIFFFDWFHAYTIK 141
DK+ ++ + + + + +F R K+++ ++ + ++NIFFFDW+ + ++
Sbjct: 292 DKDLLKRELRLQKNRPKMKRYFERVDEPFRLKIRELWHKEMREQRKNIFFFDWYESSQVR 351
Query: 142 KKIPYPYQVDVI-------------------SWQLKDGKETLSNLPPKTPFFLK-GAQSA 181
+ + ++I W+ G + S PP L A
Sbjct: 352 HFEEFFKRKNMIKKEQKSEAEDLTVIKKVSTEWETASGNKVDSVHPPFESVQLSHNGGKA 411
Query: 182 LVLASPFKTREGEGDVTSKDIKDLMEQANYTNKYLQTLGESQTNDSSVKGKGKIKIESSS 241
L S K GE + I L+EQ NY N L++LG+ QTN I +E
Sbjct: 412 CPLKSISKNTYGE-TAKVEHIGHLVEQQNYANISLRSLGQ-QTNRIET-----ILMEGYK 464
Query: 242 SSAPPDIPKGCQLEKPLFKPFQISSRSRH 270
+ P+ + +FKP ++S +R+
Sbjct: 465 TGR----PEKYDMVNKIFKPSTLTSTTRN 489
>sp|Q02964|POL_CAMVE Enzymatic polyprotein [Contains: Aspartic protease ; Endonuclease;
Reverse transcriptase ] gi|293185|gb|AAA62375.1| reverse
transcriptase
Length = 679
Score = 336 bits (861), Expect = 4e-90
Identities = 224/645 (34%), Positives = 338/645 (51%), Gaps = 41/645 (6%)
Query: 914 LETPALFDTGADSSCISEGLIPTRYFEKTTEKLSG--AEGSKLIIKY--KIPSAIIKNDS 969
+E DTGA S+ +IP ++ + A+GS + I K II +
Sbjct: 38 IELHCFVDTGASLCIASKFVIPEEHWVNAERPIMVKIADGSSITISKVCKDIDLIIAREI 97
Query: 970 LEIETSFLLVRNLTHKVIIETPFIKKLFPY-NTDEKGITVQHHGQPI-VFKFSKPPFVKT 1027
+I T + + II F + P+ ++ I ++ P+ + K ++ V T
Sbjct: 98 FKIPTVYQQESGIDF--IIGNNFCQLYEPFIQFTDRVIFTKNKSYPVHIAKLTRAVRVGT 155
Query: 1028 LNIISYKEKQINFLKEE---ISYKNIEVQLQQPSVKS------------------RIENI 1066
+ +K+ + E IS IE L++ ++ S +IE +
Sbjct: 156 EGFLESMKKRSKTQQPEPVNISTNKIENPLKEIAILSEGRRLSEEKLFITQQRMQKIEEL 215
Query: 1067 LENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREIND 1126
LE + S D PN + ++L SD K +P++ + + F ++I +
Sbjct: 216 LEKVCSENPLD-PNKTKQWMKASIKL------SDPSKAIKVKPMKYSPMDREEFDKQIKE 268
Query: 1127 LLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLA 1186
LL K+I+ SKSP AF VN +AE RG R+V+NYK +N+A Y +PNK +LL
Sbjct: 269 LLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATIGDAYNLPNKDELLT 328
Query: 1187 RLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMN 1246
+ KIFS FD KSGFWQ+ L ++ R TAFT P G YEWNV+PFGLK APS FQR M+
Sbjct: 329 LIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMD 388
Query: 1247 EIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFL 1306
E F KF VY+DD+L+FS + + H H+ + ++G+ +SK K LF+ KI FL
Sbjct: 389 EAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFL 448
Query: 1307 GHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRL 1366
G I +GT P +E +KFPD + DK QLQRFLG L Y +D+ P+L+ I KPL +L
Sbjct: 449 GLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYIPKLAQIRKPLQAKL 508
Query: 1367 KKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDK 1425
K++ P W+ T ++++K ++ P L+ P P+ I+ETDASD +GG+LK I++
Sbjct: 509 KENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINE 568
Query: 1426 ----EQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDI 1481
E I + S + A++NY + KE LA++ +I F L FL+R D K
Sbjct: 569 GTNTELICRYASGSFKAAERNYHSNDKETLAVINTIKKFSIYLTPVHFLIRTDNTHFKSF 628
Query: 1482 LQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
+ + K + RWQA LS + F++E+IKG+ N DFL+RE
Sbjct: 629 VNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSRE 673
>emb|CAA55974.1| unnamed protein product [Cauliflower mosaic virus]
Length = 680
Score = 336 bits (861), Expect = 4e-90
Identities = 224/645 (34%), Positives = 338/645 (51%), Gaps = 41/645 (6%)
Query: 914 LETPALFDTGADSSCISEGLIPTRYFEKTTEKL--SGAEGSKLIIKY--KIPSAIIKNDS 969
+E DTGA S+ +IP ++ + A+GS + I K II +
Sbjct: 39 IELHCFVDTGASLCIASKFVIPEEHWVNAERPILVKIADGSSITISKVCKDIDLIIAGEI 98
Query: 970 LEIETSFLLVRNLTHKVIIETPFIKKLFPY-NTDEKGITVQHHGQPI-VFKFSKPPFVKT 1027
+I T + + II F + P+ ++ I ++ P+ + K ++ V T
Sbjct: 99 FKIPTVYQQESGIDF--IIGNNFCQLYEPFIQFTDRVIFTKNKSYPVHIAKLTRAVRVGT 156
Query: 1028 LNIISYKEKQINFLKEE---ISYKNIEVQLQQPSVKS------------------RIENI 1066
+ +K+ + E IS IE L++ ++ S +IE +
Sbjct: 157 EGFLESMKKRSKTQQPEPVNISTNKIENPLEEIAILSEGRRLSEEKLFITQQRMQKIEEL 216
Query: 1067 LENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREIND 1126
LE + S D PN + ++L SD K +P++ + + F ++I +
Sbjct: 217 LEKVCSENPLD-PNKTKQWMKASIKL------SDPSKAIKVKPMKYSPMDREEFDKQIKE 269
Query: 1127 LLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLA 1186
LL K+I+ SKSP AF VN +AE RG R+V+NYK +N+A Y +PNK +LL
Sbjct: 270 LLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATIGDAYNLPNKDELLT 329
Query: 1187 RLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMN 1246
+ KIFS FD KSGFWQ+ L ++ R TAFT P G YEWNV+PFGLK APS FQR M+
Sbjct: 330 LIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMD 389
Query: 1247 EIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFL 1306
E F KF VY+DD+L+FS + + H H+ + ++G+ +SK K LF+ KI FL
Sbjct: 390 EAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFL 449
Query: 1307 GHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRL 1366
G I +GT P +E +KFPD + DK QLQRFLG L Y +D+ P+L+ I KPL +L
Sbjct: 450 GLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYIPKLAQIRKPLQAKL 509
Query: 1367 KKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDK 1425
K++ P W+ T ++++K ++ P L+ P P+ I+ETDASD +GG+LK I++
Sbjct: 510 KENVPWRWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINE 569
Query: 1426 ----EQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDI 1481
E I + S + A++NY + KE LA++ +I F L FL+R D K
Sbjct: 570 GTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYLTPVHFLIRTDNTHFKSF 629
Query: 1482 LQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
+ + K + RWQA LS + F++E+IKG+ N DFL+RE
Sbjct: 630 VNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSRE 674
>emb|CAA23460.1| unnamed protein product [Cauliflower mosaic virus]
gi|130592|sp|P03554|POL_CAMVS Enzymatic polyprotein
[Contains: Aspartic protease ; Endonuclease; Reverse
transcriptase ] gi|9626943|ref|NP_056728.1| reverse
transcriptase [Cauliflower mosaic virus]
Length = 679
Score = 335 bits (860), Expect = 5e-90
Identities = 224/645 (34%), Positives = 337/645 (51%), Gaps = 41/645 (6%)
Query: 914 LETPALFDTGADSSCISEGLIPTRYFEKTTEKLSG--AEGSKLIIKY--KIPSAIIKNDS 969
+E DTGA S+ +IP ++ + A+GS + I K II +
Sbjct: 38 IELHCFVDTGASLCIASKFVIPEEHWVNAERPIMVKIADGSSITISKVCKDIDLIIAGEI 97
Query: 970 LEIETSFLLVRNLTHKVIIETPFIKKLFPY-NTDEKGITVQHHGQPI-VFKFSKPPFVKT 1027
I T + + II F + P+ ++ I ++ P+ + K ++ V T
Sbjct: 98 FRIPTVYQQESGIDF--IIGNNFCQLYEPFIQFTDRVIFTKNKSYPVHIAKLTRAVRVGT 155
Query: 1028 LNIISYKEKQINFLKEE---ISYKNIEVQLQQPSVKS------------------RIENI 1066
+ +K+ + E IS IE L++ ++ S +IE +
Sbjct: 156 EGFLESMKKRSKTQQPEPVNISTNKIENPLEEIAILSEGRRLSEEKLFITQQRMQKIEEL 215
Query: 1067 LENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREIND 1126
LE + S D PN + ++L SD K +P++ + + F ++I +
Sbjct: 216 LEKVCSENPLD-PNKTKQWMKASIKL------SDPSKAIKVKPMKYSPMDREEFDKQIKE 268
Query: 1127 LLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLA 1186
LL K+I+ SKSP AF VN +AE RG R+V+NYK +N+A Y +PNK +LL
Sbjct: 269 LLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNLPNKDELLT 328
Query: 1187 RLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMN 1246
+ KIFS FD KSGFWQ+ L ++ R TAFT P G YEWNV+PFGLK APS FQR M+
Sbjct: 329 LIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMD 388
Query: 1247 EIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFL 1306
E F KF VY+DD+L+FS + + H H+ + ++G+ +SK K LF+ KI FL
Sbjct: 389 EAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFL 448
Query: 1307 GHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRL 1366
G I +GT P +E +KFPD + DK QLQRFLG L Y +D+ P+L+ I KPL +L
Sbjct: 449 GLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYIPKLAQIRKPLQAKL 508
Query: 1367 KKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDK 1425
K++ P W+ T ++++K ++ P L+ P P+ I+ETDASD +GG+LK I++
Sbjct: 509 KENVPWRWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINE 568
Query: 1426 ----EQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDI 1481
E I + S + A++NY + KE LA++ +I F L FL+R D K
Sbjct: 569 GTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYLTPVHFLIRTDNTHFKSF 628
Query: 1482 LQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
+ + K + RWQA LS + F++E+IKG+ N DFL+RE
Sbjct: 629 VNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSRE 673
>sp|P03555|POL_CAMVC Enzymatic polyprotein [Contains: Aspartic protease ; Endonuclease;
Reverse transcriptase ]
Length = 679
Score = 334 bits (856), Expect = 2e-89
Identities = 223/645 (34%), Positives = 337/645 (51%), Gaps = 41/645 (6%)
Query: 914 LETPALFDTGADSSCISEGLIPTRYFEKTTEKLSG--AEGSKLIIKY--KIPSAIIKNDS 969
+E DTGA S+ +IP ++ + A+GS + I K II +
Sbjct: 38 IELHCFVDTGASLCIASKFVIPEEHWVNAERPIMVKIADGSSITISKVCKDIDLIIAGEI 97
Query: 970 LEIETSFLLVRNLTHKVIIETPFIKKLFPY-NTDEKGITVQHHGQPI-VFKFSKPPFVKT 1027
+I T + + II F + P+ ++ I ++ P+ + K ++ V
Sbjct: 98 FKIPTVYQQESGIDF--IIGNNFCQLYEPFIQFTDRVIFTKNKSYPVHITKLTRAVRVGI 155
Query: 1028 LNIISYKEKQINFLKEE---ISYKNIEVQLQQPSVKS------------------RIENI 1066
+ +K+ + E IS IE L++ ++ S +IE +
Sbjct: 156 EGFLESMKKRSKTQQPEPVNISTNKIENPLEEIAILSEGRRLSEEKLFITQQRMQKIEEL 215
Query: 1067 LENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREIND 1126
LE + S D PN + ++L SD K +P++ + + F ++I +
Sbjct: 216 LEKVCSENPLD-PNKTKQWMKASIKL------SDPSKAIKVKPMKYSPMDREEFDKQIKE 268
Query: 1127 LLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLA 1186
LL K+I+ SKSP AF VN +AE RG R+V+NYK +N+A Y +PNK +LL
Sbjct: 269 LLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATIGDAYNLPNKDELLT 328
Query: 1187 RLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMN 1246
+ KIFS FD KSGFWQ+ L ++ R TAFT P G YEWNV+PFGLK APS FQR M+
Sbjct: 329 LIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMD 388
Query: 1247 EIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFL 1306
E F KF VY+DD+L+FS + + H H+ + ++G+ +SK K LF+ KI FL
Sbjct: 389 EAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFL 448
Query: 1307 GHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRL 1366
G I +GT P +E +KFPD + DK QLQRFLG L Y +D+ P+L+ I KPL +L
Sbjct: 449 GLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYIPKLAQIRKPLQAKL 508
Query: 1367 KKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDK 1425
K++ P W+ T ++++K ++ P L+ P P+ I+ETDASD +GG+LK I++
Sbjct: 509 KENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINE 568
Query: 1426 ----EQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDI 1481
E I + S + A++NY + KE LA++ +I F L FL+R D K
Sbjct: 569 GTNTELICRYASGSFKAAERNYHSNDKETLAVINTIKKFSIYLTPVHFLIRTDNTHFKSF 628
Query: 1482 LQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
+ + K + RWQA LS + F++E+IKG+ N DFL+RE
Sbjct: 629 VNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSRE 673
>gb|AAA21736.1| reverse transcriptase
Length = 680
Score = 332 bits (852), Expect = 5e-89
Identities = 222/645 (34%), Positives = 336/645 (51%), Gaps = 41/645 (6%)
Query: 914 LETPALFDTGADSSCISEGLIPTRYFEKTTEKLSG--AEGSKLIIKY--KIPSAIIKNDS 969
+E DTGA S+ +IP ++ + A+GS + I K II +
Sbjct: 39 IELHCFVDTGASLCIASKFVIPEEHWVNAERPIMVKIADGSSITISKVCKDIDLIIAGEI 98
Query: 970 LEIETSFLLVRNLTHKVIIETPFIKKLFPY-NTDEKGITVQHHGQPI-VFKFSKPPFVKT 1027
+I T + + II F + P+ ++ I ++ P+ + ++ V T
Sbjct: 99 FKIPTVYQQESGIDF--IIGNNFCQLYEPFIQFTDRVIFAKNKSYPVHIANVTRAVRVGT 156
Query: 1028 LNIISYKEKQINFLKEE---ISYKNIEVQLQQPSVKS------------------RIENI 1066
+ +K+ + E IS IE L++ ++ S +IE +
Sbjct: 157 EGFLESMKKRSKTQQPEPVNISTNKIENPLEEIAILSEGRRLSEEKLFITQQRMQKIEEL 216
Query: 1067 LENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREIND 1126
LE + S D PN + ++L SD K +P++ + + F ++I +
Sbjct: 217 LEKVCSENPLD-PNKTKQWMKASIKL------SDPNKAIKVKPMKYSPMDREEFDKQIKE 269
Query: 1127 LLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLA 1186
LL K+I+ SKSP AF VN +AE RG R+V+NYK +N+A Y +PNK +LL
Sbjct: 270 LLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNLPNKDELLT 329
Query: 1187 RLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMN 1246
+ KIFS FD KSGFWQ+ L ++ R TAFT P G YEWNV+PFGLK APS FQR M+
Sbjct: 330 LIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPFGLKQAPSIFQRHMD 389
Query: 1247 EIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFL 1306
E F KF VY+DD+L+FS + + H H+ + ++G+ +SK K LF+ KI FL
Sbjct: 390 EAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILSKKKAQLFKKKINFL 449
Query: 1307 GHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRL 1366
G I +GT P +E +KFPD + DK QLQRFLG L Y +D+ P+L+ I KPL +L
Sbjct: 450 GLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYIPKLAQIRKPLQAKL 509
Query: 1367 KKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILK----QK 1421
K++ P W+ T ++++K ++ P L+ P P+ I+ETDASD +GG+LK +
Sbjct: 510 KENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASDDYWGGMLKAIKINE 569
Query: 1422 IIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDI 1481
I+ E I + S + A++NY + KE LA++ +I F L FL R D K
Sbjct: 570 GINTELICRYASGSFKAAERNYHSNDKETLAVINTIKKFSIYLTPAHFLTRTDNTHFKSF 629
Query: 1482 LQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
+ + K + RWQA L+ + F++E+IKG+ N DFL+RE
Sbjct: 630 VNLNYKGDSKLGRNIRWQAWLNHYSFDVEHIKGTDNHFVDFLSRE 674
>sp|P03556|POL_CAMVD Enzymatic polyprotein [Contains: Aspartic protease ; Endonuclease;
Reverse transcriptase ]
Length = 674
Score = 331 bits (849), Expect = 1e-88
Identities = 190/470 (40%), Positives = 273/470 (57%), Gaps = 12/470 (2%)
Query: 1062 RIENILENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQ 1121
+IE +LE + S D PN + ++L SD K +P++ + + F
Sbjct: 206 KIEELLEKVCSENPLD-PNKTKQWMKASIKL------SDPSKAIKVKPMKYSPMDREEFD 258
Query: 1122 REINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 1181
++I +LL K+I+ SKSP AF VN +AE RG R+V+NYK +N+A Y PNK
Sbjct: 259 KQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNPPNK 318
Query: 1182 KDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEF 1241
+LL + KIFS FD KSGFWQ+ L ++ R TAFT P G YEWNV+PFGLK APS F
Sbjct: 319 DELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPFGLKQAPSIF 378
Query: 1242 QRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQT 1301
QR M+E F KF VY+DD+L+FS + + H H+ + ++G+ +SK K LF+
Sbjct: 379 QRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILSKKKAQLFKK 438
Query: 1302 KIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKP 1361
KI FLG I +GT P +E +KFPD + DK QLQRFLG L Y +D+ P+L+ I KP
Sbjct: 439 KINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYIPKLAQIRKP 498
Query: 1362 LHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQ 1420
L +LK++ P W+ T ++++K ++ P L+ P P+ I+ETDASD +GG+LK
Sbjct: 499 LQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASDDYWGGMLKA 558
Query: 1421 KIIDK----EQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCK 1476
I++ E I + S + A++NY + KE LA++ +I F L FL+R D
Sbjct: 559 IKINEGTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYLTPVHFLIRTDNT 618
Query: 1477 SAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
K + + K + RWQA LS + F++E+IKG+ N DFL+RE
Sbjct: 619 HFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSRE 668
>gb|AAD37341.1| reverse transcriptase [Cauliflower mosaic virus]
Length = 674
Score = 331 bits (848), Expect = 1e-88
Identities = 189/470 (40%), Positives = 273/470 (57%), Gaps = 12/470 (2%)
Query: 1062 RIENILENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQ 1121
+IE +LE + S D PN + ++L SD K +P++ + + F
Sbjct: 206 KIEELLEKVCSENPLD-PNKTKQWMKASIKL------SDPSKAIKVKPMKYSPMDREEFD 258
Query: 1122 REINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 1181
++I +LL K+I+ SKSP AF VN +AE RG R+V+NYK +N+A Y +PNK
Sbjct: 259 KQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNLPNK 318
Query: 1182 KDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEF 1241
+LL + KIFS FD KSGFWQ+ ++ R TAFT P G YEWNV+PFGLK APS F
Sbjct: 319 DELLTLIRGKKIFSSFDCKSGFWQVLPDQESRPLTAFTCPQGHYEWNVVPFGLKQAPSIF 378
Query: 1242 QRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQT 1301
QR M+E F KF VY+DD+L+FS + + H H+ + ++G+ +SK K LF+
Sbjct: 379 QRHMDEAFRVFRKFCCVYVDDILVFSNTEEDHLLHVAMILQKCNQHGIILSKKKAQLFKK 438
Query: 1302 KIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKP 1361
KI FLG I +GT P +E +KFPD + DK QLQRFLG L Y +D+ P+L+ I KP
Sbjct: 439 KINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYIPKLAQIRKP 498
Query: 1362 LHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQ 1420
L +LK++ P W+ T ++++K ++ P L+ P P+ I+ETDASD +GG+LK
Sbjct: 499 LQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHRPLPEEKLIIETDASDDYWGGMLKA 558
Query: 1421 KIIDK----EQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCK 1476
I++ E I + S + A++NY + KE LA++ +I F L FL+R D
Sbjct: 559 IQINEGTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYLTPVHFLIRTDNT 618
Query: 1477 SAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
K + + K + RWQA LS + F++E+IKG+ N DFL+RE
Sbjct: 619 HFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNRFADFLSRE 668
>emb|CAH68829.1| polyprotein [Carnation etched ring virus]
Length = 656
Score = 330 bits (846), Expect = 2e-88
Identities = 191/482 (39%), Positives = 279/482 (57%), Gaps = 14/482 (2%)
Query: 1043 EEISYKNIEVQLQQPSVKSRIENILENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQ 1102
EE+ Y E+Q+ S S IE +LE + S D P + + +EL D +
Sbjct: 179 EELCY---EIQI---SKFSAIEEMLERVSSENPID-PEKSKQWMTATIEL------IDPK 225
Query: 1103 IPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVI 1162
+ +P+ + + F ++I + L K+I+ SKSP AF V +AE RG R+V+
Sbjct: 226 TIVEVKPMSYSPSDREEFDKQIKEPLDLKVIKPSKSPHMSPAFLVENEAERRRGKKRMVV 285
Query: 1163 NYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPF 1222
NYK +N+A + +PNK +LLA + KI+S FD KSGFWQ+ L ++ + TAFT P
Sbjct: 286 NYKAMNKATKGDAHNLPNKDELLALVRGKKIYSSFDCKSGFWQVLLDKESQLLTAFTCPQ 345
Query: 1223 GQYEWNVMPFGLKNAPSEFQRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFIS 1282
G Y+WNV+PFGLK APS FQR M FN SK+ VY+DD+L+FS + ++H+ H+ +
Sbjct: 346 GHYQWNVVPFGLKQAPSIFQRHMQTAFNQHSKYCCVYVDDILVFSNTEEEHYIHVLNILR 405
Query: 1283 IIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFL 1342
+K G+ +SK K LF+ KI FLG I QGT P N +E KFPD+I DK QLQR L
Sbjct: 406 RCEKLGIILSKKKAQLFKEKINFLGLEIDQGTHCPQNHILEHIHKFPDRIEDKKQLQRSL 465
Query: 1343 GCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQA 1401
G L Y +D+ P+L++I KPL +LK+D W+D + + +IK +K+ P LY P P
Sbjct: 466 GILTYASDYIPKLASIRKPLQSKLKEDSTWTWNDTDSQYMAKIKKNLKSFPKLYHPEPND 525
Query: 1402 FKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQ 1461
++ETDAS+ +GGILK E I + S + A++NY + +KE+LA+ I F
Sbjct: 526 KLVIETDASEEFWGGILKAIHNSHEYICRYASGSFKAAERNYHSNEKELLAVFRVIYKFS 585
Query: 1462 SDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPD 1521
L +FL+R D K+ + ++K + RWQ LS +DF++E+I G+ N D
Sbjct: 586 IYLTPCRFLIRSDNKNFTHFVNINLKGDRKQGRLVRWQMWLSQYDFDVEHIAGTKNVFAD 645
Query: 1522 FL 1523
FL
Sbjct: 646 FL 647
>sp|Q00962|POL_CAMVN Enzymatic polyprotein [Contains: Aspartic protease ; Endonuclease;
Reverse transcriptase ] gi|331571|gb|AAA46358.1| reverse
transcriptase gi|445598|prf||1909346E reverse
transcriptase
Length = 680
Score = 328 bits (841), Expect = 9e-88
Identities = 219/656 (33%), Positives = 338/656 (51%), Gaps = 63/656 (9%)
Query: 914 LETPALFDTGADSSCISEGLIPTRYFEKTTEKLSG--AEGSKLIIK-------------- 957
+E DTGA S+ +IP ++ + A+GS + I
Sbjct: 39 IELHCFVDTGASLCIASKFVIPEEHWVNAERPIMVKIADGSSITISKVCKDIDLIIVGVI 98
Query: 958 YKIPSAIIKNDSLEIETSFLLVRNLTHKVIIETPFIKKLFPYNTDEKGITVQHHGQPI-V 1016
+KIP+ + ++ F++ N + PFI+ ++ I ++ P+ +
Sbjct: 99 FKIPTVYQQESGID----FIIGNNFCQ---LYEPFIQ------FTDRVIFTKNKSYPVHI 145
Query: 1017 FKFSKPPFVKTLNIISYKEKQINFLKEE---ISYKNIEVQLQQPSVKS------------ 1061
K ++ V T + +K+ + E IS IE L++ ++ S
Sbjct: 146 AKLTRAVRVGTEGFLESMKKRSKTQQPEPVNISTNKIENPLEEIAILSEGRRLSEEKLFI 205
Query: 1062 ------RIENILENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEE 1115
+ E +LE + S D PN + ++L SD K +P++ +
Sbjct: 206 TQQRMQKTEELLEKVCSENPLD-PNKTKQWMKASIKL------SDPSKAIKVKPMKYSPM 258
Query: 1116 LLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIR 1175
+ F ++I +LL K+I+ SKSP AF VN +AE RG R+V+NYK +N+A
Sbjct: 259 DREEFDKQIKELLDLKVIKPSKSPHMAPAFLVNNEAENGRGNKRMVVNYKAMNKATVGDA 318
Query: 1176 YPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLK 1235
Y +PNK +LL + KIFS FD KSGFWQ+ L ++ R TAFT P G YEWNV+PFGLK
Sbjct: 319 YNLPNKDELLTLIRGKKIFSSFDCKSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPFGLK 378
Query: 1236 NAPSEFQRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTK 1295
APS FQR M+E F KF VY+DD+++FS + + H H+ + ++G+ +SK K
Sbjct: 379 QAPSIFQRHMDEAFRVFRKFCCVYVDDIVVFSNNEEDHLLHVAMILQKCNQHGIILSKKK 438
Query: 1296 VSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQL 1355
LF+ KI FLG I +GT P +E +KFPD + DK QLQRFLG L Y +D+ P L
Sbjct: 439 AQLFKKKINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYIPNL 498
Query: 1356 STIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGF 1414
+ + +PL +LK++ P W+ T ++++K ++ P L+ P P+ I+ETDASD +
Sbjct: 499 AQMRQPLQAKLKENVPWKWTKEDTLYMQKVKKNLQGFPPLHHPLPEEKLIIETDASDDYW 558
Query: 1415 GGILKQKIIDK----EQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFL 1470
GG+LK I++ E I + S + A++NY + KE LA++ +I F L FL
Sbjct: 559 GGMLKAIKINEGTNTELICRYRSGSFKAAERNYHSNDKETLAVINTIKKFSIYLTPVHFL 618
Query: 1471 VRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
+R D K + + K + RWQA LS + F++E+IKG+ N DFL+RE
Sbjct: 619 IRTDNTHFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSRE 674
>gb|AAW56089.1| polyprotein [Horseradish latent virus]
Length = 679
Score = 328 bits (841), Expect = 9e-88
Identities = 183/435 (42%), Positives = 256/435 (58%), Gaps = 7/435 (1%)
Query: 1099 SDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTP 1158
SD K +P++ + + F+++I +LL K+IR SKSP AF VN +AE RG
Sbjct: 241 SDPTKVIKVKPMKYSPMDREEFEKQIQELLDLKVIRPSKSPHMAPAFLVNNEAEKRRGKK 300
Query: 1159 RLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAF 1218
R+V+NYK +N A Y +PNK +LL + KIFS FD KSGFWQ++L E+ + TAF
Sbjct: 301 RMVVNYKAMNDATVGDAYNLPNKDELLTLIRGKKIFSSFDCKSGFWQVRLDEESKSLTAF 360
Query: 1219 TVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLN 1278
T P G YEWNV+PFG+K APS FQR M+E F KF +Y+DD+L+FS + H H+
Sbjct: 361 TCPQGHYEWNVVPFGMKQAPSIFQRHMDEAFKVFRKFCCIYVDDILVFSDNEQNHQLHVA 420
Query: 1279 TFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQL 1338
+ ++G+ +SK K LF+ +I FLG I QGT P + +E KFPD I K QL
Sbjct: 421 MILQKCYQHGIILSKKKAQLFKERINFLGLEIDQGTHRPQSHILEHIQKFPDIIESKLQL 480
Query: 1339 QRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLP 1397
QRFLG L Y +D+ P+L+ I KPL +LK++ W+ T +K++K + P L+ P
Sbjct: 481 QRFLGVLTYASDYIPKLAQIRKPLQAKLKENVQWRWTPEDTLYMKKVKKNLNGFPPLHHP 540
Query: 1398 NPQAFKIVETDASDIGFGGILKQKIIDK------EQIIAFTSKHWNPAQQNYSTVKKEVL 1451
P+ I+ETDASD +GGILK ID E + + S + PA+QNY + KE L
Sbjct: 541 LPEEKLIIETDASDNYWGGILKAIHIDLSTNESIELVCRYASGSFKPAEQNYHSNDKETL 600
Query: 1452 AIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEY 1511
A++ +I F L +FLVR D K L + K + RWQ L + F++++
Sbjct: 601 AVIRTIQKFSIYLTPVRFLVRTDNTHFKYFLNINYKGDSKMGRNIRWQGWLQNYVFDVDH 660
Query: 1512 IKGSTNSLPDFLTRE 1526
IKG+ N L DFL+RE
Sbjct: 661 IKGTNNCLADFLSRE 675
>gb|AAA46350.1| ORF5; putative
Length = 675
Score = 326 bits (835), Expect = 4e-87
Identities = 188/470 (40%), Positives = 271/470 (57%), Gaps = 12/470 (2%)
Query: 1062 RIENILENIQSSICFDLPNAFWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQ 1121
+IE +LE + S D PN + ++L SD K +P++ + + F
Sbjct: 207 KIEELLEKVCSENPLD-PNKTKQWMKASIKL------SDPSKAIKVKPMKYSPMDREEFD 259
Query: 1122 REINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNK 1181
++I +LL K+I+ SKSP AF VN +AE RG R+V+NYK +N+A Y PNK
Sbjct: 260 KQIKELLDLKVIKPSKSPHMAPAFLVNNEAEKRRGKKRMVVNYKAMNKATVGDAYNPPNK 319
Query: 1182 KDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEF 1241
+LL + KIFS F SGFWQ+ L ++ R TAFT P G YEWNV+PFGLK APS F
Sbjct: 320 DELLTLIRGKKIFSSFHCNSGFWQVLLDQESRPLTAFTCPQGHYEWNVVPFGLKQAPSIF 379
Query: 1242 QRIMNEIFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQT 1301
QR M+E F KF VY+DD+L+FS + + H H+ + ++G+ +SK K LF+
Sbjct: 380 QRHMDEAFRVFRKFCCVYVDDILVFSNNEEDHLLHVAMILQKCNQHGIILSKKKAQLFKK 439
Query: 1302 KIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKP 1361
KI FLG I +GT P +E +KFPD + DK QLQRFLG L Y +D+ P+L+ I KP
Sbjct: 440 KINFLGLEIDEGTHKPQGHILEHINKFPDTLEDKKQLQRFLGILTYASDYIPKLAQIRKP 499
Query: 1362 LHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQ 1420
L +LK++ P W+ T ++++K ++ P L+ P P+ I+ETDASD +GG+LK
Sbjct: 500 LQAKLKENVPWKWTKEDTLYMQKVKKNLQAFPPLHHPLPEEKLIIETDASDDYWGGMLKA 559
Query: 1421 KIIDK----EQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCK 1476
I++ E I + S + A++NY + KE LA++ +I F L FL+R D
Sbjct: 560 IKINEGTNTELICRYASGSFKAAEKNYHSNDKETLAVINTIKKFSIYLTPVHFLIRTDNT 619
Query: 1477 SAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDFLTRE 1526
K + + K + RWQA LS + F++E+IKG+ N DFL+RE
Sbjct: 620 HFKSFVNLNYKGDSKLGRNIRWQAWLSHYSFDVEHIKGTDNHFADFLSRE 669
>ref|NP_619548.1| Enzymatic polyprotein [Contains: Aspartic protease; Endonuclease;
Reverse transcriptase] [Figwort mosaic virus]
gi|58813|emb|CAA29527.1| unnamed protein product [Figwort
mosaic virus] gi|130600|sp|P09523|POL_FMVD Enzymatic
polyprotein [Contains: Aspartic protease ; Endonuclease;
Reverse transcriptase ]
Length = 666
Score = 325 bits (834), Expect = 6e-87
Identities = 215/627 (34%), Positives = 328/627 (52%), Gaps = 38/627 (6%)
Query: 921 DTGADSSCISEGLIPTRYFEKTTE----KLSGAEGSKLIIKYKIPSAIIKNDSLEIETSF 976
DTGA S +IP +E + + K++ E K+ K S EI T +
Sbjct: 54 DTGASLCIASRYIIPEELWENSPKDIQVKIANQELIKITKVCKNLKVKFAGKSFEIPTVY 113
Query: 977 LLVRNLTHKVIIETPFIKKLFPYNTDEKGITVQHHGQPIV-------FKFSKPPFVKTLN 1029
+ +I F + P+ E I + ++ F S P F++ +
Sbjct: 114 QQETGIDF--LIGNNFCRLYNPFIQWEDRIAFHLKNEMVLIKKVTKAFSVSNPSFLENMK 171
Query: 1030 IISYKEKQI-------NFLKEEISYKNIEVQLQQPSVKSRIENILENIQSSICFD-LPNA 1081
S K +QI N + E Y I + Q +IE +L+ + S D + +
Sbjct: 172 KDS-KTEQIPGTNISKNIINPEERYFLITEKYQ------KIEQLLDKVCSENPIDPIKSK 224
Query: 1082 FWERKSHMVELPYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWS 1141
W + S + P + + +P+ + + + F ++I +LL LI SKS
Sbjct: 225 QWMKASIKLIDPLKV--------IRVKPMSYSPQDREGFAKQIKELLDLGLIIPSKSQHM 276
Query: 1142 CAAFYVNKQAEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKS 1201
AF V +AE RG R+V+NYK +NQA + +PN ++LL L IFS FD KS
Sbjct: 277 SPAFLVENEAERRRGKKRMVVNYKAINQATIGDSHNLPNMQELLTLLRGKSIFSSFDCKS 336
Query: 1202 GFWQIQLQEKDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQRIMNEIFNP*SKFAIVYID 1261
GFWQ+ L E+ + TAFT P G ++W V+PFGLK APS FQR M N KF +VY+D
Sbjct: 337 GFWQVVLDEESQKLTAFTCPQGHFQWKVVPFGLKQAPSIFQRHMQTALNGADKFCMVYVD 396
Query: 1262 DVLIFSQSIDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRA 1321
D+++FS S H+ H+ + I++K G+ +SK K +LF+ KI FLG I +GT P N
Sbjct: 397 DIIVFSNSELDHYNHVYAVLKIVEKYGIILSKKKANLFKEKINFLGLEIDKGTHCPQNHI 456
Query: 1322 IEFTDKFPDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNV 1380
+E KFPD++ DK LQRFLG L Y + P+L+ I KPL +LKKD W+ ++
Sbjct: 457 LENIHKFPDRLEDKKHLQRFLGVLTYAETYIPKLAEIRKPLQVKLKKDVTWNWTQSDSDY 516
Query: 1381 VKQIKLRIKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIA-FTSKHWNPA 1439
VK+IK + + P LYLP P+ I+ETDASD +GG+LK + +D ++I ++S + A
Sbjct: 517 VKKIKKNLGSFPKLYLPKPEDHLIIETDASDSFWGGVLKARALDGVELICRYSSGSFKQA 576
Query: 1440 QQNYSTVKKEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQ 1499
++NY + KE+LA+ I+ F + L +F VR D K+ L+ ++K + + RWQ
Sbjct: 577 EKNYHSNDKELLAVKQVITKFSAYLTPVRFTVRTDNKNFTYFLRINLKGDSKQGRLVRWQ 636
Query: 1500 AILSVFDFEIEYIKGSTNSLPDFLTRE 1526
S + F++E+++G N L D LTR+
Sbjct: 637 NWFSKYQFDVEHLEGVKNVLADCLTRD 663
>emb|CAA28360.1| unnamed protein product [Carnation etched ring virus]
gi|19919894|ref|NP_612577.1| Enzymatic polyprotein
[Contains: Aspartic protease; Endonuclease; Reverse
transcriptase] [Carnation etched ring virus]
gi|130593|sp|P05400|POL_CERV Enzymatic polyprotein
[Contains: Aspartic protease ; Endonuclease; Reverse
transcriptase ] gi|225356|prf||1301227E ORF 5
Length = 659
Score = 320 bits (821), Expect = 2e-85
Identities = 190/496 (38%), Positives = 281/496 (56%), Gaps = 10/496 (2%)
Query: 1031 ISYKEKQINFLKEEISYKNIEVQLQQPSVKSRIENILENIQSSICFDLPNAFWERKSHMV 1090
I+ Q FL+E ++ + + Q S S IE +LE + S D P + + +
Sbjct: 162 INITSNQHLFLEEGGNHVDEMLYEIQISKFSAIEEMLERVSSENPID-PEKSKQWMTATI 220
Query: 1091 ELPYEKDFSDKQIPTKARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQ 1150
EL D + K +P+ + + F R+I +LL+ K+I+ SKS AF V +
Sbjct: 221 EL------IDPKTVVKVKPMSYSPSDREEFDRQIKELLELKVIKPSKSTHMSPAFLVENE 274
Query: 1151 AEIERGTPRLVINYKPLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQE 1210
AE RG R+V+NYK +N+A + +PNK +LL + KI+S FD KSG WQ+ L +
Sbjct: 275 AERRRGKKRMVVNYKAMNKATKGDAHNLPNKDELLTLVRGKKIYSSFDCKSGLWQVLLDK 334
Query: 1211 KDRYKTAFTVPFGQYEWNVMPFGLKNAPSEFQR-IMNEIFNP*SKFAIVYIDDVLIFSQS 1269
+ + TAFT P G Y+WNV+PFGLK APS F + N N SK+ VY+DD+L+FS +
Sbjct: 335 ESQLLTAFTCPQGHYQWNVVPFGLKQAPSIFPKTYANSHSNQYSKYCCVYVDDILVFSNT 394
Query: 1270 -IDQHFKHLNTFISIIKKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKF 1328
+H+ H+ + +K G+ +SK K LF+ KI FLG I QGT P N +E KF
Sbjct: 395 GRKEHYIHVLNILRRCEKLGIILSKKKAQLFKEKINFLGLEIDQGTHCPQNHILEHIHKF 454
Query: 1329 PDQIIDKTQLQRFLGCLNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLR 1387
PD+I DK QLQRFLG L Y +D+ P+L++I KPL +LK+D W+D + + +IK
Sbjct: 455 PDRIEDKKQLQRFLGILTYASDYIPKLASIRKPLQSKLKEDSTWTWNDTDSQYMAKIKKN 514
Query: 1388 IKNLPCLYLPNPQAFKIVETDASDIGFGGILKQKIIDKEQIIAFTSKHWNPAQQNYSTVK 1447
+K+ P LY P P ++ETDAS+ +GGILK E I + S + A++NY + +
Sbjct: 515 LKSFPKLYHPEPNDKLVIETDASEEFWGGILKAIHNSHEYICRYASGSFKAAERNYHSNE 574
Query: 1448 KEVLAIVLSISNFQSDLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDF 1507
KE+LA++ I F L +FL+R D K+ + ++K + RWQ LS +DF
Sbjct: 575 KELLAVIRVIKKFSIYLTPSRFLIRTDNKNFTHFVNINLKGDRKQGRLVRWQMWLSQYDF 634
Query: 1508 EIEYIKGSTNSLPDFL 1523
++E+I G+ N DFL
Sbjct: 635 DVEHIAGTKNVFADFL 650
>ref|NP_043933.1| ORF V [Strawberry vein banding virus] gi|1360613|emb|CAA65970.1| ORF
V [Strawberry vein banding virus]
Length = 708
Score = 309 bits (791), Expect = 5e-82
Identities = 170/424 (40%), Positives = 258/424 (60%), Gaps = 3/424 (0%)
Query: 1106 KARPIQMNEELLQFFQREINDLLQKKLIRRSKSPWSCAAFYVNKQAEIERGTPRLVINYK 1165
K +P+Q + + + F+ +I +LL+ +IR SKSP S AF V AEI+RG R+VINYK
Sbjct: 280 KCKPMQYSPQDREEFKTQIEELLKLGIIRPSKSPHSSPAFMVRNHAEIKRGKARMVINYK 339
Query: 1166 PLNQALCWIRYPIPNKKDLLARLHDAKIFSKFDMKSGFWQIQLQEKDRYKTAFTVPFGQY 1225
LN Y +PNK+ LL R+ +S FD KSGFWQ++L + TAF+ P G Y
Sbjct: 340 KLNDHTKGDGYLLPNKEQLLQRIGGKTFYSSFDCKSGFWQVRLAPETIQLTAFSCPQGHY 399
Query: 1226 EWNVMPFGLKNAPSEFQRIMNE-IFNP*SKFAIVYIDDVLIFSQSIDQHFKHLNTFISII 1284
EW VMPFGLK AP+ FQR M+E + N +F VY+DD+++FS++ ++H H+ ++
Sbjct: 400 EWLVMPFGLKQAPAIFQRHMDESLSNMYPQFCAVYVDDIIVFSKTEEEHLGHVKIVLNRC 459
Query: 1285 KKNGMAVSKTKVSLFQTKIRFLGHNIHQGTIIPINRAIEFTDKFPDQIIDKTQLQRFLGC 1344
K G+ +SK K L +T I FLG I +G + + FPDQ+ D+ LQRFLG
Sbjct: 460 KALGIVLSKKKAQLCKTTINFLGLVIERGNLKVQSHIGLHLVAFPDQLSDRNALQRFLGL 519
Query: 1345 LNYVADFCPQLSTIIKPLHDRLKKDPP-PWSDIHTNVVKQIKLRIKNLPCLYLPNPQAFK 1403
LNY++ + P+++ + PL +LKK+ W++ T V++IK +K LP LY P+P+
Sbjct: 520 LNYISAYFPKIANLRSPLQVKLKKEITWSWTEKDTETVRKIKSLVKTLPDLYNPSPEDKP 579
Query: 1404 IVETDASDIGFGGILKQKIID-KEQIIAFTSKHWNPAQQNYSTVKKEVLAIVLSISNFQS 1462
I+E DASD +G ILK K+ + KE I + S + PA++NY + +KE+L+I+ +I F++
Sbjct: 580 IIECDASDDHWGAILKAKLPEGKEVICRYASGTFKPAEKNYHSNEKEILSIIKAIKAFRA 639
Query: 1463 DLINQKFLVRVDCKSAKDILQKDVKNLASKHIFARWQAILSVFDFEIEYIKGSTNSLPDF 1522
++ KFLVR D +A ++ ++ + RWQ L + F+IE++ G N L D
Sbjct: 640 YILPYKFLVRTDNTNAAYFVRTNIAGDYKQSRLVRWQMALREYSFDIEHVSGQKNVLADI 699
Query: 1523 LTRE 1526
+TRE
Sbjct: 700 MTRE 703
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,637,394,842
Number of Sequences: 2540612
Number of extensions: 118904696
Number of successful extensions: 704304
Number of sequences better than 10.0: 36770
Number of HSP's better than 10.0 without gapping: 3276
Number of HSP's successfully gapped in prelim test: 33521
Number of HSP's that attempted gapping in prelim test: 612480
Number of HSP's gapped (non-prelim): 87382
length of query: 1526
length of database: 863,360,394
effective HSP length: 141
effective length of query: 1385
effective length of database: 505,134,102
effective search space: 699610731270
effective search space used: 699610731270
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 82 (36.2 bits)
Lotus: description of TM0173b.7


