

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0157b.4
(77 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
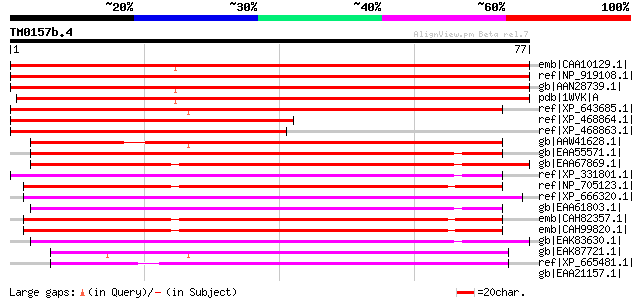
emb|CAA10129.1| hypothetical protein [Cicer arietinum] 148 3e-35
ref|NP_919108.1| unknown protein [Oryza sativa (japonica cultiva... 144 6e-34
gb|AAN28739.1| At2g23090/F21P24.15 [Arabidopsis thaliana] gi|215... 139 2e-32
pdb|1WVK|A Chain A, Nmr Solution Structure Of The Partially Diso... 137 7e-32
ref|XP_643685.1| hypothetical protein DDB0202579 [Dictyostelium ... 72 4e-12
ref|XP_468864.1| expressed protein, having alternate splicing pr... 70 1e-11
ref|XP_468863.1| expressed protein, having alternate splicing pr... 69 2e-11
gb|AAW41628.1| hypothetical protein CNB04380 [Cryptococcus neofo... 67 2e-10
gb|EAA55571.1| hypothetical protein MG01222.4 [Magnaporthe grise... 67 2e-10
gb|EAA67869.1| hypothetical protein FG00894.1 [Gibberella zeae P... 65 4e-10
ref|XP_331801.1| predicted protein [Neurospora crassa] gi|289268... 65 6e-10
ref|NP_705123.1| hypothetical protein [Plasmodium falciparum 3D7... 64 1e-09
ref|XP_666320.1| hypothetical protein Chro.60358 [Cryptosporidiu... 64 1e-09
gb|EAA61803.1| hypothetical protein AN7617.2 [Aspergillus nidula... 60 1e-08
emb|CAH82357.1| conserved hypothetical protein [Plasmodium chaba... 59 3e-08
emb|CAH99820.1| conserved hypothetical protein [Plasmodium berghei] 59 3e-08
gb|EAK83630.1| hypothetical protein UM02732.1 [Ustilago maydis 5... 58 7e-08
gb|EAK87721.1| small conserved protein [Cryptosporidium parvum] ... 54 1e-06
ref|XP_665481.1| hypothetical protein Chro.40098 [Cryptosporidiu... 52 4e-06
gb|EAA21157.1| hypothetical protein [Plasmodium yoelii yoelii] 44 0.001
>emb|CAA10129.1| hypothetical protein [Cicer arietinum]
Length = 78
Score = 148 bits (374), Expect = 3e-35
Identities = 72/78 (92%), Positives = 73/78 (93%), Gaps = 1/78 (1%)
Query: 1 MGGGNAQKAKMARERNLEKQKGA-KGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHA 59
MGGGN QKAKMARERNLEKQK A KGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHA
Sbjct: 1 MGGGNGQKAKMARERNLEKQKNAGKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHA 60
Query: 60 EAKHPKSDVVTCFPHLQK 77
EAKHPKSDV+ CFPHL K
Sbjct: 61 EAKHPKSDVLVCFPHLNK 78
>ref|NP_919108.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|22831317|dbj|BAC16171.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 77
Score = 144 bits (363), Expect = 6e-34
Identities = 64/77 (83%), Positives = 71/77 (92%)
Query: 1 MGGGNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAE 60
MGGGN QK+KMARERN+EK KGAKGSQLE NKKAM+IQCK+CMQTFICTTSE KC+EHAE
Sbjct: 1 MGGGNGQKSKMARERNMEKNKGAKGSQLEANKKAMNIQCKICMQTFICTTSETKCKEHAE 60
Query: 61 AKHPKSDVVTCFPHLQK 77
AKHPKSD+ CFPHL+K
Sbjct: 61 AKHPKSDLTACFPHLKK 77
>gb|AAN28739.1| At2g23090/F21P24.15 [Arabidopsis thaliana]
gi|21553449|gb|AAM62542.1| unknown [Arabidopsis
thaliana] gi|3169182|gb|AAC17825.1| Expressed protein
[Arabidopsis thaliana] gi|15912199|gb|AAL08233.1|
At2g23090/F21P24.15 [Arabidopsis thaliana]
gi|15450484|gb|AAK96535.1| At2g23090/F21P24.15
[Arabidopsis thaliana] gi|25412154|pir||D84620
hypothetical protein At2g23090 [imported] - Arabidopsis
thaliana gi|18400164|ref|NP_565547.1| expressed protein
[Arabidopsis thaliana]
Length = 78
Score = 139 bits (350), Expect = 2e-32
Identities = 67/78 (85%), Positives = 71/78 (90%), Gaps = 1/78 (1%)
Query: 1 MGGGNAQKAKMARERNLEKQKGA-KGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHA 59
MGGGNAQK+ MAR +NLEK K A KGSQLE NKKAMSIQCKVCMQTFICTTSEVKCREHA
Sbjct: 1 MGGGNAQKSAMARAKNLEKAKAAGKGSQLEANKKAMSIQCKVCMQTFICTTSEVKCREHA 60
Query: 60 EAKHPKSDVVTCFPHLQK 77
EAKHPK+DVV CFPHL+K
Sbjct: 61 EAKHPKADVVACFPHLKK 78
>pdb|1WVK|A Chain A, Nmr Solution Structure Of The Partially Disordered
Protein At2g23090 From Arabidopsis Thaliana
Length = 86
Score = 137 bits (345), Expect = 7e-32
Identities = 66/77 (85%), Positives = 70/77 (90%), Gaps = 1/77 (1%)
Query: 2 GGGNAQKAKMARERNLEKQKGA-KGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAE 60
GGGNAQK+ MAR +NLEK K A KGSQLE NKKAMSIQCKVCMQTFICTTSEVKCREHAE
Sbjct: 10 GGGNAQKSAMARAKNLEKAKAAGKGSQLEANKKAMSIQCKVCMQTFICTTSEVKCREHAE 69
Query: 61 AKHPKSDVVTCFPHLQK 77
AKHPK+DVV CFPHL+K
Sbjct: 70 AKHPKADVVACFPHLKK 86
>ref|XP_643685.1| hypothetical protein DDB0202579 [Dictyostelium discoideum]
gi|60471794|gb|EAL69749.1| hypothetical protein
DDB0202579 [Dictyostelium discoideum]
Length = 80
Score = 72.0 bits (175), Expect = 4e-12
Identities = 33/75 (44%), Positives = 47/75 (62%), Gaps = 2/75 (2%)
Query: 1 MGGGNAQKAKMARERNLEKQKGAKG--SQLETNKKAMSIQCKVCMQTFICTTSEVKCREH 58
M GN KA+ RERNL++ +GA SQL+TN+ A + C +C +F+CT E + R H
Sbjct: 1 MPSGNGAKAQQKRERNLKRNEGASSAKSQLKTNEAAKTTICNICRASFLCTAKETELRIH 60
Query: 59 AEAKHPKSDVVTCFP 73
++ KHPK+ CFP
Sbjct: 61 SDNKHPKNKFEECFP 75
>ref|XP_468864.1| expressed protein, having alternate splicing products [Oryza
sativa (japonica cultivar-group)]
gi|27764663|gb|AAO23088.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|40737000|gb|AAR89013.1| expressed protein, having
alternate splicing products [Oryza sativa (japonica
cultivar-group)]
Length = 42
Score = 70.5 bits (171), Expect = 1e-11
Identities = 33/42 (78%), Positives = 37/42 (87%)
Query: 1 MGGGNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVC 42
MGGGN QK++MARERN+EK KGAKGSQLETNKKAM+IQ C
Sbjct: 1 MGGGNGQKSRMARERNMEKAKGAKGSQLETNKKAMNIQVGSC 42
>ref|XP_468863.1| expressed protein, having alternate splicing products [Oryza
sativa (japonica cultivar-group)]
gi|40736999|gb|AAR89012.1| expressed protein, having
alternate splicing products [Oryza sativa (japonica
cultivar-group)]
Length = 48
Score = 69.3 bits (168), Expect = 2e-11
Identities = 33/41 (80%), Positives = 37/41 (89%)
Query: 1 MGGGNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKV 41
MGGGN QK++MARERN+EK KGAKGSQLETNKKAM+IQ V
Sbjct: 1 MGGGNGQKSRMARERNMEKAKGAKGSQLETNKKAMNIQVLV 41
>gb|AAW41628.1| hypothetical protein CNB04380 [Cryptococcus neoformans var.
neoformans JEC21] gi|50260025|gb|EAL22688.1|
hypothetical protein CNBB1370 [Cryptococcus neoformans
var. neoformans B-3501A] gi|58263050|ref|XP_568935.1|
hypothetical protein CNB04380 [Cryptococcus neoformans
var. neoformans JEC21]
Length = 77
Score = 66.6 bits (161), Expect = 2e-10
Identities = 34/74 (45%), Positives = 47/74 (62%), Gaps = 7/74 (9%)
Query: 4 GNAQKAKMARERNLEKQKGAKG----SQLETNKKAMSIQCKVCMQTFICTTSEVKCREHA 59
GN KA+ R+RN KGA G SQL++N AM+IQC C F T+ ++ ++H
Sbjct: 2 GNGAKAQAKRDRNT---KGAAGKGDQSQLKSNAAAMTIQCITCKAVFQGTSKQLVLQQHV 58
Query: 60 EAKHPKSDVVTCFP 73
++KHPKSD+ TCFP
Sbjct: 59 DSKHPKSDIKTCFP 72
>gb|EAA55571.1| hypothetical protein MG01222.4 [Magnaporthe grisea 70-15]
gi|39950524|ref|XP_363296.1| hypothetical protein
MG01222.4 [Magnaporthe grisea 70-15]
Length = 130
Score = 66.6 bits (161), Expect = 2e-10
Identities = 31/70 (44%), Positives = 44/70 (62%), Gaps = 1/70 (1%)
Query: 4 GNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAKH 63
G +A+ RERN + K A SQL+ N+ A +IQC++C TF+ TT E + +HAE KH
Sbjct: 58 GRGNRAQQKRERNAKDAKSAPKSQLKVNENAKNIQCEICKSTFLMTTREPQLLQHAENKH 117
Query: 64 PKSDVVTCFP 73
K+ + CFP
Sbjct: 118 SKT-IADCFP 126
>gb|EAA67869.1| hypothetical protein FG00894.1 [Gibberella zeae PH-1]
gi|46108024|ref|XP_381070.1| hypothetical protein
FG00894.1 [Gibberella zeae PH-1]
Length = 73
Score = 65.1 bits (157), Expect = 4e-10
Identities = 34/74 (45%), Positives = 46/74 (61%), Gaps = 2/74 (2%)
Query: 4 GNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAKH 63
GN KA+ RER + + AK SQL+ N+KA IQC++C TF+ TT +EHAE KH
Sbjct: 2 GNGAKAQQKRERAAKDKNTAK-SQLKVNEKACDIQCQICKSTFLKTTKAPALKEHAENKH 60
Query: 64 PKSDVVTCFPHLQK 77
K+ + CFP Q+
Sbjct: 61 SKT-IADCFPTYQE 73
>ref|XP_331801.1| predicted protein [Neurospora crassa] gi|28926805|gb|EAA35769.1|
predicted protein [Neurospora crassa]
Length = 80
Score = 64.7 bits (156), Expect = 6e-10
Identities = 33/73 (45%), Positives = 42/73 (57%), Gaps = 1/73 (1%)
Query: 1 MGGGNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAE 60
MGGGN KA RERN + SQL+TN AM+I C+ C TF+ T+ EHA+
Sbjct: 1 MGGGNGAKAAQKRERNAKNAAAGPKSQLKTNAAAMNIICQTCRATFLSTSRAKALDEHAQ 60
Query: 61 AKHPKSDVVTCFP 73
KH K+ + CFP
Sbjct: 61 NKHNKT-LADCFP 72
>ref|NP_705123.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23615368|emb|CAD52359.1| hypothetical protein,
conserved [Plasmodium falciparum 3D7]
Length = 78
Score = 63.5 bits (153), Expect = 1e-09
Identities = 36/71 (50%), Positives = 44/71 (61%), Gaps = 2/71 (2%)
Query: 3 GGNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAK 62
G NA K ARER + KG K SQL+ NK+A+++ CKVC F+ T S + EHAE K
Sbjct: 4 GSNACKRNQARERKNVEVKGGK-SQLKANKEALNVTCKVCYTVFMQTQSISQLAEHAENK 62
Query: 63 HPKSDVVTCFP 73
H K DV CFP
Sbjct: 63 HNK-DVKECFP 72
>ref|XP_666320.1| hypothetical protein Chro.60358 [Cryptosporidium hominis]
gi|54657292|gb|EAL36090.1| hypothetical protein
Chro.60358 [Cryptosporidium hominis]
Length = 78
Score = 63.5 bits (153), Expect = 1e-09
Identities = 29/74 (39%), Positives = 43/74 (57%)
Query: 3 GGNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAK 62
GGNA K RER + SQL+ N +AMS++C++C+Q F+ + ++H EAK
Sbjct: 4 GGNACKRNQCRERKVANGAKESKSQLKVNAEAMSLKCQICLQPFMKVQTGPLLKQHWEAK 63
Query: 63 HPKSDVVTCFPHLQ 76
HPK CFP ++
Sbjct: 64 HPKKTFQECFPGIE 77
>gb|EAA61803.1| hypothetical protein AN7617.2 [Aspergillus nidulans FGSC A4]
gi|67901260|ref|XP_680886.1| hypothetical protein
AN7617_2 [Aspergillus nidulans FGSC A4]
gi|49110613|ref|XP_411754.1| hypothetical protein
AN7617.2 [Aspergillus nidulans FGSC A4]
Length = 78
Score = 60.1 bits (144), Expect = 1e-08
Identities = 33/70 (47%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query: 4 GNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAKH 63
GN KA RERN + K SQL+TN+ A IQC VC TF+ TT EHA KH
Sbjct: 2 GNGAKAAFKRERNAKDTKSGGKSQLKTNEAAKDIQCVVCRATFLKTTRGPALTEHAANKH 61
Query: 64 PKSDVVTCFP 73
K+ + CFP
Sbjct: 62 NKT-LQDCFP 70
>emb|CAH82357.1| conserved hypothetical protein [Plasmodium chabaudi]
Length = 75
Score = 58.9 bits (141), Expect = 3e-08
Identities = 32/71 (45%), Positives = 44/71 (61%), Gaps = 2/71 (2%)
Query: 3 GGNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAK 62
G NA K ARER + KG K SQ++ N++A+++ CKVC F+ T S + EHA+ +
Sbjct: 4 GSNACKRNQARERKTVEVKGGK-SQIKANQEALNVICKVCYTAFMKTQSISQLAEHAQNR 62
Query: 63 HPKSDVVTCFP 73
H K DV CFP
Sbjct: 63 HGK-DVKECFP 72
>emb|CAH99820.1| conserved hypothetical protein [Plasmodium berghei]
Length = 75
Score = 58.9 bits (141), Expect = 3e-08
Identities = 32/71 (45%), Positives = 44/71 (61%), Gaps = 2/71 (2%)
Query: 3 GGNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAK 62
G NA K ARER + KG K SQ++ N++A+++ CKVC F+ T S + EHA+ +
Sbjct: 4 GSNACKRNQARERKTVEVKGGK-SQIKANQEALNVICKVCYTAFMKTQSISQLAEHAQNR 62
Query: 63 HPKSDVVTCFP 73
H K DV CFP
Sbjct: 63 HRK-DVKECFP 72
>gb|EAK83630.1| hypothetical protein UM02732.1 [Ustilago maydis 521]
gi|49072116|ref|XP_400347.1| hypothetical protein
UM02732.1 [Ustilago maydis 521]
Length = 74
Score = 57.8 bits (138), Expect = 7e-08
Identities = 29/74 (39%), Positives = 41/74 (55%), Gaps = 1/74 (1%)
Query: 4 GNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAKH 63
GN KA+ RERN + G SQ +TN+ A ++ C C QTF+ T E +HA +H
Sbjct: 2 GNGAKAQQKRERNAKAAGGEAKSQKKTNEAARNVMCMTCRQTFLLTVREPALIQHASDRH 61
Query: 64 PKSDVVTCFPHLQK 77
K+ + CFP+ K
Sbjct: 62 NKT-LAECFPNHAK 74
>gb|EAK87721.1| small conserved protein [Cryptosporidium parvum]
gi|66357006|ref|XP_625681.1| hypothetical protein
cgd4_790 [Cryptosporidium parvum]
Length = 82
Score = 53.5 bits (127), Expect = 1e-06
Identities = 26/80 (32%), Positives = 44/80 (54%), Gaps = 12/80 (15%)
Query: 7 QKAKMAR--ERNLEKQKGAKG----------SQLETNKKAMSIQCKVCMQTFICTTSEVK 54
++ KM+R +R++++Q+ K S +E +K +SI C +C QTF+CT +
Sbjct: 2 EEIKMSRGNQRDIDRQRAQKRQDRTAPKTGKSIIEQKEKILSIVCNICKQTFMCTANRQT 61
Query: 55 CREHAEAKHPKSDVVTCFPH 74
H + KHPK + CFP+
Sbjct: 62 LEVHVDTKHPKLEFSQCFPN 81
>ref|XP_665481.1| hypothetical protein Chro.40098 [Cryptosporidium hominis]
gi|54656190|gb|EAL35250.1| hypothetical protein
Chro.40098 [Cryptosporidium hominis]
Length = 77
Score = 52.0 bits (123), Expect = 4e-06
Identities = 24/68 (35%), Positives = 37/68 (54%), Gaps = 3/68 (4%)
Query: 7 QKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAEAKHPKS 66
Q+A+ ++R K S +E +K +SI C +C QTF+CT + H + KHPK
Sbjct: 12 QRAQKRQDRTAPK---TGKSIIEQKEKILSIVCNICKQTFMCTANRQTLEVHVDTKHPKL 68
Query: 67 DVVTCFPH 74
+ CFP+
Sbjct: 69 EFSQCFPN 76
>gb|EAA21157.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 71
Score = 43.9 bits (102), Expect = 0.001
Identities = 22/72 (30%), Positives = 34/72 (46%), Gaps = 1/72 (1%)
Query: 1 MGGGNAQKAKMARERNLEKQKGAKGSQLETNKKAMSIQCKVCMQTFICTTSEVKCREHAE 60
M GN + R +K S ++ K +++ CK+C TF+CT ++ +EH E
Sbjct: 1 MTRGNQRDVDRIRSSK-RNEKAKPNSTVKDASKNLNVICKLCRHTFMCTVNQSILKEHHE 59
Query: 61 AKHPKSDVVTCF 72
KH K CF
Sbjct: 60 KKHSKHAYEDCF 71
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.125 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 119,446,817
Number of Sequences: 2540612
Number of extensions: 3299003
Number of successful extensions: 9570
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 9539
Number of HSP's gapped (non-prelim): 37
length of query: 77
length of database: 863,360,394
effective HSP length: 53
effective length of query: 24
effective length of database: 728,707,958
effective search space: 17488990992
effective search space used: 17488990992
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0157b.4


