

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0154.7
(495 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
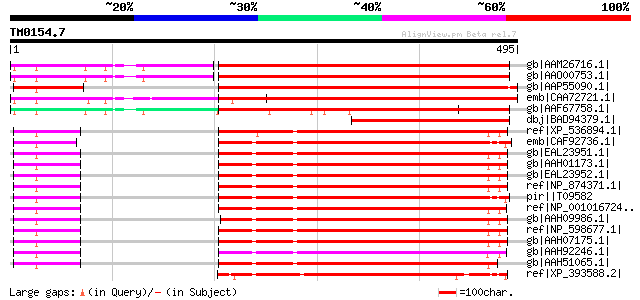
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM26716.1| AT5g27640/F15A18_100 [Arabidopsis thaliana] gi|18... 460 e-128
gb|AAO00753.1| eukaryotic translation initiation factor - like p... 454 e-126
gb|AAP55090.1| putative eukaryotic initiation factor subunit [Or... 450 e-125
emb|CAA72721.1| PRT1 protein [Nicotiana tabacum] gi|6685536|sp|P... 431 e-119
gb|AAF67758.1| eIF3b [Arabidopsis thaliana] 405 e-111
dbj|BAD94379.1| TRANSLATION INITIATION FACTOR 3 SUBUNIT 9-like p... 241 5e-62
ref|XP_536894.1| PREDICTED: similar to Eukaryotic translation in... 207 8e-52
emb|CAF92736.1| unnamed protein product [Tetraodon nigroviridis] 206 1e-51
gb|EAL23951.1| eukaryotic translation initiation factor 3, subun... 205 3e-51
gb|AAH01173.1| EIF3S9 protein [Homo sapiens] gi|2558668|gb|AAC99... 205 3e-51
gb|EAL23952.1| eukaryotic translation initiation factor 3, subun... 205 3e-51
ref|NP_874371.1| eukaryotic translation initiation factor 3, sub... 205 3e-51
pir||T09582 translation initiation factor eIF-3 Prt1 chain - hum... 204 4e-51
ref|NP_001016724.1| hypothetical protein LOC549478 [Xenopus trop... 204 4e-51
gb|AAH09986.1| Unknown (protein for IMAGE:4124553) [Homo sapiens] 204 5e-51
ref|NP_598677.1| eukaryotic translation initiation factor 3, sub... 203 9e-51
gb|AAH07175.1| Eif3s9 protein [Mus musculus] 203 9e-51
gb|AAH92246.1| Unknown (protein for MGC:99017) [Xenopus laevis] 203 1e-50
gb|AAH51065.1| Eif3s9 protein [Mus musculus] gi|23271707|gb|AAH2... 201 4e-50
ref|XP_393588.2| PREDICTED: similar to CG4878-PB, isoform B [Api... 199 1e-49
>gb|AAM26716.1| AT5g27640/F15A18_100 [Arabidopsis thaliana]
gi|18421140|ref|NP_568498.1| eukaryotic translation
initiation factor 3 subunit 9 / eIF-3 eta / eIF3b
(TIF3B1) [Arabidopsis thaliana]
gi|14190411|gb|AAK55686.1| AT5g27640/F15A18_100
[Arabidopsis thaliana] gi|23396618|sp|Q9C5Z1|IF39_ARATH
Eukaryotic translation initiation factor 3 subunit 9
(eIF-3 eta) (eIF3 p110) (eIF3b) (p82)
gi|12407662|gb|AAG53615.1| eukaryotic initiation factor
3B1 subunit [Arabidopsis thaliana]
Length = 712
Score = 460 bits (1184), Expect = e-128
Identities = 218/285 (76%), Positives = 251/285 (87%), Gaps = 1/285 (0%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R+TKTKKSTY+GFELFRIKERDIPIEVLEL+NKNDKIIAFAWEPKGHRF VIHGD +
Sbjct: 425 VDRYTKTKKSTYSGFELFRIKERDIPIEVLELDNKNDKIIAFAWEPKGHRFAVIHGDQPR 484
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
PDVS YSM+TAQ G VSKL LK KQANALFWSP G++I++AGLKGFNGQLEF+NVDEL
Sbjct: 485 PDVSFYSMKTAQNTGRVSKLATLKAKQANALFWSPTGKYIILAGLKGFNGQLEFFNVDEL 544
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVTSVHDVENGFNIWSFSGKHLYRIMKDHFFQFSW 384
ETM TAEHFM TDIEWDPTGRYV T+VTSVH++ENGF IWSF+G LYRI+KDHFFQ +W
Sbjct: 545 ETMATAEHFMATDIEWDPTGRYVATAVTSVHEMENGFTIWSFNGIMLYRILKDHFFQLAW 604
Query: 385 RARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEW 444
R RPPSFLT EKEEEIA LKKYSKKYEAEDQD S+LLSEQ++EK + LKEEW+ WV +W
Sbjct: 605 RPRPPSFLTAEKEEEIAKTLKKYSKKYEAEDQDVSLLLSEQDREKRKALKEEWEKWVMQW 664
Query: 445 KWMHEEEKLQREQHRDGETSD-EEEEYEAKDIEVEEVIDVSEEIL 488
K +HEEEKL R+ RDGE SD EE+EYEAK++E E++IDV+EEI+
Sbjct: 665 KSLHEEEKLVRQNLRDGEVSDVEEDEYEAKEVEFEDLIDVTEEIV 709
Score = 94.7 bits (234), Expect = 6e-18
Identities = 74/222 (33%), Positives = 102/222 (45%), Gaps = 35/222 (15%)
Query: 1 MGY---KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPE 46
+GY ++NTPQEAQ AKE HGYKLD +FD M V +EW+PP+ PY
Sbjct: 105 LGYCFIEFNTPQEAQNAKEKSHGYKLDKSHIFAVNMFDDFDRLMNVKEEWEPPQARPYVP 164
Query: 47 KENQQHWLTDAKARDQFVIRTDSDTE---GRFPYKIKYLIHDLSFVRRS---GNSVADFL 100
EN Q WLTD KARDQ VIR+ DTE K +H + S + + +L
Sbjct: 165 GENLQKWLTDEKARDQLVIRSGPDTEVFWNDTRQKAPEPVHKRPYWTESYVQWSPLGTYL 224
Query: 101 AKNSSKFPNVVWLEEILPKFDLLVSSDAF--IVFAQVLVIMLLSMEEWQTIIEEFPKFDG 158
K VW +D F ++ Q ++ L+ + + + +
Sbjct: 225 V-TLHKQGAAVW-----------GGADTFTRLMRYQHSMVKLVDFSPGEKYLVTYHSQEP 272
Query: 159 VDPVSWIARAEKIFEVH-NLQEQDKSGCADEFAIGGTGGVTG 199
+P K+F+V +D G ADEF+IGG GGV G
Sbjct: 273 SNPRDASKVEIKVFDVRTGRMMRDFKGSADEFSIGGPGGVAG 314
>gb|AAO00753.1| eukaryotic translation initiation factor - like protein
[Arabidopsis thaliana] gi|18420952|ref|NP_568477.1|
eukaryotic translation initiation factor 3 subunit 9,
putative / eIF-3 eta, putative / eIF3b, putative
[Arabidopsis thaliana]
Length = 714
Score = 454 bits (1167), Expect = e-126
Identities = 214/285 (75%), Positives = 248/285 (86%), Gaps = 1/285 (0%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R+TKTKKSTY+GFELFRIKERDIPIEVLEL+NKNDKIIAFAWEPKGHRF VIHGD +
Sbjct: 426 VDRYTKTKKSTYSGFELFRIKERDIPIEVLELDNKNDKIIAFAWEPKGHRFAVIHGDQPR 485
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
PDVS YSM+TAQ G VSKL LK KQANALFWSP G++I++AGLKGFNGQLEF+NVDEL
Sbjct: 486 PDVSFYSMKTAQNTGRVSKLATLKAKQANALFWSPTGKYIILAGLKGFNGQLEFFNVDEL 545
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVTSVHDVENGFNIWSFSGKHLYRIMKDHFFQFSW 384
ETM TAEHFM TDIEWDPTGRYV TSVT+VH++ENGF IWSF+G +YRI+KDHFFQ +W
Sbjct: 546 ETMATAEHFMATDIEWDPTGRYVATSVTTVHEMENGFTIWSFNGNMVYRILKDHFFQLAW 605
Query: 385 RARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEW 444
R RP SFLT EKEEEIA NL+ YSK+YEAEDQD S+LLSEQ++EK R L EEW WV +W
Sbjct: 606 RPRPASFLTAEKEEEIAKNLRNYSKRYEAEDQDVSLLLSEQDREKRRALNEEWQKWVMQW 665
Query: 445 KWMHEEEKLQREQHRDGETSD-EEEEYEAKDIEVEEVIDVSEEIL 488
K +HEEEKL R+ RDGE SD EE+EYEAK++E E++IDV+EEI+
Sbjct: 666 KSLHEEEKLVRQNLRDGEVSDVEEDEYEAKEVEFEDLIDVTEEIV 710
Score = 94.7 bits (234), Expect = 6e-18
Identities = 75/222 (33%), Positives = 102/222 (45%), Gaps = 35/222 (15%)
Query: 1 MGY---KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPE 46
+GY ++NTPQEAQ AKE HGYKLD +FD M V +EW+PP+ PY
Sbjct: 106 LGYCFIEFNTPQEAQNAKEKSHGYKLDKSHIFAVNMFDDFDRLMNVKEEWEPPQERPYVP 165
Query: 47 KENQQHWLTDAKARDQFVIRTDSDTE---GRFPYKIKYLIHDLSFVRRS---GNSVADFL 100
EN Q WLTD KARDQ VIR DTE K +H S+ S + + +L
Sbjct: 166 GENLQKWLTDDKARDQLVIRFGHDTEVYWNDARQKKPEPVHKRSYWTESYVQWSPIGTYL 225
Query: 101 AKNSSKFPNVVWLEEILPKFDLLVSSDAF--IVFAQVLVIMLLSMEEWQTIIEEFPKFDG 158
K VW +D F ++ Q ++ L+ + + + +
Sbjct: 226 V-TLHKQGAAVW-----------GGADTFTRLMRYQHSMVKLVDFSPGEKYLVTYHSQEP 273
Query: 159 VDPVSWIARAEKIFEVH-NLQEQDKSGCADEFAIGGTGGVTG 199
+P K+F+V +D G ADEF+IGG GGV G
Sbjct: 274 SNPRDASKVEIKVFDVRTGRMMRDFKGSADEFSIGGPGGVAG 315
>gb|AAP55090.1| putative eukaryotic initiation factor subunit [Oryza sativa
(japonica cultivar-group)] gi|37537002|ref|NP_922803.1|
putative eukaryotic initiation factor subunit [Oryza
sativa (japonica cultivar-group)]
gi|19224988|gb|AAL86464.1| putative eukaryotic
initiation factor subunit [Oryza sativa (japonica
cultivar-group)]
Length = 719
Score = 450 bits (1157), Expect = e-125
Identities = 212/291 (72%), Positives = 245/291 (83%), Gaps = 1/291 (0%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R+TKTKKSTYTGFELFRIKERDIPIEVLEL+NKNDKIIAFAWEPKGHRF VIHGD K
Sbjct: 430 VDRYTKTKKSTYTGFELFRIKERDIPIEVLELDNKNDKIIAFAWEPKGHRFAVIHGDGPK 489
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
PD+S YSM+T+ VSKLT LKGKQANALFWSP GRFIV AGLKGFNGQLEFYNVDEL
Sbjct: 490 PDISFYSMKTSNNISRVSKLTTLKGKQANALFWSPGGRFIVFAGLKGFNGQLEFYNVDEL 549
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVTSVHDVENGFNIWSFSGKHLYRIMKDHFFQFSW 384
ETM T EHFM TDI WDPTGRY+ ++VTSVH++ENGF IWSFSGK LY++ KDHFFQF W
Sbjct: 550 ETMATGEHFMATDIMWDPTGRYLASAVTSVHEMENGFQIWSFSGKQLYKVSKDHFFQFLW 609
Query: 385 RARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEW 444
R RPPS LTPEKE+EIA NL+KYSKKYE EDQD LSEQE+++ + L+EEW+ WV +W
Sbjct: 610 RPRPPSLLTPEKEDEIAKNLRKYSKKYEQEDQDAFNQLSEQERKRRKQLQEEWEGWVAKW 669
Query: 445 KWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVIDVSEEILHFEYGQE 495
K +HEEE+ R + RDGE SD+EEEY+ K++E+EE +DV EE+ FE QE
Sbjct: 670 KQLHEEERPYRMELRDGEASDDEEEYDTKEVEIEEEVDVQEEV-PFELDQE 719
Score = 89.7 bits (221), Expect = 2e-16
Identities = 44/80 (55%), Positives = 53/80 (66%), Gaps = 11/80 (13%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+YNTPQEA+LA+E +GYKLD +FD +M+VPDEW P E +PY EN Q
Sbjct: 116 EYNTPQEAELAREKTNGYKLDKSHIFAVNMFDDFDKYMKVPDEWMPAEIKPYTPGENLQK 175
Query: 53 WLTDAKARDQFVIRTDSDTE 72
WL D KARDQFVIR + TE
Sbjct: 176 WLADEKARDQFVIRAGTFTE 195
>emb|CAA72721.1| PRT1 protein [Nicotiana tabacum] gi|6685536|sp|P56821|IF39_TOBAC
Eukaryotic translation initiation factor 3 subunit 9
(eIF-3 eta) (eIF3 p110) (eIF3b)
Length = 719
Score = 431 bits (1107), Expect = e-119
Identities = 205/293 (69%), Positives = 247/293 (83%), Gaps = 2/293 (0%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDS- 263
++R+TKTKKSTYTGFELFRIKERDIPIEVLEL+N NDKI AF W P+G +
Sbjct: 427 VDRYTKTKKSTYTGFELFRIKERDIPIEVLELDNTNDKIPAFGWGPEGSPICCYPSVTTP 486
Query: 264 KPDVSIYSMRT-AQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVD 322
+PD+ YS+ A G VSKLT LKGKQANAL+WSP GRF+++ GLKGFNGQLEF++VD
Sbjct: 487 RPDIRFYSVPVWALTPGLVSKLTTLKGKQANALYWSPGGRFLILTGLKGFNGQLEFFDVD 546
Query: 323 ELETMVTAEHFMTTDIEWDPTGRYVTTSVTSVHDVENGFNIWSFSGKHLYRIMKDHFFQF 382
ELETM +AEHFM TD+EWDPTGRYV TSVTSVH++ENGFNIWSF+GK LYRI+KDHFFQ+
Sbjct: 547 ELETMASAEHFMATDVEWDPTGRYVATSVTSVHEMENGFNIWSFNGKLLYRILKDHFFQY 606
Query: 383 SWRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVN 442
WR RPPSFL+ EKEEEIA NLK+YSKKYEAEDQD S+ LSEQ++EK + LKEEW+ W+N
Sbjct: 607 LWRPRPPSFLSKEKEEEIAKNLKRYSKKYEAEDQDVSLQLSEQDREKRKKLKEEWEAWIN 666
Query: 443 EWKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVIDVSEEILHFEYGQE 495
EWK +HEEEK++R++ RDGE SDEEEEYEAK++EVEE+I+V+EEI+ FE Q+
Sbjct: 667 EWKRLHEEEKMERQKLRDGEASDEEEEYEAKEVEVEEIINVTEEIIPFEESQQ 719
Score = 105 bits (262), Expect = 3e-21
Identities = 88/274 (32%), Positives = 133/274 (48%), Gaps = 35/274 (12%)
Query: 1 MGY---KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPE 46
+GY +YNTPQEA+L+KE HGYKLD + + F++VPDEW PPE +PY
Sbjct: 109 LGYCFIEYNTPQEAELSKEKTHGYKLDRSHIFAVNMFDDIERFLKVPDEWAPPEIKPYVP 168
Query: 47 KENQQHWLTDAKARDQFVIRTDSDTEGRF--PYKIK-YLIHDLSFVRRS---GNSVADFL 100
EN Q WLTD KARDQFVIR +DTE + ++K L++ SF S + + +L
Sbjct: 169 GENLQQWLTDEKARDQFVIRAGNDTEVLWNDARQLKPELVYKRSFWTESFVQWSPLGTYL 228
Query: 101 AKNSSKFPNVVWLEEILPKFDLLVSSDAFIVFAQVLVIMLLSMEEWQTIIEEFPKFDGVD 160
A + +W +S+ + +A V L+ + + + + +
Sbjct: 229 A-TVHRQGGAIW--------GGATTSNRLMRYAHPQV-KLIDFSLAERYLVTYSSHEPSN 278
Query: 161 PVSWIARAEKIFEVHNLQ-EQDKSGCADEFAIGGTGGVTGGRGLFLNRHTKTKKSTY--- 216
P A I +V + +D G ADEFA+GGTGGVTG R + K+ Y
Sbjct: 279 PRDSHAVVLNISDVRTGKVMRDFQGSADEFAVGGTGGVTGVSWPVF-RWSGGKQDKYFAR 337
Query: 217 TGFELFRIKERDIPIEVLELENKNDKIIAFAWEP 250
G + + E + + + K + ++ F+W P
Sbjct: 338 IGKNVISVYETETFSLIDKKSIKVENVMDFSWSP 371
>gb|AAF67758.1| eIF3b [Arabidopsis thaliana]
Length = 716
Score = 405 bits (1042), Expect = e-111
Identities = 201/287 (70%), Positives = 233/287 (81%), Gaps = 3/287 (1%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R+TKTKKSTY+GFELFRIKERDIPIEVLEL+NKNDKIIAFAWEPKG RF IHGD
Sbjct: 426 VDRYTKTKKSTYSGFELFRIKERDIPIEVLELDNKNDKIIAFAWEPKGQRFAGIHGDQPN 485
Query: 265 PDVSIYSMRTAQKA-GWVSKLTALKGKQANA-LFWSPAGRFIVVAGLKGFNGQLEFYNVD 322
DVS YS++T K VSKL LK KQA L +P +AGLKGFNGQLEF+NVD
Sbjct: 486 RDVSFYSIKTGTKTLERVSKLATLKAKQAKCPLLVAPQASTSFLAGLKGFNGQLEFFNVD 545
Query: 323 ELETMVTAEHFMTTDIEWDPTGRYVTTSVTSVHDVENGFNIWSFSGKHLYRIMKDHFFQF 382
ELETM TAEHFM TDIEWDPTGRYV TSVT+VH++ENGF IWSF+G +YRI+KDHFFQ
Sbjct: 546 ELETMATAEHFMATDIEWDPTGRYVATSVTTVHEMENGFTIWSFNGNMVYRILKDHFFQL 605
Query: 383 SWRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVN 442
+WR RP SFLT EKEEEIA NL+ YSK+YEAEDQD S+LLSEQ++EK R L EEW WV
Sbjct: 606 AWRPRPASFLTAEKEEEIAKNLRNYSKRYEAEDQDVSLLLSEQDREKRRALNEEWQKWVM 665
Query: 443 EWKWMHEEEKLQREQHRDGETSD-EEEEYEAKDIEVEEVIDVSEEIL 488
+WK +HEEEKL R+ RDGE SD EE+EYEAK++E E++IDV+EEI+
Sbjct: 666 QWKSLHEEEKLVRQNLRDGEVSDVEEDEYEAKEVEFEDLIDVTEEIV 712
Score = 95.5 bits (236), Expect = 3e-18
Identities = 119/490 (24%), Positives = 187/490 (37%), Gaps = 83/490 (16%)
Query: 1 MGY---KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPE 46
+GY ++NTPQEAQ AKE HGYKLD +FD M V +EW+PP+ PY
Sbjct: 106 LGYCFIEFNTPQEAQNAKEKSHGYKLDKSHIFAVNMFDDFDRLMNVKEEWEPPQERPYVP 165
Query: 47 KENQQHWLTDAKARDQFVIRTDSDTE---GRFPYKIKYLIHDLSFVRRS---GNSVADFL 100
EN Q WLTD KARDQ VIR DTE K +H S+ S + + +L
Sbjct: 166 GENLQKWLTDDKARDQLVIRFGHDTEVYWNDARQKKPEPVHKRSYWTESYVQWSPIGTYL 225
Query: 101 AKNSSKFPNVVWLEEILPKFDLLVSSDAF--IVFAQVLVIMLLSMEEWQTIIEEFPKFDG 158
K VW +D F ++ Q ++ L+ + + + +
Sbjct: 226 V-TLHKQGAAVW-----------GGADTFTRLMRYQHSMVKLVDFSPGEKYLVTYHSQEP 273
Query: 159 VDPVSWIARAEKIFEVH-NLQEQDKSGCADEFAIGGTGGVTGGR--------GLFLNRHT 209
+P K+F+V +D G ADEF+IGG GGV G G
Sbjct: 274 SNPRDASKVEIKVFDVRTGRMMRDFKGSADEFSIGGPGGVAGASWPVFRWACGKDDKYFA 333
Query: 210 KTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKG---HRFVVIHGDDSKPD 266
K K+T + +E D + K D ++ W P FV G ++P
Sbjct: 334 KLSKNTISVYETETFSLID------KKSMKVDNVVDICWSPTDSILSLFVPEQGGGNQP- 386
Query: 267 VSIYSMRTAQKAGWVSKLTALKGKQAN-------ALFWSPAGRFIVV------AGLKGFN 313
K V + ++ +Q N ++W +G ++ V K
Sbjct: 387 ---------AKVALVQIPSKVELRQKNLFSVSDCKMYWQSSGEYLAVKVDRYTKTKKSTY 437
Query: 314 GQLEFYNVDELETMVTA-----EHFMTTDIEWDPTGRYVTTSVTSVHDVENGFNIWSFSG 368
E + + E + + ++ W+P G+ + + F
Sbjct: 438 SGFELFRIKERDIPIEVLELDNKNDKIIAFAWEPKGQRFAGIHGDQPNRDVSFYSIKTGT 497
Query: 369 KHLYRIMKDHFFQFSWRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQE 428
K L R+ K + + +A+ P + P+ LK ++ + E + DE ++ E
Sbjct: 498 KTLERVSKLATLK-AKQAKCPLLVAPQASTSFLAGLKGFNGQLEFFNVDELETMATAEHF 556
Query: 429 KCRMLKEEWD 438
+ EWD
Sbjct: 557 MATDI--EWD 564
>dbj|BAD94379.1| TRANSLATION INITIATION FACTOR 3 SUBUNIT 9-like protein [Arabidopsis
thaliana]
Length = 159
Score = 241 bits (614), Expect = 5e-62
Identities = 113/156 (72%), Positives = 134/156 (85%), Gaps = 1/156 (0%)
Query: 334 MTTDIEWDPTGRYVTTSVTSVHDVENGFNIWSFSGKHLYRIMKDHFFQFSWRARPPSFLT 393
M TDIEWDPTGRYV T+VTSVH++ENGF IWSF+G LYRI+KDHFFQ +WR RPPSFLT
Sbjct: 1 MATDIEWDPTGRYVATAVTSVHEMENGFTIWSFNGIMLYRILKDHFFQLAWRPRPPSFLT 60
Query: 394 PEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWKWMHEEEKL 453
EKEEEIA LKKYSKKYEAEDQD S+LLSEQ++EK + LKEEW+ WV +WK +HEEEKL
Sbjct: 61 AEKEEEIAKTLKKYSKKYEAEDQDVSLLLSEQDREKRKALKEEWEKWVMQWKSLHEEEKL 120
Query: 454 QREQHRDGETSD-EEEEYEAKDIEVEEVIDVSEEIL 488
R+ RDGE SD EE+EYEAK++E E++IDV+EEI+
Sbjct: 121 VRQNLRDGEVSDVEEDEYEAKEVEFEDLIDVTEEIV 156
>ref|XP_536894.1| PREDICTED: similar to Eukaryotic translation initiation factor 3
subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b)
[Canis familiaris]
Length = 796
Score = 207 bits (526), Expect = 8e-52
Identities = 108/295 (36%), Positives = 179/295 (60%), Gaps = 16/295 (5%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKN-----DKIIAFAWEPKGHRFVVIH 259
++R K + T FE+FR++E+ +P++V+E++ K+ + IIAFAWEP G +F V+H
Sbjct: 504 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMKGKSHGVPRETIIAFAWEPNGSKFAVLH 563
Query: 260 GDDSKPDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFY 319
G+ + VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F
Sbjct: 564 GEAPRISVSFYHVKNNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFV 620
Query: 320 NVDELETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDH 378
+ + M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD
Sbjct: 621 DTSDCTVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDR 680
Query: 379 FFQFSWRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWD 438
F Q WR RPP+ L+ ++ ++I +LKKYSK +E +D+ S++ E+ R + E++
Sbjct: 681 FCQLLWRPRPPTLLSQDQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFR 740
Query: 439 MWVNEWKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSEE 486
+ + ++ E+K +R + R G +DE E++E + IE EE+I + +
Sbjct: 741 KYRKMAQELYMEQKSERLELRGGVDTDELDSNVEDWEEETIEFFVTEEIIPLGNQ 795
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 183 EYASPAHALDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 242
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 243 WLEEAECRDQYSVIFES 259
>emb|CAF92736.1| unnamed protein product [Tetraodon nigroviridis]
Length = 630
Score = 206 bits (524), Expect = 1e-51
Identities = 112/289 (38%), Positives = 178/289 (60%), Gaps = 11/289 (3%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 347 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ESIIAFAWEPNGSKFAVLHGESPR 403
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y ++ K + L +QAN++FWSP G+F+V+AGL+ NG L F + +
Sbjct: 404 ISVSFYHVKNNGK---IDLLKMFDKQQANSIFWSPQGQFLVLAGLRSMNGALAFVDTSDC 460
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 461 TMMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 520
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPPS L+ ++ + I +LKKYSK +E +D+ S++ +K R + E++ + +E
Sbjct: 521 WRPRPPSLLSADQIKMIKKDLKKYSKIFEQKDRLSQSKASKELVDKRRAMMEDYRRYRDE 580
Query: 444 WKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVID--VSEEILHF 490
+++E+K R R G +DE + D E EE I+ ++EEI+ F
Sbjct: 581 AVKIYQEQKSIRLGLRGGVDTDELDS-NVDDWE-EETIEFFINEEIIPF 627
Score = 55.8 bits (133), Expect = 3e-06
Identities = 25/73 (34%), Positives = 39/73 (53%), Gaps = 11/73 (15%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y P +A A + GYKLD +FD +M + DEW+ PE +P+ + N +H
Sbjct: 97 EYAAPTQALEAVKNADGYKLDKQHTFRVNLFTDFDKYMNISDEWETPEKQPFKDFGNLRH 156
Query: 53 WLTDAKARDQFVI 65
W+ D RDQ+ +
Sbjct: 157 WIEDPDCRDQYSV 169
>gb|EAL23951.1| eukaryotic translation initiation factor 3, subunit 9 eta, 116kDa
[Homo sapiens] gi|33239445|ref|NP_003742.2| eukaryotic
translation initiation factor 3, subunit 9 eta, 116kDa
isoform a [Homo sapiens]
Length = 814
Score = 205 bits (521), Expect = 3e-51
Identities = 107/290 (36%), Positives = 177/290 (60%), Gaps = 14/290 (4%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 530 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPR 586
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 587 ISVSFYHVKNNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDC 643
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 644 TVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 703
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPP+ L+ E+ ++I +LKKYSK +E +D+ S++ E+ R + E++ +
Sbjct: 704 WRPRPPTLLSQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKM 763
Query: 444 WKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSEE 486
+ ++ E+K +R + R G +DE +++E + IE EE+I + +
Sbjct: 764 AQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVTEEIIPLGNQ 813
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 236 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 295
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 296 WLEEAECRDQYSVIFES 312
>gb|AAH01173.1| EIF3S9 protein [Homo sapiens] gi|2558668|gb|AAC99479.1| eukaryotic
translation initiation factor [Homo sapiens]
gi|3123230|sp|P55884|IF39_HUMAN Eukaryotic translation
initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116)
(eIF3 p110) (eIF3b)
Length = 814
Score = 205 bits (521), Expect = 3e-51
Identities = 107/290 (36%), Positives = 177/290 (60%), Gaps = 14/290 (4%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 530 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPR 586
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 587 ISVSFYHVKNNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDC 643
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 644 TVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 703
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPP+ L+ E+ ++I +LKKYSK +E +D+ S++ E+ R + E++ +
Sbjct: 704 WRPRPPTLLSQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKM 763
Query: 444 WKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSEE 486
+ ++ E+K +R + R G +DE +++E + IE EE+I + +
Sbjct: 764 AQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVTEEIIPLGNQ 813
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 236 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 295
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 296 WLEEAECRDQYSVIFES 312
>gb|EAL23952.1| eukaryotic translation initiation factor 3, subunit 9 eta, 116kDa
[Homo sapiens]
Length = 775
Score = 205 bits (521), Expect = 3e-51
Identities = 107/290 (36%), Positives = 177/290 (60%), Gaps = 14/290 (4%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 491 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPR 547
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 548 ISVSFYHVKNNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDC 604
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 605 TVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 664
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPP+ L+ E+ ++I +LKKYSK +E +D+ S++ E+ R + E++ +
Sbjct: 665 WRPRPPTLLSQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKM 724
Query: 444 WKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSEE 486
+ ++ E+K +R + R G +DE +++E + IE EE+I + +
Sbjct: 725 AQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVTEEIIPLGNQ 774
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 197 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 256
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 257 WLEEAECRDQYSVIFES 273
>ref|NP_874371.1| eukaryotic translation initiation factor 3, subunit 9 eta, 116kDa
isoform b [Homo sapiens]
Length = 775
Score = 205 bits (521), Expect = 3e-51
Identities = 107/290 (36%), Positives = 177/290 (60%), Gaps = 14/290 (4%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 491 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPR 547
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 548 ISVSFYHVKNNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDC 604
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 605 TVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 664
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPP+ L+ E+ ++I +LKKYSK +E +D+ S++ E+ R + E++ +
Sbjct: 665 WRPRPPTLLSQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKM 724
Query: 444 WKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSEE 486
+ ++ E+K +R + R G +DE +++E + IE EE+I + +
Sbjct: 725 AQELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVTEEIIPLGNQ 774
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 197 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 256
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 257 WLEEAECRDQYSVIFES 273
>pir||T09582 translation initiation factor eIF-3 Prt1 chain - human
gi|1778051|gb|AAB42010.1| Prt1 homolog [Homo sapiens]
Length = 873
Score = 204 bits (520), Expect = 4e-51
Identities = 110/287 (38%), Positives = 177/287 (61%), Gaps = 11/287 (3%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 530 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPR 586
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 587 ISVSFYHVKNNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDC 643
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 644 TVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 703
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPP+ L+ E+ ++I +LKKYSK +E +D+ S++ E+ R + E++ +
Sbjct: 704 WRPRPPTLLSQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKM 763
Query: 444 WKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVID--VSEEIL 488
+ ++ E+K +R + R G +DE + D E EE I+ V+EEI+
Sbjct: 764 AQELYMEQKNERLELRGGVDTDELDS-NVDDWE-EETIEFFVTEEII 808
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 236 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 295
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 296 WLEEAECRDQYSVIFES 312
>ref|NP_001016724.1| hypothetical protein LOC549478 [Xenopus tropicalis]
Length = 676
Score = 204 bits (520), Expect = 4e-51
Identities = 110/289 (38%), Positives = 174/289 (60%), Gaps = 14/289 (4%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ D IIAFAWEP G +F V+HG+ +
Sbjct: 394 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---DGIIAFAWEPNGSKFAVLHGEVPR 450
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 451 ISVSFYHVKNNGK---IELIKMYDKQQANTIFWSPQGQFLVLAGLRSMNGALAFVDTSDC 507
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+ +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 508 TIMNIAEHYTASDVEWDPTGRYVVTSVSWWSHKVDNAYWMWTFQGRLLQKNNKDRFCQLL 567
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPPS L+ ++ ++I +LKKYSK +E +D+ S++ E+ R + EE+ +
Sbjct: 568 WRPRPPSLLSQDQIKQIKKDLKKYSKIFEQKDRLSQSKASKELIERRRAMMEEYKTYREM 627
Query: 444 WKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSE 485
++ E+K R + R G +DE +++E + IE EE+I V E
Sbjct: 628 AAKLYMEQKAARLEIRGGVDTDELDSNVDDWEEETIEFFVTEEIIPVEE 676
Score = 51.6 bits (122), Expect = 6e-05
Identities = 27/77 (35%), Positives = 40/77 (51%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y P +AQ A + YKLD +FD +M + DEW+ PE +P+ + N +
Sbjct: 100 EYALPTQAQDAVKNADCYKLDKQHTFRVNLFTDFDKYMVIGDEWEAPEKQPFKDFGNLRS 159
Query: 53 WLTDAKARDQFVIRTDS 69
WL D RDQF + +S
Sbjct: 160 WLEDPDCRDQFSVIYES 176
>gb|AAH09986.1| Unknown (protein for IMAGE:4124553) [Homo sapiens]
Length = 622
Score = 204 bits (519), Expect = 5e-51
Identities = 107/288 (37%), Positives = 175/288 (60%), Gaps = 14/288 (4%)
Query: 207 RHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSKPD 266
R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 340 RTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPRIS 396
Query: 267 VSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDELET 326
VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 397 VSFYHVKNNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDCTV 453
Query: 327 MVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFSWR 385
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q WR
Sbjct: 454 MNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLLWR 513
Query: 386 ARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNEWK 445
RPP+ L+ E+ ++I +LKKYSK +E +D+ S++ E+ R + E++ + +
Sbjct: 514 PRPPTLLSQEQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRKYRKMAQ 573
Query: 446 WMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSEE 486
++ E+K +R + R G +DE +++E + IE EE+I + +
Sbjct: 574 ELYMEQKNERLELRGGVDTDELDSNVDDWEEETIEFFVTEEIIPLGNQ 621
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 45 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 104
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 105 WLEEAECRDQYSVIFES 121
>ref|NP_598677.1| eukaryotic translation initiation factor 3, subunit 9 [Mus
musculus] gi|21594439|gb|AAH31704.1| Eukaryotic
translation initiation factor 3, subunit 9 [Mus
musculus] gi|60687524|pir||JC7862 eukaryotic initiation
factor, eIF3 subunit, p116 protein - mouse
gi|26329413|dbj|BAC28445.1| unnamed protein product [Mus
musculus]
Length = 803
Score = 203 bits (517), Expect = 9e-51
Identities = 106/290 (36%), Positives = 178/290 (60%), Gaps = 14/290 (4%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 519 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPR 575
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y +++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 576 ISVSFYHVKSNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDC 632
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 633 TVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 692
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPP+ L+ ++ ++I +LKKYSK +E +D+ S++ E+ R + E++ +
Sbjct: 693 WRPRPPTLLSQDQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRQYRKM 752
Query: 444 WKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSEE 486
+ ++ ++K +R + R G +DE +++E + IE EEVI + +
Sbjct: 753 AQELYMKQKNERLELRGGVDTDELDSNVDDWEEETIEFFVTEEVIPLGSQ 802
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 225 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 284
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 285 WLEEAECRDQYSVIFES 301
>gb|AAH07175.1| Eif3s9 protein [Mus musculus]
Length = 641
Score = 203 bits (517), Expect = 9e-51
Identities = 106/290 (36%), Positives = 178/290 (60%), Gaps = 14/290 (4%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 357 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPR 413
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y +++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 414 ISVSFYHVKSNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDC 470
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 471 TVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 530
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPP+ L+ ++ ++I +LKKYSK +E +D+ S++ E+ R + E++ +
Sbjct: 531 WRPRPPTLLSQDQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRQYRKM 590
Query: 444 WKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE---VEEVIDVSEE 486
+ ++ ++K +R + R G +DE +++E + IE EEVI + +
Sbjct: 591 AQELYMKQKNERLELRGGVDTDELDSNVDDWEEETIEFFVTEEVIPLGSQ 640
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 63 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 122
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 123 WLEEAECRDQYSVIFES 139
>gb|AAH92246.1| Unknown (protein for MGC:99017) [Xenopus laevis]
Length = 688
Score = 203 bits (516), Expect = 1e-50
Identities = 110/289 (38%), Positives = 173/289 (59%), Gaps = 14/289 (4%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ D IIAFAWEP G +F V+HG+ +
Sbjct: 406 VDRTPKGTQGMVTNFEIFRMREKQVPVDVVEMK---DGIIAFAWEPNGSKFAVLHGEVPR 462
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y ++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 463 ISVSFYHVKNNGK---IELIKMYDKQQANTIFWSPQGQFLVLAGLRSMNGALAFVDTSDC 519
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+ +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 520 TIMNIAEHYTASDVEWDPTGRYVITSVSWWSHKVDNAYWMWTFQGRLLQKNNKDRFCQLL 579
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPPS L+ ++ ++I +LKKYSK +E +D+ S++ E+ R + EE+ +
Sbjct: 580 WRPRPPSLLSQDQIKQIKKDLKKYSKIFEQKDRLSQTKASKELIERRRAMMEEYKTYREM 639
Query: 444 WKWMHEEEKLQREQHRDGETSD----EEEEYEAKDIE---VEEVIDVSE 485
++ E+K R + R G +D E++E + IE EE+I V E
Sbjct: 640 ATKLYMEQKTARLEIRGGVDTDLLDSNVEDWEEETIEFFVTEEIIPVEE 688
Score = 56.2 bits (134), Expect = 2e-06
Identities = 29/77 (37%), Positives = 41/77 (52%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y P +AQ A + GYKLD +FD +M + DEWD PE +P+ + N +
Sbjct: 112 EYALPAQAQDAVKNADGYKLDKQHTFRVNLFTDFDKYMVIGDEWDVPEKQPFKDFGNLRS 171
Query: 53 WLTDAKARDQFVIRTDS 69
WL D RDQF + +S
Sbjct: 172 WLEDPDCRDQFSVIYES 188
>gb|AAH51065.1| Eif3s9 protein [Mus musculus] gi|23271707|gb|AAH23767.1| Eif3s9
protein [Mus musculus]
Length = 970
Score = 201 bits (511), Expect = 4e-50
Identities = 102/277 (36%), Positives = 172/277 (61%), Gaps = 11/277 (3%)
Query: 205 LNRHTKTKKSTYTGFELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIHGDDSK 264
++R K + T FE+FR++E+ +P++V+E++ + IIAFAWEP G +F V+HG+ +
Sbjct: 519 VDRTPKGTQGVVTNFEIFRMREKQVPVDVVEMK---ETIIAFAWEPNGSKFAVLHGEAPR 575
Query: 265 PDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFYNVDEL 324
VS Y +++ K + + +QAN +FWSP G+F+V+AGL+ NG L F + +
Sbjct: 576 ISVSFYHVKSNGK---IELIKMFDKQQANTIFWSPQGQFVVLAGLRSMNGALAFVDTSDC 632
Query: 325 ETMVTAEHFMTTDIEWDPTGRYVTTSVT-SVHDVENGFNIWSFSGKHLYRIMKDHFFQFS 383
M AEH+M +D+EWDPTGRYV TSV+ H V+N + +W+F G+ L + KD F Q
Sbjct: 633 TVMNIAEHYMASDVEWDPTGRYVVTSVSWWSHKVDNAYWLWTFQGRLLQKNNKDRFCQLL 692
Query: 384 WRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEKCRMLKEEWDMWVNE 443
WR RPP+ L+ ++ ++I +LKKYSK +E +D+ S++ E+ R + E++ +
Sbjct: 693 WRPRPPTLLSQDQIKQIKKDLKKYSKIFEQKDRLSQSKASKELVERRRTMMEDFRQYRKM 752
Query: 444 WKWMHEEEKLQREQHRDGETSDE----EEEYEAKDIE 476
+ ++ ++K +R + R G +DE +++E + IE
Sbjct: 753 AQELYMKQKNERLELRGGVDTDELDSNVDDWEEETIE 789
Score = 55.8 bits (133), Expect = 3e-06
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 11/77 (14%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+Y +P A A + GYKLD +FD +M + DEWD PE +P+ + N ++
Sbjct: 225 EYASPAHAVDAVKNADGYKLDKQHTFRVNLFTDFDKYMTISDEWDIPEKQPFKDLGNLRY 284
Query: 53 WLTDAKARDQFVIRTDS 69
WL +A+ RDQ+ + +S
Sbjct: 285 WLEEAECRDQYSVIFES 301
>ref|XP_393588.2| PREDICTED: similar to CG4878-PB, isoform B [Apis mellifera]
Length = 702
Score = 199 bits (507), Expect = 1e-49
Identities = 120/294 (40%), Positives = 183/294 (61%), Gaps = 24/294 (8%)
Query: 204 FLNRHTKTKKSTYTG----FELFRIKERDIPIEVLELENKNDKIIAFAWEPKGHRFVVIH 259
F R KT++ YTG FE+F ++E+ IP++ +E++ + I AFAWEP G +F +IH
Sbjct: 415 FAKRKEKTEQK-YTGMSYNFEIFHMREKQIPVDSVEIK---EPIHAFAWEPVGSKFAIIH 470
Query: 260 GDDSKPDVSIYSMRTAQKAGWVSKLTALKGKQANALFWSPAGRFIVVAGLKGFNGQLEFY 319
G+ +VS Y +R + + KL + K N LFWSP+G+FIV+AGL G LEF
Sbjct: 471 GEIPSVNVSFYEVRYGHQPALLKKL---EKKACNHLFWSPSGQFIVLAGLTTMAGALEFI 527
Query: 320 NVDELETMVTAEHFMTTDIEWDPTGRYVTTSVTS-VHDVENGFNIWSFSGKHLYRIMKDH 378
+ ++ M + +H+ T+D+EWDPTGRYV T+V+S V+NG+ IW+F G+ L R+ +
Sbjct: 528 DTNDFTIMNSTDHYQTSDVEWDPTGRYVVTAVSSWKTSVDNGYWIWTFQGRILKRVNLNQ 587
Query: 379 FFQFSWRARPPSFLTPEKEEEIANNLKKYSKKYEAEDQDESMLLSEQEQEK-CRMLK--E 435
F Q WR RPP+ LTPE+ +EI NLKKYS ++E++D+ S++ EK C +K E
Sbjct: 588 FNQLLWRPRPPTLLTPEQIKEIKKNLKKYSAQFESKDRMRLTRASKELIEKRCAAMKAFE 647
Query: 436 EW-DMWVNEWKWMHEEEKLQREQHRDGETSDEEEEYEAKDIEVEEVID--VSEE 486
E+ + EW +K +R + R+G +DE + ++K++E EEVI+ V EE
Sbjct: 648 EYRSKRIQEW----NNQKKRRLELRNGIDTDELDS-DSKNVE-EEVIEFFVKEE 695
Score = 47.0 bits (110), Expect = 0.001
Identities = 21/73 (28%), Positives = 37/73 (49%), Gaps = 11/73 (15%)
Query: 4 KYNTPQEAQLAKEMLHGYKLD-----------EFDSFMRVPDEWDPPETEPYPEKENQQH 52
+YN P A A + ++ YK+D +F F +PDEW+PP+ + + + +
Sbjct: 116 EYNNPLNALAAAQSVNNYKIDKQHTFKVNLFTDFKKFEEIPDEWEPPKPQEFKAAADLHY 175
Query: 53 WLTDAKARDQFVI 65
+L + A DQF +
Sbjct: 176 YLLEPDAYDQFCV 188
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 931,833,899
Number of Sequences: 2540612
Number of extensions: 42013392
Number of successful extensions: 210519
Number of sequences better than 10.0: 1287
Number of HSP's better than 10.0 without gapping: 385
Number of HSP's successfully gapped in prelim test: 945
Number of HSP's that attempted gapping in prelim test: 187654
Number of HSP's gapped (non-prelim): 9550
length of query: 495
length of database: 863,360,394
effective HSP length: 132
effective length of query: 363
effective length of database: 527,999,610
effective search space: 191663858430
effective search space used: 191663858430
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0154.7


