

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0162b.4
(228 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
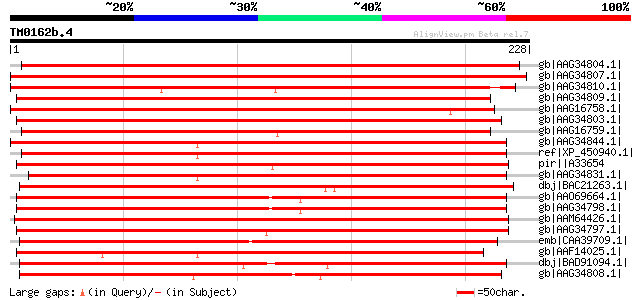
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG34804.1| glutathione S-transferase GST 14 [Glycine max] 368 e-101
gb|AAG34807.1| glutathione S-transferase GST 17 [Glycine max] 357 1e-97
gb|AAG34810.1| glutathione S-transferase GST 20 [Glycine max] 333 3e-90
gb|AAG34809.1| glutathione S-transferase GST 19 [Glycine max] 236 5e-61
gb|AAG16758.1| putative glutathione S-transferase T3 [Lycopersic... 235 8e-61
gb|AAG34803.1| glutathione S-transferase GST 13 [Glycine max] 233 2e-60
gb|AAG16759.1| putative glutathione S-transferase T4 [Lycopersic... 233 3e-60
gb|AAG34844.1| glutathione S-transferase GST 36 [Zea mays] 229 3e-59
ref|XP_450940.1| putative glutathione S-transferase [Oryza sativ... 225 8e-58
pir||A33654 heat shock protein 26A - soybean gi|417148|sp|P32110... 223 2e-57
gb|AAG34831.1| glutathione S-transferase GST 23 [Zea mays] 223 3e-57
dbj|BAC21263.1| glutathione S-transferase [Cucurbita maxima] 222 5e-57
gb|AAO69664.1| glutathione S-transferase [Phaseolus acutifolius] 221 9e-57
gb|AAG34798.1| glutathione S-transferase GST 8 [Glycine max] 221 9e-57
gb|AAM64426.1| putative glutathione S-transferase [Arabidopsis t... 219 5e-56
gb|AAG34797.1| glutathione S-transferase GST 7 [Glycine max] 218 1e-55
emb|CAA39709.1| auxin-induced protein [Nicotiana tabacum] gi|197... 216 5e-55
gb|AAF14025.1| putative glutathione transferase [Arabidopsis tha... 214 1e-54
dbj|BAD91094.1| glutathione S-transferase GST 18 [Populus alba x... 212 7e-54
gb|AAG34808.1| glutathione S-transferase GST 18 [Glycine max] 211 1e-53
>gb|AAG34804.1| glutathione S-transferase GST 14 [Glycine max]
Length = 219
Score = 368 bits (944), Expect = e-101
Identities = 169/219 (77%), Positives = 195/219 (88%)
Query: 6 VKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKTI 65
VKLLSF+ SPF KRVEWALKLKGVEYEY+E+DIFNK+SLLL+LNPVHKK+PVLVH HK I
Sbjct: 1 VKLLSFFASPFGKRVEWALKLKGVEYEYIEQDIFNKTSLLLQLNPVHKKVPVLVHAHKPI 60
Query: 66 AESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKAL 125
AESF+I+EY+DETWKQYPLLP DPY+RALARFWANF EQKLL+ ++ M +GDEQ+ A+
Sbjct: 61 AESFVIVEYVDETWKQYPLLPRDPYQRALARFWANFAEQKLLDAAWIGMYSSGDEQQNAV 120
Query: 126 NEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPATT 185
AREA+EKIEE I+GK++FGGENIGYLDIA+GWISYW+ IWEEVGSI IIDPLKFPA T
Sbjct: 121 KVAREAIEKIEEEIKGKKYFGGENIGYLDIALGWISYWLPIWEEVGSIQIIDPLKFPAIT 180
Query: 186 AWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLSETFRG 224
AW+TNFLSHPVIKD LPPRDKM+ YFH R+ LS TF+G
Sbjct: 181 AWITNFLSHPVIKDNLPPRDKMLVYFHSRRTALSSTFQG 219
>gb|AAG34807.1| glutathione S-transferase GST 17 [Glycine max]
Length = 229
Score = 357 bits (917), Expect = 1e-97
Identities = 168/227 (74%), Positives = 195/227 (85%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
MGS DVKLLSFW SPF KRVEWALKLKG+EYEY+EEDIFNKS+LLL+LNPVHKK+PVLVH
Sbjct: 2 MGSGDVKLLSFWVSPFGKRVEWALKLKGIEYEYIEEDIFNKSNLLLQLNPVHKKVPVLVH 61
Query: 61 GHKTIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDE 120
HK IAESFII+EYIDETWKQYPLLP P++RALARFWA EQKL + +VAM +G+E
Sbjct: 62 AHKPIAESFIILEYIDETWKQYPLLPCHPHQRALARFWATSVEQKLGKAGWVAMSTSGEE 121
Query: 121 QEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLK 180
QEKA+ EA E MEKIEE I+GK+FFGG+NIGYLDIA+GWI+Y + +WEEVGS+ IIDPLK
Sbjct: 122 QEKAVKEAIEMMEKIEEEIKGKKFFGGDNIGYLDIALGWIAYLVPVWEEVGSMQIIDPLK 181
Query: 181 FPATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLSETFRGRFK 227
FPATT W+TNFLSHP+IKD+LPPRDKM+ Y+H RK +L FR K
Sbjct: 182 FPATTEWITNFLSHPLIKDSLPPRDKMLVYYHNRKNNLPSVFRNLVK 228
>gb|AAG34810.1| glutathione S-transferase GST 20 [Glycine max]
Length = 224
Score = 333 bits (853), Expect = 3e-90
Identities = 162/225 (72%), Positives = 189/225 (84%), Gaps = 7/225 (3%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
MGS++VKLLSFW SPF KR+EWALKLKGVEYEY+EEDIFNKSSLLLELNPVHKK+PVLVH
Sbjct: 1 MGSEEVKLLSFWVSPFGKRIEWALKLKGVEYEYIEEDIFNKSSLLLELNPVHKKVPVLVH 60
Query: 61 GHKTI-AESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMC--IT 117
K+I AESFII+EYIDE WKQY LLPH PY+RALARFWA E+ + V++A+ +
Sbjct: 61 AEKSIIAESFIILEYIDEKWKQYSLLPHHPYQRALARFWAATAEEMFRKVVWIALRSPTS 120
Query: 118 GDEQEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIID 177
GDE+EKAL E+RE ME+IEE I GK++FGG+NIGYLDIA+GWISYW+ + EEVGS+ IID
Sbjct: 121 GDEREKALKESREVMERIEEEIRGKKYFGGDNIGYLDIALGWISYWLPVLEEVGSMQIID 180
Query: 178 PLKFPATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLSETF 222
PLKFPATTAWMTNFLS+PVIKD LPPRDKM+ Y KDL +
Sbjct: 181 PLKFPATTAWMTNFLSNPVIKDNLPPRDKMLVYL----KDLRSKY 221
>gb|AAG34809.1| glutathione S-transferase GST 19 [Glycine max]
Length = 225
Score = 236 bits (601), Expect = 5e-61
Identities = 109/208 (52%), Positives = 154/208 (73%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
++VKL++ S RVEWAL++KGVEYEY++ED+ NKSSLLL+ NPVHKK+PVL+H +K
Sbjct: 2 EEVKLIATHQSFPCARVEWALRIKGVEYEYLKEDLANKSSLLLQSNPVHKKVPVLLHNNK 61
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
IAES +I+EYIDETWK+ PLLP DPYERA ARFWA F ++K + V+ A G+E+EK
Sbjct: 62 PIAESLVILEYIDETWKKNPLLPLDPYERAQARFWARFIDEKCVLAVWGATVAQGEEKEK 121
Query: 124 ALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPA 183
A+ A E++ +E+ I+GK++FGGE IGYLDIA G +S W + EE+G + +++ +FP+
Sbjct: 122 AVGAALESLALLEKEIQGKKYFGGEKIGYLDIAAGCMSLWFSVLEELGEMELLNAERFPS 181
Query: 184 TTAWMTNFLSHPVIKDTLPPRDKMIDYF 211
W NFL +KD +P R+ +++YF
Sbjct: 182 LHEWSQNFLQTSPVKDCIPSRESVVEYF 209
>gb|AAG16758.1| putative glutathione S-transferase T3 [Lycopersicon esculentum]
Length = 225
Score = 235 bits (599), Expect = 8e-61
Identities = 111/214 (51%), Positives = 146/214 (67%), Gaps = 1/214 (0%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
M S+ VKLL +W SPF+ +V WALKLKG+EYEY EED+ NKS LLL+ NPVHKKIPVLVH
Sbjct: 1 MASEKVKLLGYWASPFALKVHWALKLKGIEYEYQEEDLSNKSPLLLQYNPVHKKIPVLVH 60
Query: 61 GHKTIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDE 120
K IAES +I+EYI+ETWK PLLP DPYERA ARFWA F + K + +F G E
Sbjct: 61 NGKPIAESLVILEYIEETWKHNPLLPEDPYERAKARFWAKFVDDKCVPGIFGTFAKVGVE 120
Query: 121 QEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLK 180
Q+K EARE ++ +E+ + K FFG IG++D+ WI W I EEV I +ID +
Sbjct: 121 QQKIAKEARENLKILEDELGKKHFFGDAKIGFMDVTSAWIICWAQIVEEVVDIRLIDAEE 180
Query: 181 FPATTAWMTNFL-SHPVIKDTLPPRDKMIDYFHG 213
P+ +W N L + P++K+ PP+DK++++ G
Sbjct: 181 MPSLVSWFQNVLEAAPILKECTPPKDKLLEHNKG 214
>gb|AAG34803.1| glutathione S-transferase GST 13 [Glycine max]
Length = 220
Score = 233 bits (595), Expect = 2e-60
Identities = 109/213 (51%), Positives = 151/213 (70%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
++V+LL W SPFS RV+ ALKLKGV Y+Y EED+ NKS+ LL+ NPVHKK+PVLVH
Sbjct: 7 EEVRLLGKWASPFSNRVDLALKLKGVPYKYSEEDLANKSADLLKYNPVHKKVPVLVHNGN 66
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
+ ES II+EYIDETWK PLLP DPYERALARFW+ + K+L ++ A + +EK
Sbjct: 67 PLPESLIIVEYIDETWKNNPLLPQDPYERALARFWSKTLDDKILPAIWNACWSDENGREK 126
Query: 124 ALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPA 183
A+ EA EA++ ++E ++ K+FFGGE+IG +DIA +I YW+ I +E+ + ++ KFP
Sbjct: 127 AVEEALEALKILQETLKDKKFFGGESIGLVDIAANFIGYWVAILQEIAGLELLTIEKFPK 186
Query: 184 TTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKK 216
W +F++HPVIK+ LPPRD++ +F K
Sbjct: 187 LYNWSQDFINHPVIKEGLPPRDELFAFFKASAK 219
>gb|AAG16759.1| putative glutathione S-transferase T4 [Lycopersicon esculentum]
Length = 226
Score = 233 bits (594), Expect = 3e-60
Identities = 108/208 (51%), Positives = 147/208 (69%), Gaps = 2/208 (0%)
Query: 6 VKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKTI 65
VKL+ S F RVEWALKLKGV+YEY++ED+ NKS LL++ NPVHKKIPVL+H K +
Sbjct: 4 VKLIGSSGSLFCTRVEWALKLKGVDYEYIQEDLLNKSELLIKSNPVHKKIPVLLHDDKPV 63
Query: 66 AESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCI--TGDEQEK 123
ES +I+EYIDETWK YPLLP DP+ERA ARFWA F + K + + AM + G+ + K
Sbjct: 64 VESLLILEYIDETWKGYPLLPQDPHERATARFWAKFVDDKCVIGSWEAMAMQDEGEAKTK 123
Query: 124 ALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPA 183
A+ +E IE+ IEGK+FFGGE IGYLD+ +GW + W+ EEVG++ ++DP KFP+
Sbjct: 124 AIESIQELYAFIEKQIEGKKFFGGEQIGYLDLVMGWKTLWLSAMEEVGNVKLLDPEKFPS 183
Query: 184 TTAWMTNFLSHPVIKDTLPPRDKMIDYF 211
W NF P+I + +P ++ +++YF
Sbjct: 184 LHQWAENFKQIPIINECMPQQETLVNYF 211
>gb|AAG34844.1| glutathione S-transferase GST 36 [Zea mays]
Length = 222
Score = 229 bits (585), Expect = 3e-59
Identities = 109/219 (49%), Positives = 148/219 (66%), Gaps = 1/219 (0%)
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
M + VK+L W SP RVEWAL+LKGVEYEYV+ED+ NKS+ LL NPV KK+PVLVH
Sbjct: 1 MAEKGVKVLGMWASPMVIRVEWALRLKGVEYEYVDEDLANKSADLLRHNPVTKKVPVLVH 60
Query: 61 GHKTIAESFIIIEYIDETWKQ-YPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGD 119
K +AES II+EYIDE WK YP++P DPYERA ARFWA F E K ++ TG+
Sbjct: 61 DGKPVAESTIIVEYIDEVWKGGYPIMPGDPYERAQARFWARFAEDKCNAALYPIFTATGE 120
Query: 120 EQEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPL 179
Q KA++EA++ ++ +E +EGK+FFGG+ +GYLDI VGW ++W+ + EEV ++
Sbjct: 121 AQRKAVHEAQQCLKTLETALEGKKFFGGDAVGYLDIVVGWFAHWLPVIEEVTGASVVTHE 180
Query: 180 KFPATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDL 218
+ P AW FL+ V+K LP RD+++ R++ L
Sbjct: 181 ELPLMKAWFGRFLALDVVKAALPDRDRLLAANKARREQL 219
>ref|XP_450940.1| putative glutathione S-transferase [Oryza sativa (japonica
cultivar-group)] gi|51979108|ref|XP_507428.1| PREDICTED
OJ1005_D12.39 gene product [Oryza sativa (japonica
cultivar-group)] gi|51963774|ref|XP_506667.1| PREDICTED
OJ1005_D12.39 gene product [Oryza sativa (japonica
cultivar-group)] gi|15430731|gb|AAK98545.1| putative
glutathione S-transferase OsGSTU17 [Oryza sativa
(japonica cultivar-group)] gi|46806331|dbj|BAD17523.1|
putative glutathione S-transferase [Oryza sativa
(japonica cultivar-group)] gi|47497667|dbj|BAD19734.1|
putative glutathione S-transferase [Oryza sativa
(japonica cultivar-group)]
Length = 223
Score = 225 bits (573), Expect = 8e-58
Identities = 101/214 (47%), Positives = 142/214 (66%), Gaps = 1/214 (0%)
Query: 6 VKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKTI 65
VK+ W SP + RVEWAL+LKGV+YEYV+ED+ NKS LL NPV KK+PVLVH K +
Sbjct: 7 VKVFGMWASPMAIRVEWALRLKGVDYEYVDEDLANKSEALLRHNPVTKKVPVLVHDGKPL 66
Query: 66 AESFIIIEYIDETWKQ-YPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
AES +I+EYIDE WK YP++P DP++RA ARFWA F E+K ++ TG+EQ K
Sbjct: 67 AESTVIVEYIDEAWKHGYPIMPSDPFDRAQARFWARFAEEKCNAALYPIFMTTGEEQRKL 126
Query: 125 LNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPAT 184
++EA++ ++ +E +EGK+FFGG+ GYLDI GW +YW+ + EE + ++ P
Sbjct: 127 VHEAQQCLKTLETALEGKKFFGGDAFGYLDIVTGWFAYWLPVIEEACGVEVVTDEALPLM 186
Query: 185 TAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDL 218
AW L+ +K LPPRDK++ R++ +
Sbjct: 187 KAWFDRVLAVDAVKAVLPPRDKLVALNKARREQI 220
>pir||A33654 heat shock protein 26A - soybean gi|417148|sp|P32110|GSTX6_SOYBN
Probable glutathione S-transferase (Heat shock protein
26A) (G2-4) gi|169981|gb|AAA33973.1| Gmhsp26-A
Length = 225
Score = 223 bits (569), Expect = 2e-57
Identities = 106/217 (48%), Positives = 149/217 (67%), Gaps = 1/217 (0%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
+DVKLL SPF RV+ ALKLKGVEY+++EE++ NKS LLL+ NPVHKK+PV VH +
Sbjct: 6 EDVKLLGIVGSPFVCRVQIALKLKGVEYKFLEENLGNKSDLLLKYNPVHKKVPVFVHNEQ 65
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAM-CITGDEQE 122
IAES +I+EYIDETWK P+LP DPY+RALARFW+ F + K++ V ++ + E+E
Sbjct: 66 PIAESLVIVEYIDETWKNNPILPSDPYQRALARFWSKFIDDKIVGAVSKSVFTVDEKERE 125
Query: 123 KALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFP 182
K + E EA++ +E ++ K+FFGGE G +DIA +I++WI I++E+ + + KFP
Sbjct: 126 KNVEETYEALQFLENELKDKKFFGGEEFGLVDIAAVFIAFWIPIFQEIAGLQLFTSEKFP 185
Query: 183 ATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLS 219
W FL+HP + + LPPRD + YF R + LS
Sbjct: 186 ILYKWSQEFLNHPFVHEVLPPRDPLFAYFKARYESLS 222
>gb|AAG34831.1| glutathione S-transferase GST 23 [Zea mays]
Length = 214
Score = 223 bits (568), Expect = 3e-57
Identities = 105/211 (49%), Positives = 143/211 (67%), Gaps = 1/211 (0%)
Query: 9 LSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKTIAES 68
L W SP RVEWAL+LKGVEYEYV+ED+ NKS+ LL NPV KK+PVLVH K +AES
Sbjct: 1 LGMWASPMVIRVEWALRLKGVEYEYVDEDLANKSADLLRHNPVTKKVPVLVHDGKPVAES 60
Query: 69 FIIIEYIDETWKQ-YPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKALNE 127
II+EYIDE WK YP++P DPYERA ARFWA F E K ++ TG+ Q KA++E
Sbjct: 61 TIIVEYIDEVWKGGYPIMPGDPYERAQARFWARFAEDKCNAALYPIFTATGEAQRKAVHE 120
Query: 128 AREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPATTAW 187
A++ ++ +E ++GK+FFGG+ +GYLDI VGW ++W+ + EEV ++ + P AW
Sbjct: 121 AQQCLKTLETALDGKKFFGGDAVGYLDIVVGWFAHWLPVIEEVTGASVVTDEELPLMKAW 180
Query: 188 MTNFLSHPVIKDTLPPRDKMIDYFHGRKKDL 218
FL+ V+K LP RD+++ R++ L
Sbjct: 181 FGRFLAVDVVKAALPDRDRLLAANKARREQL 211
>dbj|BAC21263.1| glutathione S-transferase [Cucurbita maxima]
Length = 226
Score = 222 bits (566), Expect = 5e-57
Identities = 113/223 (50%), Positives = 144/223 (63%), Gaps = 6/223 (2%)
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
+VKL WPSPFS RV WALKLKG+ YEYVEED+ NKS LLL NPVHKKIPVLVHG K
Sbjct: 3 EVKLHGTWPSPFSYRVIWALKLKGIPYEYVEEDLSNKSPLLLHYNPVHKKIPVLVHGGKP 62
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
I ES II+EYIDETW + PLLP DP+ERA ARFW F E K + + +EQE A
Sbjct: 63 ICESMIILEYIDETWPEKPLLPSDPFERATARFWIKFIEDKGPAVFMIFRAKSDEEQENA 122
Query: 125 LNEAREAMEKIEE-VIEG-----KRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDP 178
++ E + +EE G ++FFGG+ IG D+A GWI++WI + EEV ++ ++D
Sbjct: 123 KKQSLEMLRTVEEWGFRGGDGRERKFFGGDEIGMTDLAFGWIAHWIGVIEEVVNVKLLDA 182
Query: 179 LKFPATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLSET 221
+FP AW F VI+D LP R+ M F R++ L ++
Sbjct: 183 NEFPKVYAWTERFKDAAVIRDNLPDRNLMAAAFRRRREQLLQS 225
>gb|AAO69664.1| glutathione S-transferase [Phaseolus acutifolius]
Length = 225
Score = 221 bits (564), Expect = 9e-57
Identities = 103/217 (47%), Positives = 152/217 (69%), Gaps = 3/217 (1%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
++V LL SPF RV+ ALKLKG+EY+YVEE++ NKS LL+ NPVHKK+PV VHG K
Sbjct: 6 EEVTLLGATGSPFVCRVKIALKLKGIEYKYVEENLANKSEQLLKYNPVHKKVPVFVHGDK 65
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
+AES +I+EYIDETW P+LP DPY+RALARFW+ F + ++ + + ++ T DE+E+
Sbjct: 66 PLAESLVIVEYIDETWNNNPILPSDPYQRALARFWSKFIDDNIVGSTWKSV-FTADEKER 124
Query: 124 ALN--EAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
N EA E+++ +E I K+FFGGE +G +DIA ++++WI + +E+ + ++ KF
Sbjct: 125 EKNVAEASESLQFLENEIADKKFFGGEELGLVDIAAVYVAFWIPLVQEIAGLELLTSEKF 184
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDL 218
P W F+SHP++K++LPPRD + +F GR + L
Sbjct: 185 PNLYKWSQEFVSHPIVKESLPPRDPVFGFFKGRYESL 221
>gb|AAG34798.1| glutathione S-transferase GST 8 [Glycine max]
Length = 225
Score = 221 bits (564), Expect = 9e-57
Identities = 106/217 (48%), Positives = 150/217 (68%), Gaps = 3/217 (1%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
++V LL SPF RV ALKLKGV+Y+YVEE++ NKS LLL+ NPVHKK+PV +H K
Sbjct: 6 EEVTLLGATGSPFVCRVHIALKLKGVQYKYVEENLRNKSELLLKSNPVHKKVPVFIHNEK 65
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
IAES +I+EYIDETWK P+LP DPY+RALARFW+ F + K+ + ++ T DE+E+
Sbjct: 66 PIAESLVIVEYIDETWKNNPILPSDPYQRALARFWSKFIDDKVFGAAWKSV-FTADEKER 124
Query: 124 ALN--EAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
N EA EA++ +E I+ K+FFGGE IG +DIA +I++W+ + +E+ + + KF
Sbjct: 125 EKNVEEAIEALQFLENEIKDKKFFGGEEIGLVDIAAVYIAFWVPMVQEIAGLELFTSEKF 184
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDL 218
P W FL+HP++K++LPPRD + +F G + L
Sbjct: 185 PKLHNWSQEFLNHPIVKESLPPRDPVFSFFKGLYESL 221
>gb|AAM64426.1| putative glutathione S-transferase [Arabidopsis thaliana]
gi|3980393|gb|AAC95196.1| putative glutathione
S-transferase [Arabidopsis thaliana]
gi|15810097|gb|AAL06974.1| At2g29420/F16P2.20
[Arabidopsis thaliana] gi|15010756|gb|AAK74037.1|
At2g29420/F16P2.20 [Arabidopsis thaliana]
gi|15227082|ref|NP_180503.1| glutathione S-transferase,
putative [Arabidopsis thaliana]
gi|11096010|gb|AAG30137.1| glutathione S-transferase
[Arabidopsis thaliana] gi|25313229|pir||B84696 probable
glutathione S-transferase [imported] - Arabidopsis
thaliana
Length = 227
Score = 219 bits (558), Expect = 5e-56
Identities = 99/217 (45%), Positives = 147/217 (67%)
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
S++VKLL W SPFS+R+E AL LKGV YE++E+DI NKSSLLL+LNPVHK IPVLVH
Sbjct: 7 SEEVKLLGMWASPFSRRIEIALTLKGVSYEFLEQDITNKSSLLLQLNPVHKMIPVLVHNG 66
Query: 63 KTIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQE 122
K I+ES +I+EYIDETW+ P+LP DPYER +ARFW+ F ++++ T + TG E++
Sbjct: 67 KPISESLVILEYIDETWRDNPILPQDPYERTMARFWSKFVDEQIYVTAMKVVGKTGKERD 126
Query: 123 KALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFP 182
+ R+ + +E+ + GK F GG+++G++DI +++W+ EE+ + ++ KFP
Sbjct: 127 AVVEATRDLLMFLEKELVGKDFLGGKSLGFVDIVATLVAFWLMRTEEIVGVKVVPVEKFP 186
Query: 183 ATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLS 219
W+ N L + VIK +PP D+ + Y R + L+
Sbjct: 187 EIHRWVKNLLGNDVIKKCIPPEDEHLKYIRARMEKLN 223
>gb|AAG34797.1| glutathione S-transferase GST 7 [Glycine max]
Length = 225
Score = 218 bits (554), Expect = 1e-55
Identities = 104/217 (47%), Positives = 149/217 (67%), Gaps = 1/217 (0%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
+DVKLL SPF RV+ ALKLKGV+Y+++EE++ NKS LLL+ NPVHKK+PV +H K
Sbjct: 6 EDVKLLGATGSPFVCRVQIALKLKGVQYKFLEENLRNKSELLLKSNPVHKKVPVFIHNEK 65
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVF-VAMCITGDEQE 122
IAES +I+EYIDETWK P+LP DPY+RALARFW+ F + K++ + + E+E
Sbjct: 66 PIAESLVIVEYIDETWKNNPILPSDPYQRALARFWSKFIDDKVVGAAWKYIYTVDEKERE 125
Query: 123 KALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFP 182
K + E+ EA++ +E ++ K+FFGGE IG +DIA +I++WI I +EV + + KFP
Sbjct: 126 KNVEESYEALQFLENELKDKKFFGGEEIGLVDIAAVFIAFWIPIIQEVLGLKLFTSEKFP 185
Query: 183 ATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLS 219
W F++HPV+K LPPRD++ ++ + LS
Sbjct: 186 KLYKWSQEFINHPVVKQVLPPRDQLFAFYKACHESLS 222
>emb|CAA39709.1| auxin-induced protein [Nicotiana tabacum] gi|19795|emb|CAA39705.1|
auxin-induced protein [Nicotiana tabacum]
gi|416649|sp|Q03662|GSTX1_TOBAC Probable glutathione
S-transferase (Auxin-induced protein PGNT1/PCNT110)
Length = 223
Score = 216 bits (549), Expect = 5e-55
Identities = 99/210 (47%), Positives = 145/210 (68%), Gaps = 1/210 (0%)
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
+VKLL FW SPFS+RVEWALK+KGV+YEY+EED NKSSLLL+ NP+HKK+PVL+H K
Sbjct: 3 EVKLLGFWYSPFSRRVEWALKIKGVKYEYIEEDRDNKSSLLLQSNPIHKKVPVLIHNGKR 62
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
I ES +I+EYIDET++ +LP DPY+RALARFWA F + K + V G+EQEK
Sbjct: 63 IVESMVILEYIDETFEGPSILPKDPYDRALARFWAKFLDDK-VPAVVKTFLRKGEEQEKD 121
Query: 125 LNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPAT 184
E E ++ ++ ++ K+FF G+ G+ DIA +++W+ ++EE + ++ KFP
Sbjct: 122 KEEVCEMLKVLDNELKDKKFFVGDKFGFADIAANLVAFWLGVFEEASGVVLVTSEKFPNF 181
Query: 185 TAWMTNFLSHPVIKDTLPPRDKMIDYFHGR 214
W +++ IK++LPPRD+++ ++ R
Sbjct: 182 CKWRGEYINCSQIKESLPPRDELLAFYRSR 211
>gb|AAF14025.1| putative glutathione transferase [Arabidopsis thaliana]
gi|21554248|gb|AAM63323.1| putative glutathione
transferase [Arabidopsis thaliana]
gi|15232628|ref|NP_187538.1| glutathione S-transferase,
putative [Arabidopsis thaliana]
Length = 224
Score = 214 bits (545), Expect = 1e-54
Identities = 98/207 (47%), Positives = 147/207 (70%), Gaps = 2/207 (0%)
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIF-NKSSLLLELNPVHKKIPVLVHGH 62
+ VKLL W SPFSKRVE LKLKG+ YEY+EED++ N+S +LL+ NP+HKK+PVL+H
Sbjct: 5 EHVKLLGLWGSPFSKRVEMVLKLKGIPYEYIEEDVYGNRSPMLLKYNPIHKKVPVLIHNG 64
Query: 63 KTIAESFIIIEYIDETWKQ-YPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQ 121
++IAES +I+EYI++TWK + +LP DPYERA+ARFWA + ++K++ V A E+
Sbjct: 65 RSIAESLVIVEYIEDTWKTTHTILPQDPYERAMARFWAKYVDEKVMLAVKKACWGPESER 124
Query: 122 EKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
EK + EA E ++ +E+ + K FFGGE IG++DIA +I YW+ I++E + I+ +F
Sbjct: 125 EKEVKEAYEGLKCLEKELGDKLFFGGETIGFVDIAADFIGYWLGIFQEASGVTIMTAEEF 184
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMI 208
P W +F+ + IK+ LPP++K++
Sbjct: 185 PKLQRWSEDFVGNNFIKEVLPPKEKLV 211
>dbj|BAD91094.1| glutathione S-transferase GST 18 [Populus alba x Populus tremula
var. glandulosa] gi|61287176|dbj|BAD91093.1| glutathione
S-transferase GST 18 [Populus alba x Populus tremula
var. glandulosa]
Length = 219
Score = 212 bits (539), Expect = 7e-54
Identities = 106/217 (48%), Positives = 146/217 (66%), Gaps = 6/217 (2%)
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
DVKL FW SPF RV WALKLKGVE+E++EED+ NKS LLL+ NPV+KKIPVLVHG K
Sbjct: 3 DVKLHGFWASPFCYRVIWALKLKGVEFEHIEEDLTNKSELLLKYNPVYKKIPVLVHGGKP 62
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANF--TEQKLLETVFVAMCITGDEQE 122
IAES +I+EYI+ETW + PLLP DPYERA+ARFW + T+ L ++ A +G+E E
Sbjct: 63 IAESLVILEYIEETWPENPLLPTDPYERAMARFWIQYGVTKGAALVALYRA---SGEELE 119
Query: 123 KALNEAREAMEKIEEV-IEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
KA+ E E + +EE + K+FFGG++I +DI+ G + W EE + +++P
Sbjct: 120 KAVKEVVEVLRVLEEQGLGDKKFFGGDSINLVDISYGLFTCWFAAMEEAMGVKVLEPSTL 179
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDL 218
P AW NF+ P+IK+ +P DK++ + G ++ L
Sbjct: 180 PRLHAWAQNFIEVPLIKENIPDNDKLLLFMKGFREKL 216
>gb|AAG34808.1| glutathione S-transferase GST 18 [Glycine max]
Length = 220
Score = 211 bits (537), Expect = 1e-53
Identities = 107/217 (49%), Positives = 147/217 (67%), Gaps = 6/217 (2%)
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
DVK+L FW SPF RV WALKLK + YEY+E D FNKS LLL+ NPV+KK+PVL+HG K
Sbjct: 1 DVKVLGFWSSPFVHRVIWALKLKNISYEYIEVDRFNKSELLLQSNPVYKKVPVLIHGGKA 60
Query: 65 IAESFIIIEYIDETW-KQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEK 123
IAES +I+EYI+ETW + +PLLP D ++RALARFW F E + + + + DEQE+
Sbjct: 61 IAESLVILEYIEETWPENHPLLPKDNHQRALARFWIKFGEDSIASITDLFLGPSKDEQER 120
Query: 124 ALNEAREAMEKI----EEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPL 179
A + ++A E I E+ + K+FFGG NIG +DIA G +S+W+ EE+ + +I+P
Sbjct: 121 A-SAKKKAEETIMVMEEQGLGDKKFFGGNNIGMVDIAHGCLSHWLEGLEEIVGMKLIEPN 179
Query: 180 KFPATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKK 216
KFP AW NF PVIK+ LP +K++ + R++
Sbjct: 180 KFPRLHAWTQNFKQVPVIKENLPDYEKLLIHLEWRRQ 216
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 429,568,566
Number of Sequences: 2540612
Number of extensions: 18294570
Number of successful extensions: 49305
Number of sequences better than 10.0: 1056
Number of HSP's better than 10.0 without gapping: 605
Number of HSP's successfully gapped in prelim test: 451
Number of HSP's that attempted gapping in prelim test: 47878
Number of HSP's gapped (non-prelim): 1075
length of query: 228
length of database: 863,360,394
effective HSP length: 124
effective length of query: 104
effective length of database: 548,324,506
effective search space: 57025748624
effective search space used: 57025748624
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0162b.4


