

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0150.5
(168 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
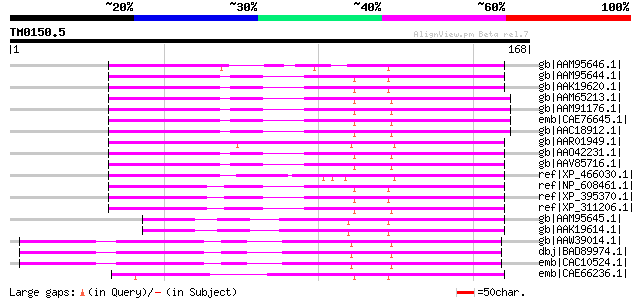
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM95646.1| WD-repeat protein GhTTG4 [Gossypium hirsutum] 107 1e-22
gb|AAM95644.1| WD-repeat protein GhTTG2 [Gossypium hirsutum] gi|... 105 4e-22
gb|AAK19620.1| WD1521 [Gossypium hirsutum] 105 4e-22
gb|AAM65213.1| flower pigmentation protein ATAN11 [Arabidopsis t... 103 1e-21
gb|AAM91176.1| WD repeat protein ATAN11 [Arabidopsis thaliana] g... 103 1e-21
emb|CAE76645.1| WD 40 protein [Matthiola incana] 103 1e-21
gb|AAC18912.1| ATAN11 [Arabidopsis thaliana] 103 1e-21
gb|AAR01949.1| WD40 repeat protein [Zea mays] 95 8e-19
gb|AAO42231.1| putative transcriptional regulator protein [Arabi... 94 2e-18
gb|AAV85716.1| At3g26640 [Arabidopsis thaliana] gi|1402884|emb|C... 93 2e-18
ref|XP_466030.1| putative WD40 repeat protein [Oryza sativa (jap... 91 2e-17
ref|NP_608461.1| CG14614-PA [Drosophila melanogaster] gi|1072635... 75 5e-13
ref|XP_395370.1| PREDICTED: similar to ENSANGP00000019078 [Apis ... 75 9e-13
ref|XP_311206.1| ENSANGP00000019078 [Anopheles gambiae str. PEST... 71 1e-11
gb|AAM95645.1| WD-repeat protein GhTTG3 [Gossypium hirsutum] 61 1e-08
gb|AAK19614.1| GHTTG1 [Gossypium hirsutum] 61 1e-08
gb|AAW39014.1| At5g24520 [Arabidopsis thaliana] gi|56121888|gb|A... 60 2e-08
dbj|BAD89974.1| mutant protein of TTG1 [Arabidopsis thaliana] 60 2e-08
emb|CAC10524.1| transparent testa glabra 1 [Arabidopsis thaliana] 60 2e-08
emb|CAE66236.1| Hypothetical protein CBG11480 [Caenorhabditis br... 60 2e-08
>gb|AAM95646.1| WD-repeat protein GhTTG4 [Gossypium hirsutum]
Length = 346
Score = 107 bits (268), Expect = 1e-22
Identities = 73/140 (52%), Positives = 82/140 (58%), Gaps = 31/140 (22%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNA--RSDPTRISPSSTRTHHQGHLHPR 90
NWSVR DKKY LAIASLLEQ PN ++IV LDDSN RSDP + HP
Sbjct: 34 NWSVRRDKKYRLAIASLLEQYPNRLQIVQLDDSNGEIRSDP-----------NLSFDHP- 81
Query: 91 *GVPAPR----PPRHLQRLPPRVADLRRR*NPTLR------RNPSVDLKSLLNGNKSSEY 140
PA + P + Q+ DL + LR VDLKSLLNGNK+SE+
Sbjct: 82 --YPATKTIFIPDKDCQK-----PDLLATSSDFLRIWRISDDGSRVDLKSLLNGNKNSEF 134
Query: 141 CGPLTSFDWNEAEPCRIGTS 160
CGPLTSFDWNEAEP RIGTS
Sbjct: 135 CGPLTSFDWNEAEPKRIGTS 154
>gb|AAM95644.1| WD-repeat protein GhTTG2 [Gossypium hirsutum]
gi|22324803|gb|AAM95643.1| WD-repeat protein GhTTG2
[Gossypium hirsutum]
Length = 346
Score = 105 bits (263), Expect = 4e-22
Identities = 69/137 (50%), Positives = 78/137 (56%), Gaps = 25/137 (18%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DKKY LAIASLLE N +EIV LDDSN R P+ + H
Sbjct: 34 NWSVRRDKKYRLAIASLLEHYNNRLEIVQLDDSNGE---IRSDPNLSFDH---------- 80
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLR------RNPSVDLKSLLNGNKSSEYCGP 143
P PP +P + DL + LR + VDLKSLLNGNK+SE+CGP
Sbjct: 81 ---PYPPTKTIFIPDKECQKPDLLATSSDFLRIWRISDDHSRVDLKSLLNGNKNSEFCGP 137
Query: 144 LTSFDWNEAEPCRIGTS 160
LTSFDWNEAEP RIGTS
Sbjct: 138 LTSFDWNEAEPKRIGTS 154
>gb|AAK19620.1| WD1521 [Gossypium hirsutum]
Length = 314
Score = 105 bits (263), Expect = 4e-22
Identities = 69/137 (50%), Positives = 78/137 (56%), Gaps = 25/137 (18%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DKKY LAIASLLE N +EIV LDDSN R P+ + H
Sbjct: 2 NWSVRRDKKYRLAIASLLEHYNNRLEIVQLDDSNGE---IRSDPNLSFDH---------- 48
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLR------RNPSVDLKSLLNGNKSSEYCGP 143
P PP +P + DL + LR + VDLKSLLNGNK+SE+CGP
Sbjct: 49 ---PYPPTKTIFIPDKECQKPDLLATSSDFLRIWRISDDHSRVDLKSLLNGNKNSEFCGP 105
Query: 144 LTSFDWNEAEPCRIGTS 160
LTSFDWNEAEP RIGTS
Sbjct: 106 LTSFDWNEAEPKRIGTS 122
>gb|AAM65213.1| flower pigmentation protein ATAN11 [Arabidopsis thaliana]
Length = 346
Score = 103 bits (258), Expect = 1e-21
Identities = 67/139 (48%), Positives = 78/139 (55%), Gaps = 25/139 (17%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DKKY LAI SLLEQ PN +EIV LD+SN R P+ + H
Sbjct: 34 NWSVRRDKKYRLAITSLLEQYPNRVEIVQLDESNGE---IRSDPNLSFEH---------- 80
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLRR------NPSVDLKSLLNGNKSSEYCGP 143
P PP +P + DL + LR + V+LKS LN NK+SE+CGP
Sbjct: 81 ---PYPPTKTIFIPDKECQRPDLLATSSDFLRLWRIADDHSRVELKSCLNSNKNSEFCGP 137
Query: 144 LTSFDWNEAEPCRIGTSLT 162
LTSFDWNEAEP RIGTS T
Sbjct: 138 LTSFDWNEAEPRRIGTSST 156
>gb|AAM91176.1| WD repeat protein ATAN11 [Arabidopsis thaliana]
gi|20260404|gb|AAM13100.1| WD repeat protein ATAN11
[Arabidopsis thaliana] gi|8698737|gb|AAF78495.1|
Identical to WD repeat protein ATAN11 from Arabidopsis
thaliana gb|U94746 and contains multiple WD domain
PF|00400 repeats. ESTs gb|H35958, gb|AA712360,
gb|R90717, gb|AW004301 come from this gene
gi|15222113|ref|NP_172751.1| flower pigmentation protein
(AN11) [Arabidopsis thaliana] gi|25518671|pir||G86262
hypothetical protein F13K23.16 - Arabidopsis thaliana
Length = 346
Score = 103 bits (258), Expect = 1e-21
Identities = 67/139 (48%), Positives = 78/139 (55%), Gaps = 25/139 (17%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DKKY LAI SLLEQ PN +EIV LD+SN R P+ + H
Sbjct: 34 NWSVRRDKKYRLAITSLLEQYPNRVEIVQLDESNGE---IRSDPNLSFEH---------- 80
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLRR------NPSVDLKSLLNGNKSSEYCGP 143
P PP +P + DL + LR + V+LKS LN NK+SE+CGP
Sbjct: 81 ---PYPPTKTIFIPDKECQRPDLLATSSDFLRLWRIADDHSRVELKSCLNSNKNSEFCGP 137
Query: 144 LTSFDWNEAEPCRIGTSLT 162
LTSFDWNEAEP RIGTS T
Sbjct: 138 LTSFDWNEAEPRRIGTSST 156
>emb|CAE76645.1| WD 40 protein [Matthiola incana]
Length = 331
Score = 103 bits (258), Expect = 1e-21
Identities = 67/139 (48%), Positives = 78/139 (55%), Gaps = 25/139 (17%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DKKY LAI SLLEQ PN +EIV LD+SN R P+ + H
Sbjct: 19 NWSVRRDKKYRLAITSLLEQYPNRVEIVQLDESNGE---IRSDPNLSFEH---------- 65
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLRR------NPSVDLKSLLNGNKSSEYCGP 143
P PP +P + DL + LR + V+LKS LN NK+SE+CGP
Sbjct: 66 ---PYPPTKTIFIPDKECQRPDLLATSSDFLRLWRIADDHSRVELKSCLNSNKNSEFCGP 122
Query: 144 LTSFDWNEAEPCRIGTSLT 162
LTSFDWNEAEP RIGTS T
Sbjct: 123 LTSFDWNEAEPRRIGTSST 141
>gb|AAC18912.1| ATAN11 [Arabidopsis thaliana]
Length = 346
Score = 103 bits (258), Expect = 1e-21
Identities = 67/139 (48%), Positives = 78/139 (55%), Gaps = 25/139 (17%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DKKY LAI SLLEQ PN +EIV LD+SN R P+ + H
Sbjct: 34 NWSVRRDKKYRLAITSLLEQYPNRVEIVQLDESNGE---IRSDPNLSFEH---------- 80
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLRR------NPSVDLKSLLNGNKSSEYCGP 143
P PP +P + DL + LR + V+LKS LN NK+SE+CGP
Sbjct: 81 ---PYPPTKTIFIPDKECQRPDLLATSSDFLRLWRIADDHSRVELKSCLNSNKNSEFCGP 137
Query: 144 LTSFDWNEAEPCRIGTSLT 162
LTSFDWNEAEP RIGTS T
Sbjct: 138 LTSFDWNEAEPRRIGTSST 156
>gb|AAR01949.1| WD40 repeat protein [Zea mays]
Length = 416
Score = 94.7 bits (234), Expect = 8e-19
Identities = 59/142 (41%), Positives = 79/142 (55%), Gaps = 14/142 (9%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPT-----RISPSSTRTHHQGHL 87
NWSVR DKKY LAIASLLEQ N +E+V LD+++ P + P+ T H
Sbjct: 49 NWSVRRDKKYRLAIASLLEQVTNRVEVVQLDEASGDIAPVLTFDHQYPPTKTMFMPDPHA 108
Query: 88 HPR*GVPAPRPPRHLQRLPPRV-----ADLRRR*NPTLRRN----PSVDLKSLLNGNKSS 138
+ + R+P A N ++R N P ++L+S LNGN++S
Sbjct: 109 LRPDLLATSADHLRIWRIPSSDDAEDGAASANNNNGSVRCNGTQQPGIELRSELNGNRNS 168
Query: 139 EYCGPLTSFDWNEAEPCRIGTS 160
+YCGPLTSFDWN+A+P RIGTS
Sbjct: 169 DYCGPLTSFDWNDADPRRIGTS 190
>gb|AAO42231.1| putative transcriptional regulator protein [Arabidopsis thaliana]
Length = 346
Score = 93.6 bits (231), Expect = 2e-18
Identities = 60/137 (43%), Positives = 74/137 (53%), Gaps = 25/137 (18%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWS+R DKKY LAI SL+EQ PN +EIV LD+SN R P+ H
Sbjct: 34 NWSIRRDKKYRLAITSLIEQYPNRVEIVQLDESNGE---VRSDPNLCFEH---------- 80
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLRR------NPSVDLKSLLNGNKSSEYCGP 143
P PP +P + DL + LR V+LKS L+ +K+SE+ GP
Sbjct: 81 ---PYPPTKTSFIPDKECQRPDLLATSSDFLRLWRISDDESRVELKSCLSSDKNSEFSGP 137
Query: 144 LTSFDWNEAEPCRIGTS 160
+TSFDWNEAEP RIGTS
Sbjct: 138 ITSFDWNEAEPRRIGTS 154
>gb|AAV85716.1| At3g26640 [Arabidopsis thaliana] gi|1402884|emb|CAA66815.1|
hypothetical protein [Arabidopsis thaliana]
gi|21554271|gb|AAM63346.1| transcriptional regulator
protein, putative [Arabidopsis thaliana]
gi|11994299|dbj|BAB01729.1| beta-transducin like protein
[Arabidopsis thaliana] gi|1495265|emb|CAA66120.1|
beta-transducin like protein [Arabidopsis thaliana]
gi|15231593|ref|NP_189298.1| transducin family protein /
WD-40 repeat family protein [Arabidopsis thaliana]
Length = 346
Score = 93.2 bits (230), Expect = 2e-18
Identities = 60/137 (43%), Positives = 74/137 (53%), Gaps = 25/137 (18%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWS+R DKKY LAI SL+EQ PN +EIV LD+SN R P+ H
Sbjct: 34 NWSIRRDKKYRLAITSLIEQYPNRVEIVQLDESNGE---IRSDPNLCFEH---------- 80
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLRR------NPSVDLKSLLNGNKSSEYCGP 143
P PP +P + DL + LR V+LKS L+ +K+SE+ GP
Sbjct: 81 ---PYPPTKTSFIPDKECQRPDLLATSSDFLRLWRISDDESRVELKSCLSSDKNSEFSGP 137
Query: 144 LTSFDWNEAEPCRIGTS 160
+TSFDWNEAEP RIGTS
Sbjct: 138 ITSFDWNEAEPRRIGTS 154
>ref|XP_466030.1| putative WD40 repeat protein [Oryza sativa (japonica
cultivar-group)] gi|49388269|dbj|BAD25387.1| putative
WD40 repeat protein [Oryza sativa (japonica
cultivar-group)]
Length = 411
Score = 90.5 bits (223), Expect = 2e-17
Identities = 64/147 (43%), Positives = 82/147 (55%), Gaps = 25/147 (17%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DKKY LAIASLLEQ N +++V LD+S+ I+P T H
Sbjct: 49 NWSVRRDKKYRLAIASLLEQHNNHVQVVQLDESSGD-----IAPVLTFDHPYPPTKTM-F 102
Query: 93 VPAPRPPR---------HLQ--RLPP----RVADLRRR*NPTLRRN----PSVDLKSLLN 133
VP P R HL+ R+P A + ++R N P V+L+ LN
Sbjct: 103 VPDPHSVRPDLLATSADHLRIWRIPSPDEAAAAAAASSNSGSVRCNGTASPDVELRCELN 162
Query: 134 GNKSSEYCGPLTSFDWNEAEPCRIGTS 160
GN++S+YCGPLTSFDWN+A+P RIGTS
Sbjct: 163 GNRNSDYCGPLTSFDWNDADPRRIGTS 189
>ref|NP_608461.1| CG14614-PA [Drosophila melanogaster] gi|10726353|gb|AAF50953.2|
CG14614-PA [Drosophila melanogaster]
gi|54643605|gb|EAL32348.1| GA13113-PA [Drosophila
pseudoobscura]
Length = 343
Score = 75.5 bits (184), Expect = 5e-13
Identities = 48/135 (35%), Positives = 67/135 (49%), Gaps = 25/135 (18%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DK++ LA+ S +E+ N ++I+ LD+ D + S ST H
Sbjct: 24 NWSVRPDKRFRLALGSFIEEYNNKVQIISLDE-----DTSEFSAKSTFDH---------- 68
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLR----RNPSVDLKSLLNGNKSSEYCGPLT 145
P P + +P DL LR P L+ +LN NK+S++C PLT
Sbjct: 69 ---PYPTTKIMWIPDSKGVYPDLLATSGDYLRVWRAGEPDTRLECVLNNNKNSDFCAPLT 125
Query: 146 SFDWNEAEPCRIGTS 160
SFDWNE +P +GTS
Sbjct: 126 SFDWNEVDPNLVGTS 140
>ref|XP_395370.1| PREDICTED: similar to ENSANGP00000019078 [Apis mellifera]
Length = 346
Score = 74.7 bits (182), Expect = 9e-13
Identities = 49/135 (36%), Positives = 67/135 (49%), Gaps = 25/135 (18%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DK++ LA+ S +E+ N ++IV LD+ + + S ST H
Sbjct: 26 NWSVRPDKRFRLALGSFVEEYNNKVQIVSLDE-----ETSEFSAKSTFDH---------- 70
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLR----RNPSVDLKSLLNGNKSSEYCGPLT 145
P P + +P DL LR P L+ +LN NK+S++C PLT
Sbjct: 71 ---PYPTTKIMWIPDSKGLFPDLLATSGDYLRVWRAAEPETRLECVLNNNKNSDFCAPLT 127
Query: 146 SFDWNEAEPCRIGTS 160
SFDWNE +P IGTS
Sbjct: 128 SFDWNEVDPNLIGTS 142
>ref|XP_311206.1| ENSANGP00000019078 [Anopheles gambiae str. PEST]
gi|30177772|gb|EAA06830.2| ENSANGP00000019078 [Anopheles
gambiae str. PEST]
Length = 344
Score = 71.2 bits (173), Expect = 1e-11
Identities = 47/135 (34%), Positives = 66/135 (48%), Gaps = 25/135 (18%)
Query: 33 NWSVRHDKKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*G 92
NWSVR DK++ LA+ S +E+ N ++I+ LD+ D + S ST H
Sbjct: 24 NWSVRPDKRFRLALGSFVEEYNNKVQIISLDE-----DTSEFSAKSTFDH---------- 68
Query: 93 VPAPRPPRHLQRLPPRVA---DLRRR*NPTLRR----NPSVDLKSLLNGNKSSEYCGPLT 145
P P + +P DL LR P L+ +LN NK+S++C PLT
Sbjct: 69 ---PYPTTKIMWIPDSKGVYPDLLATSGDYLRLWRAGEPDTRLECVLNNNKNSDFCAPLT 125
Query: 146 SFDWNEAEPCRIGTS 160
SFDWNE + +GTS
Sbjct: 126 SFDWNEVDLNLVGTS 140
>gb|AAM95645.1| WD-repeat protein GhTTG3 [Gossypium hirsutum]
Length = 345
Score = 61.2 bits (147), Expect = 1e-08
Identities = 44/129 (34%), Positives = 61/129 (47%), Gaps = 28/129 (21%)
Query: 44 LAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*GVPAPRPPRHLQ 103
+A+ S LE N ++I+ DP +S + HP+ P PP L
Sbjct: 43 IALGSFLEDYTNRVDII-------SFDPETLSFKT---------HPKLAFDHPYPPTKLM 86
Query: 104 RLPPR--------VADLRRR*NPTLR----RNPSVDLKSLLNGNKSSEYCGPLTSFDWNE 151
P R +DL LR R S++ ++LN +K+SE+C PLTSFDWN+
Sbjct: 87 FQPNRKSASSSSSCSDLLASTGDFLRLWEVRESSIEPVTVLNNSKTSEFCAPLTSFDWND 146
Query: 152 AEPCRIGTS 160
EP RIGTS
Sbjct: 147 VEPKRIGTS 155
>gb|AAK19614.1| GHTTG1 [Gossypium hirsutum]
Length = 345
Score = 61.2 bits (147), Expect = 1e-08
Identities = 44/129 (34%), Positives = 61/129 (47%), Gaps = 28/129 (21%)
Query: 44 LAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR*GVPAPRPPRHLQ 103
+A+ S LE N ++I+ DP +S + HP+ P PP L
Sbjct: 43 IALGSFLEDYTNRVDII-------SFDPETLSFKT---------HPKLAFDHPYPPTKLM 86
Query: 104 RLPPR--------VADLRRR*NPTLR----RNPSVDLKSLLNGNKSSEYCGPLTSFDWNE 151
P R +DL LR R S++ ++LN +K+SE+C PLTSFDWN+
Sbjct: 87 FQPNRKSASSSSSCSDLLASTGDFLRLWEVRESSIEPVTVLNNSKTSEFCAPLTSFDWND 146
Query: 152 AEPCRIGTS 160
EP RIGTS
Sbjct: 147 VEPKRIGTS 155
>gb|AAW39014.1| At5g24520 [Arabidopsis thaliana] gi|56121888|gb|AAV74225.1|
At5g24520 [Arabidopsis thaliana]
gi|10177852|dbj|BAB11204.1| Ttg1 protein [Arabidopsis
thaliana] gi|10636049|emb|CAC10523.1| transparent testa
glabra 1 protein [Arabidopsis thaliana]
gi|5123716|emb|CAB45372.1| Ttg1 protein [Arabidopsis
thaliana] gi|15238565|ref|NP_197840.1| transparent testa
glabra 1 protein (TTG1) [Arabidopsis thaliana]
gi|30689492|ref|NP_851070.1| transparent testa glabra 1
protein (TTG1) [Arabidopsis thaliana]
gi|30689487|ref|NP_851069.1| transparent testa glabra 1
protein (TTG1) [Arabidopsis thaliana]
gi|27151711|sp|Q9XGN1|TTG1_ARATH TRANSPARENT TESTA
GLABRA 1 protein (TTG1 protein)
Length = 341
Score = 60.5 bits (145), Expect = 2e-08
Identities = 51/167 (30%), Positives = 73/167 (43%), Gaps = 33/167 (19%)
Query: 4 PRKTSQRTTRLSETSHRPVESKAAKYQTKNWSVRHDKKYHLAIASLLEQCPNCIEIV*LD 63
P S+ T ++ S P+ + A S+R + +A+ S LE N I+I+ D
Sbjct: 6 PDSLSRSETAVTYDSPYPLYAMAFS------SLRSSSGHRIAVGSFLEDYNNRIDILSFD 59
Query: 64 DSNARSDPTRISPSSTRTHHQGHLHPR*GVPAPRPPRHLQRLPPRV-----ADLRRR*NP 118
SD + P P P PP L PP + DL
Sbjct: 60 -----SDSMTVKPL-----------PNLSFEHPYPPTKLMFSPPSLRRPSSGDLLASSGD 103
Query: 119 TLRR------NPSVDLKSLLNGNKSSEYCGPLTSFDWNEAEPCRIGT 159
LR + +V+ S+LN +K+SE+C PLTSFDWN+ EP R+GT
Sbjct: 104 FLRLWEINEDSSTVEPISVLNNSKTSEFCAPLTSFDWNDVEPKRLGT 150
>dbj|BAD89974.1| mutant protein of TTG1 [Arabidopsis thaliana]
Length = 332
Score = 60.5 bits (145), Expect = 2e-08
Identities = 51/167 (30%), Positives = 73/167 (43%), Gaps = 33/167 (19%)
Query: 4 PRKTSQRTTRLSETSHRPVESKAAKYQTKNWSVRHDKKYHLAIASLLEQCPNCIEIV*LD 63
P S+ T ++ S P+ + A S+R + +A+ S LE N I+I+ D
Sbjct: 6 PDSLSRSETAVTYDSPYPLYAMAFS------SLRSSSGHRIAVGSFLEDYNNRIDILSFD 59
Query: 64 DSNARSDPTRISPSSTRTHHQGHLHPR*GVPAPRPPRHLQRLPPRV-----ADLRRR*NP 118
SD + P P P PP L PP + DL
Sbjct: 60 -----SDSMTVKPL-----------PNLSFEHPYPPTKLMFSPPSLRRPSSGDLLASSGD 103
Query: 119 TLRR------NPSVDLKSLLNGNKSSEYCGPLTSFDWNEAEPCRIGT 159
LR + +V+ S+LN +K+SE+C PLTSFDWN+ EP R+GT
Sbjct: 104 FLRLWEINEDSSTVEPISVLNNSKTSEFCAPLTSFDWNDVEPKRLGT 150
>emb|CAC10524.1| transparent testa glabra 1 [Arabidopsis thaliana]
Length = 341
Score = 60.5 bits (145), Expect = 2e-08
Identities = 51/167 (30%), Positives = 73/167 (43%), Gaps = 33/167 (19%)
Query: 4 PRKTSQRTTRLSETSHRPVESKAAKYQTKNWSVRHDKKYHLAIASLLEQCPNCIEIV*LD 63
P S+ T ++ S P+ + A S+R + +A+ S LE N I+I+ D
Sbjct: 6 PDSLSRSETAVTYDSPYPLYAMAFS------SLRSSSGHRIAVGSFLEDYNNRIDILSFD 59
Query: 64 DSNARSDPTRISPSSTRTHHQGHLHPR*GVPAPRPPRHLQRLPPRV-----ADLRRR*NP 118
SD + P P P PP L PP + DL
Sbjct: 60 -----SDSMTVKPL-----------PNLSFEHPYPPTKLMFSPPSLRRPSSGDLLASFGD 103
Query: 119 TLRR------NPSVDLKSLLNGNKSSEYCGPLTSFDWNEAEPCRIGT 159
LR + +V+ S+LN +K+SE+C PLTSFDWN+ EP R+GT
Sbjct: 104 FLRLWEINEDSSTVEPISVLNNSKTSEFCAPLTSFDWNDVEPKRLGT 150
>emb|CAE66236.1| Hypothetical protein CBG11480 [Caenorhabditis briggsae]
Length = 477
Score = 60.1 bits (144), Expect = 2e-08
Identities = 46/137 (33%), Positives = 64/137 (46%), Gaps = 28/137 (20%)
Query: 34 WSVRHD--KKYHLAIASLLEQCPNCIEIV*LDDSNARSDPTRISPSSTRTHHQGHLHPR* 91
WS D +K+ LA++S +E+ N I IV LD+ G L PR
Sbjct: 156 WSAATDPSRKFRLAVSSFIEEYSNKITIVQLDEE------------------LGELVPRS 197
Query: 92 GVPAPRPPRHLQRLPPRVA---DLRRR*NPTLR-----RNPSVDLKSLLNGNKSSEYCGP 143
P P + +P + DL LR + + ++SLLN N+++EYC P
Sbjct: 198 QFDHPYPATKIMWIPDQKGNFPDLLATSGDYLRLWRIGTDNNARIESLLNTNRTAEYCAP 257
Query: 144 LTSFDWNEAEPCRIGTS 160
LTSFDWNE + IGTS
Sbjct: 258 LTSFDWNELDMNLIGTS 274
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.328 0.139 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 289,179,271
Number of Sequences: 2540612
Number of extensions: 11586764
Number of successful extensions: 33129
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 33031
Number of HSP's gapped (non-prelim): 90
length of query: 168
length of database: 863,360,394
effective HSP length: 118
effective length of query: 50
effective length of database: 563,568,178
effective search space: 28178408900
effective search space used: 28178408900
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0150.5


