

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0147b.4
(313 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
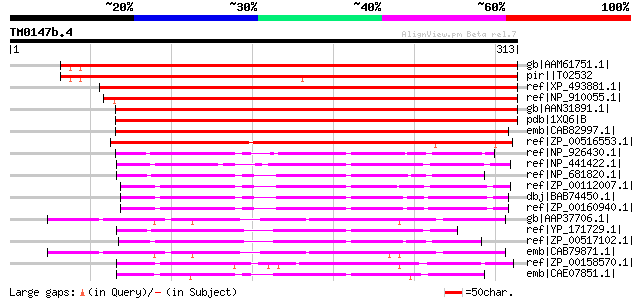
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM61751.1| putative 3-beta hydroxysteroid dehydrogenase/isom... 432 e-120
pir||T02532 hypothetical protein At2g37660 [imported] - Arabidop... 424 e-117
ref|XP_493881.1| putative 3-beta hydroxysteroid dehydrogenase/is... 417 e-115
ref|NP_910055.1| unknown protein [Oryza sativa (japonica cultiva... 413 e-114
gb|AAN31891.1| unknown protein [Arabidopsis thaliana] gi|2265543... 388 e-106
pdb|1XQ6|B Chain B, X-Ray Structure Of Gene Product From Arabido... 388 e-106
emb|CAB82997.1| putative protein [Arabidopsis thaliana] gi|11358... 380 e-104
ref|ZP_00516553.1| similar to Nucleoside-diphosphate-sugar epime... 253 4e-66
ref|NP_926430.1| hypothetical protein gll3484 [Gloeobacter viola... 130 6e-29
ref|NP_441422.1| hypothetical protein sll1218 [Synechocystis sp.... 123 6e-27
ref|NP_681820.1| hypothetical protein tll1029 [Thermosynechococc... 116 1e-24
ref|ZP_00112007.1| COG0702: Predicted nucleoside-diphosphate-sug... 114 3e-24
dbj|BAB74450.1| alr2751 [Nostoc sp. PCC 7120] gi|17230243|ref|NP... 113 6e-24
ref|ZP_00160940.1| COG0702: Predicted nucleoside-diphosphate-sug... 110 5e-23
gb|AAP37706.1| At4g31530 [Arabidopsis thaliana] gi|26449707|dbj|... 106 1e-21
ref|YP_171729.1| hypothetical protein syc1019_d [Synechococcus e... 103 6e-21
ref|ZP_00517102.1| similar to Nucleoside-diphosphate-sugar epime... 102 1e-20
emb|CAB79871.1| putative protein [Arabidopsis thaliana] gi|52627... 100 7e-20
ref|ZP_00158570.1| COG0702: Predicted nucleoside-diphosphate-sug... 99 2e-19
emb|CAE07851.1| conserved hypothetical protein [Synechococcus sp... 92 2e-17
>gb|AAM61751.1| putative 3-beta hydroxysteroid dehydrogenase/isomerase protein
[Arabidopsis thaliana] gi|20197253|gb|AAC23636.2|
expressed protein [Arabidopsis thaliana]
gi|20148255|gb|AAM10018.1| unknown protein [Arabidopsis
thaliana] gi|14596079|gb|AAK68767.1| Unknown protein
[Arabidopsis thaliana] gi|18404496|ref|NP_565868.1|
expressed protein [Arabidopsis thaliana]
gi|22096383|sp|O80934|Y230_ARATH Protein At2g37660,
chloroplast precursor
Length = 325
Score = 432 bits (1112), Expect = e-120
Identities = 216/289 (74%), Positives = 253/289 (86%), Gaps = 7/289 (2%)
Query: 32 TSLQF----SHSSSL---SLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKL 84
+SLQF S S+S+ S T K R +V A A + TVLVTGAGGRTG+IVYKKL
Sbjct: 37 SSLQFRSLVSDSTSICGPSKFTGKNRRVSVTVSAAATTEPLTVLVTGAGGRTGQIVYKKL 96
Query: 85 QERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMK 144
+ERS Q++ARGLVRT+ESK+ I D+V++GDIRDT SIAPA++GIDAL+ILTSAVP MK
Sbjct: 97 KERSEQFVARGLVRTKESKEKINGEDEVFIGDIRDTASIAPAVEGIDALVILTSAVPQMK 156
Query: 145 PGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLN 204
PGF+P+KG RPEF+F+DGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGT++N+PLN
Sbjct: 157 PGFDPSKGGRPEFFFDDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTNINHPLN 216
Query: 205 SLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKDDEILKTETRTIA 264
S+GN NILVWKRKAEQYLADSGIPYTIIRAGGLQDK+GG+REL++GKDDE+L+TETRTIA
Sbjct: 217 SIGNANILVWKRKAEQYLADSGIPYTIIRAGGLQDKDGGIRELLVGKDDELLETETRTIA 276
Query: 265 RPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFSQITTRF 313
R DVAEVC+QAL EEA+FKA DLASKPEG GTPT+DFKALF+Q+TT+F
Sbjct: 277 RADVAEVCVQALQLEEAKFKALDLASKPEGTGTPTKDFKALFTQVTTKF 325
>pir||T02532 hypothetical protein At2g37660 [imported] - Arabidopsis thaliana
Length = 337
Score = 424 bits (1089), Expect = e-117
Identities = 216/301 (71%), Positives = 253/301 (83%), Gaps = 19/301 (6%)
Query: 32 TSLQF----SHSSSL---SLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKKL 84
+SLQF S S+S+ S T K R +V A A + TVLVTGAGGRTG+IVYKKL
Sbjct: 37 SSLQFRSLVSDSTSICGPSKFTGKNRRVSVTVSAAATTEPLTVLVTGAGGRTGQIVYKKL 96
Query: 85 QERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMK 144
+ERS Q++ARGLVRT+ESK+ I D+V++GDIRDT SIAPA++GIDAL+ILTSAVP MK
Sbjct: 97 KERSEQFVARGLVRTKESKEKINGEDEVFIGDIRDTASIAPAVEGIDALVILTSAVPQMK 156
Query: 145 PGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDA------------AKAAGVKQIVLVG 192
PGF+P+KG RPEF+F+DGAYPEQVDWIGQKNQIDA AKAAGVKQIVLVG
Sbjct: 157 PGFDPSKGGRPEFFFDDGAYPEQVDWIGQKNQIDAGVDDDGLSFVVTAKAAGVKQIVLVG 216
Query: 193 SMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRELIIGKD 252
SMGGT++N+PLNS+GN NILVWKRKAEQYLADSGIPYTIIRAGGLQDK+GG+REL++GKD
Sbjct: 217 SMGGTNINHPLNSIGNANILVWKRKAEQYLADSGIPYTIIRAGGLQDKDGGIRELLVGKD 276
Query: 253 DEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFSQITTR 312
DE+L+TETRTIAR DVAEVC+QAL EEA+FKA DLASKPEG GTPT+DFKALF+Q+TT+
Sbjct: 277 DELLETETRTIARADVAEVCVQALQLEEAKFKALDLASKPEGTGTPTKDFKALFTQVTTK 336
Query: 313 F 313
F
Sbjct: 337 F 337
>ref|XP_493881.1| putative 3-beta hydroxysteroid dehydrogenase/isomerase protein
[Oryza sativa] gi|52353632|gb|AAU44198.1| putative
3-beta hydroxysteroid dehydrogenase/isomerase [Oryza
sativa (japonica cultivar-group)]
gi|14719331|gb|AAK73149.1| putative 3-beta
hydroxysteroid dehydrogenase/isomerase protein [Oryza
sativa]
Length = 293
Score = 417 bits (1073), Expect = e-115
Identities = 205/258 (79%), Positives = 230/258 (88%)
Query: 56 VVAMADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVG 115
V A A TVLVTGAGGRTG+IVYKKL+ER+ Q++ RGLVRTEESK IG + DV+VG
Sbjct: 36 VSAAAGGPLPTVLVTGAGGRTGQIVYKKLKERADQFVGRGLVRTEESKAKIGGAADVFVG 95
Query: 116 DIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKN 175
DIRD SIAPAI GIDALIILTSAVP MKPGF+P+KG RPEFYFEDG+YPEQVDWIGQ+N
Sbjct: 96 DIRDPASIAPAIDGIDALIILTSAVPKMKPGFDPSKGGRPEFYFEDGSYPEQVDWIGQRN 155
Query: 176 QIDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAG 235
QIDAAK+ GVKQ+VLVGSMGGTD+N+PLN LGN NILVWKRKAEQYLADSG+PYTIIRAG
Sbjct: 156 QIDAAKSIGVKQVVLVGSMGGTDVNHPLNKLGNANILVWKRKAEQYLADSGLPYTIIRAG 215
Query: 236 GLQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGA 295
GLQDK+GGVREL++GKDDEILKTET+TI R DVAEVC+QAL FEEA+FKAFDLASKPEG
Sbjct: 216 GLQDKDGGVRELLVGKDDEILKTETKTITRADVAEVCLQALLFEEARFKAFDLASKPEGE 275
Query: 296 GTPTRDFKALFSQITTRF 313
G PT DF+ALFSQ+ +RF
Sbjct: 276 GVPTTDFRALFSQVNSRF 293
>ref|NP_910055.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|27545035|gb|AAO18441.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 257
Score = 413 bits (1061), Expect = e-114
Identities = 200/257 (77%), Positives = 228/257 (87%), Gaps = 2/257 (0%)
Query: 59 MADSS--RSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGD 116
MADSS R TVLVTGAGGRTG IVY KL+ERS Q++ RGLVRTEESKQ IG +DVY+ D
Sbjct: 1 MADSSAARPTVLVTGAGGRTGNIVYNKLKERSDQFVVRGLVRTEESKQKIGGGNDVYIAD 60
Query: 117 IRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQ 176
IRD + PA+QG+DALIILTSAVP MKPGF+P+KG RPEFY+EDG YPEQVDWIGQKNQ
Sbjct: 61 IRDRDHLVPAVQGVDALIILTSAVPKMKPGFDPSKGGRPEFYYEDGMYPEQVDWIGQKNQ 120
Query: 177 IDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGG 236
ID AKAAGVK IVLVGSMGGT+ N+PLNSLGNGNILVWKRK+EQYLADSG+PYTIIR GG
Sbjct: 121 IDTAKAAGVKHIVLVGSMGGTNPNHPLNSLGNGNILVWKRKSEQYLADSGVPYTIIRPGG 180
Query: 237 LQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAG 296
LQDK+GGVRELI+G DDE+L+T+T++I R DVAEVC+QAL +EE +FKAFDLASKPEG G
Sbjct: 181 LQDKDGGVRELIVGNDDELLQTDTKSIPRADVAEVCVQALQYEETKFKAFDLASKPEGTG 240
Query: 297 TPTRDFKALFSQITTRF 313
TPT+DFK+LFSQ+T RF
Sbjct: 241 TPTKDFKSLFSQVTARF 257
>gb|AAN31891.1| unknown protein [Arabidopsis thaliana] gi|22655434|gb|AAM98309.1|
At5g02240/T7H20_290 [Arabidopsis thaliana]
gi|62320775|dbj|BAD95439.1| hypothetical protein
[Arabidopsis thaliana] gi|18413869|ref|NP_568098.1|
expressed protein [Arabidopsis thaliana]
gi|15294290|gb|AAK95322.1| AT5g02240/T7H20_290
[Arabidopsis thaliana]
Length = 253
Score = 388 bits (996), Expect = e-106
Identities = 189/248 (76%), Positives = 217/248 (87%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAP 125
TVLVTGA GRTG+IVYKKL+E S +++A+GLVR+ + K+ IG DV++GDI D SI P
Sbjct: 6 TVLVTGASGRTGQIVYKKLKEGSDKFVAKGLVRSAQGKEKIGGEADVFIGDITDADSINP 65
Query: 126 AIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGV 185
A QGIDAL+ILTSAVP MKPGF+PTKG RPEF FEDG YPEQVDWIGQKNQIDAAK AGV
Sbjct: 66 AFQGIDALVILTSAVPKMKPGFDPTKGGRPEFIFEDGQYPEQVDWIGQKNQIDAAKVAGV 125
Query: 186 KQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVR 245
K IV+VGSMGGT+ ++PLN LGNGNILVWKRKAEQYLADSG PYTIIRAGGL DKEGGVR
Sbjct: 126 KHIVVVGSMGGTNPDHPLNKLGNGNILVWKRKAEQYLADSGTPYTIIRAGGLLDKEGGVR 185
Query: 246 ELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKAL 305
EL++GKDDE+L+T+T+T+ R DVAEVCIQAL FEEA+ KAFDL SKPEG TPT+DFKAL
Sbjct: 186 ELLVGKDDELLQTDTKTVPRADVAEVCIQALLFEEAKNKAFDLGSKPEGTSTPTKDFKAL 245
Query: 306 FSQITTRF 313
FSQ+T+RF
Sbjct: 246 FSQVTSRF 253
>pdb|1XQ6|B Chain B, X-Ray Structure Of Gene Product From Arabidopsis Thaliana
At5g02240 gi|56554664|pdb|1XQ6|A Chain A, X-Ray
Structure Of Gene Product From Arabidopsis Thaliana
At5g02240 gi|60594215|pdb|1YBM|B Chain B, X-Ray
Structure Of Selenomethionyl Gene Product From
Arabidopsis Thaliana At5g02240 In Space Group P21212
gi|60594214|pdb|1YBM|A Chain A, X-Ray Structure Of
Selenomethionyl Gene Product From Arabidopsis Thaliana
At5g02240 In Space Group P21212
Length = 253
Score = 388 bits (996), Expect = e-106
Identities = 189/248 (76%), Positives = 217/248 (87%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAP 125
TVLVTGA GRTG+IVYKKL+E S +++A+GLVR+ + K+ IG DV++GDI D SI P
Sbjct: 6 TVLVTGASGRTGQIVYKKLKEGSDKFVAKGLVRSAQGKEKIGGEADVFIGDITDADSINP 65
Query: 126 AIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGV 185
A QGIDAL+ILTSAVP MKPGF+PTKG RPEF FEDG YPEQVDWIGQKNQIDAAK AGV
Sbjct: 66 AFQGIDALVILTSAVPKMKPGFDPTKGGRPEFIFEDGQYPEQVDWIGQKNQIDAAKVAGV 125
Query: 186 KQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVR 245
K IV+VGSMGGT+ ++PLN LGNGNILVWKRKAEQYLADSG PYTIIRAGGL DKEGGVR
Sbjct: 126 KHIVVVGSMGGTNPDHPLNKLGNGNILVWKRKAEQYLADSGTPYTIIRAGGLLDKEGGVR 185
Query: 246 ELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKAL 305
EL++GKDDE+L+T+T+T+ R DVAEVCIQAL FEEA+ KAFDL SKPEG TPT+DFKAL
Sbjct: 186 ELLVGKDDELLQTDTKTVPRADVAEVCIQALLFEEAKNKAFDLGSKPEGTSTPTKDFKAL 245
Query: 306 FSQITTRF 313
FSQ+T+RF
Sbjct: 246 FSQVTSRF 253
>emb|CAB82997.1| putative protein [Arabidopsis thaliana] gi|11358410|pir||T48245
hypothetical protein T7H20.290 - Arabidopsis thaliana
Length = 376
Score = 380 bits (976), Expect = e-104
Identities = 186/243 (76%), Positives = 212/243 (86%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAP 125
TVLVTGA GRTG+IVYKKL+E S +++A+GLVR+ + K+ IG DV++GDI D SI P
Sbjct: 6 TVLVTGASGRTGQIVYKKLKEGSDKFVAKGLVRSAQGKEKIGGEADVFIGDITDADSINP 65
Query: 126 AIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGV 185
A QGIDAL+ILTSAVP MKPGF+PTKG RPEF FEDG YPEQVDWIGQKNQIDAAK AGV
Sbjct: 66 AFQGIDALVILTSAVPKMKPGFDPTKGGRPEFIFEDGQYPEQVDWIGQKNQIDAAKVAGV 125
Query: 186 KQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVR 245
K IV+VGSMGGT+ ++PLN LGNGNILVWKRKAEQYLADSG PYTIIRAGGL DKEGGVR
Sbjct: 126 KHIVVVGSMGGTNPDHPLNKLGNGNILVWKRKAEQYLADSGTPYTIIRAGGLLDKEGGVR 185
Query: 246 ELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKAL 305
EL++GKDDE+L+T+T+T+ R DVAEVCIQAL FEEA+ KAFDL SKPEG TPT+DFKAL
Sbjct: 186 ELLVGKDDELLQTDTKTVPRADVAEVCIQALLFEEAKNKAFDLGSKPEGTSTPTKDFKAL 245
Query: 306 FSQ 308
FSQ
Sbjct: 246 FSQ 248
>ref|ZP_00516553.1| similar to Nucleoside-diphosphate-sugar epimerases [Crocosphaera
watsonii WH 8501] gi|67855074|gb|EAM50341.1| similar to
Nucleoside-diphosphate-sugar epimerases [Crocosphaera
watsonii WH 8501]
Length = 257
Score = 253 bits (647), Expect = 4e-66
Identities = 136/255 (53%), Positives = 176/255 (68%), Gaps = 9/255 (3%)
Query: 63 SRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEES-KQTIGASDDVYVGDIRDTG 121
S+ VLVTGA GRTG V KKL++ S ++ G R E K+ G+++ +VGDI +
Sbjct: 2 SKKRVLVTGATGRTGLFVVKKLRQTSDKFEVFGFARDNEKVKELFGSTEGFFVGDITNKS 61
Query: 122 SIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAK 181
S+ PA++G D+L+I+TS+ P MK +G+RPEF FE G PE+VDWIGQKNQID AK
Sbjct: 62 SLEPALKGCDSLVIVTSSFPKMKAPAQ--EGQRPEFEFEPGGMPEEVDWIGQKNQIDLAK 119
Query: 182 AAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKE 241
G+ +IVLVGSMGGT+ +PLN +GNGN+L+WKRKAE+YL DSGI YTIIRAGGL ++
Sbjct: 120 ELGINKIVLVGSMGGTNREHPLNKMGNGNVLIWKRKAEEYLIDSGIDYTIIRAGGLINEP 179
Query: 242 GGVRELIIGKDDEILKTETR----TIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGT 297
GG RELI+GK+D L+ I R DVAE+ +QAL A+ KAFD+ SKPE T
Sbjct: 180 GGKRELIVGKNDTFLENPPNGIPTVIPREDVAELVVQALIESTAKNKAFDVISKPEDDST 239
Query: 298 P--TRDFKALFSQIT 310
T+DF +LF Q T
Sbjct: 240 ANITKDFASLFGQTT 254
>ref|NP_926430.1| hypothetical protein gll3484 [Gloeobacter violaceus PCC 7421]
gi|35214056|dbj|BAC91425.1| gll3484 [Gloeobacter
violaceus PCC 7421]
Length = 228
Score = 130 bits (326), Expect = 6e-29
Identities = 83/235 (35%), Positives = 135/235 (57%), Gaps = 27/235 (11%)
Query: 66 TVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTE-ESKQTIGASDDVYVGDIRDTGSIA 124
++LV GA G+TG+ + KKL R+ R L R+ ++++ G +V GD+ T S+
Sbjct: 2 SILVVGATGQTGQQIVKKL--RAQSMAPRVLARSRAKAREVFGDGTEVVEGDVLKTDSLG 59
Query: 125 PAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAG 184
PA+ G++ + T + GF +GA +QVD+ G +N + AA+ AG
Sbjct: 60 PALNGVETIFCATGT----RTGFGA-----------NGA--QQVDYEGTRNLVYAARRAG 102
Query: 185 VKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGV 244
V +++LV S+ + L +PLN G +L WK++AE YL DSG+ +TI+R GGL+D GG
Sbjct: 103 VGRLILVSSLCVSRLIHPLNLFGG--VLFWKKRAEDYLLDSGLNFTIVRPGGLRDGAGGA 160
Query: 245 RELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPT 299
E+++ D + + TI R DVA VC++AL E+++K ++ + P GA P+
Sbjct: 161 -EIVVRPADTLFE---GTIDRADVARVCVEALGSAESEYKIVEIVAGP-GAAQPS 210
>ref|NP_441422.1| hypothetical protein sll1218 [Synechocystis sp. PCC 6803]
gi|1653186|dbj|BAA18102.1| ycf39 [Synechocystis sp. PCC
6803] gi|7450531|pir||S75541 hypothetical protein
sll1218 - Synechocystis sp. (strain PCC 6803)
Length = 219
Score = 123 bits (309), Expect = 6e-27
Identities = 87/244 (35%), Positives = 131/244 (53%), Gaps = 29/244 (11%)
Query: 67 VLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEES-KQTIGASDDVYVGDIRDTGSIAP 125
VLV GA G TGK V L +R Q R LVR +S K + ++ VGD+ + +I
Sbjct: 3 VLVIGATGETGKRVVNTLTDR--QIAVRALVRNYDSAKAVLPPGTEIMVGDLLEPETIKA 60
Query: 126 AIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGV 185
AI G +++ +A G RP D P +VD++G +N +D AKA G+
Sbjct: 61 AIAG--CTVVINAA------------GARPS---ADLTGPFKVDYLGTRNLVDIAKANGI 103
Query: 186 KQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVR 245
+Q+VLV S+ ++L +PLN G ILVWK+ E YL SG+PYTI+R GGL++++
Sbjct: 104 EQLVLVSSLCVSNLFHPLNLFGL--ILVWKQWGENYLRQSGVPYTIVRPGGLKNEDNDNA 161
Query: 246 ELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKAL 305
++ G D +I R VAE C+++L A+ K ++ SKP+ P + F L
Sbjct: 162 IVMAGADTLF----DGSIPRQKVAEACVESLFSPSAKNKIVEIVSKPD---IPVQSFDEL 214
Query: 306 FSQI 309
F+ +
Sbjct: 215 FAMV 218
>ref|NP_681820.1| hypothetical protein tll1029 [Thermosynechococcus elongatus BP-1]
gi|22294753|dbj|BAC08582.1| ycf39 [Thermosynechococcus
elongatus BP-1]
Length = 228
Score = 116 bits (290), Expect = 1e-24
Identities = 78/227 (34%), Positives = 123/227 (53%), Gaps = 25/227 (11%)
Query: 67 VLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPA 126
V V GA GRTG+ + LQ SS++ A +VR Q + ++ + D+ ++ PA
Sbjct: 11 VAVVGATGRTGQRIVSALQ--SSEHQAIAVVRNPAKAQGRWPTVEIRIADVTQPQTLPPA 68
Query: 127 IQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVK 186
++ +A+I T A P NP + P VD++G KN +DAAKA V+
Sbjct: 69 LKDCEAVICATGA----SPNLNPLE-------------PLSVDYLGTKNLVDAAKATQVQ 111
Query: 187 QIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRE 246
Q +LV S+ + +PLN IL WK++AE+YL +SG+ YTI+R GGL++ + G
Sbjct: 112 QFILVSSLCVSQFFHPLNLFWL--ILYWKQQAERYLQESGLTYTIVRPGGLKETDDGGFP 169
Query: 247 LIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPE 293
II + D + + +I R VAE+C+ AL A K F++ ++P+
Sbjct: 170 -IIARADTLFE---GSIPRSRVAEICVAALGEPSAYNKIFEVVNRPD 212
>ref|ZP_00112007.1| COG0702: Predicted nucleoside-diphosphate-sugar epimerases [Nostoc
punctiforme PCC 73102]
Length = 219
Score = 114 bits (286), Expect = 3e-24
Identities = 82/242 (33%), Positives = 131/242 (53%), Gaps = 29/242 (11%)
Query: 69 VTGAGGRTGKIVYKKLQERSSQYIARGLVRT-EESKQTIGASDDVYVGDIRDTGSIAPAI 127
V GA G TG+ + ++L R+ R LVR E++K + ++ VGD+ SI A+
Sbjct: 5 VAGATGETGRRIVQELIARNIP--VRALVRDIEKAKGILSPEAELVVGDVLQPESITAAL 62
Query: 128 QGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQ 187
L++ T A KP F+PT P +VD+ G KN +DAAKA G++
Sbjct: 63 GDSTVLLVATGA----KPSFDPTG-------------PYKVDFEGTKNLVDAAKAKGIEH 105
Query: 188 IVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVREL 247
VLV S+ + +PLN ILVWK++AE+Y+ SG+ YTI+R GGL++ E + +
Sbjct: 106 FVLVSSLCTSQFFHPLNLFWL--ILVWKKQAEEYIQKSGLTYTIVRPGGLKN-EDNLDAI 162
Query: 248 IIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFS 307
++ D + +I R VA+V ++AL +A+ K ++ +KPE A ++ F LF+
Sbjct: 163 VMQSADTLF---DGSIPRQKVAQVAVEALFEADARNKIVEIVAKPEAA---SKSFGELFA 216
Query: 308 QI 309
+
Sbjct: 217 NV 218
>dbj|BAB74450.1| alr2751 [Nostoc sp. PCC 7120] gi|17230243|ref|NP_486791.1|
hypothetical protein alr2751 [Nostoc sp. PCC 7120]
gi|25327951|pir||AH2149 hypothetical protein alr2751
[imported] - Nostoc sp. (strain PCC 7120)
Length = 218
Score = 113 bits (283), Expect = 6e-24
Identities = 81/241 (33%), Positives = 132/241 (54%), Gaps = 29/241 (12%)
Query: 69 VTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASD-DVYVGDIRDTGSIAPAI 127
V GA G TG+ + ++L R+ R LVR E++ + I D ++ VGD+ + S+ A+
Sbjct: 5 VAGATGETGRRIVQELIARNIP--VRALVRDEQTARAILPPDAELVVGDVLNPASLTAAL 62
Query: 128 QGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQ 187
++ T A KP F+PT P +VD+ G KN +D AKA G++
Sbjct: 63 GDSTVVLCATGA----KPSFDPTG-------------PYKVDFEGTKNLVDVAKAKGIEN 105
Query: 188 IVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVREL 247
VLV S+ + +PLN ILVWK++AE+YL SG+ YTI+R GGL++++ +
Sbjct: 106 FVLVTSLCVSQFFHPLNLFWL--ILVWKKQAEEYLQKSGLTYTIVRPGGLKNEDNS--DA 161
Query: 248 IIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFS 307
I+ + + L +I R VA+VC+++L +A+ K ++ +KPE + ++ F LF
Sbjct: 162 IVMQSSDTL--FDGSIPRQKVAQVCVESLFEPDARNKIVEIVAKPEAS---SKTFTELFQ 216
Query: 308 Q 308
Q
Sbjct: 217 Q 217
>ref|ZP_00160940.1| COG0702: Predicted nucleoside-diphosphate-sugar epimerases
[Anabaena variabilis ATCC 29413]
Length = 218
Score = 110 bits (275), Expect = 5e-23
Identities = 81/241 (33%), Positives = 130/241 (53%), Gaps = 29/241 (12%)
Query: 69 VTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASD-DVYVGDIRDTGSIAPAI 127
V GA G TG+ + ++L R+ R LVR E + + I D ++ VGD+ + S+ A+
Sbjct: 5 VAGATGETGRRIVQELIARNIP--VRALVRDEHTARAILPPDTELVVGDVLNPASLTAAL 62
Query: 128 QGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQ 187
++ T A KP F+PT P +VD+ G KN +D AKA G++
Sbjct: 63 GDSTVVLCATGA----KPSFDPTG-------------PYKVDFEGTKNLVDVAKAKGIEN 105
Query: 188 IVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVREL 247
VLV S+ + +PLN ILVWK++AE+YL SG+ YTI+R GGL++++ +
Sbjct: 106 FVLVTSLCVSQFFHPLNLFWL--ILVWKKQAEEYLQKSGLTYTIVRPGGLKNEDNS--DA 161
Query: 248 IIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALFS 307
I+ + + L +I R VA+VC+++L A+ K ++ +KPE + ++ F LF
Sbjct: 162 IVMQSADTL--FDGSIPRQKVAQVCVESLFEPGARNKIVEIVAKPEAS---SKTFTELFQ 216
Query: 308 Q 308
Q
Sbjct: 217 Q 217
>gb|AAP37706.1| At4g31530 [Arabidopsis thaliana] gi|26449707|dbj|BAC41977.1|
unknown protein [Arabidopsis thaliana]
gi|30689062|ref|NP_194881.2| expressed protein
[Arabidopsis thaliana]
Length = 324
Score = 106 bits (264), Expect = 1e-21
Identities = 94/306 (30%), Positives = 148/306 (47%), Gaps = 45/306 (14%)
Query: 24 LPSSLKLLTSLQFSHSSSLSLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKK 83
LPSS L+ + SS + TS +A SS VLV G G G++V
Sbjct: 35 LPSSFFLVRNEASLSSSITPVQAFTEEAVDTS--DLASSSSKLVLVVGGTGGVGQLVVAS 92
Query: 84 LQERS--SQYIARGLVRTEESKQTIGASDD----VYVGDIRDTGSIAPAI-QGIDALIIL 136
L +R+ S+ + R L +++ + G D+ V GD R+ + P++ +G+ +I
Sbjct: 93 LLKRNIRSRLLLRDL---DKATKLFGKQDEYSLQVVKGDTRNAEDLDPSMFEGVTHVICT 149
Query: 137 TSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGG 196
T P + + PE+VDW G KN I A ++ VK++VLV S+G
Sbjct: 150 TGTTAF------------PSKRWNEENTPEKVDWEGVKNLISALPSS-VKRVVLVSSVGV 196
Query: 197 TDLNN-PLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQD--------------KE 241
T N P + + +L +K+ E +L DSG+P+TIIR G L D
Sbjct: 197 TKSNELPWSIMNLFGVLKYKKMGEDFLRDSGLPFTIIRPGRLTDGPYTSYDLNTLLKATA 256
Query: 242 GGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLAS-KPEGAGTPTR 300
G R ++IG+ D ++ +R + VAE CIQAL+ E Q KA+++ S K +G G+ +
Sbjct: 257 GERRAVVIGQGDNLVGEVSRLV----VAEACIQALDIEFTQGKAYEINSVKGDGPGSDPQ 312
Query: 301 DFKALF 306
++ LF
Sbjct: 313 QWRELF 318
>ref|YP_171729.1| hypothetical protein syc1019_d [Synechococcus elongatus PCC 6301]
gi|56685987|dbj|BAD79209.1| hypothetical protein
[Synechococcus elongatus PCC 6301]
gi|45511858|ref|ZP_00163425.1| COG0702: Predicted
nucleoside-diphosphate-sugar epimerases [Synechococcus
elongatus PCC 7942]
Length = 216
Score = 103 bits (257), Expect = 6e-21
Identities = 72/210 (34%), Positives = 109/210 (51%), Gaps = 25/210 (11%)
Query: 67 VLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIAPA 126
VLV GA GRTG+ V + ++ + R LVR+ + + ++ VGD+ D S+ A
Sbjct: 3 VLVVGATGRTGRCVVETAI--AAGHSVRALVRSANPQPPLPEGVELVVGDLSDRASLEAA 60
Query: 127 IQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVK 186
+ G+DA+I A P + P P +VD++G ID A AAG++
Sbjct: 61 LAGMDAVISAAGATPNLDP-----------------LGPFKVDYLGTTQLIDLAGAAGIQ 103
Query: 187 QIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRE 246
+ VLV S+ + L +PLN +L WKR+AE+YL SG+ YTI+R GGL+ V
Sbjct: 104 RFVLVSSLCVSRLLHPLNLFWL--VLFWKRRAERYLQSSGLSYTIVRPGGLRSDRTRVPL 161
Query: 247 LIIGKDDEILKTETRTIARPDVAEVCIQAL 276
+ G D+ ++ R VAEV ++AL
Sbjct: 162 KLTGPDELF----DGSLPRLQVAEVAVEAL 187
>ref|ZP_00517102.1| similar to Nucleoside-diphosphate-sugar epimerases [Crocosphaera
watsonii WH 8501] gi|67854514|gb|EAM49803.1| similar to
Nucleoside-diphosphate-sugar epimerases [Crocosphaera
watsonii WH 8501]
Length = 207
Score = 102 bits (255), Expect = 1e-20
Identities = 68/225 (30%), Positives = 119/225 (52%), Gaps = 26/225 (11%)
Query: 68 LVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEE-SKQTIGASDDVYVGDIRDTGSIAPA 126
LV GA G TG+ + ++L R Q + LVR ++ +K + ++ VGD+ D S+ A
Sbjct: 4 LVAGATGETGRRIVQELVNR--QIPVKALVRDQDRAKSILSPEAELVVGDVLDVDSLTKA 61
Query: 127 IQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVK 186
+ LI T A P + P + P QVD+ G KN ++ AK G++
Sbjct: 62 MTECTVLICATGARPSLDP-----------------SGPYQVDYEGTKNLVNVAKGQGIE 104
Query: 187 QIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRE 246
+ VLV S+ + +PLN +L WK++AE YL +SG+ YTI+R GGL++++
Sbjct: 105 KFVLVSSLCVSQFFHPLNLFWL--VLYWKKQAENYLENSGLKYTIVRPGGLKNEDNS-DP 161
Query: 247 LIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASK 291
+++ D + + +I R VA+VC+ +L ++ Q + ++ ++
Sbjct: 162 IVVSSADTLFE---GSIPRKKVAQVCVDSLFKDDYQQRILEIVAQ 203
>emb|CAB79871.1| putative protein [Arabidopsis thaliana] gi|5262764|emb|CAB45912.1|
putative protein [Arabidopsis thaliana]
gi|7486379|pir||T10683 hypothetical protein F3L17.100 -
Arabidopsis thaliana
Length = 329
Score = 100 bits (248), Expect = 7e-20
Identities = 94/311 (30%), Positives = 148/311 (47%), Gaps = 50/311 (16%)
Query: 24 LPSSLKLLTSLQFSHSSSLSLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTGKIVYKK 83
LPSS L+ + SS + TS +A SS VLV G G G++V
Sbjct: 35 LPSSFFLVRNEASLSSSITPVQAFTEEAVDTS--DLASSSSKLVLVVGGTGGVGQLVVAS 92
Query: 84 LQERS--SQYIARGLVRTEESKQTIGASDD----VYVGDIRDTGSIAPAI-QGIDALIIL 136
L +R+ S+ + R L +++ + G D+ V GD R+ + P++ +G+ +I
Sbjct: 93 LLKRNIRSRLLLRDL---DKATKLFGKQDEYSLQVVKGDTRNAEDLDPSMFEGVTHVICT 149
Query: 137 TSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGG 196
T P + + PE+VDW G KN I A ++ VK++VLV S+G
Sbjct: 150 TGTTAF------------PSKRWNEENTPEKVDWEGVKNLISALPSS-VKRVVLVSSVGV 196
Query: 197 TDLNN-PLNSLGNGNILVWKRKAEQYLADSGIPYTIIR-----AGGLQD----------- 239
T N P + + +L +K+ E +L DSG+P+TIIR G L D
Sbjct: 197 TKSNELPWSIMNLFGVLKYKKMGEDFLRDSGLPFTIIRFRTKEPGRLTDGPYTSYDLNTL 256
Query: 240 ---KEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLAS-KPEGA 295
G R ++IG+ D ++ +R + VAE CIQAL+ E Q KA+++ S K +G
Sbjct: 257 LKATAGERRAVVIGQGDNLVGEVSRLV----VAEACIQALDIEFTQGKAYEINSVKGDGP 312
Query: 296 GTPTRDFKALF 306
G+ + ++ LF
Sbjct: 313 GSDPQQWRELF 323
>ref|ZP_00158570.1| COG0702: Predicted nucleoside-diphosphate-sugar epimerases
[Anabaena variabilis ATCC 29413]
Length = 272
Score = 98.6 bits (244), Expect = 2e-19
Identities = 88/276 (31%), Positives = 132/276 (46%), Gaps = 44/276 (15%)
Query: 67 VLVTGAGGRTGKIVYKKLQERSSQYIARGLVRT-EESKQTIGASDDVYVGDIRDTGSIAP 125
VLV GA G G+IV KL E+ ++ R L R E++KQ +V+VGDIR ++
Sbjct: 10 VLVVGATGGVGQIVVGKLLEKGAK--VRILTRNAEKAKQLFNEKVEVFVGDIRQPNTLPA 67
Query: 126 AIQGIDALIILT--SAVPLMKPGFNPTKGERPEFY-----FEDGAY--------PEQVDW 170
A+ + +I T +A P + F+P P F+ D Y P +VD
Sbjct: 68 AVDHVTHIICCTGTTAFPSARWEFDP----EPNFFEWGKILLDSDYREATAKNTPAKVDA 123
Query: 171 IGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNP-LNSLGNGNILVWKRKAEQYLADSGIPY 229
G N + AA + + V V S+G + P N L +L K+K E+ + SG+PY
Sbjct: 124 EGVSNLV-AAAPKDLSRFVFVSSVGILRKDQPPFNILNAFGVLDAKKKGEEAIIHSGLPY 182
Query: 230 TIIRAGGLQD--------------KEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQA 275
TIIR G L D GG ++IGK D T +R DVA C+++
Sbjct: 183 TIIRPGRLIDGPYTSYDLNTLLKATTGGKLNVVIGKGD----TLAGDASRIDVAAACVES 238
Query: 276 LNFEEAQFKAFDLASKPEGAGTPTRDFKALFSQITT 311
+ + ++ + F+L +K G PT D++ LFSQ+ T
Sbjct: 239 IFYSASEGQVFELVNK--GTRPPTIDWETLFSQLPT 272
>emb|CAE07851.1| conserved hypothetical protein [Synechococcus sp. WH 8102]
gi|33865870|ref|NP_897429.1| hypothetical protein
SYNW1336 [Synechococcus sp. WH 8102]
Length = 234
Score = 92.0 bits (227), Expect = 2e-17
Identities = 75/237 (31%), Positives = 119/237 (49%), Gaps = 36/237 (15%)
Query: 67 VLVTGAGGRTG-KIVYKKLQERSSQYIARGLVRTEES-KQTIGASD-----DVYVGDIRD 119
V V+GA G+TG ++V + LQ S R ++R E + + A++ DV D+
Sbjct: 4 VAVSGASGKTGWRVVEEALQRGMS---VRAIMRPESTLPPALAAAERDQRLDVQRLDLNS 60
Query: 120 TGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDA 179
++ A++G AL+I T A +P N A P QVD G ++Q+ A
Sbjct: 61 GEALLHALKGCTALVIATGA----RPSINL-------------AGPLQVDAAGVQSQVQA 103
Query: 180 AKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQD 239
+A G++++VLV S+ +PLN G ILVWKR E++L SG+ +T+IR GGL +
Sbjct: 104 CRAVGLQRVVLVSSLCAGRWLHPLNLFGL--ILVWKRLGERWLERSGLDWTVIRPGGLSE 161
Query: 240 KEGGVRE---LIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPE 293
+G + G D + + +I R VA VC+ AL A + ++ S P+
Sbjct: 162 DDGRAEAEGVVFTGADQQ----QNSSIPRRLVARVCLDALESPAASGRIIEITSSPD 214
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 509,269,645
Number of Sequences: 2540612
Number of extensions: 20993061
Number of successful extensions: 52107
Number of sequences better than 10.0: 557
Number of HSP's better than 10.0 without gapping: 69
Number of HSP's successfully gapped in prelim test: 488
Number of HSP's that attempted gapping in prelim test: 51642
Number of HSP's gapped (non-prelim): 604
length of query: 313
length of database: 863,360,394
effective HSP length: 128
effective length of query: 185
effective length of database: 538,162,058
effective search space: 99559980730
effective search space used: 99559980730
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0147b.4


