

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.9
(746 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
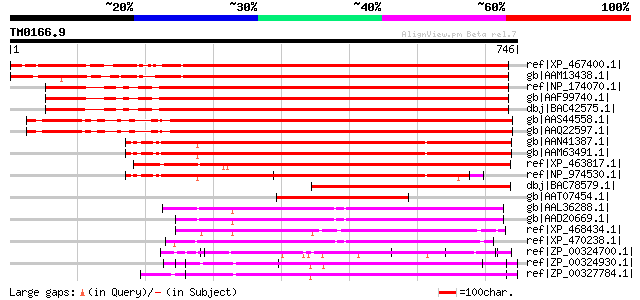
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_467400.1| putative kinesin light chain [Oryza sativa (jap... 833 0.0
gb|AAM13438.1| similar to A. thaliana protein BAB01483 similar ... 816 0.0
ref|NP_174070.1| kinesin light chain-related [Arabidopsis thaliana] 745 0.0
gb|AAF99740.1| F17L21.29 [Arabidopsis thaliana] 745 0.0
dbj|BAC42575.1| unknown protein [Arabidopsis thaliana] gi|290289... 743 0.0
gb|AAS44558.1| kinesin light chain-like protein [Arabidopsis tha... 717 0.0
gb|AAQ22597.1| At3g27960 [Arabidopsis thaliana] 715 0.0
gb|AAN41387.1| unknown protein [Arabidopsis thaliana] gi|7267782... 642 0.0
gb|AAM63491.1| putative kinesin light chain [Arabidopsis thaliana] 642 0.0
ref|XP_463817.1| kinesin light chain-like protein [Oryza sativa ... 637 0.0
ref|NP_974530.1| kinesin light chain-related [Arabidopsis thaliana] 561 e-158
dbj|BAC78579.1| hypothetical protein [Oryza sativa (japonica cul... 392 e-107
gb|AAT07454.1| kinesin-like protein [Mirabilis jalapa] 306 2e-81
gb|AAL36288.1| putative kinesin light chain [Arabidopsis thalian... 172 3e-41
gb|AAD20669.1| putative kinesin light chain [Arabidopsis thalian... 172 4e-41
ref|XP_468434.1| kinesin light chain-like [Oryza sativa (japonic... 155 3e-36
ref|XP_470238.1| Hypothetical protein [Oryza sativa (japonica cu... 137 1e-30
ref|ZP_00324700.1| COG0457: FOG: TPR repeat [Trichodesmium eryth... 127 1e-27
ref|ZP_00324930.1| COG0457: FOG: TPR repeat [Trichodesmium eryth... 121 9e-26
ref|ZP_00327784.1| COG0457: FOG: TPR repeat [Trichodesmium eryth... 119 3e-25
>ref|XP_467400.1| putative kinesin light chain [Oryza sativa (japonica
cultivar-group)] gi|41053168|dbj|BAD08110.1| putative
kinesin light chain [Oryza sativa (japonica
cultivar-group)] gi|19387245|gb|AAL87157.1| putative
kinesin light chain gene [Oryza sativa (japonica
cultivar-group)]
Length = 711
Score = 833 bits (2152), Expect = 0.0
Identities = 449/737 (60%), Positives = 554/737 (74%), Gaps = 41/737 (5%)
Query: 1 MPGIVTNGVC-DGAVNEMNGNHSPSKETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPS 59
MPGI+ +GV + A NE+N S +K+ + +SP S S + + G EPS
Sbjct: 1 MPGIIVDGVVTEEAQNEVNS--SQNKDNSSAPRSPVASKS-MHSEALEMHVEGSGAGEPS 57
Query: 60 IEQLYENVCDMQSSDQ--SPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEEVEVEV 117
IEQLY NVC+M+SS + SPSR+SFGSDG+ESRIDSELR LV G M ++++EEE
Sbjct: 58 IEQLYNNVCEMESSSEGGSPSRESFGSDGEESRIDSELRHLVAGEMEAMKVIEEE----- 112
Query: 118 EKERGGSSSGEISSGVGGLSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDASPKSK 177
E E G+++ +++ G A +S+ S + S KA SQL++
Sbjct: 113 EGEGSGNAANAVTAAENGTPVK------------AMSSNSSKKSSKKAAKSQLESESSVG 160
Query: 178 PKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVENSAESALDKP 237
P GK+ + E + KP G G + K+SP N + ++ L+ P
Sbjct: 161 PNGKASTEEG--EAEVSKP-------GSRVG-RRRKASP-------NPHNGTEDAGLNNP 203
Query: 238 ERAPILLKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLELVMCLHVTAAIYCNL 297
+ P LLK ARDLI+S DNP++AL+ AL+A EK GKPSL LVM LHV AAIYCNL
Sbjct: 204 DLGPFLLKHARDLIAS-DNPRRALKYALRATKSFEKCAGGKPSLNLVMSLHVVAAIYCNL 262
Query: 298 GQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQ 357
G+Y+EA+P+L+RS+EIPVI E Q+HALAKF+G MQLGDTY MLGQ S+ Y++G E+Q
Sbjct: 263 GKYDEAVPVLQRSLEIPVIEEGQEHALAKFSGCMQLGDTYGMLGQTALSLQWYAAGLEIQ 322
Query: 358 RQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRRLM 417
+Q LGE DPRVGETCRY+AEA+VQALQ DEA+RLCQ ALDIH+ N SLEE ADRRLM
Sbjct: 323 KQTLGEQDPRVGETCRYLAEAHVQALQLDEAQRLCQKALDIHRENGEPASLEETADRRLM 382
Query: 418 GLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKA 477
GLI DTKG+HEAALEHLV+ASMAMVANGQE EVASVDCSIGD YLS+ RYDEAVF+Y+KA
Sbjct: 383 GLICDTKGDHEAALEHLVMASMAMVANGQETEVASVDCSIGDIYLSLGRYDEAVFSYQKA 442
Query: 478 LTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIAS 537
LTVFKT KGENH V SVFVRLADL+N+TGK++ESKSYC++AL+IY+ P+PG + EEIA+
Sbjct: 443 LTVFKTSKGENHATVASVFVRLADLYNKTGKLRESKSYCENALKIYQKPIPGTSLEEIAT 502
Query: 538 GLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYMLGNYTESY 597
GLT+VSAIYE+MNE E+ALKLLQKAL +YN+S GQQS+IAGIEAQMGV++Y+LGNY E+Y
Sbjct: 503 GLTDVSAIYETMNEHEQALKLLQKALKMYNNSAGQQSTIAGIEAQMGVLHYILGNYGEAY 562
Query: 598 NTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYH 657
++ K+AI KLR GEKK++F G+ALNQMGLACVQ Y+ EA ELFEEA+++LE EYGPYH
Sbjct: 563 DSFKSAIAKLRTCGEKKTAFFGVALNQMGLACVQRYSINEAAELFEEARAVLEQEYGPYH 622
Query: 658 PETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAG 717
P+TLGVYSNLAGTYDA+GRLDEAI+ILE+VV +REEKLGTANPDVDDEKRRL+ELLKEAG
Sbjct: 623 PDTLGVYSNLAGTYDAMGRLDEAIEILEHVVGMREEKLGTANPDVDDEKRRLAELLKEAG 682
Query: 718 RVRSRKVMSLENLLDAN 734
R RSRK SLENLL+ N
Sbjct: 683 RGRSRKAKSLENLLETN 699
>gb|AAM13438.1| similar to A. thaliana protein BAB01483 similar to kinesin light
chain [Hordeum vulgare subsp. vulgare]
Length = 708
Score = 816 bits (2107), Expect = 0.0
Identities = 440/739 (59%), Positives = 541/739 (72%), Gaps = 48/739 (6%)
Query: 1 MPGIVTNGVCDGAVNEMNGNHSPSKETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPSI 60
MPGI+ +GV + + + AP +P + + + G EPSI
Sbjct: 1 MPGIIVDGVVTEEIQHEVSSSQNKENLTAPTMAPS-----MQSEALEMHVEDSGAGEPSI 55
Query: 61 EQLYENVCDMQSSDQ---SPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEEVEVEV 117
EQLY NVC+M+SS + SPSR+SFGSDG+ESRIDSELR LV G M ++++EEE E
Sbjct: 56 EQLYNNVCEMESSSEGGGSPSRESFGSDGEESRIDSELRHLVAGEMEAMKVIEEEEE--- 112
Query: 118 EKERGGSSSGEISSGVGGLSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDASPKSK 177
KE+G S + G + + SS S++K KA SQL++
Sbjct: 113 -KEKGSGSVTNAAPTAG------------NGTPARPQSSNSSKK--KAAKSQLESDASVG 157
Query: 178 PKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLN-RVENSAESA-LD 235
P GK+ P + E + KP S +G+ N + +N E A LD
Sbjct: 158 PNGKASPEEG--ESEVSKP-----------------GSRVGRRRKANAKSQNGTEDAGLD 198
Query: 236 KPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLELVMCLHVTAAIYC 295
P+ P LLK ARDLI+S DNP++AL+ AL+A EK GKPSL LVM LHV AAI+C
Sbjct: 199 NPDLGPFLLKHARDLIAS-DNPRRALKYALRATKSFEKCAGGKPSLNLVMSLHVVAAIHC 257
Query: 296 NLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFE 355
N+G+Y EA+P+L+RS+EIPV E Q+HALAKF+G MQLGDTY MLGQ+ S+ Y+ G +
Sbjct: 258 NMGKYEEAVPVLQRSLEIPVTEEGQEHALAKFSGCMQLGDTYGMLGQIALSLQWYAKGLD 317
Query: 356 VQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRR 415
+Q+Q LGE DPRVGETCRY+AEA+VQALQ DEA++LCQMALDIH+ N SLEE ADRR
Sbjct: 318 IQKQTLGEQDPRVGETCRYLAEAHVQALQLDEAQKLCQMALDIHRDNGQPASLEETADRR 377
Query: 416 LMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYE 475
LMGLI DTKG+HEAALEHLV+ASMAMVANGQE EVASVDCSIGD YLS+ RYDEAV AY+
Sbjct: 378 LMGLICDTKGDHEAALEHLVMASMAMVANGQETEVASVDCSIGDIYLSLGRYDEAVCAYQ 437
Query: 476 KALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEI 535
KALTVFKT KGENH V SVF+RLADL+N+TGK++ESKSYC++AL+IY+ P+PG + EEI
Sbjct: 438 KALTVFKTSKGENHATVASVFLRLADLYNKTGKLRESKSYCENALKIYQKPIPGTSLEEI 497
Query: 536 ASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYMLGNYTE 595
A+GLT+VSAIYE+MNE ++ALKLLQKAL +YN+S GQQS+IAGIEAQ+GV++Y+ GNY +
Sbjct: 498 ATGLTDVSAIYETMNEHDQALKLLQKALKMYNNSAGQQSTIAGIEAQIGVLHYISGNYGD 557
Query: 596 SYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGP 655
+Y++ K+AITKLR GEKKS+F GIALNQMGLACVQ Y+ EA ELFEEA+++LE EYGP
Sbjct: 558 AYDSFKSAITKLRTCGEKKSAFFGIALNQMGLACVQRYSINEAAELFEEARTVLEQEYGP 617
Query: 656 YHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKE 715
YH +TLGVYSNLAGTYDA+GRLDEAI+ILEYVV +REEKLGTANPDVDDEKRRL+ELLKE
Sbjct: 618 YHADTLGVYSNLAGTYDAMGRLDEAIEILEYVVEMREEKLGTANPDVDDEKRRLAELLKE 677
Query: 716 AGRVRSRKVMSLENLLDAN 734
AGR RSRK SLENLL N
Sbjct: 678 AGRGRSRKAKSLENLLGTN 696
>ref|NP_174070.1| kinesin light chain-related [Arabidopsis thaliana]
Length = 650
Score = 745 bits (1923), Expect = 0.0
Identities = 395/684 (57%), Positives = 503/684 (72%), Gaps = 57/684 (8%)
Query: 53 DGVIEPSIEQLYENVCDMQSSDQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEE 112
D + + +IE+L +N+C++QSS+QSPSR+SFGS GDES+IDS+L+ L G MR+++I+E+E
Sbjct: 14 DQMFDTTIEELCKNLCELQSSNQSPSRQSFGSYGDESKIDSDLQHLALGEMRDIDILEDE 73
Query: 113 VEVEVEKERGGSSSGEISSGVGGLSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDA 172
+E ++ K E + SS N L+
Sbjct: 74 -------------------------GDEDEVAKPEEFDVKSNSS----------NLDLEV 98
Query: 173 SPKSKPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVEN-SAE 231
P+ +E++ K + K GV K K V +++N + E
Sbjct: 99 MPRD------------MEKQTGKKNVTKSNVGVGGMRK--------KKVGGTKLQNGNEE 138
Query: 232 SALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLE-KLGNGKPSLELVMCLHVT 290
+ + E A LL QAR+L+SSGD+ KALEL +A L E NGKP LE +MCLHVT
Sbjct: 139 PSSENVELARFLLNQARNLVSSGDSTHKALELTHRAAKLFEASAENGKPCLEWIMCLHVT 198
Query: 291 AAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCY 350
AA++C L +YNEAIP+L+RS+EIPV+ E ++HALAKFAG MQLGDTYAM+GQLE+SI CY
Sbjct: 199 AAVHCKLKEYNEAIPVLQRSVEIPVVEEGEEHALAKFAGLMQLGDTYAMVGQLESSISCY 258
Query: 351 SSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEE 410
+ G +Q++VLGE DPRVGETCRY+AEA VQAL+FDEA+++C+ AL IH+ + S+ E
Sbjct: 259 TEGLNIQKKVLGENDPRVGETCRYLAEALVQALRFDEAQQVCETALSIHRESGLPGSIAE 318
Query: 411 AADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEA 470
AADRRLMGLI +TKG+HE ALEHLVLASMAM ANGQE EVA VD SIGD+YLS+SR+DEA
Sbjct: 319 AADRRLMGLICETKGDHENALEHLVLASMAMAANGQESEVAFVDTSIGDSYLSLSRFDEA 378
Query: 471 VFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGV 530
+ AY+K+LT KT KGENHPAVGSV++RLADL+NRTGK++E+KSYC++ALRIYE+ +
Sbjct: 379 ICAYQKSLTALKTAKGENHPAVGSVYIRLADLYNRTGKVREAKSYCENALRIYESHNLEI 438
Query: 531 NPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYML 590
+PEEIASGLT++S I ESMNE+E+A+ LLQKAL IY DSPGQ+ IAGIEAQMGV+YYM+
Sbjct: 439 SPEEIASGLTDISVICESMNEVEQAITLLQKALKIYADSPGQKIMIAGIEAQMGVLYYMM 498
Query: 591 GNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILE 650
G Y ESYNT K+AI+KLRA G+K+S+F GIALNQMGLAC+QL A EA ELFEEAK ILE
Sbjct: 499 GKYMESYNTFKSAISKLRATGKKQSTFFGIALNQMGLACIQLDAIEEAVELFEEAKCILE 558
Query: 651 HEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLS 710
E GPYHPETLG+YSNLAG YDAIGRLD+AI++L +VV VREEKLGTANP +DEKRRL+
Sbjct: 559 QECGPYHPETLGLYSNLAGAYDAIGRLDDAIKLLGHVVGVREEKLGTANPVTEDEKRRLA 618
Query: 711 ELLKEAGRVRSRKVMSLENLLDAN 734
+LLKEAG V RK SL+ L+D++
Sbjct: 619 QLLKEAGNVTGRKAKSLKTLIDSD 642
>gb|AAF99740.1| F17L21.29 [Arabidopsis thaliana]
Length = 684
Score = 745 bits (1923), Expect = 0.0
Identities = 395/684 (57%), Positives = 503/684 (72%), Gaps = 57/684 (8%)
Query: 53 DGVIEPSIEQLYENVCDMQSSDQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEE 112
D + + +IE+L +N+C++QSS+QSPSR+SFGS GDES+IDS+L+ L G MR+++I+E+E
Sbjct: 48 DQMFDTTIEELCKNLCELQSSNQSPSRQSFGSYGDESKIDSDLQHLALGEMRDIDILEDE 107
Query: 113 VEVEVEKERGGSSSGEISSGVGGLSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDA 172
+E ++ K E + SS N L+
Sbjct: 108 -------------------------GDEDEVAKPEEFDVKSNSS----------NLDLEV 132
Query: 173 SPKSKPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVEN-SAE 231
P+ +E++ K + K GV K K V +++N + E
Sbjct: 133 MPRD------------MEKQTGKKNVTKSNVGVGGMRK--------KKVGGTKLQNGNEE 172
Query: 232 SALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLE-KLGNGKPSLELVMCLHVT 290
+ + E A LL QAR+L+SSGD+ KALEL +A L E NGKP LE +MCLHVT
Sbjct: 173 PSSENVELARFLLNQARNLVSSGDSTHKALELTHRAAKLFEASAENGKPCLEWIMCLHVT 232
Query: 291 AAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCY 350
AA++C L +YNEAIP+L+RS+EIPV+ E ++HALAKFAG MQLGDTYAM+GQLE+SI CY
Sbjct: 233 AAVHCKLKEYNEAIPVLQRSVEIPVVEEGEEHALAKFAGLMQLGDTYAMVGQLESSISCY 292
Query: 351 SSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEE 410
+ G +Q++VLGE DPRVGETCRY+AEA VQAL+FDEA+++C+ AL IH+ + S+ E
Sbjct: 293 TEGLNIQKKVLGENDPRVGETCRYLAEALVQALRFDEAQQVCETALSIHRESGLPGSIAE 352
Query: 411 AADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEA 470
AADRRLMGLI +TKG+HE ALEHLVLASMAM ANGQE EVA VD SIGD+YLS+SR+DEA
Sbjct: 353 AADRRLMGLICETKGDHENALEHLVLASMAMAANGQESEVAFVDTSIGDSYLSLSRFDEA 412
Query: 471 VFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGV 530
+ AY+K+LT KT KGENHPAVGSV++RLADL+NRTGK++E+KSYC++ALRIYE+ +
Sbjct: 413 ICAYQKSLTALKTAKGENHPAVGSVYIRLADLYNRTGKVREAKSYCENALRIYESHNLEI 472
Query: 531 NPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYML 590
+PEEIASGLT++S I ESMNE+E+A+ LLQKAL IY DSPGQ+ IAGIEAQMGV+YYM+
Sbjct: 473 SPEEIASGLTDISVICESMNEVEQAITLLQKALKIYADSPGQKIMIAGIEAQMGVLYYMM 532
Query: 591 GNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILE 650
G Y ESYNT K+AI+KLRA G+K+S+F GIALNQMGLAC+QL A EA ELFEEAK ILE
Sbjct: 533 GKYMESYNTFKSAISKLRATGKKQSTFFGIALNQMGLACIQLDAIEEAVELFEEAKCILE 592
Query: 651 HEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLS 710
E GPYHPETLG+YSNLAG YDAIGRLD+AI++L +VV VREEKLGTANP +DEKRRL+
Sbjct: 593 QECGPYHPETLGLYSNLAGAYDAIGRLDDAIKLLGHVVGVREEKLGTANPVTEDEKRRLA 652
Query: 711 ELLKEAGRVRSRKVMSLENLLDAN 734
+LLKEAG V RK SL+ L+D++
Sbjct: 653 QLLKEAGNVTGRKAKSLKTLIDSD 676
>dbj|BAC42575.1| unknown protein [Arabidopsis thaliana] gi|29028990|gb|AAO64874.1|
At1g27500 [Arabidopsis thaliana]
Length = 650
Score = 743 bits (1919), Expect = 0.0
Identities = 396/684 (57%), Positives = 504/684 (72%), Gaps = 57/684 (8%)
Query: 53 DGVIEPSIEQLYENVCDMQSSDQSPSRKSFGSDGDESRIDSELRQLVGGRMREVEIMEEE 112
D + + +IE+L +N+C++QSS+QSPSR+SFGS GDES+IDS+L+ L G MR+++I+E+
Sbjct: 14 DQMFDTTIEELCKNLCELQSSNQSPSRQSFGSYGDESKIDSDLQHLALGEMRDIDILED- 72
Query: 113 VEVEVEKERGGSSSGEISSGVGGLSSNEKKLDKVHEIQSATTSSVSTEKSVKALNSQLDA 172
GG +E ++ K E + SS N L+
Sbjct: 73 ---------GG---------------DEDEVAKPEEFDVKSNSS----------NLDLEV 98
Query: 173 SPKSKPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVEN-SAE 231
P+ +E++ K + K GV K K V +++N + E
Sbjct: 99 MPRD------------MEKQTGKKNVTKSNVGVGGMRK--------KKVGGTKLQNGNEE 138
Query: 232 SALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLE-KLGNGKPSLELVMCLHVT 290
+ + E A LL QAR+L+SSGD+ KALEL +A L E NGKP LE +MCLHVT
Sbjct: 139 PSSENVELARFLLNQARNLVSSGDSTHKALELTHRAAKLFEASAENGKPCLEWIMCLHVT 198
Query: 291 AAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCY 350
AA++C L +YNEAIP+L+RS+EIPV+ E ++HALAKFAG MQLGDTYAM+GQLE+SI CY
Sbjct: 199 AAVHCKLKEYNEAIPVLQRSVEIPVVEEGEEHALAKFAGLMQLGDTYAMVGQLESSISCY 258
Query: 351 SSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEE 410
+ G +Q++VLGE DPRVGETCRY+AEA VQAL+FDEA+++C+ AL IH+ + S+ E
Sbjct: 259 TEGLNIQKKVLGENDPRVGETCRYLAEALVQALRFDEAQQVCETALSIHRESGLPGSIAE 318
Query: 411 AADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEA 470
AADRRLMGLI +TKG+HE ALEHLVLASMAM ANGQE EVA VD SIGD+YLS+SR+DEA
Sbjct: 319 AADRRLMGLICETKGDHENALEHLVLASMAMAANGQESEVAFVDTSIGDSYLSLSRFDEA 378
Query: 471 VFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGV 530
+ AY+K+LT KT KGENHPAVGSV++RLADL+NRTGK++E+KSYC++ALRIYE+ +
Sbjct: 379 ICAYQKSLTALKTAKGENHPAVGSVYIRLADLYNRTGKVREAKSYCENALRIYESHNLEI 438
Query: 531 NPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYML 590
+PEEIASGLT++S I ESMNE+E+A+ LLQKAL IY DSPGQ+ IAGIEAQMGV+YYM+
Sbjct: 439 SPEEIASGLTDISVICESMNEVEQAITLLQKALKIYADSPGQKIMIAGIEAQMGVLYYMM 498
Query: 591 GNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILE 650
G Y ESYNT K+AI+KLRA G+K+S+F GIALNQMGLAC+QL A EA ELFEEAK ILE
Sbjct: 499 GKYMESYNTFKSAISKLRATGKKQSTFFGIALNQMGLACIQLDAIEEAVELFEEAKCILE 558
Query: 651 HEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLS 710
E GPYHPETLG+YSNLAG YDAIGRLD+AI++L +VV VREEKLGTANP +DEKRRL+
Sbjct: 559 QECGPYHPETLGLYSNLAGAYDAIGRLDDAIKLLGHVVGVREEKLGTANPVTEDEKRRLA 618
Query: 711 ELLKEAGRVRSRKVMSLENLLDAN 734
+LLKEAG V RK SL+ L+D++
Sbjct: 619 QLLKEAGNVTGRKAKSLKTLIDSD 642
>gb|AAS44558.1| kinesin light chain-like protein [Arabidopsis thaliana]
gi|9294312|dbj|BAB01483.1| unnamed protein product
[Arabidopsis thaliana] gi|15232873|ref|NP_189435.1|
kinesin light chain-related [Arabidopsis thaliana]
Length = 663
Score = 717 bits (1852), Expect = 0.0
Identities = 401/720 (55%), Positives = 503/720 (69%), Gaps = 78/720 (10%)
Query: 25 KETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPSIEQLYENVCDMQSSD-QSPSRKSFG 83
K+ A SPR LS + +DG + SIEQLY NVC+M+SSD QSPSR SF
Sbjct: 11 KDDSALQASPRSPLS-------SIDLAIDGAMNASIEQLYHNVCEMESSDDQSPSRASFI 63
Query: 84 SDGDESRIDSELRQLVGGRMREVEIMEEEVEVEVEKERGGSSSGEISSGVGGLSSNEKKL 143
S G ESRID ELR LVG E E + E+ +EK+ E S+G G LS +
Sbjct: 64 SYGAESRIDLELRHLVGDVGEEGE---SKKEIILEKK-------EESNGEGSLSQKKP-- 111
Query: 144 DKVHEIQSATTSSVSTEKSVKALNSQLDASPKSKPKGKSPPAKAPLERKNDKPLLRKQTK 203
+S K V K+ P P
Sbjct: 112 -------------LSNGKKVA----------KTSPNNPKMPG------------------ 130
Query: 204 GVASGVKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARDLISSGDNPQKALEL 263
S + S KS LGK VS++ + PE +LLKQAR+L+SSG+N KAL+L
Sbjct: 131 ---SRISSRKSPDLGK-VSVDE---------ESPELGVVLLKQARELVSSGENLNKALDL 177
Query: 264 ALQAMNLLEKLGNGKPSL--ELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQ 321
AL+A+ + EK G G+ L LVM LH+ AAIY LG+YN+A+P+LERSIEIP+I + +
Sbjct: 178 ALRAVKVFEKCGEGEKQLGLNLVMSLHILAAIYAGLGRYNDAVPVLERSIEIPMIEDGED 237
Query: 322 HALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQ 381
HALAKFAG MQLGD Y ++GQ+ENSIM Y++G E+QRQVLGE+D RVGETCRY+AEA+VQ
Sbjct: 238 HALAKFAGCMQLGDMYGLMGQVENSIMLYTAGLEIQRQVLGESDARVGETCRYLAEAHVQ 297
Query: 382 ALQFDEAERLCQMALDIHKANSSAP--SLEEAADRRLMGLILDTKGNHEAALEHLVLASM 439
A+QF+EA RLCQMALDIHK N +A S+EEAADR+LMGLI D KG++E ALEH VLASM
Sbjct: 298 AMQFEEASRLCQMALDIHKENGAAATASIEEAADRKLMGLICDAKGDYEVALEHYVLASM 357
Query: 440 AMVANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRL 499
AM + +VA+VDCSIGD Y+S++R+DEA+FAY+KAL VFK GKGE H +V V+VRL
Sbjct: 358 AMSSQNHREDVAAVDCSIGDAYMSLARFDEAIFAYQKALAVFKQGKGETHSSVALVYVRL 417
Query: 500 ADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLL 559
ADL+N+ GK ++SKSYC++AL+IY P PG EE+A+G +SAIY+SMNEL++ALKLL
Sbjct: 418 ADLYNKIGKTRDSKSYCENALKIYLKPTPGTPMEEVATGFIEISAIYQSMNELDQALKLL 477
Query: 560 QKALLIYNDSPGQQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIG 619
++AL IY ++PGQQ++IAGIEAQMGV+ YM+GNY+ESY+ K+AI+K R GEKK++ G
Sbjct: 478 RRALKIYANAPGQQNTIAGIEAQMGVVTYMMGNYSESYDIFKSAISKFRNSGEKKTALFG 537
Query: 620 IALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDE 679
IALNQMGLACVQ YA EA +LFEEAK+ILE E GPYHP+TL VYSNLAGTYDA+GRLD+
Sbjct: 538 IALNQMGLACVQRYAINEAADLFEEAKTILEKECGPYHPDTLAVYSNLAGTYDAMGRLDD 597
Query: 680 AIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAGRVRSRKVMSLENLLDANARIPN 739
AI+ILEYVV REEKLGTANP+V+DEK+RL+ LLKEAGR RS++ +L LLD N I N
Sbjct: 598 AIEILEYVVGTREEKLGTANPEVEDEKQRLAALLKEAGRGRSKRNRALLTLLDNNPEIAN 657
>gb|AAQ22597.1| At3g27960 [Arabidopsis thaliana]
Length = 663
Score = 715 bits (1845), Expect = 0.0
Identities = 400/720 (55%), Positives = 502/720 (69%), Gaps = 78/720 (10%)
Query: 25 KETLAPVKSPRGSLSPQRGQNGGPNFPVDGVIEPSIEQLYENVCDMQSSD-QSPSRKSFG 83
K+ A SPR LS + +DG + SIEQLY NVC+M+SSD QSPSR SF
Sbjct: 11 KDDSALQASPRSPLS-------SIDLAIDGAMNASIEQLYHNVCEMESSDDQSPSRASFI 63
Query: 84 SDGDESRIDSELRQLVGGRMREVEIMEEEVEVEVEKERGGSSSGEISSGVGGLSSNEKKL 143
S G ESRID ELR LVG E E + E+ +EK+ E S+G G LS +
Sbjct: 64 SYGAESRIDLELRHLVGDVGEEGE---SKKEIILEKK-------EESNGEGSLSQKKP-- 111
Query: 144 DKVHEIQSATTSSVSTEKSVKALNSQLDASPKSKPKGKSPPAKAPLERKNDKPLLRKQTK 203
+S K V K+ P P
Sbjct: 112 -------------LSNGKKVA----------KTSPNNPKMPG------------------ 130
Query: 204 GVASGVKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARDLISSGDNPQKALEL 263
S + S KS LGK VS++ + PE +LLKQAR+L+SSG+N KAL+L
Sbjct: 131 ---SRISSRKSPDLGK-VSVDE---------ESPELGVVLLKQARELVSSGENLNKALDL 177
Query: 264 ALQAMNLLEKLGNGKPSL--ELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQ 321
AL+A+ + EK G G+ L LVM LH+ AAIY LG+YN+A+P+L RSIEIP+I + +
Sbjct: 178 ALRAVKVFEKCGEGEKQLGLNLVMSLHILAAIYAGLGRYNDAVPVLGRSIEIPMIEDGED 237
Query: 322 HALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQ 381
HALAKFAG MQLGD Y ++GQ+ENSIM Y++G E+QRQVLGE+D RVGETCRY+AEA+VQ
Sbjct: 238 HALAKFAGCMQLGDMYGLMGQVENSIMLYTAGLEIQRQVLGESDARVGETCRYLAEAHVQ 297
Query: 382 ALQFDEAERLCQMALDIHKANSSAP--SLEEAADRRLMGLILDTKGNHEAALEHLVLASM 439
A+QF+EA RLCQMALDIHK N +A S+EEAADR+LMGLI D KG++E ALEH VLASM
Sbjct: 298 AMQFEEASRLCQMALDIHKENGAAATASIEEAADRKLMGLICDAKGDYEVALEHYVLASM 357
Query: 440 AMVANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRL 499
AM + +VA+VDCSIGD Y+S++R+DEA+FAY+KAL VFK GKGE H +V V+VRL
Sbjct: 358 AMSSQNHREDVAAVDCSIGDAYMSLARFDEAIFAYQKALAVFKQGKGETHSSVALVYVRL 417
Query: 500 ADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLL 559
ADL+N+ GK ++SKSYC++AL+IY P PG EE+A+G +SAIY+SMNEL++ALKLL
Sbjct: 418 ADLYNKIGKTRDSKSYCENALKIYLKPTPGTPMEEVATGFIEISAIYQSMNELDQALKLL 477
Query: 560 QKALLIYNDSPGQQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIG 619
++AL IY ++PGQQ++IAGIEAQMGV+ YM+GNY+ESY+ K+AI+K R GEKK++ G
Sbjct: 478 RRALKIYANAPGQQNTIAGIEAQMGVVTYMMGNYSESYDIFKSAISKFRNSGEKKTALFG 537
Query: 620 IALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDE 679
IALNQMGLACVQ YA EA +LFEEAK+ILE E GPYHP+TL VYSNLAGTYDA+GRLD+
Sbjct: 538 IALNQMGLACVQRYAINEAADLFEEAKTILEKECGPYHPDTLAVYSNLAGTYDAMGRLDD 597
Query: 680 AIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAGRVRSRKVMSLENLLDANARIPN 739
AI+ILEYVV REEKLGTANP+V+DEK+RL+ LLKEAGR RS++ +L LLD N I N
Sbjct: 598 AIEILEYVVGTREEKLGTANPEVEDEKQRLAALLKEAGRGRSKRNRALLTLLDNNPEIAN 657
>gb|AAN41387.1| unknown protein [Arabidopsis thaliana] gi|7267782|emb|CAB81185.1|
putative protein [Arabidopsis thaliana]
gi|4539358|emb|CAB40052.1| putative protein [Arabidopsis
thaliana] gi|18176148|gb|AAL59992.1| unknown protein
[Arabidopsis thaliana] gi|19699309|gb|AAL91265.1|
AT4g10840/F25I24_50 [Arabidopsis thaliana]
gi|3513727|gb|AAC33943.1| contains similarity to TPR
domains (Pfam: TPR.hmm: score: 11.15) and kinesin motor
domains (Pfam: kinesin2.hmm, score: 17.49, 20.52 and
10.94) [Arabidopsis thaliana]
gi|15236944|ref|NP_192822.1| kinesin light chain-related
[Arabidopsis thaliana] gi|7486710|pir||T01892
hypothetical protein F8M12.21 - Arabidopsis thaliana
Length = 609
Score = 642 bits (1656), Expect = 0.0
Identities = 334/579 (57%), Positives = 431/579 (73%), Gaps = 21/579 (3%)
Query: 171 DASPKSKPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVENSA 230
D P S P ++P K P + P K + ++G K SP S S V +
Sbjct: 25 DTQPLSNPP-RTPMKKTP----SSTPSRSKPSPNRSTGKKD---SPTVSS-STAAVIDVD 75
Query: 231 ESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKL-----------GNGKP 279
+ +LD P+ P LLK ARD I+SG+ P KAL+ A++A E+ +G P
Sbjct: 76 DPSLDNPDLGPFLLKLARDAIASGEGPNKALDYAIRATKSFERCCAAVAPPIPGGSDGGP 135
Query: 280 SLELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAM 339
L+L M LHV AAIYC+LG+++EA+P LER+I++P H+LA F+GHMQLGDT +M
Sbjct: 136 VLDLAMSLHVLAAIYCSLGRFDEAVPPLERAIQVPDPTRGPDHSLAAFSGHMQLGDTLSM 195
Query: 340 LGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIH 399
LGQ++ SI CY G ++Q Q LG+TDPRVGETCRY+AEA VQA+QF++AE LC+ L+IH
Sbjct: 196 LGQIDRSIACYEEGLKIQIQTLGDTDPRVGETCRYLAEAYVQAMQFNKAEELCKKTLEIH 255
Query: 400 KANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGD 459
+A+S SLEEAADRRLM +I + KG++E ALEHLVLASMAM+A+GQE EVAS+D SIG+
Sbjct: 256 RAHSEPASLEEAADRRLMAIICEAKGDYENALEHLVLASMAMIASGQESEVASIDVSIGN 315
Query: 460 TYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSA 519
Y+S+ R+DEAVF+Y+KALTVFK KGE HP V SVFVRLA+L++RTGK++ESKSYC++A
Sbjct: 316 IYMSLCRFDEAVFSYQKALTVFKASKGETHPTVASVFVRLAELYHRTGKLRESKSYCENA 375
Query: 520 LRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGI 579
LRIY P+PG EEIA GLT +SAIYES++E E+ALKLLQK++ + D PGQQS+IAG+
Sbjct: 376 LRIYNKPVPGTTVEEIAGGLTEISAIYESVDEPEEALKLLQKSMKLLEDKPGQQSAIAGL 435
Query: 580 EAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEAT 639
EA+MGVMYY +G Y ++ N ++A+TKLRA GE KS+F G+ LNQMGLACVQL+ EA
Sbjct: 436 EARMGVMYYTVGRYEDARNAFESAVTKLRAAGE-KSAFFGVVLNQMGLACVQLFKIDEAG 494
Query: 640 ELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTAN 699
ELFEEA+ ILE E GP +TLGVYSNLA TYDA+GR+++AI+ILE V+ +REEKLGTAN
Sbjct: 495 ELFEEARGILEQERGPCDQDTLGVYSNLAATYDAMGRIEDAIEILEQVLKLREEKLGTAN 554
Query: 700 PDVDDEKRRLSELLKEAGRVRSRKVMSLENLLDANARIP 738
PD +DEK+RL+ELLKEAGR R+ K SL+NL+D NAR P
Sbjct: 555 PDFEDEKKRLAELLKEAGRSRNYKAKSLQNLIDPNARPP 593
>gb|AAM63491.1| putative kinesin light chain [Arabidopsis thaliana]
Length = 606
Score = 642 bits (1656), Expect = 0.0
Identities = 334/579 (57%), Positives = 431/579 (73%), Gaps = 21/579 (3%)
Query: 171 DASPKSKPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVENSA 230
D P S P ++P K P + P K + ++G K SP S S V +
Sbjct: 22 DTQPLSNPP-RTPMKKTP----SSTPSRSKPSPNRSTGKKD---SPTVSS-STAAVIDVD 72
Query: 231 ESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKL-----------GNGKP 279
+ +LD P+ P LLK ARD I+SG+ P KAL+ A++A E+ +G P
Sbjct: 73 DPSLDNPDLGPFLLKLARDAIASGEGPNKALDYAIRATKSFERCCAAVAPPIPGGSDGGP 132
Query: 280 SLELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAM 339
L+L M LHV AAIYC+LG+++EA+P LER+I++P H+LA F+GHMQLGDT +M
Sbjct: 133 VLDLAMSLHVLAAIYCSLGRFDEAVPPLERAIQVPDPTRGPDHSLAAFSGHMQLGDTLSM 192
Query: 340 LGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIH 399
LGQ++ SI CY G ++Q Q LG+TDPRVGETCRY+AEA VQA+QF++AE LC+ L+IH
Sbjct: 193 LGQIDRSIACYEEGLKIQIQTLGDTDPRVGETCRYLAEAYVQAMQFNKAEELCKKTLEIH 252
Query: 400 KANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGD 459
+A+S SLEEAADRRLM +I + KG++E ALEHLVLASMAM+A+GQE EVAS+D SIG+
Sbjct: 253 RAHSEPASLEEAADRRLMAIICEAKGDYENALEHLVLASMAMIASGQESEVASIDVSIGN 312
Query: 460 TYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSA 519
Y+S+ R+DEAVF+Y+KALTVFK KGE HP V SVFVRLA+L++RTGK++ESKSYC++A
Sbjct: 313 IYMSLCRFDEAVFSYQKALTVFKASKGETHPTVASVFVRLAELYHRTGKLRESKSYCENA 372
Query: 520 LRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGI 579
LRIY P+PG EEIA GLT +SAIYES++E E+ALKLLQK++ + D PGQQS+IAG+
Sbjct: 373 LRIYNKPVPGTTVEEIAGGLTEISAIYESVDEPEEALKLLQKSMKLLEDKPGQQSAIAGL 432
Query: 580 EAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEAT 639
EA+MGVMYY +G Y ++ N ++A+TKLRA GE KS+F G+ LNQMGLACVQL+ EA
Sbjct: 433 EARMGVMYYTVGRYEDARNAFESAVTKLRAAGE-KSAFFGVVLNQMGLACVQLFKIDEAG 491
Query: 640 ELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTAN 699
ELFEEA+ ILE E GP +TLGVYSNLA TYDA+GR+++AI+ILE V+ +REEKLGTAN
Sbjct: 492 ELFEEARGILEQERGPCDQDTLGVYSNLAATYDAMGRIEDAIEILEQVLKLREEKLGTAN 551
Query: 700 PDVDDEKRRLSELLKEAGRVRSRKVMSLENLLDANARIP 738
PD +DEK+RL+ELLKEAGR R+ K SL+NL+D NAR P
Sbjct: 552 PDFEDEKKRLAELLKEAGRSRNYKAKSLQNLIDPNARPP 590
>ref|XP_463817.1| kinesin light chain-like protein [Oryza sativa (japonica
cultivar-group)] gi|41052919|dbj|BAD07830.1| kinesin
light chain-like protein [Oryza sativa (japonica
cultivar-group)] gi|50252012|dbj|BAD27945.1| kinesin
light chain-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 599
Score = 637 bits (1642), Expect = 0.0
Identities = 326/578 (56%), Positives = 433/578 (74%), Gaps = 28/578 (4%)
Query: 182 SPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVENSAESALDKPERAP 241
SPP AP R+ PL R + S S ++ P + + E+ALD P+ P
Sbjct: 10 SPPVAAPPPRRLSSPLPRSRAPPSPSPSTSSRAKPRKVAAQ----PETDEAALDNPDLGP 65
Query: 242 ILLKQARDLISSGDN--PQKALELALQAMNLLEKLGNGKPSLELVMCLHVTAAIYCNLGQ 299
LLKQARD + SG+ +ALE A +A LE+ G G LEL M LHV AAI+C LG+
Sbjct: 66 FLLKQARDAMVSGEGGGAARALEFAERAARALERRGEGA-ELELAMSLHVAAAIHCGLGR 124
Query: 300 YNEAIPILERSIEI--------PVIGES-------------QQHALAKFAGHMQLGDTYA 338
+ +AIP+LER++ + P GES ++ ALA F+G MQLGDT+A
Sbjct: 125 HADAIPVLERAVAVVTPPPPPPPPEGESSAEEQPPEDQQKGEEWALAAFSGWMQLGDTHA 184
Query: 339 MLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDI 398
MLG+++ SI CY G E+Q LGE DPRV ETCRY+AEA+VQALQFDEAE+LC+ AL+I
Sbjct: 185 MLGRMDESIACYGKGLEIQMAALGERDPRVAETCRYLAEAHVQALQFDEAEKLCRKALEI 244
Query: 399 HKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIG 458
H+ +S+ SLEEA+DRRLM LILD KG+++ ALEHLVLASM MVANG+++EVA++D +IG
Sbjct: 245 HREHSAPASLEEASDRRLMALILDAKGDYDGALEHLVLASMTMVANGRDIEVATIDVAIG 304
Query: 459 DTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDS 518
+TYL+++R+DEAVF+Y+KALTV K+ +G++HP+V SVFVRLADL++RTG+++ESKSYC++
Sbjct: 305 NTYLALARFDEAVFSYQKALTVLKSARGDDHPSVASVFVRLADLYHRTGRLRESKSYCEN 364
Query: 519 ALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAG 578
ALR+Y P PG P+E+A GL ++AIYE++ +L++ALKLLQ+AL + DSPGQ S++AG
Sbjct: 365 ALRVYAKPAPGAAPDEVAGGLMEIAAIYEALGDLDEALKLLQRALKLLEDSPGQWSTVAG 424
Query: 579 IEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEA 638
IEAQMGV+YYM+G Y +S N+ +NA+ KLRA GE+KS+F G+ LNQMGLACVQL+ EA
Sbjct: 425 IEAQMGVLYYMVGRYADSRNSFENAVAKLRASGERKSAFFGVLLNQMGLACVQLFKIDEA 484
Query: 639 TELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTA 698
+LFEEA+++LE E G HP+TLGVYSNLA YDA+GR+++AI+ILE+V+ VREEKLGTA
Sbjct: 485 AQLFEEARAVLEQECGASHPDTLGVYSNLAAIYDAMGRVEDAIEILEHVLKVREEKLGTA 544
Query: 699 NPDVDDEKRRLSELLKEAGRVRSRKVMSLENLLDANAR 736
NPDV+DEK RL+ELLKEAGR R+RK SLENL N++
Sbjct: 545 NPDVEDEKLRLAELLKEAGRSRNRKQKSLENLFVTNSQ 582
>ref|NP_974530.1| kinesin light chain-related [Arabidopsis thaliana]
Length = 531
Score = 561 bits (1445), Expect = e-158
Identities = 293/517 (56%), Positives = 378/517 (72%), Gaps = 21/517 (4%)
Query: 171 DASPKSKPKGKSPPAKAPLERKNDKPLLRKQTKGVASGVKSLKSSPLGKSVSLNRVENSA 230
D P S P ++P K P + P K + ++G K SP S S V +
Sbjct: 25 DTQPLSNPP-RTPMKKTP----SSTPSRSKPSPNRSTGKKD---SPTVSS-STAAVIDVD 75
Query: 231 ESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKL-----------GNGKP 279
+ +LD P+ P LLK ARD I+SG+ P KAL+ A++A E+ +G P
Sbjct: 76 DPSLDNPDLGPFLLKLARDAIASGEGPNKALDYAIRATKSFERCCAAVAPPIPGGSDGGP 135
Query: 280 SLELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAM 339
L+L M LHV AAIYC+LG+++EA+P LER+I++P H+LA F+GHMQLGDT +M
Sbjct: 136 VLDLAMSLHVLAAIYCSLGRFDEAVPPLERAIQVPDPTRGPDHSLAAFSGHMQLGDTLSM 195
Query: 340 LGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIH 399
LGQ++ SI CY G ++Q Q LG+TDPRVGETCRY+AEA VQA+QF++AE LC+ L+IH
Sbjct: 196 LGQIDRSIACYEEGLKIQIQTLGDTDPRVGETCRYLAEAYVQAMQFNKAEELCKKTLEIH 255
Query: 400 KANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGD 459
+A+S SLEEAADRRLM +I + KG++E ALEHLVLASMAM+A+GQE EVAS+D SIG+
Sbjct: 256 RAHSEPASLEEAADRRLMAIICEAKGDYENALEHLVLASMAMIASGQESEVASIDVSIGN 315
Query: 460 TYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSA 519
Y+S+ R+DEAVF+Y+KALTVFK KGE HP V SVFVRLA+L++RTGK++ESKSYC++A
Sbjct: 316 IYMSLCRFDEAVFSYQKALTVFKASKGETHPTVASVFVRLAELYHRTGKLRESKSYCENA 375
Query: 520 LRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGI 579
LRIY P+PG EEIA GLT +SAIYES++E E+ALKLLQK++ + D PGQQS+IAG+
Sbjct: 376 LRIYNKPVPGTTVEEIAGGLTEISAIYESVDEPEEALKLLQKSMKLLEDKPGQQSAIAGL 435
Query: 580 EAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEAT 639
EA+MGVMYY +G Y ++ N ++A+TKLRA GE KS+F G+ LNQMGLACVQL+ EA
Sbjct: 436 EARMGVMYYTVGRYEDARNAFESAVTKLRAAGE-KSAFFGVVLNQMGLACVQLFKIDEAG 494
Query: 640 ELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGR 676
ELFEEA+ ILE E GP +TLGVYSNLA TYDA+GR
Sbjct: 495 ELFEEARGILEQERGPCDQDTLGVYSNLAATYDAMGR 531
Score = 68.9 bits (167), Expect = 6e-10
Identities = 73/321 (22%), Positives = 139/321 (42%), Gaps = 23/321 (7%)
Query: 389 ERLCQ-MALDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLA-SMAMVANGQ 446
ER C +A I + P L+ A ++ I + G + A+ L A + G
Sbjct: 117 ERCCAAVAPPIPGGSDGGPVLDLAMSLHVLAAIYCSLGRFDEAVPPLERAIQVPDPTRGP 176
Query: 447 EVEVASVD--CSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHN 504
+ +A+ +GDT + + D ++ YE+ L + G+ P VG LA+ +
Sbjct: 177 DHSLAAFSGHMQLGDTLSMLGQIDRSIACYEEGLKIQIQTLGDTDPRVGETCRYLAEAYV 236
Query: 505 RTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALL 564
+ + +++ C L I+ + EE A ++ I E+ + E AL+ L A +
Sbjct: 237 QAMQFNKAEELCKKTLEIHRAHSEPASLEEAADRRL-MAIICEAKGDYENALEHLVLASM 295
Query: 565 IYNDSPGQQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAI-GEKKSSFIGIALN 623
S GQ+S +A I+ +G +Y L + E+ + + A+T +A GE + +
Sbjct: 296 AMIAS-GQESEVASIDVSIGNIYMSLCRFDEAVFSYQKALTVFKASKGETHPTVASVF-- 352
Query: 624 QMGLACVQLYAFGEATELFEEAKSILEHEYGPYHP--------ETLGVYSNLAGTYDAIG 675
V+L T E+KS E+ Y+ E G + ++ Y+++
Sbjct: 353 ------VRLAELYHRTGKLRESKSYCENALRIYNKPVPGTTVEEIAGGLTEISAIYESVD 406
Query: 676 RLDEAIQILEYVVSVREEKLG 696
+EA+++L+ + + E+K G
Sbjct: 407 EPEEALKLLQKSMKLLEDKPG 427
>dbj|BAC78579.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 309
Score = 392 bits (1007), Expect = e-107
Identities = 183/292 (62%), Positives = 249/292 (84%)
Query: 445 GQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHN 504
G+++EVA++D +IG+TYL+++R+DEAVF+Y+KALTV K+ +G++HP+V SVFVRLADL++
Sbjct: 1 GRDIEVATIDVAIGNTYLALARFDEAVFSYQKALTVLKSARGDDHPSVASVFVRLADLYH 60
Query: 505 RTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALL 564
RTG+++ESKSYC++ALR+Y P PG P+E+A GL ++AIYE++ +L++ALKLLQ+AL
Sbjct: 61 RTGRLRESKSYCENALRVYAKPAPGAAPDEVAGGLMEIAAIYEALGDLDEALKLLQRALK 120
Query: 565 IYNDSPGQQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQ 624
+ DSPGQ S++AGIEAQMGV+YYM+G Y +S N+ +NA+ KLRA GE+KS+F G+ LNQ
Sbjct: 121 LLEDSPGQWSTVAGIEAQMGVLYYMVGRYADSRNSFENAVAKLRASGERKSAFFGVLLNQ 180
Query: 625 MGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQIL 684
MGLACVQL+ EA +LFEEA+++LE E G HP+TLGVYSNLA YDA+GR+++AI+IL
Sbjct: 181 MGLACVQLFKIDEAAQLFEEARAVLEQECGASHPDTLGVYSNLAAIYDAMGRVEDAIEIL 240
Query: 685 EYVVSVREEKLGTANPDVDDEKRRLSELLKEAGRVRSRKVMSLENLLDANAR 736
E+V+ VREEKLGTANPDV+DEK RL+ELLKEAGR R+RK SLENL N++
Sbjct: 241 EHVLKVREEKLGTANPDVEDEKLRLAELLKEAGRSRNRKQKSLENLFVTNSQ 292
Score = 42.0 bits (97), Expect = 0.073
Identities = 47/200 (23%), Positives = 80/200 (39%), Gaps = 3/200 (1%)
Query: 235 DKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLELVMCLHVTAAIY 294
D P A + ++ A DL +++ A+ + K G E+ L AAIY
Sbjct: 44 DHPSVASVFVRLA-DLYHRTGRLRESKSYCENALRVYAKPAPGAAPDEVAGGLMEIAAIY 102
Query: 295 CNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCYSSGF 354
LG +EA+ +L+R++++ Q +A Q+G Y M+G+ +S + +
Sbjct: 103 EALGDLDEALKLLQRALKLLEDSPGQWSTVAGI--EAQMGVLYYMVGRYADSRNSFENAV 160
Query: 355 EVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADR 414
R G + A VQ + DEA +L + A + + A +
Sbjct: 161 AKLRASGERKSAFFGVLLNQMGLACVQLFKIDEAAQLFEEARAVLEQECGASHPDTLGVY 220
Query: 415 RLMGLILDTKGNHEAALEHL 434
+ I D G E A+E L
Sbjct: 221 SNLAAIYDAMGRVEDAIEIL 240
>gb|AAT07454.1| kinesin-like protein [Mirabilis jalapa]
Length = 194
Score = 306 bits (784), Expect = 2e-81
Identities = 150/194 (77%), Positives = 177/194 (90%)
Query: 393 QMALDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVAS 452
QMAL+IH+ N S S+EE ADRRLMGLI +TKG+HE ALEHLVLASMAMVANGQE EVAS
Sbjct: 1 QMALEIHRENGSPGSVEETADRRLMGLIYETKGDHEKALEHLVLASMAMVANGQEKEVAS 60
Query: 453 VDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKES 512
VDCSIGDTYLS+SRYDEAVFAYEKALT FK KGENHP++ SV+VRLADL+N+TGK++ES
Sbjct: 61 VDCSIGDTYLSLSRYDEAVFAYEKALTSFKASKGENHPSIASVYVRLADLYNKTGKLRES 120
Query: 513 KSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQ 572
KSYC++ALRIY P+PG+ PE+IASG T+VSAIYESMNELE+A+KLLQKAL IY+++PGQ
Sbjct: 121 KSYCENALRIYSKPVPGIPPEDIASGFTDVSAIYESMNELEQAIKLLQKALKIYDNAPGQ 180
Query: 573 QSSIAGIEAQMGVM 586
Q++IAGI+AQMGVM
Sbjct: 181 QNTIAGIQAQMGVM 194
Score = 42.0 bits (97), Expect = 0.073
Identities = 37/144 (25%), Positives = 63/144 (43%), Gaps = 5/144 (3%)
Query: 560 QKALLIY--NDSPGQQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSF 617
Q AL I+ N SPG A MG++Y G++ ++ L A + A G++K
Sbjct: 1 QMALEIHRENGSPGSVEETAD-RRLMGLIYETKGDHEKALEHLVLASMAMVANGQEKE-- 57
Query: 618 IGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRL 677
+ +G + L + EA +E+A + + G HP VY LA Y+ G+L
Sbjct: 58 VASVDCSIGDTYLSLSRYDEAVFAYEKALTSFKASKGENHPSIASVYVRLADLYNKTGKL 117
Query: 678 DEAIQILEYVVSVREEKLGTANPD 701
E+ E + + + + P+
Sbjct: 118 RESKSYCENALRIYSKPVPGIPPE 141
Score = 37.4 bits (85), Expect = 1.8
Identities = 40/186 (21%), Positives = 78/186 (41%), Gaps = 6/186 (3%)
Query: 276 NGKP-SLELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQLG 334
NG P S+E + IY G + +A+ L + + ++ Q+ +A +G
Sbjct: 10 NGSPGSVEETADRRLMGLIYETKGDHEKALEHLVLA-SMAMVANGQEKEVASV--DCSIG 66
Query: 335 DTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQM 394
DTY L + + ++ Y + GE P + +A+ + + E++ C+
Sbjct: 67 DTYLSLSRYDEAVFAYEKALTSFKASKGENHPSIASVYVRLADLYNKTGKLRESKSYCEN 126
Query: 395 ALDIH-KANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLA-SMAMVANGQEVEVAS 452
AL I+ K P + A+ + I ++ E A++ L A + A GQ+ +A
Sbjct: 127 ALRIYSKPVPGIPPEDIASGFTDVSAIYESMNELEQAIKLLQKALKIYDNAPGQQNTIAG 186
Query: 453 VDCSIG 458
+ +G
Sbjct: 187 IQAQMG 192
>gb|AAL36288.1| putative kinesin light chain [Arabidopsis thaliana]
gi|30684882|ref|NP_850163.1| tetratricopeptide repeat
(TPR)-containing protein [Arabidopsis thaliana]
Length = 617
Score = 172 bits (437), Expect = 3e-41
Identities = 134/509 (26%), Positives = 238/509 (46%), Gaps = 13/509 (2%)
Query: 225 RVENSAESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLELV 284
R+ E + + E LK L G++P+K L A +A+ + GN KP+L +
Sbjct: 103 RLFKEMELSFEGNELGLSALKLGLHLDREGEDPEKVLSYADKALKSFDGDGN-KPNLLVA 161
Query: 285 MCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAK------FAGHMQLGDTYA 338
M + + L ++++++ L R+ I V E+ + + A ++L +
Sbjct: 162 MASQLMGSANYGLKRFSDSLGYLNRANRILVKLEADGDCVVEDVRPVLHAVQLELANVKN 221
Query: 339 MLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDI 398
+G+ E +I E++ E +G R +A+A V L F+EA AL+I
Sbjct: 222 AMGRREEAIENLKKSLEIKEMTFDEDSKEMGVANRSLADAYVAVLNFNEALPYALKALEI 281
Query: 399 HKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIG 458
HK S E A DRRL+G+I H+ ALE L+ + G ++E+ +
Sbjct: 282 HKKELGNNSAEVAQDRRLLGVIYSGLEQHDKALEQNRLSQRVLKNWGMKLELIRAEIDAA 341
Query: 459 DTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDS 518
+ +++ +Y+EA+ + V +T K A+ VF+ ++ K ESK +
Sbjct: 342 NMKVALGKYEEAIDILKSV--VQQTDKDSEMRAM--VFISMSKALVNQQKFAESKRCLEF 397
Query: 519 ALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAG 578
A I E + P E+A + V+ YESMNE E A+ LLQK L I P +Q S
Sbjct: 398 ACEILEKKETAL-PVEVAEAYSEVAMQYESMNEFETAISLLQKTLGILEKLPQEQHSEGS 456
Query: 579 IEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEA 638
+ A++G + G +++ L++A +L+ K +G N +G A ++L A
Sbjct: 457 VSARIGWLLLFSGRVSQAVPYLESAAERLKESFGAKHFGVGYVYNNLGAAYLELGRPQSA 516
Query: 639 TELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTA 698
++F AK I++ GP H +++ NL+ Y +G A++ + V++ + +A
Sbjct: 517 AQMFAVAKDIMDVSLGPNHVDSIDACQNLSKAYAGMGNYSLAVEFQQRVINAWDNHGDSA 576
Query: 699 NPDVDDEKRRLSEL-LKEAGRVRSRKVMS 726
++ + KR L +L LK G V + K+++
Sbjct: 577 KDEMKEAKRLLEDLRLKARGGVSTNKLLN 605
>gb|AAD20669.1| putative kinesin light chain [Arabidopsis thaliana]
gi|25408142|pir||C84718 probable kinesin light chain
[imported] - Arabidopsis thaliana
Length = 510
Score = 172 bits (436), Expect = 4e-41
Identities = 131/490 (26%), Positives = 232/490 (46%), Gaps = 13/490 (2%)
Query: 244 LKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLELVMCLHVTAAIYCNLGQYNEA 303
LK L G++P+K L A +A+ + GN KP+L + M + + L +++++
Sbjct: 15 LKLGLHLDREGEDPEKVLSYADKALKSFDGDGN-KPNLLVAMASQLMGSANYGLKRFSDS 73
Query: 304 IPILERSIEIPVIGESQQHALAK------FAGHMQLGDTYAMLGQLENSIMCYSSGFEVQ 357
+ L R+ I V E+ + + A ++L + +G+ E +I E++
Sbjct: 74 LGYLNRANRILVKLEADGDCVVEDVRPVLHAVQLELANVKNAMGRREEAIENLKKSLEIK 133
Query: 358 RQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRRLM 417
E +G R +A+A V L F+EA AL+IHK S E A DRRL+
Sbjct: 134 EMTFDEDSKEMGVANRSLADAYVAVLNFNEALPYALKALEIHKKELGNNSAEVAQDRRLL 193
Query: 418 GLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKA 477
G+I H+ ALE L+ + G ++E+ + + +++ +Y+EA+ +
Sbjct: 194 GVIYSGLEQHDKALEQNRLSQRVLKNWGMKLELIRAEIDAANMKVALGKYEEAIDILKSV 253
Query: 478 LTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIAS 537
V +T K A+ VF+ ++ K ESK + A I E + P E+A
Sbjct: 254 --VQQTDKDSEMRAM--VFISMSKALVNQQKFAESKRCLEFACEILEKKETAL-PVEVAE 308
Query: 538 GLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGIEAQMGVMYYMLGNYTESY 597
+ V+ YESMNE E A+ LLQK L I P +Q S + A++G + G +++
Sbjct: 309 AYSEVAMQYESMNEFETAISLLQKTLGILEKLPQEQHSEGSVSARIGWLLLFSGRVSQAV 368
Query: 598 NTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYH 657
L++A +L+ K +G N +G A ++L A ++F AK I++ GP H
Sbjct: 369 PYLESAAERLKESFGAKHFGVGYVYNNLGAAYLELGRPQSAAQMFAVAKDIMDVSLGPNH 428
Query: 658 PETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSEL-LKEA 716
+++ NL+ Y +G A++ + V++ + +A ++ + KR L +L LK
Sbjct: 429 VDSIDACQNLSKAYAGMGNYSLAVEFQQRVINAWDNHGDSAKDEMKEAKRLLEDLRLKAR 488
Query: 717 GRVRSRKVMS 726
G V + K+++
Sbjct: 489 GGVSTNKLLN 498
>ref|XP_468434.1| kinesin light chain-like [Oryza sativa (japonica cultivar-group)]
gi|48716499|dbj|BAD23104.1| kinesin light chain-like
[Oryza sativa (japonica cultivar-group)]
gi|48716364|dbj|BAD22975.1| kinesin light chain-like
[Oryza sativa (japonica cultivar-group)]
Length = 621
Score = 155 bits (393), Expect = 3e-36
Identities = 126/514 (24%), Positives = 241/514 (46%), Gaps = 40/514 (7%)
Query: 244 LKQARDLISSGD-NPQKALELALQAMNLLEKLGNGKPS--------LELVMCLHVTAAIY 294
+K + L +SG +P + L LAL+++ +LE N S + L M LH+ +
Sbjct: 97 IKLGQHLEASGAADPSRVLALALRSLGILEATPNSATSTTASHSDAVSLAMALHLAGSAS 156
Query: 295 CNLGQYNEAIPILERSIEI--PVIGESQQHALAK--------------FAGHMQLGDTYA 338
+L ++++A+ L RS+ + P++ S A A A +QL +
Sbjct: 157 FDLSRFHDALSFLTRSLRLVSPLLPSSSSAAAASGDDDAQGFDVRPVAHAVRLQLANVKT 216
Query: 339 MLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDI 398
LG+ E ++ + +++ +L +G R +AEA L F EA LC+ AL++
Sbjct: 217 ALGRREEALADMRACLDLKESILPPGSRELGAAYRDLAEAYSTVLDFKEALPLCEKALEL 276
Query: 399 HKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVA---NGQEVEVASVDC 455
H++ S+E A DRRL+G+I HE AL+ ++ M + G E+ A +D
Sbjct: 277 HQSTLGKNSVEVAHDRRLLGVIYTGLEQHEQALQQNEMSQKVMKSWGVAGDELLHAEIDA 336
Query: 456 SIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSY 515
+ + +++ + DEAV + ++ VF+ +A K ++K
Sbjct: 337 A--NIKIALGKCDEAVTVLRNVSKQVE----KDSEIRALVFISMAKALANQEKAGDTKRC 390
Query: 516 CDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSS 575
+ A I E P+++A VS++YE +NE +KA+ LL+++L + P Q
Sbjct: 391 LEIACDILEKKELAA-PDKVAEAYVEVSSLYEMVNEFDKAISLLKRSLGMLERIPQAQHM 449
Query: 576 IAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAF 635
+ A++G + + G +E+ L++A+ +++ K +G N +G A +++
Sbjct: 450 EGNVAARIGWLLLLTGKVSEAVPYLEDAVERMKDSFGPKHYGVGYVYNNLGAAYMEIGRP 509
Query: 636 GEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKL 695
A ++F AK +++ GP+H +T+ +LA Y+A+G A++ + VV +
Sbjct: 510 QSAAQMFALAKEVMDVSLGPHHSDTIEACQSLANAYNAMGSYALAMEFQKRVV----DSW 565
Query: 696 GTANPDVDDEKRRLSELLKEAGRVRSRKVMSLEN 729
P DE + L ++ ++++ +S EN
Sbjct: 566 RNHGPSARDELKEAIRLYEQI-KIKALSCLSPEN 598
>ref|XP_470238.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|27261475|gb|AAN87741.1| Hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 556
Score = 137 bits (345), Expect = 1e-30
Identities = 127/491 (25%), Positives = 217/491 (43%), Gaps = 18/491 (3%)
Query: 230 AESALDKPERAP------ILLKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLEL 283
A A PE AP L +A L + D+ Q+AL LA++A+ L+ +G SL +
Sbjct: 47 AAPAAPPPEAAPAAQSDGFALLEAAQLREAADDHQEALALAIKALEPLQA-SHGGWSLPV 105
Query: 284 VMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKF--AGHMQLGDTYAMLG 341
L + A LG+ +++ L + +I E+ +A A H QL T +G
Sbjct: 106 ARTLRLAGAAASRLGRLTDSLDSLNAAADIIDSLEAGDAEVAAVGAAVHEQLARTKTAMG 165
Query: 342 QLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKA 401
+ ++ E++ L + +G + VAEA L D+A LC AL+I +
Sbjct: 166 RRWDAASDLMRAMELKAVFLEKGSLELGNAYKDVAEAYRGVLACDKALPLCLEALEIARN 225
Query: 402 NSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDTY 461
+ S E A R+L+ I G +E ALE + M G +VE++ + +
Sbjct: 226 HFGGDSQEVAKVRQLLATIYAGSGRNEEALEQYEIVRMVYERLGLDVELSLAETDVAMVL 285
Query: 462 LSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALR 521
+ + R +EA+ ++ + + GK A+ FV +A++ + +SK + A
Sbjct: 286 VLLGRSEEAMDVLKRVIN--RAGKESEERAL--AFVAMANILCIQDRKADSKRCLEIARE 341
Query: 522 IYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSSIAGIEA 581
I + + V+P ++A +S +YE+M E E AL L++K L+ + Q I A
Sbjct: 342 ILDTKI-SVSPLQVAQVYAEMSMLYETMIEFEVALCLMKKTLVFLDGVSEMQHIQGSISA 400
Query: 582 QMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATEL 641
+MG + E+ L++AI KL+ +G A +G A + + A +
Sbjct: 401 RMGWLLLKTERVDEAVPYLQSAIEKLKNCFGPLHFGLGFAYKHLGDAYLAMNQSESAIKY 460
Query: 642 FEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPD 701
F AK I+ YGP H +T+ ++A Y +G +A+ E V+ E + P
Sbjct: 461 FTIAKDIINATYGPKHEDTIDTIQSIANAYGVMGSYKQAMDYQEQVIDAYE----SCGPG 516
Query: 702 VDDEKRRLSEL 712
+E R L
Sbjct: 517 AFEELREAQRL 527
>ref|ZP_00324700.1| COG0457: FOG: TPR repeat [Trichodesmium erythraeum IMS101]
Length = 1507
Score = 127 bits (320), Expect = 1e-27
Identities = 125/525 (23%), Positives = 236/525 (44%), Gaps = 22/525 (4%)
Query: 223 LNRVENSAESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLE 282
L + N ++ E+ + Q +L ++G + A+ +A Q + L +L G+ E
Sbjct: 65 LQGLANELAEMMEAKEKLDV---QVMELYNAGKYDE-AIPIAEQGVTLARQLW-GESHAE 119
Query: 283 LVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQH-ALAKFAGHMQLGDTYAMLG 341
+ L+ +Y + G+++EA P+ +++IEI + H +LA + L + Y G
Sbjct: 120 VATNLNNLTLLYESQGRHSEAEPLYKQAIEIHKVALPANHPSLATNLNN--LANLYRAQG 177
Query: 342 QLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHK- 400
+ + Y E+ + L P + +A ++ EAE L + A++I K
Sbjct: 178 RYSEAEPLYKQAIEIFKIALPANHPSLATNLNNLAGLYESQGRYSEAEPLYKQAIEIFKI 237
Query: 401 ---AN--SSAPSLEEAADR-RLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVD 454
AN S A +L A+ R G + + ++ A+E + ++A+ AN +A
Sbjct: 238 ALPANHPSLATNLNNLANLYRAQGRYSEAEPLYKQAIE---IDNIALPAN--HPSLARDL 292
Query: 455 CSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKS 514
++ + Y + RY EA Y++A+ + K NHP++ + LA+L+ G+ E++
Sbjct: 293 NNLAELYRAQGRYSEAEPLYKQAIEIHKVALPANHPSLATNLNNLANLYRAQGRYSEAEP 352
Query: 515 YCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDS-PGQQ 573
A+ I++ +P +P +A+ L N++ +YES +A L +KA+ I N + P
Sbjct: 353 LYKQAIEIFKIALPANHPS-LATNLNNLAGLYESQGRYSEAEPLFKKAIEIDNIALPANH 411
Query: 574 SSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLY 633
S+A + +Y G Y+E+ K AI + + LN +
Sbjct: 412 PSLATDLNNLAGLYSSQGRYSEAEPLYKQAIEIDKIALPANHPDLATHLNNLAGLYKSQG 471
Query: 634 AFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREE 693
+ EA L+++A I + HP+ +NLAG Y A GR EA +L+ + + +
Sbjct: 472 RYSEAEPLYKQAIEIHKVALPANHPQRASGLNNLAGLYRAQGRYSEAEPLLKQAIEIYKV 531
Query: 694 KLGTANPDVDDEKRRLSELLKEAGRVRSRKVMSLENLLDANARIP 738
L +P + L+EL + GR + + + + N +P
Sbjct: 532 ALPANHPFLATNLNNLAELYRAQGRYSEAEPLYKQAIEIDNIALP 576
Score = 109 bits (273), Expect = 3e-22
Identities = 103/438 (23%), Positives = 186/438 (41%), Gaps = 11/438 (2%)
Query: 287 LHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQH-ALAKFAGHMQLGDTYAMLGQLEN 345
L+ A +Y G+Y+EA P+ +++IEI + H +LA + L + Y G+
Sbjct: 292 LNNLAELYRAQGRYSEAEPLYKQAIEIHKVALPANHPSLATNLNN--LANLYRAQGRYSE 349
Query: 346 SIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSA 405
+ Y E+ + L P + +A ++ EAE L + A++I A
Sbjct: 350 AEPLYKQAIEIFKIALPANHPSLATNLNNLAGLYESQGRYSEAEPLFKKAIEIDNIALPA 409
Query: 406 PSLEEAADRRLMGLILDTKGNHEAA----LEHLVLASMAMVANGQEVEVASVDCSIGDTY 461
A D + + ++G + A + + + +A+ AN ++A+ ++ Y
Sbjct: 410 NHPSLATDLNNLAGLYSSQGRYSEAEPLYKQAIEIDKIALPAN--HPDLATHLNNLAGLY 467
Query: 462 LSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALR 521
S RY EA Y++A+ + K NHP S LA L+ G+ E++ A+
Sbjct: 468 KSQGRYSEAEPLYKQAIEIHKVALPANHPQRASGLNNLAGLYRAQGRYSEAEPLLKQAIE 527
Query: 522 IYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDS-PGQQSSIAGIE 580
IY+ +P +P +A+ L N++ +Y + +A L ++A+ I N + P +A
Sbjct: 528 IYKVALPANHP-FLATNLNNLAELYRAQGRYSEAEPLYKQAIEIDNIALPANHPELATNL 586
Query: 581 AQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATE 640
+ +Y G Y+E+ K AI + + LN + + EA
Sbjct: 587 NNLAELYRAQGRYSEAEPLYKQAIEVDKIALPANHPSLATNLNNLAELYRAQGRYSEAEP 646
Query: 641 LFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANP 700
L+++A + + HP +NLA Y A GR EA + + + V + L +P
Sbjct: 647 LYKQAIEVDKIALPANHPSLATHLNNLAVLYSAQGRYSEAEPLYKQAIEVDKIALPANHP 706
Query: 701 DVDDEKRRLSELLKEAGR 718
+ L+EL GR
Sbjct: 707 SLATNLNNLAELYHAQGR 724
Score = 109 bits (272), Expect = 4e-22
Identities = 106/438 (24%), Positives = 198/438 (45%), Gaps = 18/438 (4%)
Query: 287 LHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQHALAKFAGHMQ-LGDTYAMLGQLEN 345
L+ A +Y + G+Y+EA P+ +++IEI I H A H+ L Y G+
Sbjct: 418 LNNLAGLYSSQGRYSEAEPLYKQAIEIDKIALPANHP--DLATHLNNLAGLYKSQGRYSE 475
Query: 346 SIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHK----A 401
+ Y E+ + L P+ +A ++ EAE L + A++I+K A
Sbjct: 476 AEPLYKQAIEIHKVALPANHPQRASGLNNLAGLYRAQGRYSEAEPLLKQAIEIYKVALPA 535
Query: 402 NSS--APSLEEAADR-RLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIG 458
N A +L A+ R G + + ++ A+E + ++A+ AN E+A+ ++
Sbjct: 536 NHPFLATNLNNLAELYRAQGRYSEAEPLYKQAIE---IDNIALPAN--HPELATNLNNLA 590
Query: 459 DTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDS 518
+ Y + RY EA Y++A+ V K NHP++ + LA+L+ G+ E++
Sbjct: 591 ELYRAQGRYSEAEPLYKQAIEVDKIALPANHPSLATNLNNLAELYRAQGRYSEAEPLYKQ 650
Query: 519 ALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDS-PGQQSSIA 577
A+ + + +P +P +A+ L N++ +Y + +A L ++A+ + + P S+A
Sbjct: 651 AIEVDKIALPANHPS-LATHLNNLAVLYSAQGRYSEAEPLYKQAIEVDKIALPANHPSLA 709
Query: 578 GIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGE 637
+ +Y+ G Y+E+ K AI + + LN + + E
Sbjct: 710 TNLNNLAELYHAQGRYSEAEPLYKQAIEIHKVALPANHPSLATNLNNLAELYRAQGRYSE 769
Query: 638 ATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGT 697
A L+++A I + HP+ +NLA Y + GR EA++++ S E+K
Sbjct: 770 AEPLYKQAIEIHKVALPANHPQRASCLNNLALLYASQGRYSEALKLM-LKASKIEDKTIQ 828
Query: 698 ANPDVDDEKRRLSELLKE 715
A E RL+ + +E
Sbjct: 829 AIFASSSESDRLAHIKQE 846
Score = 106 bits (265), Expect = 2e-21
Identities = 99/438 (22%), Positives = 188/438 (42%), Gaps = 11/438 (2%)
Query: 287 LHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQH-ALAKFAGHMQLGDTYAMLGQLEN 345
L+ A +Y + G+Y+EA P+ +++IEI I H +LA + L Y+ G+
Sbjct: 376 LNNLAGLYESQGRYSEAEPLFKKAIEIDNIALPANHPSLATDLNN--LAGLYSSQGRYSE 433
Query: 346 SIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSA 405
+ Y E+ + L P + +A ++ EAE L + A++IHK A
Sbjct: 434 AEPLYKQAIEIDKIALPANHPDLATHLNNLAGLYKSQGRYSEAEPLYKQAIEIHKVALPA 493
Query: 406 PSLEEAADRRLMGLILDTKGNHEAA----LEHLVLASMAMVANGQEVEVASVDCSIGDTY 461
+ A+ + + +G + A + + + +A+ AN +A+ ++ + Y
Sbjct: 494 NHPQRASGLNNLAGLYRAQGRYSEAEPLLKQAIEIYKVALPAN--HPFLATNLNNLAELY 551
Query: 462 LSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALR 521
+ RY EA Y++A+ + NHP + + LA+L+ G+ E++ A+
Sbjct: 552 RAQGRYSEAEPLYKQAIEIDNIALPANHPELATNLNNLAELYRAQGRYSEAEPLYKQAIE 611
Query: 522 IYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDS-PGQQSSIAGIE 580
+ + +P +P +A+ L N++ +Y + +A L ++A+ + + P S+A
Sbjct: 612 VDKIALPANHPS-LATNLNNLAELYRAQGRYSEAEPLYKQAIEVDKIALPANHPSLATHL 670
Query: 581 AQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATE 640
+ V+Y G Y+E+ K AI + + LN + + EA
Sbjct: 671 NNLAVLYSAQGRYSEAEPLYKQAIEVDKIALPANHPSLATNLNNLAELYHAQGRYSEAEP 730
Query: 641 LFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANP 700
L+++A I + HP +NLA Y A GR EA + + + + + L +P
Sbjct: 731 LYKQAIEIHKVALPANHPSLATNLNNLAELYRAQGRYSEAEPLYKQAIEIHKVALPANHP 790
Query: 701 DVDDEKRRLSELLKEAGR 718
L+ L GR
Sbjct: 791 QRASCLNNLALLYASQGR 808
Score = 105 bits (263), Expect = 4e-21
Identities = 99/438 (22%), Positives = 187/438 (42%), Gaps = 11/438 (2%)
Query: 287 LHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQH-ALAKFAGHMQLGDTYAMLGQLEN 345
L+ A +Y G+Y+EA P+ +++IEI I H +LA + L Y G+
Sbjct: 334 LNNLANLYRAQGRYSEAEPLYKQAIEIFKIALPANHPSLATNLNN--LAGLYESQGRYSE 391
Query: 346 SIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSA 405
+ + E+ L P + +A ++ EAE L + A++I K A
Sbjct: 392 AEPLFKKAIEIDNIALPANHPSLATDLNNLAGLYSSQGRYSEAEPLYKQAIEIDKIALPA 451
Query: 406 PSLEEAADRRLMGLILDTKGNHEAA----LEHLVLASMAMVANGQEVEVASVDCSIGDTY 461
+ A + + ++G + A + + + +A+ AN + AS ++ Y
Sbjct: 452 NHPDLATHLNNLAGLYKSQGRYSEAEPLYKQAIEIHKVALPAN--HPQRASGLNNLAGLY 509
Query: 462 LSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALR 521
+ RY EA ++A+ ++K NHP + + LA+L+ G+ E++ A+
Sbjct: 510 RAQGRYSEAEPLLKQAIEIYKVALPANHPFLATNLNNLAELYRAQGRYSEAEPLYKQAIE 569
Query: 522 IYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDS-PGQQSSIAGIE 580
I +P +PE +A+ L N++ +Y + +A L ++A+ + + P S+A
Sbjct: 570 IDNIALPANHPE-LATNLNNLAELYRAQGRYSEAEPLYKQAIEVDKIALPANHPSLATNL 628
Query: 581 AQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATE 640
+ +Y G Y+E+ K AI + + LN + + + EA
Sbjct: 629 NNLAELYRAQGRYSEAEPLYKQAIEVDKIALPANHPSLATHLNNLAVLYSAQGRYSEAEP 688
Query: 641 LFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANP 700
L+++A + + HP +NLA Y A GR EA + + + + + L +P
Sbjct: 689 LYKQAIEVDKIALPANHPSLATNLNNLAELYHAQGRYSEAEPLYKQAIEIHKVALPANHP 748
Query: 701 DVDDEKRRLSELLKEAGR 718
+ L+EL + GR
Sbjct: 749 SLATNLNNLAELYRAQGR 766
Score = 76.3 bits (186), Expect = 4e-12
Identities = 66/282 (23%), Positives = 119/282 (41%), Gaps = 44/282 (15%)
Query: 282 ELVMCLHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQH-ALAKFAGHMQLGDTYAML 340
EL L+ A +Y G+Y+EA P+ +++IE+ I H +LA + L + Y
Sbjct: 581 ELATNLNNLAELYRAQGRYSEAEPLYKQAIEVDKIALPANHPSLATNLNN--LAELYRAQ 638
Query: 341 GQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHK 400
G+ + Y EV + L P + +A ++ EAE L + A+++ K
Sbjct: 639 GRYSEAEPLYKQAIEVDKIALPANHPSLATHLNNLAVLYSAQGRYSEAEPLYKQAIEVDK 698
Query: 401 ANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDT 460
+ NH + +L ++ +
Sbjct: 699 --------------------IALPANHPSLATNL--------------------NNLAEL 718
Query: 461 YLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSAL 520
Y + RY EA Y++A+ + K NHP++ + LA+L+ G+ E++ A+
Sbjct: 719 YHAQGRYSEAEPLYKQAIEIHKVALPANHPSLATNLNNLAELYRAQGRYSEAEPLYKQAI 778
Query: 521 RIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKA 562
I++ +P +P+ AS L N++ +Y S +ALKL+ KA
Sbjct: 779 EIHKVALPANHPQR-ASCLNNLALLYASQGRYSEALKLMLKA 819
Score = 56.2 bits (134), Expect = 4e-06
Identities = 83/377 (22%), Positives = 157/377 (41%), Gaps = 44/377 (11%)
Query: 287 LHVTAAIYCNLGQYNEAIPILERSIEIPVIGESQQH-ALAKFAGHMQLGDTYAMLGQLEN 345
L+ A +Y G+Y+EA P+ +++IE+ I H +LA + L + Y G+
Sbjct: 670 LNNLAVLYSAQGRYSEAEPLYKQAIEVDKIALPANHPSLATNLNN--LAELYHAQGRYSE 727
Query: 346 SIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSA 405
+ Y E+ + L P + +AE ++ EAE L + A++IHK A
Sbjct: 728 AEPLYKQAIEIHKVALPANHPSLATNLNNLAELYRAQGRYSEAEPLYKQAIEIHKVALPA 787
Query: 406 PSLEEAADRRLMGLILDTKGNHEAALEHLVLAS-------MAMVANGQEVE-VASVDCSI 457
+ A+ + L+ ++G + AL+ ++ AS A+ A+ E + +A + +
Sbjct: 788 NHPQRASCLNNLALLYASQGRYSEALKLMLKASKIEDKTIQAIFASSSESDRLAHIKQEV 847
Query: 458 G---DTYLSM--SRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKES 512
+T+LS+ + + A + AL + K ++ A A R +++E
Sbjct: 848 RPNLETFLSLVYQHFPHSPEAKQAALNLVL--KRKSLTATAMAAQNQAVFSGRYPQLQEI 905
Query: 513 ----KSYCDSALRI-YENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYN 567
+ CD L + YE P PE+ + ++ + + NELE+ L
Sbjct: 906 FPQWRRKCDQLLTLTYEVP----RPEKRETHKQQLAQLQKECNELERKLT---------- 951
Query: 568 DSPGQQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEK---KSSFIGIALNQ 624
QQ I+ QMG + + +E L+ + + EK K+ ++ L
Sbjct: 952 ----QQMPEIEIQQQMGDIRAVASELSEGEKLLEFVQFDVYDVQEKSWGKARYLAFVLAG 1007
Query: 625 MGLACVQLYAFGEATEL 641
+ V++ GEA +
Sbjct: 1008 GKPSEVEMIDLGEAENI 1024
>ref|ZP_00324930.1| COG0457: FOG: TPR repeat [Trichodesmium erythraeum IMS101]
Length = 1363
Score = 121 bits (303), Expect = 9e-26
Identities = 122/511 (23%), Positives = 222/511 (42%), Gaps = 22/511 (4%)
Query: 227 ENSAESALDKPERAPILLKQARDLISSGDNPQKALELALQAMNLLEK-LGNGKPSLELVM 285
+ S SA + ++A L KQ + +A+ L Q++ + ++ LG P ++
Sbjct: 193 DQSLASATELNDKAIELYKQGKY--------DEAVPLLEQSLKIRQQALGAEHP--DVAA 242
Query: 286 CLHVTAAIYCNLGQYNEAIPILERSIEI--PVIGESQQHALAKFAGHMQLGDTYAMLGQL 343
L+ A +Y G+Y EA P+ +++E+ ++G + L Y G+
Sbjct: 243 SLNNLAFLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNN---LAFLYKAQGRY 299
Query: 344 ENSIMCYSSGFEVQRQVLGETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANS 403
+ Y E+++++LG P V + +A ++ EAE L AL+I K
Sbjct: 300 TEAEPLYIQALEMRKKLLGAEHPDVATSLNNLASLYESQGRYTEAEPLYIQALEIFKKLL 359
Query: 404 SAPSLEEAADRRLMGLILDTKGNHEAALEHLVLASMAMVANGQEVEVASVDCSIGDT--- 460
A + A + + + +G + A E L + ++ M E SV S+ +
Sbjct: 360 GAEHPDVATSLNNLAFLYNAQGRYTEA-EPLYIQALDMTKKLLGAEHPSVATSLNNLALL 418
Query: 461 YLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSAL 520
Y RY EA Y +AL + K G HP V + LA L+N G+ E++ AL
Sbjct: 419 YEDQGRYTEAEPLYIQALEMRKKLLGAEHPDVATSLNNLAGLYNAQGRYTEAEPLYIQAL 478
Query: 521 RIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPG-QQSSIAGI 579
+ + + +P+ +A+ L N++++YES +A L +AL I+ G + +A
Sbjct: 479 EMRKKLLGAEHPD-VATSLNNLASLYESQGRYTEAEPLYIQALEIFKKLLGAEHPDVASS 537
Query: 580 EAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEAT 639
+ +Y G YTE+ A+ + + + + +LN + + EA
Sbjct: 538 LNNLAGLYKDQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAALYKDQGRYTEAE 597
Query: 640 ELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTAN 699
L+ +A + + G HP+ +NLAG Y+A GR EA + + +R++ LG +
Sbjct: 598 PLYIQALEMRKKLLGAEHPDVASSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEH 657
Query: 700 PDVDDEKRRLSELLKEAGRVRSRKVMSLENL 730
P V L+ L GR + + ++ L
Sbjct: 658 PLVASSLNNLAGLYNAQGRYTEAEPLYIQAL 688
Score = 119 bits (298), Expect = 4e-25
Identities = 115/478 (24%), Positives = 209/478 (43%), Gaps = 12/478 (2%)
Query: 259 KALELALQAMNLLEKLGNGKPSLELVMCLHVTAAIYCNLGQYNEAIPILERSIEI--PVI 316
+A L +QA+ + +KL G ++ L+ A++Y + G+Y EA P+ +++EI ++
Sbjct: 301 EAEPLYIQALEMRKKL-LGAEHPDVATSLNNLASLYESQGRYTEAEPLYIQALEIFKKLL 359
Query: 317 GESQQHALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVA 376
G L Y G+ + Y ++ +++LG P V + +A
Sbjct: 360 GAEHPDVATSLNN---LAFLYNAQGRYTEAEPLYIQALDMTKKLLGAEHPSVATSLNNLA 416
Query: 377 EANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVL 436
++ EAE L AL++ K A + A + + + +G + A E L +
Sbjct: 417 LLYEDQGRYTEAEPLYIQALEMRKKLLGAEHPDVATSLNNLAGLYNAQGRYTEA-EPLYI 475
Query: 437 ASMAM---VANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVG 493
++ M + + +VA+ ++ Y S RY EA Y +AL +FK G HP V
Sbjct: 476 QALEMRKKLLGAEHPDVATSLNNLASLYESQGRYTEAEPLYIQALEIFKKLLGAEHPDVA 535
Query: 494 SVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELE 553
S LA L+ G+ E++ AL + + + +P+ +AS L N++A+Y+
Sbjct: 536 SSLNNLAGLYKDQGRYTEAEPLYIQALEMRKKLLGAEHPD-VASSLNNLAALYKDQGRYT 594
Query: 554 KALKLLQKALLIYNDSPG-QQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGE 612
+A L +AL + G + +A + +Y G YTE+ A+ + +
Sbjct: 595 EAEPLYIQALEMRKKLLGAEHPDVASSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLG 654
Query: 613 KKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYD 672
+ + +LN + + EA L+ +A I + G HP +NLAG Y+
Sbjct: 655 AEHPLVASSLNNLAGLYNAQGRYTEAEPLYIQALEIFKKLLGAEHPLVATSLNNLAGLYN 714
Query: 673 AIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAGRVRSRKVMSLENL 730
A GR EA + + +R++ LG +P V L+ L GR + + ++ L
Sbjct: 715 AQGRYTEAEPLYIQALEMRKKLLGAEHPYVATSLNNLALLYYAQGRYTEAEPLYIQAL 772
Score = 114 bits (285), Expect = 1e-23
Identities = 108/444 (24%), Positives = 197/444 (44%), Gaps = 14/444 (3%)
Query: 259 KALELALQAMNLLEKL-GNGKPSLELVMCLHVTAAIYCNLGQYNEAIPILERSIEI--PV 315
+A L +QA+++ +KL G PS+ L+ A +Y + G+Y EA P+ +++E+ +
Sbjct: 385 EAEPLYIQALDMTKKLLGAEHPSV--ATSLNNLALLYEDQGRYTEAEPLYIQALEMRKKL 442
Query: 316 IGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYV 375
+G L Y G+ + Y E+++++LG P V + +
Sbjct: 443 LGAEHPDVATSLNN---LAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPDVATSLNNL 499
Query: 376 AEANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLV 435
A ++ EAE L AL+I K A + A+ + + +G + A E L
Sbjct: 500 ASLYESQGRYTEAEPLYIQALEIFKKLLGAEHPDVASSLNNLAGLYKDQGRYTEA-EPLY 558
Query: 436 LASMAM---VANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAV 492
+ ++ M + + +VAS ++ Y RY EA Y +AL + K G HP V
Sbjct: 559 IQALEMRKKLLGAEHPDVASSLNNLAALYKDQGRYTEAEPLYIQALEMRKKLLGAEHPDV 618
Query: 493 GSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNEL 552
S LA L+N G+ E++ AL + + + +P +AS L N++ +Y +
Sbjct: 619 ASSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPL-VASSLNNLAGLYNAQGRY 677
Query: 553 EKALKLLQKALLIYNDSPGQQSSIAGIEAQ-MGVMYYMLGNYTESYNTLKNAITKLRAIG 611
+A L +AL I+ G + + + +Y G YTE+ A+ + +
Sbjct: 678 TEAEPLYIQALEIFKKLLGAEHPLVATSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLL 737
Query: 612 EKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTY 671
+ ++ +LN + L + EA L+ +A + + G HP+ +NLA Y
Sbjct: 738 GAEHPYVATSLNNLALLYYAQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLATLY 797
Query: 672 DAIGRLDEAIQILEYVVSVREEKL 695
+ G + A+Q L+ + V+E+ L
Sbjct: 798 NVQGDIASAVQYLKRGLEVQEKNL 821
Score = 108 bits (271), Expect = 5e-22
Identities = 119/508 (23%), Positives = 217/508 (42%), Gaps = 11/508 (2%)
Query: 244 LKQARDLISSGDNPQKALELALQAMNLLEKLGNGKPSLELVMCLHVTAAIYCNLGQYNEA 303
L L S +A L +QA+ + +KL G ++ L+ A +Y G+Y EA
Sbjct: 328 LNNLASLYESQGRYTEAEPLYIQALEIFKKL-LGAEHPDVATSLNNLAFLYNAQGRYTEA 386
Query: 304 IPILERSIEIPVIGESQQH-ALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQRQVLG 362
P+ +++++ +H ++A ++ L Y G+ + Y E+++++LG
Sbjct: 387 EPLYIQALDMTKKLLGAEHPSVATSLNNLAL--LYEDQGRYTEAEPLYIQALEMRKKLLG 444
Query: 363 ETDPRVGETCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRRLMGLILD 422
P V + +A ++ EAE L AL++ K A + A + + +
Sbjct: 445 AEHPDVATSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPDVATSLNNLASLYE 504
Query: 423 TKGNHEAALEHLVLASMAM---VANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALT 479
++G + A E L + ++ + + + +VAS ++ Y RY EA Y +AL
Sbjct: 505 SQGRYTEA-EPLYIQALEIFKKLLGAEHPDVASSLNNLAGLYKDQGRYTEAEPLYIQALE 563
Query: 480 VFKTGKGENHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGL 539
+ K G HP V S LA L+ G+ E++ AL + + + +P+ +AS L
Sbjct: 564 MRKKLLGAEHPDVASSLNNLAALYKDQGRYTEAEPLYIQALEMRKKLLGAEHPD-VASSL 622
Query: 540 TNVSAIYESMNELEKALKLLQKALLIYNDSPGQQSS-IAGIEAQMGVMYYMLGNYTESYN 598
N++ +Y + +A L +AL + G + +A + +Y G YTE+
Sbjct: 623 NNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPLVASSLNNLAGLYNAQGRYTEAEP 682
Query: 599 TLKNAITKLRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHP 658
A+ + + + + +LN + + EA L+ +A + + G HP
Sbjct: 683 LYIQALEIFKKLLGAEHPLVATSLNNLAGLYNAQGRYTEAEPLYIQALEMRKKLLGAEHP 742
Query: 659 ETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAGR 718
+NLA Y A GR EA + + +R++ LG +PDV L+ L G
Sbjct: 743 YVATSLNNLALLYYAQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLATLYNVQGD 802
Query: 719 VRSRKVMSLENLLDANARIPNINLVIKA 746
+ S V L+ L+ + NL A
Sbjct: 803 IAS-AVQYLKRGLEVQEKNLTYNLAAGA 829
Score = 100 bits (249), Expect = 2e-19
Identities = 84/338 (24%), Positives = 151/338 (43%), Gaps = 4/338 (1%)
Query: 396 LDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLV--LASMAMVANGQEVEVASV 453
L +A++ SL A + + L +G ++ A+ L L + +VA+
Sbjct: 184 LSWRQASAKDQSLASATELNDKAIELYKQGKYDEAVPLLEQSLKIRQQALGAEHPDVAAS 243
Query: 454 DCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVGSVFVRLADLHNRTGKIKESK 513
++ Y + RY EA Y +AL + K G HP V S LA L+ G+ E++
Sbjct: 244 LNNLAFLYNAQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAFLYKAQGRYTEAE 303
Query: 514 SYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELEKALKLLQKALLIYNDSPG-Q 572
AL + + + +P+ +A+ L N++++YES +A L +AL I+ G +
Sbjct: 304 PLYIQALEMRKKLLGAEHPD-VATSLNNLASLYESQGRYTEAEPLYIQALEIFKKLLGAE 362
Query: 573 QSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGEKKSSFIGIALNQMGLACVQL 632
+A + +Y G YTE+ A+ + + + + +LN + L
Sbjct: 363 HPDVATSLNNLAFLYNAQGRYTEAEPLYIQALDMTKKLLGAEHPSVATSLNNLALLYEDQ 422
Query: 633 YAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYDAIGRLDEAIQILEYVVSVRE 692
+ EA L+ +A + + G HP+ +NLAG Y+A GR EA + + +R+
Sbjct: 423 GRYTEAEPLYIQALEMRKKLLGAEHPDVATSLNNLAGLYNAQGRYTEAEPLYIQALEMRK 482
Query: 693 EKLGTANPDVDDEKRRLSELLKEAGRVRSRKVMSLENL 730
+ LG +PDV L+ L + GR + + ++ L
Sbjct: 483 KLLGAEHPDVATSLNNLASLYESQGRYTEAEPLYIQAL 520
>ref|ZP_00327784.1| COG0457: FOG: TPR repeat [Trichodesmium erythraeum IMS101]
Length = 1215
Score = 119 bits (299), Expect = 3e-25
Identities = 125/544 (22%), Positives = 239/544 (42%), Gaps = 16/544 (2%)
Query: 193 NDKPLLRKQTKG-VASGVKSLKSSPLGKSVSLNRVENSAESALDKPERAPILLKQARDLI 251
+ +PL + T+ + +G ++ K +S + ES E L +A +L
Sbjct: 41 SQQPLTTEATEEQIINGTRNEKGKQGSYKLSWRQASAKEESLTFATE----LNDEAFELY 96
Query: 252 SSGDNPQKALELALQAMNL-LEKLGNGKPSLELVMCLHVTAAIYCNLGQYNEAIPILERS 310
G + A+ L Q++ + L+ LG P ++ L+ A +Y G+Y EA P+ ++
Sbjct: 97 KQGKYDE-AVPLLEQSLKIRLQLLGAEHP--DVATSLNNLAFLYQRQGRYTEAEPLYIQA 153
Query: 311 IEIPVIGESQQHALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGE 370
+++ +H L + + L + Y + G+ + Y ++++++LG P V
Sbjct: 154 LDMIKKLLGAEHPLVATSLN-NLAELYRVQGRYTEAEPLYQQALKMRKKLLGAKHPDVAT 212
Query: 371 TCRYVAEANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAA 430
+ +A ++ EAE L AL++ K A + A+ + + ++G + A
Sbjct: 213 SLNSLALLYKDQGRYTEAEPLYIQALEMRKKLLGAEHPDVASSLNNLAELYRSQGRYTEA 272
Query: 431 LEHLVLASMAM---VANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGE 487
E L L ++ M + + +VAS ++ Y RY EA Y +AL + K G
Sbjct: 273 -EPLYLQALEMTKKLLGAEHPDVASSLNNLAGLYKDQGRYTEAEPLYLQALEMRKKLLGA 331
Query: 488 NHPAVGSVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYE 547
H V S LA L+ G+ E+++ AL + + + G ++A+ L N++ +Y
Sbjct: 332 EHRYVASSLNNLALLYRVQGRYTEAEALYIQALEM-DKKLLGAEHPDVATSLNNLALLYS 390
Query: 548 SMNELEKALKLLQKALLIYNDSPGQQSS-IAGIEAQMGVMYYMLGNYTESYNTLKNAITK 606
+A L +AL I+ G + +A + +Y + G YTE+ A+
Sbjct: 391 DQGRYTEAEPLYIQALEIFKKLLGAEHRYVASSLNNLAGLYRVQGRYTEAEPLYVQALKM 450
Query: 607 LRAIGEKKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSN 666
+ + + + +LN + L + EA L+++A + + G HP+ +N
Sbjct: 451 WKKLLGAEHPDVATSLNNLALLYKDQGRYTEAEPLYQQALDMRKKLLGAEHPDVATSLNN 510
Query: 667 LAGTYDAIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAGRVRSRKVMS 726
LAG Y GR EA + + + ++ LG +PDV L+ L K GR + +
Sbjct: 511 LAGLYRVQGRYTEAEPLYVQALKMWKKLLGAEHPDVATSLNNLALLYKAQGRYTEAEPLY 570
Query: 727 LENL 730
+++L
Sbjct: 571 IQSL 574
Score = 119 bits (297), Expect = 5e-25
Identities = 116/494 (23%), Positives = 215/494 (43%), Gaps = 13/494 (2%)
Query: 259 KALELALQAMNLLEKLGNGKPSLELVMCLHVTAAIYCNLGQYNEAIPILERSIEI--PVI 316
+A L QA+ + +KL K ++ L+ A +Y + G+Y EA P+ +++E+ ++
Sbjct: 187 EAEPLYQQALKMRKKLLGAKHP-DVATSLNSLALLYKDQGRYTEAEPLYIQALEMRKKLL 245
Query: 317 GESQQHALAKFAGHMQLGDTYAMLGQLENSIMCYSSGFEVQRQVLGETDPRVGETCRYVA 376
G + L + Y G+ + Y E+ +++LG P V + +A
Sbjct: 246 GAEHPDVASSLNN---LAELYRSQGRYTEAEPLYLQALEMTKKLLGAEHPDVASSLNNLA 302
Query: 377 EANVQALQFDEAERLCQMALDIHKANSSAPSLEEAADRRLMGLILDTKGNHEAALEHLVL 436
++ EAE L AL++ K A A+ + L+ +G + A E L +
Sbjct: 303 GLYKDQGRYTEAEPLYLQALEMRKKLLGAEHRYVASSLNNLALLYRVQGRYTEA-EALYI 361
Query: 437 ASMAM---VANGQEVEVASVDCSIGDTYLSMSRYDEAVFAYEKALTVFKTGKGENHPAVG 493
++ M + + +VA+ ++ Y RY EA Y +AL +FK G H V
Sbjct: 362 QALEMDKKLLGAEHPDVATSLNNLALLYSDQGRYTEAEPLYIQALEIFKKLLGAEHRYVA 421
Query: 494 SVFVRLADLHNRTGKIKESKSYCDSALRIYENPMPGVNPEEIASGLTNVSAIYESMNELE 553
S LA L+ G+ E++ AL++++ + +P+ +A+ L N++ +Y+
Sbjct: 422 SSLNNLAGLYRVQGRYTEAEPLYVQALKMWKKLLGAEHPD-VATSLNNLALLYKDQGRYT 480
Query: 554 KALKLLQKALLIYNDSPG-QQSSIAGIEAQMGVMYYMLGNYTESYNTLKNAITKLRAIGE 612
+A L Q+AL + G + +A + +Y + G YTE+ A+ + +
Sbjct: 481 EAEPLYQQALDMRKKLLGAEHPDVATSLNNLAGLYRVQGRYTEAEPLYVQALKMWKKLLG 540
Query: 613 KKSSFIGIALNQMGLACVQLYAFGEATELFEEAKSILEHEYGPYHPETLGVYSNLAGTYD 672
+ + +LN + L + EA L+ ++ + + G HP +NLA Y
Sbjct: 541 AEHPDVATSLNNLALLYKAQGRYTEAEPLYIQSLEMRKKLLGAEHPSVAQSLNNLAALYY 600
Query: 673 AIGRLDEAIQILEYVVSVREEKLGTANPDVDDEKRRLSELLKEAGRVRSRKVMSLENLLD 732
GR +A + + + +R++ LG +PDV L+ L G + S V LE L+
Sbjct: 601 YQGRYTDAEPLYQQALEMRKKLLGAEHPDVAISLNNLALLYSAQGNIAS-AVQYLERGLE 659
Query: 733 ANARIPNINLVIKA 746
+ NL A
Sbjct: 660 VQEKNLTYNLAAGA 673
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.311 0.130 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,189,227,142
Number of Sequences: 2540612
Number of extensions: 50378609
Number of successful extensions: 162969
Number of sequences better than 10.0: 1415
Number of HSP's better than 10.0 without gapping: 345
Number of HSP's successfully gapped in prelim test: 1122
Number of HSP's that attempted gapping in prelim test: 156319
Number of HSP's gapped (non-prelim): 4725
length of query: 746
length of database: 863,360,394
effective HSP length: 136
effective length of query: 610
effective length of database: 517,837,162
effective search space: 315880668820
effective search space used: 315880668820
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0166.9


