

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
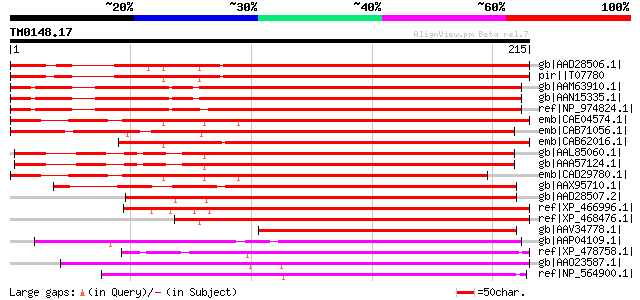
Query= TM0148.17
(215 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAD28506.1| remorin 1 [Lycopersicon esculentum] 234 9e-61
pir||T07780 remorin - potato gi|1881585|gb|AAB49425.1| remorin [... 233 2e-60
gb|AAM63910.1| remorin [Arabidopsis thaliana] 227 2e-58
gb|AAN15335.1| Unknown protein [Arabidopsis thaliana] gi|1017684... 226 3e-58
ref|NP_974824.1| remorin family protein [Arabidopsis thaliana] 225 7e-58
emb|CAE04574.1| OSJNBb0039L24.13 [Oryza sativa (japonica cultiva... 223 4e-57
emb|CAB71056.1| putative DNA-binding protein [Arabidopsis thalia... 216 5e-55
emb|CAB62016.1| remorin-like protein [Arabidopsis thaliana] gi|1... 200 3e-50
gb|AAL85060.1| putative remorin protein [Arabidopsis thaliana] g... 184 1e-45
gb|AAA57124.1| DNA-binding protein 182 4e-45
emb|CAD29780.1| putative remorin 1 protein [Oryza sativa] 179 4e-44
gb|AAX95710.1| Remorin, C-terminal region, putative [Oryza sativ... 178 8e-44
gb|AAD28507.2| remorin 2 [Lycopersicon esculentum] 177 2e-43
ref|XP_466996.1| putative remorin 1 [Oryza sativa (japonica cult... 166 4e-40
ref|XP_468476.1| putative DNA-binding protein [Oryza sativa (jap... 148 1e-34
gb|AAV34778.1| At4g00670 [Arabidopsis thaliana] gi|48310044|gb|A... 92 1e-17
gb|AAP04109.1| unknown protein [Arabidopsis thaliana] gi|2645263... 80 4e-14
ref|XP_478758.1| remorin-like protein [Oryza sativa (japonica cu... 79 7e-14
gb|AAO23587.1| At2g02170/F5O4.6 [Arabidopsis thaliana] gi|403803... 79 9e-14
ref|NP_564900.1| remorin family protein [Arabidopsis thaliana] g... 77 3e-13
>gb|AAD28506.1| remorin 1 [Lycopersicon esculentum]
Length = 197
Score = 234 bits (598), Expect = 9e-61
Identities = 139/219 (63%), Positives = 159/219 (72%), Gaps = 26/219 (11%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSII-PQP 59
M E + KKVE P+ PA P E APKE VA+EK+I+ P P
Sbjct: 1 MAELEAKKVEIVDPA----PAQEPVE-----------------APKEVVADEKAIVEPAP 39
Query: 60 SPP--ESKPVDDSKAIVKVE-KTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEES 116
PP E + DDSKA+V VE K +EAA+EK EGSI+RDAVL RVATEKRLSLIKAWEES
Sbjct: 40 PPPAEEKEKPDDSKALVVVENKAEEAADEKK-EGSIDRDAVLARVATEKRLSLIKAWEES 98
Query: 117 EKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREA 176
EKS A+NKA KK+S I AWENSK A E EL+K+EE LEKKKA+Y EK+KNKIA++H+EA
Sbjct: 99 EKSKAENKAQKKVSAIGAWENSKKANLESELKKMEEQLEKKKAIYTEKMKNKIALLHKEA 158
Query: 177 EEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
EEKRA IEAK+GEDLLKAEELAAKYRATGTAPKK F
Sbjct: 159 EEKRAMIEAKRGEDLLKAEELAAKYRATGTAPKKILGIF 197
>pir||T07780 remorin - potato gi|1881585|gb|AAB49425.1| remorin [Solanum
tuberosum] gi|34925093|sp|P93788|REMO_SOLTU Remorin
(pp34)
Length = 198
Score = 233 bits (595), Expect = 2e-60
Identities = 137/219 (62%), Positives = 156/219 (70%), Gaps = 25/219 (11%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M E + KKVE P+ PPAP P E APKE VA+EK+I+
Sbjct: 1 MAELEAKKVEIVDPA---PPAPGPVE-----------------APKEVVADEKAIVAPAL 40
Query: 61 PP---ESKPVDDSKAIVKVE-KTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEES 116
PP E + DDSKA+V VE K E A+EK EGSI+RDAVL RVATEKR+SLIKAWEES
Sbjct: 41 PPPAEEKEKPDDSKALVVVETKAPEPADEKK-EGSIDRDAVLARVATEKRVSLIKAWEES 99
Query: 117 EKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREA 176
EKS A+NKA KK+S I AWENSK A E EL+K+EE LEKKKA Y EK+KNKIA++H+EA
Sbjct: 100 EKSKAENKAQKKVSAIGAWENSKKANLEAELKKMEEQLEKKKAEYTEKMKNKIALLHKEA 159
Query: 177 EEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
EEKRA IEAK+GEDLLKAEELAAKYRATGTAPKK F
Sbjct: 160 EEKRAMIEAKRGEDLLKAEELAAKYRATGTAPKKILGIF 198
>gb|AAM63910.1| remorin [Arabidopsis thaliana]
Length = 202
Score = 227 bits (578), Expect = 2e-58
Identities = 125/212 (58%), Positives = 151/212 (70%), Gaps = 13/212 (6%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M EE+PKKV +E+ S P P P E KP AP+E ++P P+
Sbjct: 1 MAEEEPKKV-TETVSEPTPTPEVPVE---------KPAAAADVAPQEKPVAPPPVLPSPA 50
Query: 61 PPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSI 120
P E K +DSKAIV V + EE+ EGS+NRDAVL RV TEKR+SLIKAWEE+EK
Sbjct: 51 PAEEKQ-EDSKAIVPV--VPKEVEEEKKEGSVNRDAVLARVETEKRMSLIKAWEEAEKCK 107
Query: 121 ADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKR 180
+NKA KKLS I +WEN+K AA E EL+K+EE+LEKKKA YVE++KNKIA +H+EAEEKR
Sbjct: 108 VENKAEKKLSSIGSWENNKKAAVEAELKKMEEHLEKKKAEYVEQMKNKIAQIHKEAEEKR 167
Query: 181 AFIEAKKGEDLLKAEELAAKYRATGTAPKKPF 212
A IEAK+GE++LKAEELAAKYRATGTAPKK F
Sbjct: 168 AMIEAKRGEEILKAEELAAKYRATGTAPKKLF 199
>gb|AAN15335.1| Unknown protein [Arabidopsis thaliana] gi|10176842|dbj|BAB10048.1|
unnamed protein product [Arabidopsis thaliana]
gi|15237822|ref|NP_197764.1| remorin family protein
[Arabidopsis thaliana] gi|14423538|gb|AAK62451.1|
Unknown protein [Arabidopsis thaliana]
Length = 202
Score = 226 bits (577), Expect = 3e-58
Identities = 125/212 (58%), Positives = 150/212 (69%), Gaps = 13/212 (6%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M EE+PKKV +E+ S P P P E KP AP+E ++P P+
Sbjct: 1 MAEEEPKKV-TETVSEPTPTPEVPVE---------KPAAAADVAPQEKPVAPPPVLPSPA 50
Query: 61 PPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSI 120
P E K +DSKAIV V + EE+ EGS+NRDAVL RV TEKR+SLIKAWEE+EK
Sbjct: 51 PAEEKQ-EDSKAIVPV--VPKEVEEEKKEGSVNRDAVLARVETEKRMSLIKAWEEAEKCK 107
Query: 121 ADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKR 180
+NKA KKLS I +WEN+K AA E EL+K+EE LEKKKA YVE++KNKIA +H+EAEEKR
Sbjct: 108 VENKAEKKLSSIGSWENNKKAAVEAELKKMEEQLEKKKAEYVEQMKNKIAQIHKEAEEKR 167
Query: 181 AFIEAKKGEDLLKAEELAAKYRATGTAPKKPF 212
A IEAK+GE++LKAEELAAKYRATGTAPKK F
Sbjct: 168 AMIEAKRGEEILKAEELAAKYRATGTAPKKLF 199
>ref|NP_974824.1| remorin family protein [Arabidopsis thaliana]
Length = 201
Score = 225 bits (573), Expect = 7e-58
Identities = 125/212 (58%), Positives = 149/212 (69%), Gaps = 14/212 (6%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M EE+PKKV +E+ S P P P E KP AP+E ++P P+
Sbjct: 1 MAEEEPKKV-TETVSEPTPTPEVPVE---------KPAAAADVAPQEKPVAPPPVLPSPA 50
Query: 61 PPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSI 120
P E K +DSKAIV V EE+ EGS+NRDAVL RV TEKR+SLIKAWEE+EK
Sbjct: 51 PAEEKQ-EDSKAIVPVVPK---VEEEKKEGSVNRDAVLARVETEKRMSLIKAWEEAEKCK 106
Query: 121 ADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKR 180
+NKA KKLS I +WEN+K AA E EL+K+EE LEKKKA YVE++KNKIA +H+EAEEKR
Sbjct: 107 VENKAEKKLSSIGSWENNKKAAVEAELKKMEEQLEKKKAEYVEQMKNKIAQIHKEAEEKR 166
Query: 181 AFIEAKKGEDLLKAEELAAKYRATGTAPKKPF 212
A IEAK+GE++LKAEELAAKYRATGTAPKK F
Sbjct: 167 AMIEAKRGEEILKAEELAAKYRATGTAPKKLF 198
>emb|CAE04574.1| OSJNBb0039L24.13 [Oryza sativa (japonica cultivar-group)]
gi|50926698|ref|XP_473296.1| OSJNBb0039L24.13 [Oryza
sativa (japonica cultivar-group)]
Length = 206
Score = 223 bits (567), Expect = 4e-57
Identities = 128/220 (58%), Positives = 153/220 (69%), Gaps = 21/220 (9%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M EE+ KKVE E TE+ PA A A+ EP +DVAEEK++IP P+
Sbjct: 1 MAEEEAKKVEVEV-----------TEAPPAAAAAAETEPAA-----KDVAEEKAVIPAPA 44
Query: 61 PP---ESKPVDDSKAIVKVEKT-QEAAEEKPLEGSIN-RDAVLTRVATEKRLSLIKAWEE 115
PP E PVDDSKA+ VEK E EKP +G N RD L RV TEKR SLIKAWEE
Sbjct: 45 PPAEEEKPPVDDSKALAIVEKVADEPPAEKPAQGGSNDRDVALARVETEKRNSLIKAWEE 104
Query: 116 SEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHRE 175
+EK+ A+NKA KKLS I +WEN+K A E +L+KIEE LEKKKA Y EK+KNK+A+VH+E
Sbjct: 105 NEKTKAENKASKKLSAILSWENTKKANIEAQLKKIEEQLEKKKAEYAEKMKNKVAIVHKE 164
Query: 176 AEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
AEEKRA +EAK+GE++LKAEE+AAKYRATG APKK F
Sbjct: 165 AEEKRAMVEAKRGEEVLKAEEMAAKYRATGHAPKKLIGCF 204
>emb|CAB71056.1| putative DNA-binding protein [Arabidopsis thaliana]
gi|21593640|gb|AAM65607.1| putative DNA-binding protein
[Arabidopsis thaliana] gi|17104525|gb|AAL34151.1|
putative DNA-binding protein [Arabidopsis thaliana]
gi|13878167|gb|AAK44161.1| putative DNA-binding protein
[Arabidopsis thaliana] gi|15233068|ref|NP_191685.1|
DNA-binding family protein / remorin family protein
[Arabidopsis thaliana] gi|11281997|pir||T47918 probable
DNA-binding protein - Arabidopsis thaliana
Length = 212
Score = 216 bits (549), Expect = 5e-55
Identities = 119/213 (55%), Positives = 146/213 (67%), Gaps = 11/213 (5%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKE---DVAEEKSIIP 57
M EEQ +ESESP+ PAPA TPAP P P P + DVAEEK
Sbjct: 1 MAEEQKIALESESPAKVTTPAPA---DTPAPAPAEIPAPAPAPTPADVTKDVAEEKI--- 54
Query: 58 QPSPPESKPVDDSKAIVKVEK-TQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEES 116
+PP + DDSKA+ VEK +E A KP S++RD L ++ EKRLS ++AWEES
Sbjct: 55 -QNPPPEQIFDDSKALTVVEKPVEEPAPAKPASASLDRDVKLADLSKEKRLSFVRAWEES 113
Query: 117 EKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREA 176
EKS A+NKA KK++D+ AWENSK AA E +L+KIEE LEKKKA Y E++KNK+A +H+EA
Sbjct: 114 EKSKAENKAEKKIADVHAWENSKKAAVEAQLKKIEEQLEKKKAEYAERMKNKVAAIHKEA 173
Query: 177 EEKRAFIEAKKGEDLLKAEELAAKYRATGTAPK 209
EE+RA IEAK+GED+LKAEE AAKYRATG PK
Sbjct: 174 EERRAMIEAKRGEDVLKAEETAAKYRATGIVPK 206
>emb|CAB62016.1| remorin-like protein [Arabidopsis thaliana]
gi|15229057|ref|NP_190463.1| remorin family protein
[Arabidopsis thaliana] gi|11281998|pir||T46136
remorin-like protein - Arabidopsis thaliana
Length = 175
Score = 200 bits (508), Expect = 3e-50
Identities = 103/171 (60%), Positives = 132/171 (76%), Gaps = 2/171 (1%)
Query: 46 KEDVAEEKSIIPQPSPP-ESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATE 104
++ V +++ +PSPP + + DDSKAIV V +E E+K + GS++RDAVL R+ +
Sbjct: 6 QKKVIMPEAVASEPSPPSKEEKSDDSKAIVLVVAAKEPTEDKKV-GSVHRDAVLVRLEQD 64
Query: 105 KRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEK 164
KR+SLIKAWEE+EKS +NKA KK+S + AWENSK A+ E EL+KIEE L KKKA Y E+
Sbjct: 65 KRISLIKAWEEAEKSKVENKAQKKISSVGAWENSKKASVEAELKKIEEQLNKKKAHYTEQ 124
Query: 165 LKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
+KNKIA +H+EAEEKRA EAK+GED+LKAEE+AAKYRATGTAP K F FF
Sbjct: 125 MKNKIAQIHKEAEEKRAMTEAKRGEDVLKAEEMAAKYRATGTAPTKLFGFF 175
>gb|AAL85060.1| putative remorin protein [Arabidopsis thaliana]
gi|15028387|gb|AAK76670.1| putative remorin protein
[Arabidopsis thaliana] gi|3386612|gb|AAC28542.1| remorin
[Arabidopsis thaliana] gi|15225899|ref|NP_182106.1|
DNA-binding protein, putative [Arabidopsis thaliana]
gi|7484929|pir||T02465 remorin [imported] - Arabidopsis
thaliana
Length = 190
Score = 184 bits (467), Expect = 1e-45
Identities = 109/208 (52%), Positives = 134/208 (64%), Gaps = 28/208 (13%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
E++ KV+ ESP+ AP E P PV +VA+EK I P P
Sbjct: 4 EQKTSKVDVESPA------------VLAPAKEPTPAPV-------EVADEK--IHNPPPV 42
Query: 63 ESKPVDDSKAIVKVEKT-QEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIA 121
ESK A+ VEK +E +K GS +RD +L + EK+ S IKAWEESEKS A
Sbjct: 43 ESK------ALAVVEKPIEEHTPKKASSGSADRDVILADLEKEKKTSFIKAWEESEKSKA 96
Query: 122 DNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRA 181
+N+A KK+SD+ AWENSK AA E +LRKIEE LEKKKA Y EK+KNK+A +H+ AEEKRA
Sbjct: 97 ENRAQKKISDVHAWENSKKAAVEAQLRKIEEKLEKKKAQYGEKMKNKVAAIHKLAEEKRA 156
Query: 182 FIEAKKGEDLLKAEELAAKYRATGTAPK 209
+EAKKGE+LLKAEE+ AKYRATG PK
Sbjct: 157 MVEAKKGEELLKAEEMGAKYRATGVVPK 184
>gb|AAA57124.1| DNA-binding protein
Length = 190
Score = 182 bits (463), Expect = 4e-45
Identities = 108/208 (51%), Positives = 134/208 (63%), Gaps = 28/208 (13%)
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
E++ KV+ ESP+ AP E P PV +VA+EK I P P
Sbjct: 4 EQKTSKVDVESPA------------VLAPAKEPTPAPV-------EVADEK--IHNPPPV 42
Query: 63 ESKPVDDSKAIVKVEKT-QEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIA 121
ESK A+ VEK +E +K GS +RD +L + EK+ S IKAWEESEKS A
Sbjct: 43 ESK------ALAVVEKPIEEHTPKKASSGSADRDVILADLEKEKKTSFIKAWEESEKSKA 96
Query: 122 DNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRA 181
+N+A KK+SD+ AWENSK AA E +LRKIEE LEKKKA Y EK+KNK+A +H+ AEEKRA
Sbjct: 97 ENRAQKKISDVHAWENSKKAAVEAQLRKIEEKLEKKKAQYGEKMKNKVAAIHKLAEEKRA 156
Query: 182 FIEAKKGEDLLKAEELAAKYRATGTAPK 209
+EAKKGE+LL+AEE+ AKYRATG PK
Sbjct: 157 MVEAKKGEELLEAEEMGAKYRATGVVPK 184
>emb|CAD29780.1| putative remorin 1 protein [Oryza sativa]
Length = 195
Score = 179 bits (455), Expect = 4e-44
Identities = 108/203 (53%), Positives = 131/203 (64%), Gaps = 21/203 (10%)
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
M EE+ KKVE E TE+ PA A A+ EP +DVAEEK++IP P+
Sbjct: 1 MAEEEAKKVEVEV-----------TEAPPAAAAAAETEPAA-----KDVAEEKAVIPAPA 44
Query: 61 PP---ESKPVDDSKAIVKVEKT-QEAAEEKPLEGSIN-RDAVLTRVATEKRLSLIKAWEE 115
PP E PVDDSKA+ VEK E EKP +G N RD L RV TEKR SLIKAWEE
Sbjct: 45 PPAEEEKPPVDDSKALAIVEKVADEPPAEKPAQGGSNDRDVALARVETEKRNSLIKAWEE 104
Query: 116 SEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHRE 175
+EK+ A+NKA KKLS I +WEN+K A E +L+KIEE LEKKKA Y EK+ NK+A+VH E
Sbjct: 105 NEKTKAENKASKKLSAILSWENTKKANIEAQLKKIEEQLEKKKAEYSEKMXNKVAIVHXE 164
Query: 176 AEEKRAFIEAKKGEDLLKAEELA 198
EKRA +EA ++LKA +A
Sbjct: 165 XXEKRAMVEAXXRXEVLKAXXIA 187
>gb|AAX95710.1| Remorin, C-terminal region, putative [Oryza sativa (japonica
cultivar-group)]
Length = 171
Score = 178 bits (452), Expect = 8e-44
Identities = 100/192 (52%), Positives = 129/192 (67%), Gaps = 30/192 (15%)
Query: 19 PPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEK 78
PPAPAP P +D+AEE++ +P P ++SKA+ V+
Sbjct: 6 PPAPAPE-------------------PTKDIAEERAAVPAP--------EESKAMTVVDD 38
Query: 79 TQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENS 138
++AA GS RDA+LT VATEKR+SLIKAWEE+EK+ ADNKA KKL+DI++WENS
Sbjct: 39 AEKAAATG---GSHERDALLTTVATEKRISLIKAWEENEKAKADNKAAKKLADIASWENS 95
Query: 139 KIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELA 198
K+A E E++K +E LE+KKA VEKL N +A VHR AEEKRA EA++GE+++KAEE A
Sbjct: 96 KVAEIEAEIKKYQEYLERKKAEQVEKLMNGVAKVHRAAEEKRAATEARRGEEVVKAEEAA 155
Query: 199 AKYRATGTAPKK 210
AKYRA G PKK
Sbjct: 156 AKYRAKGEPPKK 167
>gb|AAD28507.2| remorin 2 [Lycopersicon esculentum]
Length = 174
Score = 177 bits (449), Expect = 2e-43
Identities = 93/168 (55%), Positives = 120/168 (71%), Gaps = 6/168 (3%)
Query: 49 VAEEKSIIPQPSPPESKPV-----DDSKAIVKVEKTQ-EAAEEKPLEGSINRDAVLTRVA 102
+AE ++ QP P + P DDSKAI + T+ +++ +K +GS +RD L +
Sbjct: 1 MAEATAVAAQPQPESTTPPPMAKSDDSKAIATLPPTKPDSSTKKSSKGSFDRDVALAHLE 60
Query: 103 TEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYV 162
EKR S IKAWEESEKS +NKA KKLS + WEN+K A E +L+K+EE LE+KKA Y
Sbjct: 61 EEKRNSYIKAWEESEKSKVNNKAEKKLSSVGTWENTKKANIEAKLKKLEEQLEQKKAEYA 120
Query: 163 EKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKK 210
EK+KNK+A VH EAEEKRA +EA++GE+LLKAEE+AAKYRATG APKK
Sbjct: 121 EKIKNKVAAVHMEAEEKRAMVEARRGEELLKAEEIAAKYRATGQAPKK 168
>ref|XP_466996.1| putative remorin 1 [Oryza sativa (japonica cultivar-group)]
gi|49388098|dbj|BAD25231.1| putative remorin 1 [Oryza
sativa (japonica cultivar-group)]
Length = 203
Score = 166 bits (420), Expect = 4e-40
Identities = 96/185 (51%), Positives = 123/185 (65%), Gaps = 17/185 (9%)
Query: 48 DVAEEKSIIP--QPSPPESK-------PVDDSKAIVK-VEKTQE-------AAEEKPLEG 90
+ EEK++IP SP SK P DDSKA+V VEK + A P
Sbjct: 17 NAGEEKAVIPAASTSPVISKTDDDTEPPADDSKALVVFVEKVADKPHAEKATATATPTRT 76
Query: 91 SINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKI 150
S +RD L +V T+KR SLIKAWEE+EK+ A+N+A KKL DI +WEN+K A + +L+K
Sbjct: 77 SNDRDIALAKVETDKRESLIKAWEENEKAKAENRASKKLLDIISWENTKKAVIKTQLKKK 136
Query: 151 EENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKK 210
EE LE+KKA Y EK KNK A+VH+EAEEKRA + A++GE+++KAEE+AAKYRATG PKK
Sbjct: 137 EEELERKKAEYAEKAKNKEAIVHKEAEEKRAMVMARRGEEVIKAEEIAAKYRATGVTPKK 196
Query: 211 PFSFF 215
F
Sbjct: 197 HIGCF 201
>ref|XP_468476.1| putative DNA-binding protein [Oryza sativa (japonica
cultivar-group)] gi|48717092|dbj|BAD22865.1| putative
DNA-binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 179
Score = 148 bits (373), Expect = 1e-34
Identities = 75/148 (50%), Positives = 106/148 (70%), Gaps = 1/148 (0%)
Query: 69 DSKAIVKVE-KTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHK 127
D+ A+ K + A + GS++RDA+L V E++LS+IKAWEESEKS A+NKA K
Sbjct: 30 DAGAVTKTNGPSAPAGKAATPTGSVDRDAILANVELERKLSMIKAWEESEKSKAENKAQK 89
Query: 128 KLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKK 187
K+S I +WEN++ AA E +LR EE LE+KKA Y EK++N++A +H+ AEEKRA +EA +
Sbjct: 90 KMSSILSWENTRKAAIEAKLRTQEEKLERKKAEYAEKMRNQVAAIHKAAEEKRATVEATR 149
Query: 188 GEDLLKAEELAAKYRATGTAPKKPFSFF 215
E+++K EE+AAK+R+ GT P K S F
Sbjct: 150 HEEIIKYEEMAAKHRSKGTTPTKFLSCF 177
>gb|AAV34778.1| At4g00670 [Arabidopsis thaliana] gi|48310044|gb|AAT41742.1|
At4g00670 [Arabidopsis thaliana]
Length = 123
Score = 92.0 bits (227), Expect = 1e-17
Identities = 43/107 (40%), Positives = 71/107 (66%)
Query: 104 EKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVE 163
+++ +IKAW+E + + +NK KKL DIS WE K E EL +I+ ++ KK E
Sbjct: 11 DEQSKVIKAWKELKITKVNNKTQKKLLDISGWEKKKTTKIESELARIQRKMDSKKMEKSE 70
Query: 164 KLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKK 210
KL+N+ A VH +A++K+A ++ ++ +++L AEE AA+++A G PKK
Sbjct: 71 KLRNEKAAVHAKAQKKKADVQTRRAQEILDAEEAAARFQAAGKIPKK 117
>gb|AAP04109.1| unknown protein [Arabidopsis thaliana] gi|26452636|dbj|BAC43401.1|
unknown protein [Arabidopsis thaliana]
gi|15220725|ref|NP_174322.1| remorin family protein
[Arabidopsis thaliana] gi|25518593|pir||E86427
hypothetical protein T4K22.7 - Arabidopsis thaliana
gi|12322121|gb|AAG51095.1| hypothetical protein
[Arabidopsis thaliana]
Length = 509
Score = 80.1 bits (196), Expect = 4e-14
Identities = 60/203 (29%), Positives = 94/203 (45%), Gaps = 7/203 (3%)
Query: 11 SESPSNPPPPAPAPTESTPAPEAEAKPEPV-VHDAPKEDVAEEKSIIPQPSPPESKPVDD 69
S +P P +PT S P+ +PE + + +++EE+ + V
Sbjct: 305 SVTPVGATTPLRSPTSSLPSTPRGGQPEESSMSKNTRRELSEEEEKAKTRREIVALGVQL 364
Query: 70 SKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKL 129
K + ++E E K G ++ EKR + AWEE+EKS + + ++
Sbjct: 365 GKMNIAAWASKEEEENKKNNGDAEE---AQKIEFEKRAT---AWEEAEKSKHNARYKREE 418
Query: 130 SDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGE 189
I AWE+ + A E E+R+IE +E+ KA K+ KIA+ + +EEKRA EA+K
Sbjct: 419 IRIQAWESQEKAKLEAEMRRIEAKVEQMKAEAEAKIMKKIALAKQRSEEKRALAEARKTR 478
Query: 190 DLLKAEELAAKYRATGTAPKKPF 212
D KA A R TG P +
Sbjct: 479 DAEKAVAEAQYIRETGRIPASSY 501
>ref|XP_478758.1| remorin-like protein [Oryza sativa (japonica cultivar-group)]
gi|33146770|dbj|BAC79688.1| remorin-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 316
Score = 79.3 bits (194), Expect = 7e-14
Identities = 57/170 (33%), Positives = 88/170 (51%), Gaps = 7/170 (4%)
Query: 47 EDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAV-LTRVATEK 105
ED EE + P P+S P+ + + A + G + D V + +V E+
Sbjct: 153 EDELEETN--PLAIVPDSNPIPSPR---RAHLALPAPGDVSSAGGGHGDEVSVGQVKKEE 207
Query: 106 RLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKL 165
S I AW+ +E + +N+ ++ I+ WE ++ L+K E LE+K+A +EK
Sbjct: 208 VESKIAAWQIAEVAKVNNRFKREEVVINGWEGDQVEKANAWLKKYERKLEEKRAKAMEKA 267
Query: 166 KNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
+N++A R+AEEKRA EAK+G + + ELA RA G AP K SFF
Sbjct: 268 QNEVAKARRKAEEKRASAEAKRGTKVARVLELANFMRAVGRAPSKR-SFF 316
>gb|AAO23587.1| At2g02170/F5O4.6 [Arabidopsis thaliana] gi|4038035|gb|AAC97217.1|
expressed protein [Arabidopsis thaliana]
gi|14326558|gb|AAK60323.1| At2g02170/F5O4.6 [Arabidopsis
thaliana] gi|25410968|pir||G84433 hypothetical protein
At2g02170 [imported] - Arabidopsis thaliana
gi|18395321|ref|NP_027421.1| remorin family protein
[Arabidopsis thaliana]
Length = 486
Score = 79.0 bits (193), Expect = 9e-14
Identities = 59/226 (26%), Positives = 93/226 (41%), Gaps = 32/226 (14%)
Query: 22 PAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQE 81
P+ S + + P+ P + ++ P SP S+P + + +
Sbjct: 257 PSTARSVSMRDMGTEMTPIASQEPSRNGTPIRATTPIRSPISSEPSSPGRQASASPMSNK 316
Query: 82 AAEEKPLEGSINRDAVL------------------------TRVATEKRLSLIK------ 111
EK L+ R+ ++ T + T+ L K
Sbjct: 317 ELSEKELQMKTRREIMVLGTQLGKFNIAAWASKEDEDKDASTSLKTKASLQTSKSVSEAR 376
Query: 112 --AWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKI 169
AWEE+EK+ + ++ I AWEN + A E E++K E +E+ K ++L K+
Sbjct: 377 ATAWEEAEKAKHMARFRREEMKIQAWENHQKAKSEAEMKKTEVKVERIKGRAQDRLMKKL 436
Query: 170 AMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
A + R+AEEKRA EAKK K E+ A + R TG P FS F
Sbjct: 437 ATIERKAEEKRAAAEAKKDHQAAKTEKQAEQIRRTGKVPSLLFSCF 482
>ref|NP_564900.1| remorin family protein [Arabidopsis thaliana]
gi|12324673|gb|AAG52296.1| unknown protein [Arabidopsis
thaliana] gi|11072019|gb|AAG28898.1| F12A21.28
[Arabidopsis thaliana]
Length = 347
Score = 77.0 bits (188), Expect = 3e-13
Identities = 57/180 (31%), Positives = 85/180 (46%), Gaps = 5/180 (2%)
Query: 39 PVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVL 98
P+ P ++ P P + PV S+ V E E S N + V
Sbjct: 158 PIGSQEPSRTATPVRATTPVGRSPVTSPVRASQRGEAVGVVMETVTEVRRVESNNSEKVN 217
Query: 99 TRVATEKRLSLIKA----WEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENL 154
V ++K +S ++A W+E+E++ + ++ I AWEN + E+E++K+E
Sbjct: 218 GFVESKKAMSAMEARAMAWDEAERAKFMARYKREEVKIQAWENHEKRKAEMEMKKMEVKA 277
Query: 155 EKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSF 214
E+ KA EKL NK+A R AEE+RA EAK E +K E A R +G P FSF
Sbjct: 278 ERMKARAEEKLANKLAATKRIAEERRANAEAKLNEKAVKTSEKADYIRRSGHLPSS-FSF 336
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.302 0.122 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 385,489,438
Number of Sequences: 2540612
Number of extensions: 18174154
Number of successful extensions: 284646
Number of sequences better than 10.0: 9541
Number of HSP's better than 10.0 without gapping: 1948
Number of HSP's successfully gapped in prelim test: 7978
Number of HSP's that attempted gapping in prelim test: 197275
Number of HSP's gapped (non-prelim): 47438
length of query: 215
length of database: 863,360,394
effective HSP length: 123
effective length of query: 92
effective length of database: 550,865,118
effective search space: 50679590856
effective search space used: 50679590856
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0148.17


