

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
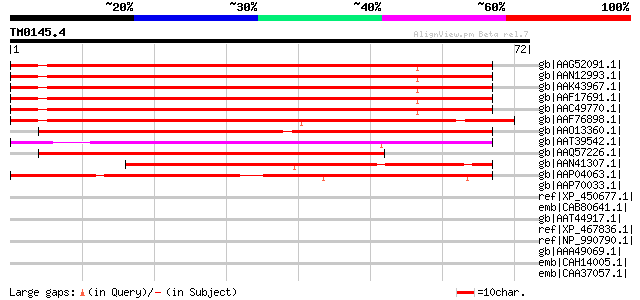
Query= TM0145.4
(72 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG52091.1| putative AP2 domain transcriptional regulator, 5'... 83 2e-15
gb|AAN12993.1| putative AP2 domain containing protein [Arabidops... 83 2e-15
gb|AAK43967.1| putative AP2 domain-containing protein [Arabidops... 83 2e-15
gb|AAF17691.1| F28K19.29 [Arabidopsis thaliana] 83 2e-15
gb|AAC49770.1| AP2 domain containing protein RAP2.4 [Arabidopsis... 80 1e-14
gb|AAF76898.1| apetala2 domain-containing protein [Atriplex hort... 73 2e-12
gb|AAO13360.1| dehydration-responsive element binding protein 3 ... 63 2e-09
gb|AAT39542.1| transcription factor DRE-binding factor 2 [Gossyp... 59 4e-08
gb|AAQ57226.1| DREB2 [Glycine max] 56 3e-07
gb|AAN41307.1| putative AP2 domain containing protein RAP2 [Arab... 49 4e-05
gb|AAP04063.1| putative AP2 domain transcription factor RAP2 [Ar... 47 2e-04
gb|AAP70033.1| DRE binding factor 2 [Oryza sativa (japonica cul... 39 0.046
ref|XP_450677.1| DRE binding factor 2 [Oryza sativa (japonica cu... 39 0.046
emb|CAB80641.1| putative protein [Arabidopsis thaliana] gi|30804... 37 0.10
gb|AAT44917.1| putative AP2/EREBP transcription factor [Arabidop... 37 0.10
ref|XP_467836.1| putative dehydration-responsive element binding... 35 0.39
ref|NP_990790.1| smooth muscle myosin light chain kinase (61-kDa... 34 1.1
gb|AAA49069.1| smooth muscle myosin light chain kinase precursor... 34 1.1
emb|CAH14005.1| hypothetical protein [Legionella pneumophila str... 34 1.1
emb|CAA37057.1| myosin light chain kinase [Gallus gallus] 34 1.1
>gb|AAG52091.1| putative AP2 domain transcriptional regulator, 5' partial; 1-558
[Arabidopsis thaliana]
Length = 185
Score = 82.8 bits (203), Expect = 2e-15
Identities = 43/69 (62%), Positives = 52/69 (75%), Gaps = 3/69 (4%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKK--SKA 58
PNLR H GS +GGDFGEYKPLHSSVDAKL+AIC+ +AE QKQ K+ K S+K +KK S
Sbjct: 59 PNLR-HNGSHIGGDFGEYKPLHSSVDAKLEAICKSMAETQKQDKSTKSSKKREKKVSSPD 117
Query: 59 TASKVAPQE 67
+ KV +E
Sbjct: 118 LSEKVKAEE 126
>gb|AAN12993.1| putative AP2 domain containing protein [Arabidopsis thaliana]
gi|15218204|ref|NP_177931.1| AP2 domain-containing
transcription factor RAP2.4 [Arabidopsis thaliana]
Length = 334
Score = 82.8 bits (203), Expect = 2e-15
Identities = 43/69 (62%), Positives = 52/69 (75%), Gaps = 3/69 (4%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKK--SKA 58
PNLR H GS +GGDFGEYKPLHSSVDAKL+AIC+ +AE QKQ K+ K S+K +KK S
Sbjct: 208 PNLR-HNGSHIGGDFGEYKPLHSSVDAKLEAICKSMAETQKQDKSTKSSKKREKKVSSPD 266
Query: 59 TASKVAPQE 67
+ KV +E
Sbjct: 267 LSEKVKAEE 275
>gb|AAK43967.1| putative AP2 domain-containing protein [Arabidopsis thaliana]
Length = 334
Score = 82.8 bits (203), Expect = 2e-15
Identities = 43/69 (62%), Positives = 52/69 (75%), Gaps = 3/69 (4%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKK--SKA 58
PNLR H GS +GGDFGEYKPLHSSVDAKL+AIC+ +AE QKQ K+ K S+K +KK S
Sbjct: 208 PNLR-HNGSHIGGDFGEYKPLHSSVDAKLEAICKSMAETQKQDKSTKSSKKREKKVSSPD 266
Query: 59 TASKVAPQE 67
+ KV +E
Sbjct: 267 LSEKVKAEE 275
>gb|AAF17691.1| F28K19.29 [Arabidopsis thaliana]
Length = 330
Score = 82.8 bits (203), Expect = 2e-15
Identities = 43/69 (62%), Positives = 52/69 (75%), Gaps = 3/69 (4%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKK--SKA 58
PNLR H GS +GGDFGEYKPLHSSVDAKL+AIC+ +AE QKQ K+ K S+K +KK S
Sbjct: 204 PNLR-HNGSHIGGDFGEYKPLHSSVDAKLEAICKSMAETQKQDKSTKSSKKREKKVSSPD 262
Query: 59 TASKVAPQE 67
+ KV +E
Sbjct: 263 LSEKVKAEE 271
>gb|AAC49770.1| AP2 domain containing protein RAP2.4 [Arabidopsis thaliana]
Length = 229
Score = 80.5 bits (197), Expect = 1e-14
Identities = 42/69 (60%), Positives = 51/69 (73%), Gaps = 3/69 (4%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKK--SKA 58
PNLR H G +GGDFGEYKPLHSSVDAKL+AIC+ +AE QKQ K+ K S+K +KK S
Sbjct: 103 PNLR-HNGFHIGGDFGEYKPLHSSVDAKLEAICKSMAETQKQDKSTKSSKKREKKVSSPD 161
Query: 59 TASKVAPQE 67
+ KV +E
Sbjct: 162 LSEKVKAEE 170
>gb|AAF76898.1| apetala2 domain-containing protein [Atriplex hortensis]
Length = 240
Score = 73.2 bits (178), Expect = 2e-12
Identities = 38/72 (52%), Positives = 49/72 (67%), Gaps = 4/72 (5%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEM--QKQGKTEKKSEKPQKKSKA 58
PNLR H+GS +GG+FGEYKPLHSSV+AKL+AICE LA+ +KQGK+ K +K +
Sbjct: 94 PNLR-HEGSHIGGEFGEYKPLHSSVNAKLEAICESLAKQGNEKQGKSGKSKKKDVANNNN 152
Query: 59 TASKVAPQEGCC 70
S + CC
Sbjct: 153 NTSS-SSSSSCC 163
>gb|AAO13360.1| dehydration-responsive element binding protein 3 [Lycopersicon
esculentum]
Length = 264
Score = 63.2 bits (152), Expect = 2e-09
Identities = 32/63 (50%), Positives = 41/63 (64%), Gaps = 1/63 (1%)
Query: 5 RHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKATASKVA 64
RH G+ +GGDFGEY PLHSSVDAKL+ IC+ LA+ K ++KK K KA K+
Sbjct: 144 RHNGNLIGGDFGEYNPLHSSVDAKLKDICQSLAQ-GKSIDSKKKKTKGLSAEKAAVVKME 202
Query: 65 PQE 67
+E
Sbjct: 203 EEE 205
>gb|AAT39542.1| transcription factor DRE-binding factor 2 [Gossypium hirsutum]
Length = 350
Score = 58.5 bits (140), Expect = 4e-08
Identities = 33/72 (45%), Positives = 42/72 (57%), Gaps = 10/72 (13%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSE-----KPQKK 55
PNLR H G G+YKPL SSVDAKLQAICE L + KQG +K S+ K +
Sbjct: 217 PNLRHH-----GSHVGDYKPLPSSVDAKLQAICESLVQNPKQGSKKKSSKVTADTKSRNN 271
Query: 56 SKATASKVAPQE 67
K+ ++ P+E
Sbjct: 272 KKSDMAEPKPEE 283
>gb|AAQ57226.1| DREB2 [Glycine max]
Length = 312
Score = 55.8 bits (133), Expect = 3e-07
Identities = 26/48 (54%), Positives = 35/48 (72%)
Query: 5 RHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKP 52
RH G+ V G+FG+YKPL SSVD+KLQAICE LA+ +++ + KP
Sbjct: 180 RHHGAFVFGEFGDYKPLPSSVDSKLQAICESLAKQEEKPCCSVEDVKP 227
>gb|AAN41307.1| putative AP2 domain containing protein RAP2 [Arabidopsis thaliana]
gi|15219839|ref|NP_173638.1| AP2 domain-containing
transcription factor, putative [Arabidopsis thaliana]
gi|25518572|pir||E86354 hypothetical protein F16L1.8
[imported] - Arabidopsis thaliana
gi|9454531|gb|AAF87854.1| Contains similarity to a
cadmium-imduced protein AS30 from Arabidopsis thaliana
gi|1168862 and contains an AP2 PF|00847 domain. EST
gb|AI099641 comes from this gene
Length = 261
Score = 48.5 bits (114), Expect = 4e-05
Identities = 29/52 (55%), Positives = 38/52 (72%), Gaps = 3/52 (5%)
Query: 17 EYKPLHSSVDAKLQAICEGLAE-MQKQGKTEKKSEKPQKKSKATASKVAPQE 67
EY+PL SSVDAKL+AIC+ LAE QKQ ++ KKS +K+S A K+ P+E
Sbjct: 146 EYQPLQSSVDAKLEAICQNLAETTQKQVRSTKKSSS-RKRSSTVAVKL-PEE 195
>gb|AAP04063.1| putative AP2 domain transcription factor RAP2 [Arabidopsis
thaliana] gi|28973692|gb|AAO64163.1| putative AP2 domain
transcription factor RAP2 [Arabidopsis thaliana]
gi|18400001|ref|NP_564468.1| AP2 domain-containing
transcription factor, putative [Arabidopsis thaliana]
gi|6598593|gb|AAF18648.1| F5J5.5 [Arabidopsis thaliana]
gi|12324713|gb|AAG52316.1| putative AP2
domain-containing transcription factor; 19304-20248
[Arabidopsis thaliana] gi|25350213|pir||E86482 protein
F5J5.5 [imported] - Arabidopsis thaliana
Length = 314
Score = 46.6 bits (109), Expect = 2e-04
Identities = 33/69 (47%), Positives = 44/69 (62%), Gaps = 6/69 (8%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQ-GKTEKKSEKPQKKSKAT 59
P LR GSS D GEY P+ ++VDAKL+AI LAE + Q GKTE+ S K K + ++
Sbjct: 199 PALRYQTGSSPS-DTGEYGPIQAAVDAKLEAI---LAEPKNQPGKTERTSRKRAKAAASS 254
Query: 60 ASK-VAPQE 67
A + APQ+
Sbjct: 255 AEQPSAPQQ 263
>gb|AAP70033.1| DRE binding factor 2 [Oryza sativa (japonica cultivar-group)]
Length = 291
Score = 38.5 bits (88), Expect = 0.046
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 7/50 (14%)
Query: 20 PLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKATASKVAPQEGC 69
PLH+SVDAKLQ +C+ +A K ++K + A A+ AP C
Sbjct: 175 PLHASVDAKLQTLCQNIA-------AAKNAKKSSVSASAAATSSAPTSNC 217
>ref|XP_450677.1| DRE binding factor 2 [Oryza sativa (japonica cultivar-group)]
gi|49388786|dbj|BAD25981.1| DRE binding factor 2 [Oryza
sativa (japonica cultivar-group)]
gi|49387678|dbj|BAD25924.1| DRE binding factor 2 [Oryza
sativa (japonica cultivar-group)]
Length = 291
Score = 38.5 bits (88), Expect = 0.046
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 7/50 (14%)
Query: 20 PLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKATASKVAPQEGC 69
PLH+SVDAKLQ +C+ +A K ++K + A A+ AP C
Sbjct: 175 PLHASVDAKLQTLCQNIA-------AAKNAKKSSVSASAAATSSAPTSNC 217
>emb|CAB80641.1| putative protein [Arabidopsis thaliana] gi|3080447|emb|CAA18764.1|
putative protein [Arabidopsis thaliana]
gi|15236020|ref|NP_195688.1| AP2 domain-containing
transcription factor, putative [Arabidopsis thaliana]
gi|7487213|pir||T05015 hypothetical protein T19P19.170 -
Arabidopsis thaliana
Length = 272
Score = 37.4 bits (85), Expect = 0.10
Identities = 19/40 (47%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQ 40
P R G GG + PLHSSVDAKLQ IC+ L + +
Sbjct: 149 PQFRHEDGYYGGGSC--FNPLHSSVDAKLQEICQSLRKTE 186
>gb|AAT44917.1| putative AP2/EREBP transcription factor [Arabidopsis thaliana]
Length = 272
Score = 37.4 bits (85), Expect = 0.10
Identities = 19/40 (47%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQ 40
P R G GG + PLHSSVDAKLQ IC+ L + +
Sbjct: 149 PQFRHEDGYYGGGSC--FNPLHSSVDAKLQEICQSLRKTE 186
>ref|XP_467836.1| putative dehydration-responsive element binding protein 3 [Oryza
sativa (japonica cultivar-group)]
gi|51964380|ref|XP_506975.1| PREDICTED OJ1288_G09.10
gene product [Oryza sativa (japonica cultivar-group)]
gi|46390126|dbj|BAD15561.1| putative
dehydration-responsive element binding protein 3 [Oryza
sativa (japonica cultivar-group)]
Length = 338
Score = 35.4 bits (80), Expect = 0.39
Identities = 24/66 (36%), Positives = 33/66 (49%), Gaps = 14/66 (21%)
Query: 1 PNLRRHQGSSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKATA 60
P LRR G+ + G PLH+SVDAKL AIC+ LA T P + + + A
Sbjct: 225 PTLRRG-GAHLAG------PLHASVDAKLTAICQSLA-------TSSSKNTPAESAASAA 270
Query: 61 SKVAPQ 66
+P+
Sbjct: 271 EPESPK 276
>ref|NP_990790.1| smooth muscle myosin light chain kinase (61-kDa active fragment)
[Gallus gallus] gi|992993|emb|CAA37056.1| myosin light
chain kinase [Gallus gallus]
gi|2851396|sp|P11799|MYLK_CHICK Myosin light chain
kinase, smooth muscle and non-muscle isozymes (MLCK)
[Contains: Telokin] gi|3403202|gb|AAC29031.1| smooth
muscle/non-muscle myosin light chain kinase [Gallus
gallus]
Length = 1906
Score = 33.9 bits (76), Expect = 1.1
Identities = 19/54 (35%), Positives = 24/54 (44%)
Query: 14 DFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKATASKVAPQE 67
D GEYK + + K + C+ L E K K +EK KK K T V E
Sbjct: 1150 DGGEYKCIAENAAGKAECACKVLVEDTSSTKAAKPAEKKTKKPKTTLPPVLSTE 1203
>gb|AAA49069.1| smooth muscle myosin light chain kinase precursor (EC 2.7.2.37)
Length = 972
Score = 33.9 bits (76), Expect = 1.1
Identities = 19/54 (35%), Positives = 24/54 (44%)
Query: 14 DFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKATASKVAPQE 67
D GEYK + + K + C+ L E K K +EK KK K T V E
Sbjct: 216 DGGEYKCIAENAAGKAECACKVLVEDTSSTKAAKPAEKKTKKPKTTLPPVLSTE 269
>emb|CAH14005.1| hypothetical protein [Legionella pneumophila str. Paris]
gi|54298788|ref|YP_125157.1| hypothetical protein
lpp2852 [Legionella pneumophila str. Paris]
Length = 460
Score = 33.9 bits (76), Expect = 1.1
Identities = 15/42 (35%), Positives = 25/42 (58%)
Query: 9 SSVGGDFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSE 50
+S+ D EYKP+H S + L + + +++Q +TEKK E
Sbjct: 368 TSISEDSKEYKPVHESTEDVLVRLSVNIDRLKEQVRTEKKEE 409
>emb|CAA37057.1| myosin light chain kinase [Gallus gallus]
Length = 972
Score = 33.9 bits (76), Expect = 1.1
Identities = 19/54 (35%), Positives = 24/54 (44%)
Query: 14 DFGEYKPLHSSVDAKLQAICEGLAEMQKQGKTEKKSEKPQKKSKATASKVAPQE 67
D GEYK + + K + C+ L E K K +EK KK K T V E
Sbjct: 216 DGGEYKCIAENAAGKAECACKVLVEDTSSTKAAKPAEKKTKKPKTTLPPVLSTE 269
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.131 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 112,452,166
Number of Sequences: 2540612
Number of extensions: 3394622
Number of successful extensions: 42181
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 42148
Number of HSP's gapped (non-prelim): 40
length of query: 72
length of database: 863,360,394
effective HSP length: 48
effective length of query: 24
effective length of database: 741,411,018
effective search space: 17793864432
effective search space used: 17793864432
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0145.4


