

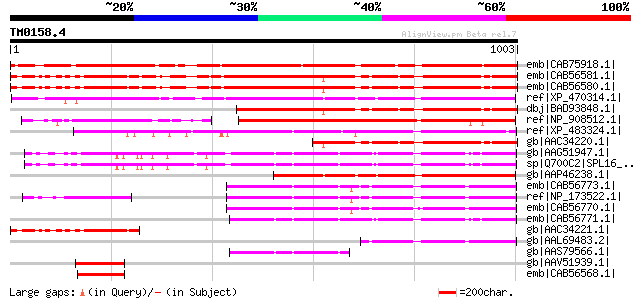
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0158.4
(1003 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB75918.1| squamosa promoter binding protein-like 12 [Arabi... 980 0.0
emb|CAB56581.1| squamosa promoter binding protein-like 1 [Arabid... 979 0.0
emb|CAB56580.1| squamosa promoter binding protein-like 1 [Arabid... 977 0.0
ref|XP_470314.1| putative SBP-domain protein [Oryza sativa (japo... 756 0.0
dbj|BAD93848.1| squamosa promoter binding protein-like 1 [Arabid... 637 0.0
ref|NP_908512.1| unnamed protein product [Oryza sativa (japonica... 458 e-127
ref|XP_483324.1| putative SPL1-Related2 protein [Oryza sativa (j... 449 e-124
gb|AAC34220.1| unknown protein [Arabidopsis thaliana] 428 e-118
gb|AAG51947.1| unknown protein; 70902-74753 [Arabidopsis thaliana] 382 e-104
sp|Q700C2|SPL16_ARATH Squamosa-promoter binding-like protein 16 ... 381 e-104
gb|AAP46238.1| unknown protein, 5'-partial [Oryza sativa (japoni... 369 e-100
emb|CAB56773.1| Spl1-Related2 protein [Arabidopsis thaliana] 301 7e-80
ref|NP_173522.1| SPL1-Related2 protein (SPL1R2) [Arabidopsis tha... 301 7e-80
emb|CAB56770.1| SPL1-Related2 protein [Arabidopsis thaliana] 300 1e-79
emb|CAB56771.1| SPL1-Related3 protein [Arabidopsis thaliana] 281 8e-74
gb|AAC34221.1| putative squamosa-promoter binding protein [Arabi... 248 9e-64
gb|AAL69483.2| putative SPL1-related protein [Arabidopsis thaliana] 171 9e-41
gb|AAS79566.1| At1g76580 [Arabidopsis thaliana] gi|46367500|emb|... 138 8e-31
gb|AAV51939.1| SBP transcription factor [Gossypium hirsutum] 131 1e-28
emb|CAB56568.1| squamosa promoter binding protein-homologue 3 [A... 128 1e-27
>emb|CAB75918.1| squamosa promoter binding protein-like 12 [Arabidopsis thaliana]
gi|6006403|emb|CAB56769.1| squamosa promoter binding
protein-like 12 [Arabidopsis thaliana]
gi|6006395|emb|CAB56768.1| squamosa promoter binding
protein-like 12 [Arabidopsis thaliana]
gi|28392866|gb|AAO41870.1| putative squamosa promoter
binding protein 12 [Arabidopsis thaliana]
gi|67461584|sp|Q9S7P5|SPL12_ARATH Squamosa-promoter
binding-like protein 12 gi|15232241|ref|NP_191562.1|
squamosa promoter-binding protein-like 12 (SPL12)
[Arabidopsis thaliana]
Length = 927
Score = 980 bits (2534), Expect = 0.0
Identities = 552/1022 (54%), Positives = 703/1022 (68%), Gaps = 114/1022 (11%)
Query: 1 MEARFGAEAYHYYGVGASSDMRGMGKRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQQL 60
MEAR E G S + GKRS EWDLN+W+W+GDLF+A+++N
Sbjct: 1 MEARIEGEVE-----GHSLEYGFSGKRSVEWDLNDWKWNGDLFVATQLNH---------- 45
Query: 61 FPLVSGIPASNSSSSCSEEVDLRDPMGSR---EGERKRR---VIVLEDDGLNEEGG-TLS 113
+SNSSS+CS+E ++ R E ++KRR V+ +E+D L ++ L+
Sbjct: 46 -------GSSNSSSTCSDEGNVEIMERRRIEMEKKKKRRAVTVVAMEEDNLKDDDAHRLT 98
Query: 114 LKLGGHAADREVASWDGGNGKKSRVAGGGASNRAVC-QVEDCCADLSRAKDYHRRHKVCE 172
L LGG+ + GNG K GGG +RA+C QV++C ADLS+ KDYHRRHKVCE
Sbjct: 99 LNLGGNNIE--------GNGVKKTKLGGGIPSRAICCQVDNCGADLSKVKDYHRRHKVCE 150
Query: 173 MHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNGN 232
+HSKAT ALVG MQRFCQQCSRFH+L+EFDEGKRSCRRRLAGHNKRRRK N + + NG
Sbjct: 151 IHSKATTALVGGIMQRFCQQCSRFHVLEEFDEGKRSCRRRLAGHNKRRRKANPDTIGNGT 210
Query: 233 PLNDDQTSSYLLISLLKILSNMNSDRSDQTTDQDLLTHLLRSLASRNGEQGGNNLSNILR 292
++DDQTS+Y+LI+LLKILSN++S++SDQT DQDLL+HLL+SL S+ GE G NL
Sbjct: 211 SMSDDQTSNYMLITLLKILSNIHSNQSDQTGDQDLLSHLLKSLVSQAGEHIGRNL----- 265
Query: 293 EPENLLREGGSSRESEMVPTL--LSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATD 350
LL+ GG + S+ + L L + Q DI+ H S+++ + V+A+ A+
Sbjct: 266 --VGLLQGGGGLQASQNIGNLSALLSLEQAPREDIKHH---SVSETPWQEVYANSAQE-- 318
Query: 351 QQLMSYIKPSISNTPPAYAEARDSTAGQIKMNNFDLNDIYIDSDDGIEDLERLPGTTNHV 410
A D + Q+K+N+FDLNDIYIDSDD + P TN
Sbjct: 319 ------------------RVAPDRSEKQVKVNDFDLNDIYIDSDDTTDIERSSPPPTNPA 360
Query: 411 TSSLDYPWTQQDSHQSSPPQTS-RNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDF 469
TSSLDY QDS QSSPPQTS RNS+S S SPSSS+G+AQ RTDRIV KLFGKEP+DF
Sbjct: 361 TSSLDY---HQDSRQSSPPQTSRRNSDSASDQSPSSSSGDAQSRTDRIVFKLFGKEPNDF 417
Query: 470 PLILRAQILDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSD 529
P+ LR QIL+WL+H+PTD+ESYIRPGCIVLTIYLRQ EA WEE+C DL+ SL RLLD+SD
Sbjct: 418 PVALRGQILNWLAHTPTDMESYIRPGCIVLTIYLRQDEASWEELCCDLSFSLRRLLDLSD 477
Query: 530 DTFWRTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVK 589
D W GW+++RVQ+Q+AF FNGQVV+DTSLP RS++YS+I+TV P+A +K+ QF+VK
Sbjct: 478 DPLWTDGWLYLRVQNQLAFAFNGQVVLDTSLPLRSHDYSQIITVRPLA--VTKKAQFTVK 535
Query: 590 GVNMIRPATRLMCALEGKYLVCEDAHESMDQYSE--ELDGLHCIQFSCAVPVTNGRGFIE 647
G+N+ RP TRL+C +EG +LV E M++ + E + + + FSC +P+ +GRGF+E
Sbjct: 536 GINLRRPGTRLLCTVEGTHLVQEATQGGMEERDDLKENNEIDFVNFSCEMPIASGRGFME 595
Query: 648 IEDQ-GLSSSFFPFIVAE-EDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIHE 705
IEDQ GLSSSFFPFIV+E ED+CS+I LE LE + TD + QAMDFIHE
Sbjct: 596 IEDQGGLSSSFFPFIVSEDEDICSEIRRLESTLEFTGTD----------SAMQAMDFIHE 645
Query: 706 IGWLLHGSQLKSRMVHLS-SSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLL-EGTVS 763
IGWLLH S+LKSR+ + DLF L RFK+L+EFSMD +WC V+KKLLN+L EGTV
Sbjct: 646 IGWLLHRSELKSRLAASDHNPEDLFSLIRFKFLIEFSMDREWCCVMKKLLNILFEEGTVD 705
Query: 764 TGDHPSLDLALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFL 823
PS D ALSE+ LLHRAVR+NSK +VE+LL + P+ + + + L
Sbjct: 706 ----PSPDAALSELCLLHRAVRKNSKPMVEMLLRFSPKK-------------KNQTLAGL 748
Query: 824 FRPDAAGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLR 883
FRPDAAGP GLTPLHIAAGKDGSEDVLDALT DP M GI+AWKN+RD+TG TPEDYARLR
Sbjct: 749 FRPDAAGPGGLTPLHIAAGKDGSEDVLDALTEDPGMTGIQAWKNSRDNTGFTPEDYARLR 808
Query: 884 GHYTYIHLVQKKINKRQGA-PHVVVEIPTNLT-ENDTNQKQNESSTTFEIAKTRDQGHCK 941
GH++YIHLVQ+K++++ A HVVV IP + E+ ++ S++ EI + CK
Sbjct: 809 GHFSYIHLVQRKLSRKPIAKEHVVVNIPESFNIEHKQEKRSPMDSSSLEITQI---NQCK 865
Query: 942 LCDIKLSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFG 1001
LCD K T +S+ Y+PAMLSMVA+AAVCVCVALLFKS PEVLY+F+PF WE L++G
Sbjct: 866 LCDHKRVFVTTHHKSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLEYG 925
Query: 1002 TS 1003
TS
Sbjct: 926 TS 927
>emb|CAB56581.1| squamosa promoter binding protein-like 1 [Arabidopsis thaliana]
gi|3650274|emb|CAA09698.1| squamosa-promoter binding
protein-like 1 [Arabidopsis thaliana]
gi|67461551|sp|Q9SMX9|SPL1_ARATH Squamosa-promoter
binding-like protein 1 gi|30690591|ref|NP_850468.1|
squamosa promoter-binding protein-like 1 (SPL1)
[Arabidopsis thaliana]
Length = 881
Score = 979 bits (2530), Expect = 0.0
Identities = 547/1013 (53%), Positives = 682/1013 (66%), Gaps = 142/1013 (14%)
Query: 1 MEARF--GAEAYHYYGVGASSDMRGMGKRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQ 58
MEAR G EA +YG +GKRS EWDLN+W+WDGDLF+A++ G+
Sbjct: 1 MEARIDEGGEAQQFYG--------SVGKRSVEWDLNDWKWDGDLFLATQTTR------GR 46
Query: 59 QLFPLVSGIPASNSSSSCSEEVDLRDPMGSREGERKRRVIVLEDDGLNEEGGTLSLKLGG 118
Q FPL + +SNSSSSCS+E + ++KRR + ++ D G L+L L G
Sbjct: 47 QFFPLGN---SSNSSSSCSDEGN----------DKKRRAVAIQGD----TNGALTLNLNG 89
Query: 119 HAADREVASWDGGNGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKAT 178
+ DG K +G AVCQVE+C ADLS+ KDYHRRHKVCEMHSKAT
Sbjct: 90 ES--------DGLFPAKKTKSG------AVCQVENCEADLSKVKDYHRRHKVCEMHSKAT 135
Query: 179 RALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNGNPLNDDQ 238
A VG +QRFCQQCSRFH+LQEFDEGKRSCRRRLAGHNKRRRKTN E NGNP +DD
Sbjct: 136 SATVGGILQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGNP-SDDH 194
Query: 239 TSSYLLISLLKILSNMNSDRSDQTTDQDLLTHLLRSLASRNGEQGGNNLSNILREPENLL 298
+S+YLLI+LLKILSNM++ T DQDL++HLL+SL S GEQ G NL LL
Sbjct: 195 SSNYLLITLLKILSNMHN----HTGDQDLMSHLLKSLVSHAGEQLGKNLVE-------LL 243
Query: 299 REGGSSRESEMVPTLLSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIK 358
+GG GSQGS +I + + + QE + AR
Sbjct: 244 LQGG--------------GSQGS-LNIGNSALLGIEQAPQEELKQFSARQDG-------- 280
Query: 359 PSISNTPPAYAEARDSTAGQIKMNNFDLNDIYIDSDDGIEDLERLPGTTNHVTSSLDYP- 417
+ + Q+KMN+FDLNDIYIDSDD D+ER P TN TSSLDYP
Sbjct: 281 ----------TATENRSEKQVKMNDFDLNDIYIDSDD--TDVERSPPPTNPATSSLDYPS 328
Query: 418 WTQQDSHQSSPPQTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQI 477
W HQSSPPQTSRNS+S S SPSSS+ +AQ RT RIV KLFGKEP++FP++LR QI
Sbjct: 329 WI----HQSSPPQTSRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEPNEFPIVLRGQI 384
Query: 478 LDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGW 537
LDWLSHSPTD+ESYIRPGCIVLTIYLRQAE WEE+ DL SLG+LLD+SDD W TGW
Sbjct: 385 LDWLSHSPTDMESYIRPGCIVLTIYLRQAETAWEELSDDLGFSLGKLLDLSDDPLWTTGW 444
Query: 538 VHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPA 597
+++RVQ+Q+AF++NGQVV+DTSL +S +YS I++V P+A A+++ QF+VKG+N+ +
Sbjct: 445 IYVRVQNQLAFVYNGQVVVDTSLSLKSRDYSHIISVKPLAIAATEKAQFTVKGMNLRQRG 504
Query: 598 TRLMCALEGKYLVCEDAHESM----DQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGL 653
TRL+C++EGKYL+ E H+S D + + + + C+ FSC +P+ +GRGF+EIEDQGL
Sbjct: 505 TRLLCSVEGKYLIQETTHDSTTREDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGL 564
Query: 654 SSSFFPFIVAE-EDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHG 712
SSSFFPF+V E +DVCS+I LE LE + TD + QAMDFIHEIGWLLH
Sbjct: 565 SSSFFPFLVVEDDDVCSEIRILETTLEFTGTD----------SAKQAMDFIHEIGWLLH- 613
Query: 713 SQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGDHPSLDL 772
+S++ + +FPL RF+WL+EFSMD +WCAV++KLLN+ +G V S +
Sbjct: 614 ---RSKLGESDPNPGVFPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAVGEFSSSS-NA 669
Query: 773 ALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPA 832
LSE+ LLHRAVR+NSK +VE+LL Y+P+ ++++ LFRPDAAGPA
Sbjct: 670 TLSELCLLHRAVRKNSKPMVEMLLRYIPK----------------QQRNSLFRPDAAGPA 713
Query: 833 GLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLV 892
GLTPLHIAAGKDGSEDVLDALT DP MVGIEAWK RDSTG TPEDYARLRGH++YIHL+
Sbjct: 714 GLTPLHIAAGKDGSEDVLDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLI 773
Query: 893 QKKINKRQGA-PHVVVEIPTNLTENDTNQ-KQNESSTTFEIAKTRDQGHCKLCDIKLSCR 950
Q+KINK+ HVVV IP + ++ + + K ++ EI Q CKLCD KL
Sbjct: 774 QRKINKKSTTEDHVVVNIPVSFSDREQKEPKSGPMASALEIT----QIPCKLCDHKLVYG 829
Query: 951 TAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGTS 1003
T RS+ Y+PAMLSMVA+AAVCVCVALLFKS PEVLY+F+PF WE LD+GTS
Sbjct: 830 T-TRRSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGTS 881
>emb|CAB56580.1| squamosa promoter binding protein-like 1 [Arabidopsis thaliana]
Length = 881
Score = 977 bits (2526), Expect = 0.0
Identities = 547/1013 (53%), Positives = 681/1013 (66%), Gaps = 142/1013 (14%)
Query: 1 MEARF--GAEAYHYYGVGASSDMRGMGKRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQ 58
MEAR G EA +YG +GKRS EWDLN+W+WDGDLF+A++ G+
Sbjct: 1 MEARIDEGGEAQQFYG--------SVGKRSVEWDLNDWKWDGDLFLATQTTR------GR 46
Query: 59 QLFPLVSGIPASNSSSSCSEEVDLRDPMGSREGERKRRVIVLEDDGLNEEGGTLSLKLGG 118
Q FPL + +SNSSSSCS+E + ++KRR + ++ D G L+L L G
Sbjct: 47 QFFPLGN---SSNSSSSCSDEGN----------DKKRRAVAIQGD----TNGALTLNLNG 89
Query: 119 HAADREVASWDGGNGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKAT 178
+ DG K +G AVCQVE+C ADLS+ KDYHRRHKVCEMHSKAT
Sbjct: 90 ES--------DGLFPAKKTKSG------AVCQVENCEADLSKVKDYHRRHKVCEMHSKAT 135
Query: 179 RALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNGNPLNDDQ 238
A V +QRFCQQCSRFH+LQEFDEGKRSCRRRLAGHNKRRRKTN E NGNP +DD
Sbjct: 136 SATVEGILQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGNP-SDDH 194
Query: 239 TSSYLLISLLKILSNMNSDRSDQTTDQDLLTHLLRSLASRNGEQGGNNLSNILREPENLL 298
+S+YLLI+LLKILSNM++ T DQDL++HLL+SL S GEQ G NL LL
Sbjct: 195 SSNYLLITLLKILSNMHN----HTGDQDLMSHLLKSLVSHAGEQLGKNLVE-------LL 243
Query: 299 REGGSSRESEMVPTLLSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIK 358
+GG GSQGS +I + + + QE + AR
Sbjct: 244 LQGG--------------GSQGS-LNIGNSALLGIEQAPQEELKQFSARQDG-------- 280
Query: 359 PSISNTPPAYAEARDSTAGQIKMNNFDLNDIYIDSDDGIEDLERLPGTTNHVTSSLDYP- 417
+ + Q+KMN+FDLNDIYIDSDD D+ER P TN TSSLDYP
Sbjct: 281 ----------TATENRSEKQVKMNDFDLNDIYIDSDD--TDVERSPPPTNPATSSLDYPS 328
Query: 418 WTQQDSHQSSPPQTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQI 477
W HQSSPPQTSRNS+S S SPSSS+ +AQ RT RIV KLFGKEP++FP++LR QI
Sbjct: 329 WI----HQSSPPQTSRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEPNEFPIVLRGQI 384
Query: 478 LDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGW 537
LDWLSHSPTD+ESYIRPGCIVLTIYLRQAE WEE+ DL SLG+LLD+SDD W TGW
Sbjct: 385 LDWLSHSPTDMESYIRPGCIVLTIYLRQAETAWEELSDDLGFSLGKLLDLSDDPLWTTGW 444
Query: 538 VHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPA 597
+++RVQ+Q+AF++NGQVV+DTSL +S +YS I++V P+A A+++ QF+VKG+N+ R
Sbjct: 445 IYVRVQNQLAFVYNGQVVVDTSLSLKSRDYSHIISVKPLAIAATEKAQFTVKGMNLRRRG 504
Query: 598 TRLMCALEGKYLVCEDAHESM----DQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGL 653
TRL+C++EGKYL+ E H+S D + + + + C+ FSC +P+ +GRGF+EIEDQGL
Sbjct: 505 TRLLCSVEGKYLIQETTHDSTTREDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGL 564
Query: 654 SSSFFPFIVAE-EDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHG 712
SSSFFPF+V E +DVCS+I LE LE + TD + QAMDFIHEIGWLLH
Sbjct: 565 SSSFFPFLVVEDDDVCSEIRILETTLEFTGTD----------SAKQAMDFIHEIGWLLH- 613
Query: 713 SQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGDHPSLDL 772
+S++ + +FPL RF+WL+EFSMD +WCAV++KLLN+ +G V S +
Sbjct: 614 ---RSKLGESDPNPGVFPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAVGEFSSSS-NA 669
Query: 773 ALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPA 832
LSE+ LLHRAVR+NSK +VE+LL Y+P+ ++++ LFRPDAAGPA
Sbjct: 670 TLSELCLLHRAVRKNSKPMVEMLLRYIPK----------------QQRNSLFRPDAAGPA 713
Query: 833 GLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLV 892
GLTPLHIAAGKDGSEDVLDALT DP MVGIEAWK RDSTG TPEDYARLRGH++YIHL+
Sbjct: 714 GLTPLHIAAGKDGSEDVLDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLI 773
Query: 893 QKKINKRQGA-PHVVVEIPTNLTENDTNQ-KQNESSTTFEIAKTRDQGHCKLCDIKLSCR 950
Q+KINK+ HVVV IP + ++ + + K ++ EI Q CKLCD KL
Sbjct: 774 QRKINKKSTTEDHVVVNIPVSFSDREQKEPKSGPMASALEIT----QIPCKLCDHKLVYG 829
Query: 951 TAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGTS 1003
T RS+ Y+PAMLSMVA+AAVCVCVALLFKS PEVLY+F+PF WE LD+GTS
Sbjct: 830 T-TRRSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGTS 881
>ref|XP_470314.1| putative SBP-domain protein [Oryza sativa (japonica cultivar-group)]
gi|40714694|gb|AAR88600.1| putative SBP-domain protein
[Oryza sativa (japonica cultivar-group)]
Length = 969
Score = 756 bits (1951), Expect = 0.0
Identities = 441/1021 (43%), Positives = 610/1021 (59%), Gaps = 79/1021 (7%)
Query: 3 ARFGAEAYHYYGVGASS-DMRGMGKRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQQLF 61
AR GA++ H YG G DM KR WDLN+WRWD D F+A+ V + G+
Sbjct: 4 ARVGAQSRHLYGGGLGEPDMDRRDKRLFGWDLNDWRWDSDRFVATPVPAAEASGL----- 58
Query: 62 PLVSGIPASNSSSSCSEEV---DLRDPMGSREGERKRRVIVLEDDGLNEE------GGTL 112
A NSS S SEE +R+ + ++++RV+V++DD + ++ GG+L
Sbjct: 59 -------ALNSSPSSSEEAGAASVRNVNARGDSDKRKRVVVIDDDDVEDDELVENGGGSL 111
Query: 113 SLKLGGHAADREVASWDGG-----NGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRR 167
SL++GG A G NGKK RV GG S A CQVE C ADL+ +DYHRR
Sbjct: 112 SLRIGGDAVAHGAGVGGGADEEDRNGKKIRVQGGSPSGPA-CQVEGCTADLTGVRDYHRR 170
Query: 168 HKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEA 227
HKVCEMH+KAT A+VGN +QRFCQQCSRFH LQEFDEGKRSCRRRLAGHN+RRRKT E
Sbjct: 171 HKVCEMHAKATTAVVGNTVQRFCQQCSRFHPLQEFDEGKRSCRRRLAGHNRRRRKTRPEV 230
Query: 228 VPNGNPLNDDQTSSYLLISLLKILSNMNSDRSDQTTDQDLLTHLLRSLASRNGEQGGNNL 287
G+ +D+ SSYLL+ LL + +N+N+D ++ Q+L++ LLR+L + L
Sbjct: 231 AVGGSAFTEDKISSYLLLGLLGVCANLNADNAEHLRGQELISGLLRNLGAVAKSLDPKEL 290
Query: 288 SNILREPENLLREGGSSRESEMVPTLLSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDAR 347
+L ++ +++G ++ SE L++ T + + S +KM
Sbjct: 291 CKLLEACQS-MQDGSNAGTSETANALVN-------TAVAEAAGPSNSKMPFVNGDQCGLA 342
Query: 348 ATDQQLMSYIKPSISN-TPPAYAEARDSTAGQIKMNNFDLNDIYIDSDDGIED-LERLPG 405
++ + P+++ PPA K +FDLND Y +G ED E P
Sbjct: 343 SSSVVPVQSKSPTVATPDPPA-----------CKFKDFDLNDTY-GGMEGFEDGYEGSPT 390
Query: 406 TTNHVTSSLDYP-WTQQDSHQSSPPQTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGK 464
T S + P W QDS Q SPPQTS NS+S SA S SSS G+AQCRTD+IV KLF K
Sbjct: 391 PAFKTTDSPNCPSWMHQDSTQ-SPPQTSGNSDSTSAQSLSSSNGDAQCRTDKIVFKLFEK 449
Query: 465 EPSDFPLILRAQILDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRL 524
PSD P +LR+QIL WLS SPTDIESYIRPGCI+LT+YLR E+ W+E+ +++S L +L
Sbjct: 450 VPSDLPPVLRSQILGWLSSSPTDIESYIRPGCIILTVYLRLVESAWKELSDNMSSYLDKL 509
Query: 525 LDVSDDTFWRTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRV 584
L+ S FW +G V + V+HQ+AF+ NGQ+++D L +++Y KIL V PIAAP S +V
Sbjct: 510 LNSSTGNFWASGLVFVMVRHQIAFMHNGQLMLDRPLANSAHHYCKILCVRPIAAPFSTKV 569
Query: 585 QFSVKGVNMIRPATRLMCALEGKYLVCEDAHESMDQYSEELDGLHCIQFSCAVPVTNGRG 644
F V+G+N++ ++RL+C+ EG + ED +D E D + + F C +P + GRG
Sbjct: 570 NFRVEGLNLVSDSSRLICSFEGSCIFQEDTDNIVDDV--EHDDIEYLNFCCPLPSSRGRG 627
Query: 645 FIEIEDQGLSSSFFPFIVAEEDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIH 704
F+E+ED G S+ FFPFI+AE+D+CS++C LE + E S E A++QA++F++
Sbjct: 628 FVEVEDGGFSNGFFPFIIAEQDICSEVCELESIFESSSH----EQADDDNARNQALEFLN 683
Query: 705 EIGWLLHGSQLKSRM--VHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTV 762
E+GWLLH + + S+ V L+S F + RF+ L F+M+ +WCAV K LL+ L G V
Sbjct: 684 ELGWLLHRANIISKQDKVPLAS----FNIWRFRNLGIFAMEREWCAVTKLLLDFLFTGLV 739
Query: 763 STGDHPSLDLALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSF 822
G ++ LSE LLH AVR S Q+V LL Y P ++ ++F
Sbjct: 740 DIGSQSPEEVVLSE-NLLHAAVRMKSAQMVRFLLGYKPNE-----------SLKRTAETF 787
Query: 823 LFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARL 882
LFRPDA GP+ TPLHIAA D +EDVLDALTNDP +VGI W+NARD G TPEDYAR
Sbjct: 788 LFRPDAQGPSKFTPLHIAAATDDAEDVLDALTNDPGLVGINTWRNARDGAGFTPEDYARQ 847
Query: 883 RGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTENDTN-QKQNESSTTFEIAKTRDQGHCK 941
RG+ Y+++V+KKINK G HVV+ +P+++ T+ K E S + C
Sbjct: 848 RGNDAYLNMVEKKINKHLGKGHVVLGVPSSIHPVITDGVKPGEVSLEIGMTVPPPAPSCN 907
Query: 942 LCDIK-LSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDF 1000
C + L + R+ +Y+PAML+++ +A +CVCV LL + P+V Y F WE L+
Sbjct: 908 ACSRQALMYPNSTARTFLYRPAMLTVMGIAVICVCVGLLLHTCPKV-YAAPTFRWELLER 966
Query: 1001 G 1001
G
Sbjct: 967 G 967
>dbj|BAD93848.1| squamosa promoter binding protein-like 1 [Arabidopsis thaliana]
Length = 528
Score = 637 bits (1642), Expect = 0.0
Identities = 325/561 (57%), Positives = 414/561 (72%), Gaps = 43/561 (7%)
Query: 450 AQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIESYIRPGCIVLTIYLRQAEAV 509
+Q RT RIV KLFGKEP++FP++LR QILDWLSHSPTD+ESYIRPGCIVLTIYLRQAE
Sbjct: 4 SQMRTGRIVFKLFGKEPNEFPIVLRGQILDWLSHSPTDMESYIRPGCIVLTIYLRQAETA 63
Query: 510 WEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSK 569
WEE+ DL SLG+LLD+SDD W TGW+++RVQ+Q+AF++NGQVV+DTSL +S +YS
Sbjct: 64 WEELSDDLGFSLGKLLDLSDDPLWTTGWIYVRVQNQLAFVYNGQVVVDTSLSLKSRDYSH 123
Query: 570 ILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYLVCEDAHESM----DQYSEEL 625
I++V P+A A+++ QF+VKG+N+ + TRL+C++EGKYL+ E H+S D + +
Sbjct: 124 IISVKPLAIAATEKAQFTVKGMNLRQRGTRLLCSVEGKYLIQETTHDSTTREDDDFKDNS 183
Query: 626 DGLHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIVAE-EDVCSDICGLEPLLELSETD 684
+ + C+ FSC +P+ +GRGF+EIEDQGLSSSFFPF+V E +DVCS+I LE LE + TD
Sbjct: 184 EIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFLVVEDDDVCSEIRILETTLEFTGTD 243
Query: 685 PDIEGTGKIKAKSQAMDFIHEIGWLLHGSQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDH 744
+ QAMDFIHEIGWLLH +S++ + +FPL RF+WL+EFSMD
Sbjct: 244 ----------SAKQAMDFIHEIGWLLH----RSKLGESDPNPGVFPLIRFQWLIEFSMDR 289
Query: 745 DWCAVVKKLLNLLLEGTVSTGDHPSLDLALSEMGLLHRAVRRNSKQLVELLLTYVPENIS 804
+WCAV++KLLN+ +G V S + LSE+ LLHRAVR+NSK +VE+LL Y+P+
Sbjct: 290 EWCAVIRKLLNMFFDGAVGEFSSSS-NATLSELCLLHRAVRKNSKPMVEMLLRYIPK--- 345
Query: 805 NEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEA 864
++++ LFRPDAAGPAGLTPLHIAAGKDGSEDVLDALT DP MVGIEA
Sbjct: 346 -------------QQRNSLFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTEDPAMVGIEA 392
Query: 865 WKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGA-PHVVVEIPTNLTENDTNQ-KQ 922
WK RDSTG TPEDYARLRGH++YIHL+Q+KINK+ HVVV IP + ++ + + K
Sbjct: 393 WKTCRDSTGFTPEDYARLRGHFSYIHLIQRKINKKSTTEDHVVVNIPVSFSDREQKEPKS 452
Query: 923 NESSTTFEIAKTRDQGHCKLCDIKLSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKS 982
++ EI Q CKLCD KL T RS+ Y+PAMLSMVA+AAVCVCVALLFKS
Sbjct: 453 GPMASALEIT----QIPCKLCDHKLVYGT-TRRSVAYRPAMLSMVAIAAVCVCVALLFKS 507
Query: 983 SPEVLYMFRPFSWESLDFGTS 1003
PEVLY+F+PF WE LD+GTS
Sbjct: 508 CPEVLYVFQPFRWELLDYGTS 528
>ref|NP_908512.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|8468036|dbj|BAA96636.1| putative squamosa promoter
binding protein-like 1 [Oryza sativa (japonica
cultivar-group)]
Length = 862
Score = 458 bits (1178), Expect = e-127
Identities = 246/560 (43%), Positives = 344/560 (60%), Gaps = 19/560 (3%)
Query: 453 RTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEE 512
RTD+IV KLFGKEP+DFP LRAQIL WLS+ P+DIESYIRPGCI+LTIY+R +W++
Sbjct: 309 RTDKIVFKLFGKEPNDFPSDLRAQILSWLSNCPSDIESYIRPGCIILTIYMRLPNWMWDK 368
Query: 513 ICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILT 572
+ D + +L+ +S DT WRTGW++ RVQ + NG +++ + N +IL
Sbjct: 369 LAADPAHWIQKLISLSTDTLWRTGWMYARVQDYLTLSCNGNLMLASPWQPAIGNKHQILF 428
Query: 573 VSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYLVCEDAHESMDQYSEELDGLHCIQ 632
++PIA S FSVKG+N+ +P T+L+C GKYL+ E + +D ++ G C+
Sbjct: 429 ITPIAVACSSTANFSVKGLNIAQPTTKLLCIFGGKYLIQEATEKLLDD-TKMQRGPQCLT 487
Query: 633 FSCAVPVTNGRGFIEIEDQGLSSSFFPFIVAEEDVCSDICGLEPLLEL-SETDPDIEGTG 691
FSC+ P T+GRGFIE+ED SS FPF+VAEEDVCS+I LE LL L S D +E
Sbjct: 488 FSCSFPSTSGRGFIEVEDLDQSSLSFPFVVAEEDVCSEIRTLEHLLNLVSFDDTLVEKND 547
Query: 692 KIKAKSQAMDFIHEIGWLLHGSQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVK 751
+ ++ +A++F+HE GW L S +++ T+ FP RF+WL+ F++D ++CAV+K
Sbjct: 548 LLASRDRALNFLHEFGWFLQRSHIRATSETPKDCTEGFPAARFRWLLSFAVDREFCAVIK 607
Query: 752 KLLNLLLEGTVSTGDHPSLDLALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEG 811
KLL+ L +G V +++ L + L+ AV + SK L++ LLTY + +
Sbjct: 608 KLLDTLFQGGVDLDVQSTVEFVLKQ-DLVFVAVNKRSKPLIDFLLTYTTSSAPMD----- 661
Query: 812 KALVEGEKQSFLFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDS 871
FLF PD AGP+ +TPLHIAA + VLDALT+DP +GI+AWKNARD+
Sbjct: 662 -GTESAAPAQFLFTPDIAGPSDITPLHIAATYSDTAGVLDALTDDPQQLGIKAWKNARDA 720
Query: 872 TGSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEI-----PTNLTENDTNQKQNESS 926
TG TPEDYAR RGH +YI +VQ KI+ R HV V I T+ TE +Q +
Sbjct: 721 TGLTPEDYARKRGHESYIEMVQNKIDSRLPKAHVSVTISSTTSTTDFTEKHASQSKTTDQ 780
Query: 927 TTFEIAK-----TRDQGHCKLCDIKLSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFK 981
T F++ K T+ C+ C +L+ R + R L +PA+LS+VA+AAVCVCV L+ +
Sbjct: 781 TAFDVEKGQQISTKPPLSCRQCLPELAYRHHLNRFLSTRPAVLSLVAIAAVCVCVGLIMQ 840
Query: 982 SSPEVLYMFRPFSWESLDFG 1001
P + M PF W SL G
Sbjct: 841 GPPHIGGMRGPFRWNSLRSG 860
Score = 174 bits (442), Expect = 1e-41
Identities = 124/386 (32%), Positives = 187/386 (48%), Gaps = 87/386 (22%)
Query: 23 GMGKRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQQLFPLVSGIPASNSSSSCSEEVDL 82
G+ K+ EWDLN+WRWD +LF+A+ P++ S S CS
Sbjct: 4 GLKKKGLEWDLNDWRWDSNLFLAT---------------------PSNASPSKCS----- 37
Query: 83 RDPMGSREGE-------RKRRVIVLEDDGLNEEGGTLSLKLGGHAADREVASWDGGNGKK 135
R +G EGE ++RRV +DDG EE + G D +++ G + +
Sbjct: 38 RRELGRAEGEIDFGVVDKRRRVSPEDDDG--EECINAATTNGD---DGQISGQRGRSSED 92
Query: 136 SRVAGGGASNRA-VCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCS 194
G S+ CQV+ C +LS A+DY++RHKVCE+H+K+ + N RFCQQCS
Sbjct: 93 EMPRQGTCSSSGPCCQVDGCTVNLSSARDYNKRHKVCEVHTKSGVVRIKNVEHRFCQQCS 152
Query: 195 RFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNGNPLNDDQTSSYLLISLLKILSNM 254
RFH LQEFDEGK+SCR RLA HN+RRRK +A + N L+++ + S L+ LLK LS +
Sbjct: 153 RFHFLQEFDEGKKSCRSRLAQHNRRRRKVQVQAGVDVNSLHENHSLSNTLLLLLKQLSGL 212
Query: 255 NSD-RSDQTTDQDLLTHLLRSLASRNGEQGGNNLSNILREPENLLREGGSSRESEMVPTL 313
+S S+Q + LT+L+++LA+ G Q +++L+ S+ +
Sbjct: 213 DSSGPSEQINGPNYLTNLVKNLAALAGTQ----------RNQDMLKNANSAAIASHTGNY 262
Query: 314 LSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIKPSISNTPPAYAEARD 373
++ G+ S D R H V +E P++
Sbjct: 263 VAKGN--SLHDSRPHIPVGTESTAEE-------------------PTVER---------- 291
Query: 374 STAGQIKMNNFDLNDIYIDSDDGIED 399
++ NFDLND Y++ D+ D
Sbjct: 292 ------RVQNFDLNDAYVEGDENRTD 311
>ref|XP_483324.1| putative SPL1-Related2 protein [Oryza sativa (japonica
cultivar-group)] gi|42408812|dbj|BAD10073.1| putative
SPL1-Related2 protein [Oryza sativa (japonica
cultivar-group)]
Length = 1140
Score = 449 bits (1156), Expect = e-124
Identities = 315/997 (31%), Positives = 478/997 (47%), Gaps = 141/997 (14%)
Query: 126 ASWDGGNGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNA 185
AS GG G +GGG + +CQV+DC ADL+ AKDYHRRHKVCE+H K T+ALVGN
Sbjct: 164 ASGGGGGGGGGGNSGGGGGSYPMCQVDDCRADLTNAKDYHRRHKVCEIHGKTTKALVGNQ 223
Query: 186 MQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNG--------NPLNDD 237
MQRFCQQCSRFH L EFDEGKRSCRRRLAGHN+RRRKT V + N N
Sbjct: 224 MQRFCQQCSRFHPLSEFDEGKRSCRRRLAGHNRRRRKTQPTDVASQLLLPGNQENAANRT 283
Query: 238 QTSSYLLI-----------------------SLLKILSNMNSDRSDQTTDQDLLTHLLRS 274
Q L+ +L++I+S +NS + + + + +
Sbjct: 284 QDIVNLITVIARLQGSNVGKLPSIPPIPDKDNLVQIISKINSINNGNSASKSPPSEAVDL 343
Query: 275 LASRNGEQ--------GGNNLSNILREPENLLREGGSSRESEMVPTLL------------ 314
AS + +Q G +N L + N + + + VP+ +
Sbjct: 344 NASHSQQQDSVQRTTNGFEKQTNGLDKQTNGFDKQADGFDKQAVPSTMDLLAVLSTALAT 403
Query: 315 ------SNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDAR-----------ATDQQLMSYI 357
++ SQGS +++ S + VV++H+ A ++ Y
Sbjct: 404 SNPDSNTSQSQGSSDSSGNNKSKSQSTEPANVVNSHEKSIRVFSATRKNDALERSPEMYK 463
Query: 358 KPSISNTPPAYAEARDSTAGQIKMNNFDLNDIYIDSDDGIEDLERLPGTTNHVTSSL--- 414
+P TPP + + + D + Y+ S+ ER P ++ VT
Sbjct: 464 QPD-QETPPYLSLRLFGSTEEDVPCKMDTANKYLSSESSNPLDERSPSSSPPVTHKFFPI 522
Query: 415 ----------DY-------------PW----------TQQDSHQSSPP----QTSRNSES 437
DY W +++ SPP Q+ S S
Sbjct: 523 RSVDEDARIADYGEDIATVEVSTSRAWRAPPLELFKDSERPIENGSPPNPAYQSCYTSTS 582
Query: 438 GSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIESYIRPGCI 497
S HSPS+S + Q RT RI+ KLFGKEPS P LR +I++WL HSP ++E YIRPGC+
Sbjct: 583 CSDHSPSTSNSDGQDRTGRIIFKLFGKEPSTIPGNLRGEIVNWLKHSPNEMEGYIRPGCL 642
Query: 498 VLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFIFNGQVVID 557
VL++YL W+E+ +L + L+ SD FWR G +R Q+ +G +
Sbjct: 643 VLSMYLSMPAIAWDELEENLLQRVNTLVQGSDLDFWRKGRFLVRTDAQLVSYKDGATRLS 702
Query: 558 TSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYLVCEDAHES 617
S +R+ N ++ VSPIA ++ +KG N+ P T++ C GKY+ E +
Sbjct: 703 KS--WRTWNTPELTFVSPIAVVGGRKTSLILKGRNLTIPGTQIHCTSTGKYISKEVLCSA 760
Query: 618 MDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIVAEEDVCSDICGLEPL 677
+ G+ + GR FIE+E++ +S FP I+A VC ++ LE
Sbjct: 761 YPGTIYDDSGVETFDLPGEPHLILGRYFIEVENRFRGNS-FPVIIANSSVCQELRSLEAE 819
Query: 678 LELSE-----TDPDIEGTGKIKAKSQAMDFIHEIGWLLH-GSQLKSRMVHLSSSTDL--F 729
LE S+ +D ++K K + + F++E+GWL + S SS DL F
Sbjct: 820 LEGSQFVDGSSDDQAHDARRLKPKDEVLHFLNELGWLFQKAAASTSAEKSDSSGLDLMYF 879
Query: 730 PLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGDHPSLDL-ALSEMGLLHRAVRRNS 788
RF++L+ FS + DWC++ K LL +L + ++++ + L LSE+ LL+RAV+R S
Sbjct: 880 STARFRYLLLFSSERDWCSLTKTLLEILAKRSLASDELSQETLEMLSEIHLLNRAVKRKS 939
Query: 789 KQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAGKDGSED 848
+ LL+ +V + + + + F P+ AGP GLTPLH+AA + + D
Sbjct: 940 SHMARLLVQFV-------------VVCPDDSKLYPFLPNVAGPGGLTPLHLAASIEDAVD 986
Query: 849 VLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVE 908
++DALT+DP +G+ W +A D G +PE YA+LR + Y LV +K+ R+ ++
Sbjct: 987 IVDALTDDPQQIGLSCWHSALDDDGQSPETYAKLRNNNAYNELVAQKLVDRKNNQVTIMV 1046
Query: 909 IPTNLTENDTNQKQNESSTTFEIAKTRDQGHCKLCDIKLSCRTAVGRSLVYKPAMLSMVA 968
+ + + ++ + + + R C + D L R R L+ +P + SM+A
Sbjct: 1047 GKEEIHMDQSGNVGEKNKSAIQALQIRSCNQCAILDAGLLRRPMHSRGLLARPYIHSMLA 1106
Query: 969 VAAVCVCVALLFKSSPEVLYMF---RPFSWESLDFGT 1002
+AAVCVCV + ++ L F R F WE LDFGT
Sbjct: 1107 IAAVCVCVCVFMRA----LLRFNSGRSFKWERLDFGT 1139
>gb|AAC34220.1| unknown protein [Arabidopsis thaliana]
Length = 383
Score = 428 bits (1101), Expect = e-118
Identities = 229/412 (55%), Positives = 289/412 (69%), Gaps = 43/412 (10%)
Query: 599 RLMCALEGKYLVCEDAHESM----DQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLS 654
RL+C++EGKYL+ E H+S D + + + + C+ FSC +P+ +GRGF+EIEDQGLS
Sbjct: 8 RLLCSVEGKYLIQETTHDSTTREDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGLS 67
Query: 655 SSFFPFIVAEED-VCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHGS 713
SSFFPF+V E+D VCS+I LE LE + TD + QAMDFIHEIGWLLH
Sbjct: 68 SSFFPFLVVEDDDVCSEIRILETTLEFTGTD----------SAKQAMDFIHEIGWLLH-- 115
Query: 714 QLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGDHPSLDLA 773
+S++ + +FPL RF+WL+EFSMD +WCAV++KLLN+ +G V S +
Sbjct: 116 --RSKLGESDPNPGVFPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAVGEFSSSS-NAT 172
Query: 774 LSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAG 833
LSE+ LLHRAVR+NSK +VE+LL Y+P+ ++++ LFRPDAAGPAG
Sbjct: 173 LSELCLLHRAVRKNSKPMVEMLLRYIPK----------------QQRNSLFRPDAAGPAG 216
Query: 834 LTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQ 893
LTPLHIAAGKDGSEDVLDALT DP MVGIEAWK RDSTG TPEDYARLRGH++YIHL+Q
Sbjct: 217 LTPLHIAAGKDGSEDVLDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLIQ 276
Query: 894 KKINKRQGA-PHVVVEIPTNLTENDTNQ-KQNESSTTFEIAKTRDQGHCKLCDIKLSCRT 951
+KINK+ HVVV IP + ++ + + K ++ EI Q CKLCD KL T
Sbjct: 277 RKINKKSTTEDHVVVNIPVSFSDREQKEPKSGPMASALEIT----QIPCKLCDHKLVYGT 332
Query: 952 AVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGTS 1003
RS+ Y+PAMLSMVA+AAVCVCVALLFKS PEVLY+F+PF WE LD+GTS
Sbjct: 333 -TRRSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGTS 383
>gb|AAG51947.1| unknown protein; 70902-74753 [Arabidopsis thaliana]
Length = 1020
Score = 382 bits (980), Expect = e-104
Identities = 318/1074 (29%), Positives = 480/1074 (44%), Gaps = 195/1074 (18%)
Query: 30 EWDLNNWRWDGDLFIASRVNPVQEDGVGQQLFPLVSGIPASNSSSSCSEEVDLRDPMGSR 89
+W +N W+WDG F A ++ G QL S + +DL P G
Sbjct: 40 DWQMNRWKWDGQRFEA-----IELQGESLQL--------------SNKKGLDLNLPCGFN 80
Query: 90 EGERKRRVIVLEDDGLNEEGGTLSLKLGGHAADREVASWDGGNGKKSRVAGGGASNRAVC 149
+ EG + L ++V S G+G GGG N C
Sbjct: 81 D----------------VEGTPVDLT----RPSKKVRSGSPGSG------GGGGGNYPKC 114
Query: 150 QVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSC 209
QV++C DLS AKDYHRRHKVCE+HSKAT+ALVG MQRFCQQCSRFH+L EFDEGKRSC
Sbjct: 115 QVDNCKEDLSIAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSC 174
Query: 210 ---------RRR-------------LAGHNKRRRKTN----------------NEAVPNG 231
RRR L + TN NEA NG
Sbjct: 175 RRRLDGHNRRRRKTQPDAITSQVVALENRDNTSNNTNMDVMALLTALVCAQGRNEATTNG 234
Query: 232 NPLNDDQTSSYLLISLLKIL------------------------SNMNSDRS----DQTT 263
+P + +++ +K L S MN S +
Sbjct: 235 SPGVPQREQLLQILNKIKALPLPMNLTSKLNNIGILARKNPEQPSPMNPQNSMNGASSPS 294
Query: 264 DQDLLTHLLRSLASRNGE------QGG--NNLSNILREPENLLREGGSSRESEMVP---- 311
DLL L SL S E QGG N SN + + +S E + +
Sbjct: 295 TMDLLAALSASLGSSAPEAIAFLSQGGFGNKESNDRTKLTSSDHSATTSLEKKTLEFPSF 354
Query: 312 -----TLLSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIKPSISNTPP 366
T +N S +D R T S +Q + + S S+ P
Sbjct: 355 GGGERTSSTNHSPSQYSDSRGQDTRSSLSLQLFTSSPEEESRPKVASSTKYYSSASSNP- 413
Query: 367 AYAEARDSTAGQIKMNNFDLN------------DIYIDSDDGIEDLERLPGTTNHVTSSL 414
E R ++ + F L+ D LE + T++
Sbjct: 414 --VEDRSPSSSPVMQELFPLHTSPETRRYNNYKDTSTSPRTSCLPLELFGASNRGATANP 471
Query: 415 DYPWTQQDSHQSSPPQTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILR 474
+Y + S +S SGS +SP S AQ RT +I KLF K+PS P LR
Sbjct: 472 NYNVLRHQSGYAS---------SGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLPNTLR 522
Query: 475 AQILDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWR 534
+I WLS P+D+ES+IRPGC++L++Y+ + + WE++ +L + L V D FW
Sbjct: 523 TEIFRWLSSFPSDMESFIRPGCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDSEFWS 580
Query: 535 TGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMI 594
+ Q+A +G++ + S +R+ N +++TVSP+A A + V+G N+
Sbjct: 581 NSRFLVNAGRQLASHKHGEIRLSKS--WRTLNLPELITVSPLAVVAGEETALIVRGRNLT 638
Query: 595 RPATRLMCALEGKYLVCE-DAHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGL 653
RL CA G Y E E +EL+ + Q A V+ GR FIE+E+ GL
Sbjct: 639 NDGMRLRCAHMGNYASMEVTGREHRLTKVDELN-VSSFQVQSASSVSLGRCFIELEN-GL 696
Query: 654 SSSFFPFIVAEEDVCSDICGLEPLLELSET-DPDIEGTGKIKAKSQAMDFIHEIGWLLHG 712
FP I+A +C ++ LE + + I+ + +++ + + F++E+GWL
Sbjct: 697 RGDNFPLIIANATICKELNRLEEEFHPKDVIEEQIQNLDRPRSREEVLCFLNELGWLFQR 756
Query: 713 SQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLE---GTVSTGDHPS 769
+H F L RFK+L+ S++ D+C++++ +L++++E G + S
Sbjct: 757 KWTSD--IHGEPD---FSLPRFKFLLVCSVERDYCSLIRTVLDMMVERNLGKDGLLNKES 811
Query: 770 LDLALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAA 829
LD+ L+++ LL+RA++R + ++ E L+ Y V ++F+F P A
Sbjct: 812 LDM-LADIQLLNRAIKRRNTKMAETLIHY---------------SVNPSTRNFIFLPSIA 855
Query: 830 GPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYI 889
GP +TPLH+AA S+D++DALTNDP +G+ W D+TG TP YA +R +++Y
Sbjct: 856 GPGDITPLHLAASTSSSDDMIDALTNDPQEIGLSCWNTLVDATGQTPFSYAAMRDNHSYN 915
Query: 890 HLVQKKI-NKRQGAPHVVVEIPTNLTENDTNQKQNESSTTFEIAKTRDQGHCKLCDIKLS 948
LV +K+ +KR G +I N+ EN +Q + E+ R C +K
Sbjct: 916 TLVARKLADKRNG------QISLNI-ENGIDQIGLSKRLSSEL--KRSCNTCASVALKYQ 966
Query: 949 CRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGT 1002
+ + R L P + SM+AVA VCVCV + + P ++ FSW LD+G+
Sbjct: 967 RKVSGSRRLFPTPIIHSMLAVATVCVCVCVFMHAFP-MVRQGSHFSWGGLDYGS 1019
>sp|Q700C2|SPL16_ARATH Squamosa-promoter binding-like protein 16 (SPL1-related protein 3)
Length = 988
Score = 381 bits (979), Expect = e-104
Identities = 318/1074 (29%), Positives = 480/1074 (44%), Gaps = 195/1074 (18%)
Query: 30 EWDLNNWRWDGDLFIASRVNPVQEDGVGQQLFPLVSGIPASNSSSSCSEEVDLRDPMGSR 89
+W +N W+WDG F A ++ G QL S + +DL P G
Sbjct: 8 DWQMNRWKWDGQRFEA-----IELQGESLQL--------------SNKKGLDLNLPCGFN 48
Query: 90 EGERKRRVIVLEDDGLNEEGGTLSLKLGGHAADREVASWDGGNGKKSRVAGGGASNRAVC 149
+ EG + L ++V S G+G GGG N C
Sbjct: 49 D----------------VEGTPVDLT----RPSKKVRSGSPGSG------GGGGGNYPKC 82
Query: 150 QVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEGKRSC 209
QV++C DLS AKDYHRRHKVCE+HSKAT+ALVG MQRFCQQCSRFH+L EFDEGKRSC
Sbjct: 83 QVDNCKEDLSIAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSC 142
Query: 210 ---------RRR-------------LAGHNKRRRKTN----------------NEAVPNG 231
RRR L + TN NEA NG
Sbjct: 143 RRRLDGHNRRRRKTQPDAITSQVVALENRDNTSNNTNMDVMALLTALVCAQGRNEATTNG 202
Query: 232 NPLNDDQTSSYLLISLLKIL------------------------SNMNSDRS----DQTT 263
+P + +++ +K L S MN S +
Sbjct: 203 SPGVPQREQLLQILNKIKALPLPMNLTSKLNNIGILARKNPEQPSPMNPQNSMNGASSPS 262
Query: 264 DQDLLTHLLRSLASRNGE------QGG--NNLSNILREPENLLREGGSSRESEMVP---- 311
DLL L SL S E QGG N SN + + +S E + +
Sbjct: 263 TMDLLAALSASLGSSAPEAIAFLSQGGFGNKESNDRTKLTSSDHSATTSLEKKTLEFPSF 322
Query: 312 -----TLLSNGSQGSPTDIRQHQTVSMNKMQQEVVHAHDARATDQQLMSYIKPSISNTPP 366
T +N S +D R T S +Q + + S S+ P
Sbjct: 323 GGGERTSSTNHSPSQYSDSRGQDTRSSLSLQLFTSSPEEESRPKVASSTKYYSSASSNP- 381
Query: 367 AYAEARDSTAGQIKMNNFDLN------------DIYIDSDDGIEDLERLPGTTNHVTSSL 414
E R ++ + F L+ D LE + T++
Sbjct: 382 --VEDRSPSSSPVMQELFPLHTSPETRRYNNYKDTSTSPRTSCLPLELFGASNRGATANP 439
Query: 415 DYPWTQQDSHQSSPPQTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILR 474
+Y + S +S SGS +SP S AQ RT +I KLF K+PS P LR
Sbjct: 440 NYNVLRHQSGYAS---------SGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLPNTLR 490
Query: 475 AQILDWLSHSPTDIESYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWR 534
+I WLS P+D+ES+IRPGC++L++Y+ + + WE++ +L + L V D FW
Sbjct: 491 TEIFRWLSSFPSDMESFIRPGCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDSEFWS 548
Query: 535 TGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMI 594
+ Q+A +G++ + S +R+ N +++TVSP+A A + V+G N+
Sbjct: 549 NSRFLVNAGRQLASHKHGRIRLSKS--WRTLNLPELITVSPLAVVAGEETALIVRGRNLT 606
Query: 595 RPATRLMCALEGKYLVCE-DAHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGL 653
RL CA G Y E E +EL+ + Q A V+ GR FIE+E+ GL
Sbjct: 607 NDGMRLRCAHMGNYASMEVTGREHRLTKVDELN-VSSFQVQSASSVSLGRCFIELEN-GL 664
Query: 654 SSSFFPFIVAEEDVCSDICGLEPLLELSET-DPDIEGTGKIKAKSQAMDFIHEIGWLLHG 712
FP I+A +C ++ LE + + I+ + +++ + + F++E+GWL
Sbjct: 665 RGDNFPLIIANATICKELNRLEEEFHPKDVIEEQIQNLDRPRSREEVLCFLNELGWLFQR 724
Query: 713 SQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLE---GTVSTGDHPS 769
+H F L RFK+L+ S++ D+C++++ +L++++E G + S
Sbjct: 725 KWTSD--IHGEPD---FSLPRFKFLLVCSVERDYCSLIRTVLDMMVERNLGKDGLLNKES 779
Query: 770 LDLALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAA 829
LD+ L+++ LL+RA++R + ++ E L+ Y V ++F+F P A
Sbjct: 780 LDM-LADIQLLNRAIKRRNTKMAETLIHY---------------SVNPSTRNFIFLPSIA 823
Query: 830 GPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYI 889
GP +TPLH+AA S+D++DALTNDP +G+ W D+TG TP YA +R +++Y
Sbjct: 824 GPGDITPLHLAASTSSSDDMIDALTNDPQEIGLSCWNTLVDATGQTPFSYAAMRDNHSYN 883
Query: 890 HLVQKKI-NKRQGAPHVVVEIPTNLTENDTNQKQNESSTTFEIAKTRDQGHCKLCDIKLS 948
LV +K+ +KR G +I N+ EN +Q + E+ R C +K
Sbjct: 884 TLVARKLADKRNG------QISLNI-ENGIDQIGLSKRLSSEL--KRSCNTCASVALKYQ 934
Query: 949 CRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGT 1002
+ + R L P + SM+AVA VCVCV + + P ++ FSW LD+G+
Sbjct: 935 RKVSGSRRLFPTPIIHSMLAVATVCVCVCVFMHAFP-MVRQGSHFSWGGLDYGS 987
>gb|AAP46238.1| unknown protein, 5'-partial [Oryza sativa (japonica cultivar-group)]
gi|50919619|ref|XP_470170.1| unknown protein, 5'-partial
[Oryza sativa (japonica cultivar-group)]
Length = 462
Score = 369 bits (947), Expect = e-100
Identities = 201/481 (41%), Positives = 293/481 (60%), Gaps = 23/481 (4%)
Query: 523 RLLDVSDDTFWRTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKILTVSPIAAPASK 582
+LL+ S FW +G V + V+HQ+AF+ NGQ+++D L +++Y KIL V PIAAP S
Sbjct: 1 KLLNSSTGNFWASGLVFVMVRHQIAFMHNGQLMLDRPLANSAHHYCKILCVRPIAAPFST 60
Query: 583 RVQFSVKGVNMIRPATRLMCALEGKYLVCEDAHESMDQYSEELDGLHCIQFSCAVPVTNG 642
+V F V+G+N++ ++RL+C+ EG + ED +D E D + + F C +P + G
Sbjct: 61 KVNFRVEGLNLVSDSSRLICSFEGSCIFQEDTDNIVDDV--EHDDIEYLNFCCPLPSSRG 118
Query: 643 RGFIEIEDQGLSSSFFPFIVAEEDVCSDICGLEPLLELSETDPDIEGTGKIKAKSQAMDF 702
RGF+E+ED G S+ FFPFI+AE+D+CS++C LE + E S E A++QA++F
Sbjct: 119 RGFVEVEDGGFSNGFFPFIIAEQDICSEVCELESIFESSSH----EQADDDNARNQALEF 174
Query: 703 IHEIGWLLHGSQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTV 762
++E+GWLLH + + S+ + ++ F + RF+ L F+M+ +WCAV K LL+ L G V
Sbjct: 175 LNELGWLLHRANIISKQDKVPLAS--FNIWRFRNLGIFAMEREWCAVTKLLLDFLFTGLV 232
Query: 763 STGDHPSLDLALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSF 822
G ++ LSE LLH AVR S Q+V LL Y P ++ ++F
Sbjct: 233 DIGSQSPEEVVLSE-NLLHAAVRMKSAQMVRFLLGYKPNES-----------LKRTAETF 280
Query: 823 LFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARL 882
LFRPDA GP+ TPLHIAA D +EDVLDALTNDP +VGI W+NARD G TPEDYAR
Sbjct: 281 LFRPDAQGPSKFTPLHIAAATDDAEDVLDALTNDPGLVGINTWRNARDGAGFTPEDYARQ 340
Query: 883 RGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTENDTN-QKQNESSTTFEIAKTRDQGHCK 941
RG+ Y+++V+KKINK G HVV+ +P+++ T+ K E S + C
Sbjct: 341 RGNDAYLNMVEKKINKHLGKGHVVLGVPSSIHPVITDGVKPGEVSLEIGMTVPPPAPSCN 400
Query: 942 LCDIK-LSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDF 1000
C + L + R+ +Y+PAML+++ +A +CVCV LL + P+V Y F WE L+
Sbjct: 401 ACSRQALMYPNSTARTFLYRPAMLTVMGIAVICVCVGLLLHTCPKV-YAAPTFRWELLER 459
Query: 1001 G 1001
G
Sbjct: 460 G 460
>emb|CAB56773.1| Spl1-Related2 protein [Arabidopsis thaliana]
Length = 712
Score = 301 bits (771), Expect = 7e-80
Identities = 189/581 (32%), Positives = 307/581 (52%), Gaps = 35/581 (6%)
Query: 430 QTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIE 489
Q S + SGS +SP S +AQ RT +IV KL K+PS P LR++I +WLS+ P+++E
Sbjct: 158 QQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQLPGTLRSEIYNWLSNIPSEME 217
Query: 490 SYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFI 549
SYIRPGC+VL++Y+ + A WE++ L LG LL S FWR + Q+A
Sbjct: 218 SYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNSPSDFWRNARFIVNTGRQLASH 277
Query: 550 FNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYL 609
NG+V S +R+ N ++++VSP+A A + V+G ++ + C G Y+
Sbjct: 278 KNGKVRCSKS--WRTWNSPELISVSPVAVVAGEETSLVVRGRSLTNDGISIRCTHMGSYM 335
Query: 610 VCEDAHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIVAEEDVCS 669
E Q + ++ + P GR FIE+E+ G FP I+A +C
Sbjct: 336 AMEVTRAVCRQTIFDELNVNSFKVQNVHPGFLGRCFIEVEN-GFRGDSFPLIIANASICK 394
Query: 670 DICGL-----EPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHGSQLKSRMVHLSS 724
++ L +++E G ++ + + F++E+GWL K++ L
Sbjct: 395 ELNRLGEEFHPKSQDMTEEQAQSSNRGP-TSREEVLCFLNELGWLFQ----KNQTSELRE 449
Query: 725 STDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGD--HPSLDLALSEMGLLHR 782
+D F L RFK+L+ S++ D+CA+++ LL++L+E + + +LD+ L+E+ LL+R
Sbjct: 450 QSD-FSLARFKFLLVCSVERDYCALIRTLLDMLVERNLVNDELNREALDM-LAEIQLLNR 507
Query: 783 AVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAG 842
AV+R S ++VELL+ Y+ V P L + F+F P+ GP G+TPLH+AA
Sbjct: 508 AVKRKSTKMVELLIHYL-------VNP----LTLSSSRKFVFLPNITGPGGITPLHLAAC 556
Query: 843 KDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGA 902
GS+D++D LTNDP +G+ +W RD+TG TP YA +R ++ Y LV +K+ ++
Sbjct: 557 TSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAAIRNNHNYNSLVARKLADKRNK 616
Query: 903 PHVVVEIPTNLTENDTNQKQNESSTTFEIAKTRDQ-GHCKLCDIKLSCRTAVGRSLVYKP 961
++ N+ +Q + E+ K+ C +K R + + L P
Sbjct: 617 -----QVSLNIEHEVVDQTGLSKRLSLEMNKSSSSCASCATVALKYQRRVSGSQRLFPTP 671
Query: 962 AMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGT 1002
+ SM+AVA VCVCV + + P ++ FSW LD+G+
Sbjct: 672 IIHSMLAVATVCVCVCVFMHAFP-IVRQGSHFSWGGLDYGS 711
>ref|NP_173522.1| SPL1-Related2 protein (SPL1R2) [Arabidopsis thaliana]
gi|67461574|sp|Q8RY95|SPL14_ARATH Squamosa-promoter
binding-like protein 14 (SPL1-related protein 2)
gi|4836890|gb|AAD30593.1| Unknown protein [Arabidopsis
thaliana]
Length = 1035
Score = 301 bits (771), Expect = 7e-80
Identities = 189/581 (32%), Positives = 307/581 (52%), Gaps = 35/581 (6%)
Query: 430 QTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIE 489
Q S + SGS +SP S +AQ RT +IV KL K+PS P LR++I +WLS+ P+++E
Sbjct: 481 QQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQLPGTLRSEIYNWLSNIPSEME 540
Query: 490 SYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFI 549
SYIRPGC+VL++Y+ + A WE++ L LG LL S FWR + Q+A
Sbjct: 541 SYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNSPSDFWRNARFIVNTGRQLASH 600
Query: 550 FNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYL 609
NG+V S +R+ N ++++VSP+A A + V+G ++ + C G Y+
Sbjct: 601 KNGKVRCSKS--WRTWNSPELISVSPVAVVAGEETSLVVRGRSLTNDGISIRCTHMGSYM 658
Query: 610 VCEDAHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIVAEEDVCS 669
E Q + ++ + P GR FIE+E+ G FP I+A +C
Sbjct: 659 AMEVTRAVCRQTIFDELNVNSFKVQNVHPGFLGRCFIEVEN-GFRGDSFPLIIANASICK 717
Query: 670 DICGL-----EPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHGSQLKSRMVHLSS 724
++ L +++E G ++ + + F++E+GWL K++ L
Sbjct: 718 ELNRLGEEFHPKSQDMTEEQAQSSNRGP-TSREEVLCFLNELGWLFQ----KNQTSELRE 772
Query: 725 STDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGD--HPSLDLALSEMGLLHR 782
+D F L RFK+L+ S++ D+CA+++ LL++L+E + + +LD+ L+E+ LL+R
Sbjct: 773 QSD-FSLARFKFLLVCSVERDYCALIRTLLDMLVERNLVNDELNREALDM-LAEIQLLNR 830
Query: 783 AVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAG 842
AV+R S ++VELL+ Y+ V P L + F+F P+ GP G+TPLH+AA
Sbjct: 831 AVKRKSTKMVELLIHYL-------VNP----LTLSSSRKFVFLPNITGPGGITPLHLAAC 879
Query: 843 KDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGA 902
GS+D++D LTNDP +G+ +W RD+TG TP YA +R ++ Y LV +K+ ++
Sbjct: 880 TSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAAIRNNHNYNSLVARKLADKRNK 939
Query: 903 PHVVVEIPTNLTENDTNQKQNESSTTFEIAKTRDQ-GHCKLCDIKLSCRTAVGRSLVYKP 961
++ N+ +Q + E+ K+ C +K R + + L P
Sbjct: 940 -----QVSLNIEHEVVDQTGLSKRLSLEMNKSSSSCASCATVALKYQRRVSGSQRLFPTP 994
Query: 962 AMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGT 1002
+ SM+AVA VCVCV + + P ++ FSW LD+G+
Sbjct: 995 IIHSMLAVATVCVCVCVFMHAFP-IVRQGSHFSWGGLDYGS 1034
Score = 152 bits (384), Expect = 5e-35
Identities = 95/219 (43%), Positives = 114/219 (51%), Gaps = 44/219 (20%)
Query: 26 KRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQQLFPLVSGIPASNSSSSCSEEVDLRDP 85
+R EW+ W WD F A V+ V Q F L
Sbjct: 38 QRRDEWNSKMWDWDSRRFEAKPVD------VEVQEFDLT--------------------- 70
Query: 86 MGSREGERKRRVIVLEDDGLNEEGGTLSLKLGGHAADREVASWDGGNGKKSRVAGGGASN 145
+ +R GE E G L+L G A + + K +G N
Sbjct: 71 LRNRSGE--------------ERGLDLNLGSGLTAVEETTTTTQNVRPNKKVRSGSPGGN 116
Query: 146 RAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQCSRFHMLQEFDEG 205
+CQV++C DLS AKDYHRRHKVCE+HSKAT+ALVG MQRFCQQCSRFH+L EFDEG
Sbjct: 117 YPMCQVDNCTEDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEG 176
Query: 206 KRSCRRRLAGHNKRRRKTNN-EAVPNG--NPLNDDQTSS 241
KRSCRRRLAGHN+RRRKT E V +G P N D T++
Sbjct: 177 KRSCRRRLAGHNRRRRKTTQPEEVASGVVVPGNHDTTNN 215
>emb|CAB56770.1| SPL1-Related2 protein [Arabidopsis thaliana]
Length = 812
Score = 300 bits (769), Expect = 1e-79
Identities = 188/581 (32%), Positives = 307/581 (52%), Gaps = 35/581 (6%)
Query: 430 QTSRNSESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIE 489
Q S + SGS +SP S +AQ RT +IV KL K+PS P LR++I +WLS+ P+++E
Sbjct: 258 QQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQLPGTLRSEIYNWLSNIPSEME 317
Query: 490 SYIRPGCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFI 549
SYIRPGC+VL++Y+ + A WE++ L LG LL S FWR + Q+A
Sbjct: 318 SYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNSPSDFWRNARFIVNTGRQLASH 377
Query: 550 FNGQVVIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYL 609
NG+V S +R+ N ++++VSP+A A + V+G ++ + C G Y+
Sbjct: 378 KNGKVRCSKS--WRTWNSPELISVSPVAVVAGEETSLVVRGRSLTNDGISIRCTHMGSYM 435
Query: 610 VCEDAHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIVAEEDVCS 669
E Q + ++ + P GR FIE+E+ G FP I+A +C
Sbjct: 436 AMEVTRAVCRQTIFDELNVNSFKVQNVHPGFLGRCFIEVEN-GFRGDSFPLIIANASICK 494
Query: 670 DICGL-----EPLLELSETDPDIEGTGKIKAKSQAMDFIHEIGWLLHGSQLKSRMVHLSS 724
++ L +++E G ++ + + F++E+GWL K++ L
Sbjct: 495 ELNRLGEEFHPKSQDMTEEQAQSSNRGP-TSREEVLCFLNELGWLFQ----KNQTSELRE 549
Query: 725 STDLFPLKRFKWLMEFSMDHDWCAVVKKLLNLLLEGTVSTGD--HPSLDLALSEMGLLHR 782
+D F L RFK+L+ S++ D+CA+++ LL++L+E + + +LD+ L+E+ LL+R
Sbjct: 550 QSD-FSLARFKFLLVCSVERDYCALIRTLLDMLVERNLVNDELNREALDM-LAEIQLLNR 607
Query: 783 AVRRNSKQLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAG 842
AV+R S ++VELL+ Y+ V P + + F+F P+ GP G+TPLH+AA
Sbjct: 608 AVKRKSTKMVELLIHYL-------VNPSALS----SSRKFVFLPNITGPGGITPLHLAAC 656
Query: 843 KDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKRQGA 902
GS+D++D LTNDP +G+ +W RD+TG TP YA +R ++ Y LV +K+ ++
Sbjct: 657 TSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAAIRNNHNYNSLVARKLADKRNK 716
Query: 903 PHVVVEIPTNLTENDTNQKQNESSTTFEIAKTRDQ-GHCKLCDIKLSCRTAVGRSLVYKP 961
++ N+ +Q + E+ K+ C +K R + + L P
Sbjct: 717 -----QVSLNIEHEVVDQTGLSKRLSLEMNKSSSSCASCATVALKYQRRVSGSQRLFPTP 771
Query: 962 AMLSMVAVAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGT 1002
+ SM+AVA VCVCV + + P ++ FSW LD+G+
Sbjct: 772 IIHSMLAVATVCVCVCVFMHAFP-IVRQGSHFSWGGLDYGS 811
>emb|CAB56771.1| SPL1-Related3 protein [Arabidopsis thaliana]
Length = 647
Score = 281 bits (719), Expect = 8e-74
Identities = 185/574 (32%), Positives = 302/574 (52%), Gaps = 43/574 (7%)
Query: 435 SESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIESYIRP 494
+ SGS +SP S AQ RT +I KLF K+PS P LR +I WLS P+D+ES+IRP
Sbjct: 110 ASSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLPNTLRTEIFRWLSSFPSDMESFIRP 169
Query: 495 GCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFIFNGQV 554
GC++L++Y+ + + WE++ +L + L V D FW + Q+A +G++
Sbjct: 170 GCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDSEFWSNSRFLVNAGRQLASHKHGRI 227
Query: 555 VIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYLVCE-D 613
+ S +R+ N +++TVSP+A A + V+G N+ RL CA G Y E
Sbjct: 228 RLSKS--WRTLNLPELITVSPLAVVAGEETALIVRGRNLTNDGMRLRCAHMGNYASMEVT 285
Query: 614 AHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIVAEEDVCSDICG 673
E +EL+ + Q A V+ GR FIE+E+ GL FP I+A +C ++
Sbjct: 286 GREHRLTKVDELN-VSSFQVQSASSVSLGRCFIELEN-GLRGDNFPLIIANATICKELNR 343
Query: 674 LEPLLELSET-DPDIEGTGKIKAKSQAMDFIHEIGWLLHGSQLKSRMVHLSSSTDLFPLK 732
LE + + I+ + +++ + + F++E+GWL +H F L
Sbjct: 344 LEEEFHPKDVIEEQIQNLDRPRSREEVLCFLNELGWLFQRKWTSD--IHGEPD---FSLP 398
Query: 733 RFKWLMEFSMDHDWCAVVKKLLNLLLE---GTVSTGDHPSLDLALSEMGLLHRAVRRNSK 789
RFK+L+ S++ D+C++++ +L++++E G + SLD+ L+++ LL+RA++R +
Sbjct: 399 RFKFLLVCSVERDYCSLIRTVLDMMVERNLGKDGLLNKESLDM-LADIQLLNRAIKRRNT 457
Query: 790 QLVELLLTYVPENISNEVRPEGKALVEGEKQSFLFRPDAAGPAGLTPLHIAAGKDGSEDV 849
++ E L+ Y V ++F+F P AGP +TPLH+AA S+D+
Sbjct: 458 KMAETLIHY---------------SVNPSTRNFIFLPSIAGPGDITPLHLAASTSSSDDM 502
Query: 850 LDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKI-NKRQGAPHVVVE 908
+DALTNDP +G+ W D+TG TP YA +R +++Y LV +K+ +KR G +
Sbjct: 503 IDALTNDPQEIGLSCWNTLVDATGQTPFSYAAMRDNHSYNTLVARKLADKRNG------Q 556
Query: 909 IPTNLTENDTNQKQNESSTTFEIAKTRDQGHCKLCDIKLSCRTAVGRSLVYKPAMLSMVA 968
I N+ EN +Q + E+ R C +K + + R L P + SM+A
Sbjct: 557 ISLNI-ENGIDQIGLSKRLSSEL--KRSCNTCASVALKYQRKVSGSRRLFPTPIIHSMLA 613
Query: 969 VAAVCVCVALLFKSSPEVLYMFRPFSWESLDFGT 1002
VA VCVCV + + P ++ FSW LD+G+
Sbjct: 614 VATVCVCVCVFMHAFP-MVRQGSHFSWGGLDYGS 646
>gb|AAC34221.1| putative squamosa-promoter binding protein [Arabidopsis thaliana]
Length = 240
Score = 248 bits (632), Expect = 9e-64
Identities = 142/258 (55%), Positives = 171/258 (66%), Gaps = 48/258 (18%)
Query: 1 MEARF--GAEAYHYYGVGASSDMRGMGKRSAEWDLNNWRWDGDLFIASRVNPVQEDGVGQ 58
MEAR G EA +YG +GKRS EWDLN+W+WDGDLF+A++ G+
Sbjct: 1 MEARIDEGGEAQQFYG--------SVGKRSVEWDLNDWKWDGDLFLATQTTR------GR 46
Query: 59 QLFPLVSGIPASNSSSSCSEEVDLRDPMGSREGERKRRVIVLEDDGLNEEGGTLSLKLGG 118
Q FPL + +SNSSSSCS+E + ++KRR + ++ D G L+L L G
Sbjct: 47 QFFPLGN---SSNSSSSCSDEGN----------DKKRRAVAIQGD----TNGALTLNLNG 89
Query: 119 HAADREVASWDGGNGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKAT 178
+ DG K +G AVCQVE+C ADLS+ KDYHRRHKVCEMHSKAT
Sbjct: 90 ES--------DGLFPAKKTKSG------AVCQVENCEADLSKVKDYHRRHKVCEMHSKAT 135
Query: 179 RALVGNAMQRFCQQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEAVPNGNPLNDDQ 238
A VG +QRFCQQCSRFH+LQEFDEGKRSCRRRLAGHNKRRRKTN E NGNP +DD
Sbjct: 136 SATVGGILQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGNP-SDDH 194
Query: 239 TSSYLLISLLKILSNMNS 256
+S+YLLI+LLKILSNM+S
Sbjct: 195 SSNYLLITLLKILSNMHS 212
>gb|AAL69483.2| putative SPL1-related protein [Arabidopsis thaliana]
Length = 314
Score = 171 bits (434), Expect = 9e-41
Identities = 103/311 (33%), Positives = 173/311 (55%), Gaps = 26/311 (8%)
Query: 695 AKSQAMDFIHEIGWLLHGSQLKSRMVHLSSSTDLFPLKRFKWLMEFSMDHDWCAVVKKLL 754
++ + + F++E+GWL K++ L +D F L RFK+L+ S++ D+CA+++ LL
Sbjct: 26 SREEVLCFLNELGWLFQ----KNQTSELREQSD-FSLARFKFLLVCSVERDYCALIRTLL 80
Query: 755 NLLLEGTVSTGD--HPSLDLALSEMGLLHRAVRRNSKQLVELLLTYVPENISNEVRPEGK 812
++L+E + + +LD+ L+E+ LL+RAV+R S ++VELL+ Y+ V P
Sbjct: 81 DMLVERNLVNDELNREALDM-LAEIQLLNRAVKRKSTKMVELLIHYL-------VNP--- 129
Query: 813 ALVEGEKQSFLFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDST 872
L + F+F P+ GP G+TPLH+AA GS+D++D LTNDP +G+ +W RD+T
Sbjct: 130 -LTLSSSRKFVFLPNITGPGGITPLHLAACTSGSDDMIDLLTNDPQEIGLSSWNTLRDAT 188
Query: 873 GSTPEDYARLRGHYTYIHLVQKKINKRQGAPHVVVEIPTNLTENDTNQKQNESSTTFEIA 932
G TP YA +R ++ Y LV +K+ ++ ++ N+ +Q + E+
Sbjct: 189 GQTPYSYAAIRNNHNYNSLVARKLADKRNK-----QVSLNIEHEVVDQTGLSKRLSLEMN 243
Query: 933 KTRDQ-GHCKLCDIKLSCRTAVGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYMFR 991
K+ C +K R + + L P + SM+AVA VCVCV + + P ++
Sbjct: 244 KSSSSCASCATVALKYQRRVSGSQRLFPTPIIHSMLAVATVCVCVCVFMHAFP-IVRQGS 302
Query: 992 PFSWESLDFGT 1002
FSW LD+G+
Sbjct: 303 HFSWGGLDYGS 313
>gb|AAS79566.1| At1g76580 [Arabidopsis thaliana] gi|46367500|emb|CAG25876.1|
hypothetical protein [Arabidopsis thaliana]
Length = 503
Score = 138 bits (348), Expect = 8e-31
Identities = 84/238 (35%), Positives = 128/238 (53%), Gaps = 7/238 (2%)
Query: 435 SESGSAHSPSSSTGEAQCRTDRIVIKLFGKEPSDFPLILRAQILDWLSHSPTDIESYIRP 494
+ SGS +SP S AQ RT +I KLF K+PS P LR +I WLS P+D+ES+IRP
Sbjct: 272 ASSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLPNTLRTEIFRWLSSFPSDMESFIRP 331
Query: 495 GCIVLTIYLRQAEAVWEEICYDLTSSLGRLLDVSDDTFWRTGWVHIRVQHQMAFIFNGQV 554
GC++L++Y+ + + WE++ +L + L V D FW + Q+A +G++
Sbjct: 332 GCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDSEFWSNSRFLVNAGRQLASHKHGRI 389
Query: 555 VIDTSLPFRSNNYSKILTVSPIAAPASKRVQFSVKGVNMIRPATRLMCALEGKYLVCE-D 613
+ S +R+ N +++TVSP+A A + V+G N+ RL CA G Y E
Sbjct: 390 RLSKS--WRTLNLPELITVSPLAVVAGEETALIVRGRNLTNDGMRLRCAHMGNYASMEVT 447
Query: 614 AHESMDQYSEELDGLHCIQFSCAVPVTNGRGFIEIEDQGLSSSFFPFIVAEEDVCSDI 671
E +EL+ + Q A V+ GR FIE+E+ GL FP I+A +C ++
Sbjct: 448 GREHRLTKVDELN-VSSFQVQSASSVSLGRCFIELEN-GLRGDNFPLIIANATICKEL 503
>gb|AAV51939.1| SBP transcription factor [Gossypium hirsutum]
Length = 169
Score = 131 bits (330), Expect = 1e-28
Identities = 61/97 (62%), Positives = 71/97 (72%)
Query: 131 GNGKKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFC 190
G G++ GGG + CQVE C DLS AK YHRRHKVCE+H+KA +V QRFC
Sbjct: 24 GYGRRGAAGGGGGVSPPACQVEKCGLDLSDAKRYHRRHKVCEIHAKAPFVVVAGLRQRFC 83
Query: 191 QQCSRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEA 227
QQCSRFH L EFDE KRSCRRRLAGHN+RRRK++ E+
Sbjct: 84 QQCSRFHELPEFDEAKRSCRRRLAGHNERRRKSSAES 120
>emb|CAB56568.1| squamosa promoter binding protein-homologue 3 [Antirrhinum majus]
gi|25458128|pir||T52299 squamosa promoter binding
protein-homolog 3 [imported] - garden snapdragon
Length = 305
Score = 128 bits (321), Expect = 1e-27
Identities = 58/94 (61%), Positives = 68/94 (71%)
Query: 134 KKSRVAGGGASNRAVCQVEDCCADLSRAKDYHRRHKVCEMHSKATRALVGNAMQRFCQQC 193
++SR+ G+ N CQ E C ADLS K YHRRHKVCE HSKA + QRFCQQC
Sbjct: 133 RRSRLLEPGSINAPRCQAEGCNADLSHCKHYHRRHKVCEFHSKAANVVAAGLTQRFCQQC 192
Query: 194 SRFHMLQEFDEGKRSCRRRLAGHNKRRRKTNNEA 227
SRFH+L EFD GKRSCRRRLA HN+RRRK++ +A
Sbjct: 193 SRFHVLSEFDNGKRSCRRRLADHNRRRRKSSQQA 226
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,759,034,604
Number of Sequences: 2540612
Number of extensions: 77696142
Number of successful extensions: 210011
Number of sequences better than 10.0: 288
Number of HSP's better than 10.0 without gapping: 109
Number of HSP's successfully gapped in prelim test: 192
Number of HSP's that attempted gapping in prelim test: 209379
Number of HSP's gapped (non-prelim): 668
length of query: 1003
length of database: 863,360,394
effective HSP length: 138
effective length of query: 865
effective length of database: 512,755,938
effective search space: 443533886370
effective search space used: 443533886370
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0158.4


