

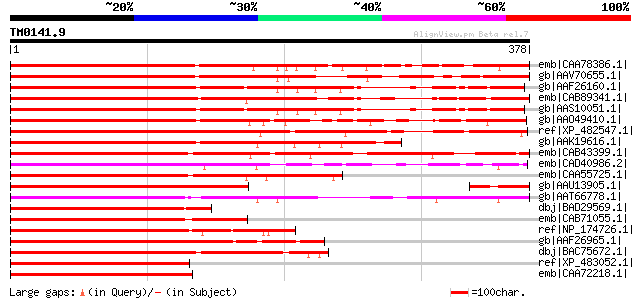
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0141.9
(378 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA78386.1| protein 1 [Petunia x hybrida] gi|282964|pir||S26... 424 e-117
gb|AAV70655.1| MYB transcription factor MIXTA-like 2 [Antirrhinu... 386 e-106
gb|AAF26160.1| putative Myb-related transcription factor [Arabid... 368 e-100
emb|CAB89341.1| myb-related protein-like [Arabidopsis thaliana] ... 367 e-100
gb|AAS10051.1| MYB transcription factor [Arabidopsis thaliana] 364 3e-99
gb|AAO49410.1| MYB1 [Dendrobium sp. XMW-2002-1] 363 4e-99
ref|XP_482547.1| MYB27 protein [Oryza sativa (japonica cultivar-... 357 3e-97
gb|AAK19616.1| GHMYB25 [Gossypium hirsutum] 335 2e-90
emb|CAB43399.1| Myb-related transcription factor mixta-like 1 [A... 334 2e-90
emb|CAD40986.2| OSJNBa0072F16.11 [Oryza sativa (japonica cultiva... 332 9e-90
emb|CAA55725.1| mixta [Antirrhinum majus] gi|629737|pir||S45338 ... 303 4e-81
gb|AAU13905.1| MYB transcription factor MYBML3 [Antirrhinum majus] 302 1e-80
gb|AAT66778.1| putative MYB related protein [Solanum demissum] 285 1e-75
dbj|BAD29569.1| MYB27 protein-like [Oryza sativa (japonica culti... 254 2e-66
emb|CAB71055.1| putative transcription factor (MYB17) [Arabidops... 243 7e-63
ref|NP_174726.1| myb family transcription factor [Arabidopsis th... 239 8e-62
gb|AAF26965.1| putative MYB family transcription factor [Arabido... 239 1e-61
dbj|BAC75672.1| transcription factor MYB102 [Lotus corniculatus ... 237 4e-61
ref|XP_483052.1| putative transcription factor Myb protein [Oryz... 235 1e-60
emb|CAA72218.1| myb [Oryza sativa (japonica cultivar-group)] gi|... 234 4e-60
>emb|CAA78386.1| protein 1 [Petunia x hybrida] gi|282964|pir||S26605 myb-related
protein 1 - garden petunia
Length = 421
Score = 424 bits (1089), Expect = e-117
Identities = 257/442 (58%), Positives = 292/442 (65%), Gaps = 85/442 (19%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC+KVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKF+LQEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRL+KMG
Sbjct: 61 LRPDIKRGKFTLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLVKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLG---- 176
IDPVTHKPKNDAL S +G SK AANLSHMAQWESARLEAEARLVR+SKLRS+
Sbjct: 121 IDPVTHKPKNDALLS--HDGQSKNAANLSHMAQWESARLEAEARLVRQSKLRSNSFQNPL 178
Query: 177 TSSSILPSPS-SSSLNKH--------------DTVKAWN-------------NGSSVVG- 207
S + SP+ SS L+K D +KAWN +GS+
Sbjct: 179 ASHELFTSPTPSSPLHKPIVTPTKAPGSPRCLDVLKAWNGVWTKPMNDVLHADGSTSASA 238
Query: 208 ---------DLESPKSTLSFSEN----NNNNHNDNNVAPIMIEFVGT---SLERGIVKEE 251
DLESP STLS+ EN + +N+ + + EFVG S E GI+ EE
Sbjct: 239 TVSVNALGLDLESPTSTLSYFENAQHISTGMIQENSTS--LFEFVGNSSGSSEGGIMNEE 296
Query: 252 GEQEWKG-----------YKDGM-ENSMPFTSTTFHELTMTMEATWPPSDESLRTITTAA 299
E++WKG YKDG+ ENSM TS T +LTM M+ TW + ESLR
Sbjct: 297 SEEDWKGFGNSSTGHLPEYKDGINENSMSLTS-TLQDLTMPMDTTW--TAESLR------ 347
Query: 300 TTAARAHVLGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGESNSGDGGDGCGSDFYE---D 356
+ A G+ V E TDLLL+++ D S G D G G G+D E D
Sbjct: 348 -SNAEDISHGNNFV-ETFTDLLLSTSGDGGLS------GNGTDSDNGGGSGNDPSETCGD 399
Query: 357 NKNYWNNILNLVNSSPTNSPMF 378
NKNYWN+I NLVNSSP++S MF
Sbjct: 400 NKNYWNSIFNLVNSSPSDSAMF 421
>gb|AAV70655.1| MYB transcription factor MIXTA-like 2 [Antirrhinum majus]
Length = 352
Score = 386 bits (991), Expect = e-106
Identities = 223/401 (55%), Positives = 258/401 (63%), Gaps = 72/401 (17%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC+KVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPA+AGLQRCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPARAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGK SLQEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRL KMG
Sbjct: 61 LRPDIKRGKISLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLAKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTS-S 179
IDPVTHKPK+D L S ++G SK AANLSHMAQWESARLEAEARLVR+SKL+ S
Sbjct: 121 IDPVTHKPKSDTLMS--NDGQSKNAANLSHMAQWESARLEAEARLVRQSKLQPPSANSFQ 178
Query: 180 SILPSPSSSSLNKH---DTVKAWNN----------------GSSVVGDLESPKSTLSFSE 220
+ +P S+ + D +KAWN + +VG+L SP STLS +
Sbjct: 179 ASTSNPVQKSMGQARCLDVLKAWNGVVWGKNDEAAGVSVTVTTGIVGELGSPTSTLSSAA 238
Query: 221 NNNNNHNDNNVAPIMIEFVGTSLERGIVKEEGEQEWKGY---KDGMENSMPFTSTTFHEL 277
N GI+KEE E+EWK +D + N+ PFTS
Sbjct: 239 GTN----------------------GIMKEESEEEWKQMPENRDEIGNATPFTS------ 270
Query: 278 TMTMEATWPPSDESLRTITTAATTAARAHVLGDPVVQEGLTDLLLNSNSDEDRSLSPEGG 337
W +L + ++ R + G +T+ L R
Sbjct: 271 ------NWESEQVALNSSEACSSREFRGKLYG------LVTECFL------CRWRGMRIV 312
Query: 338 GESNSGDGGDGCGSDFYEDNKNYWNNILNLVNSSPTNSPMF 378
G +NSG G D SD+YEDNKNY N+ILNLVNSSP++SP+F
Sbjct: 313 GYTNSG-GNDVGVSDYYEDNKNYGNSILNLVNSSPSHSPIF 352
>gb|AAF26160.1| putative Myb-related transcription factor [Arabidopsis thaliana]
gi|7644368|gb|AAF65559.1| putative transcription factor
[Arabidopsis thaliana] gi|15232045|ref|NP_186763.1| myb
family transcription factor (MYB106) [Arabidopsis
thaliana]
Length = 345
Score = 368 bits (944), Expect = e-100
Identities = 217/395 (54%), Positives = 250/395 (62%), Gaps = 74/395 (18%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC+K GLKKGPWTPEEDQKLLAYIEEHGHGSWR+LP KAGLQRCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDKAGLKKGPWTPEEDQKLLAYIEEHGHGSWRSLPEKAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKF++QEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRLIKMG
Sbjct: 61 LRPDIKRGKFTVQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLIKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSS 180
IDPVTHK KN+ L SS G SK AA LSHMAQWESARLEAEARL RESKL HL +
Sbjct: 121 IDPVTHKHKNETLSSS--TGQSKNAATLSHMAQWESARLEAEARLARESKL-LHLQHYQN 177
Query: 181 ILPSPSSSSLNKH----DTVKAWNNGSSVVGD--LESPKSTLSFSEN-----------NN 223
S++ +H T W + GD LESP ST++FSEN +
Sbjct: 178 NNNLNKSAAPQQHCFTQKTSTNWTKPNQGNGDQQLESPTSTVTFSENLLMPLGIPTDSSR 237
Query: 224 NNHNDNNVAPIMIEFV---GTSLERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELTMT 280
N +N+NN + MIE TS + +VKE E +W
Sbjct: 238 NRNNNNNESSAMIELAVSSSTSSDVSLVKEH-EHDW------------------------ 272
Query: 281 MEATWPPSDESLRTITTAATTAARAHVLGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGES 340
+R I G + EG T LL+ + R L P G E+
Sbjct: 273 -----------IRQIN-----------CGSGGIGEGFTSLLIGDS--VGRGL-PTGKNEA 307
Query: 341 NSGDGGDGCGSDFYEDNKNYWNNILNLVNSSPTNS 375
+G G + ++YEDNKNYWN+ILNLV+SSP++S
Sbjct: 308 TAGVGNES-EYNYYEDNKNYWNSILNLVDSSPSDS 341
>emb|CAB89341.1| myb-related protein-like [Arabidopsis thaliana]
gi|15242272|ref|NP_197035.1| myb family transcription
factor [Arabidopsis thaliana] gi|13877709|gb|AAK43932.1|
myb-related protein-like [Arabidopsis thaliana]
gi|41619402|gb|AAS10094.1| MYB transcription factor
[Arabidopsis thaliana] gi|11274013|pir||T49966
myb-related protein-like - Arabidopsis thaliana
Length = 326
Score = 367 bits (943), Expect = e-100
Identities = 211/380 (55%), Positives = 243/380 (63%), Gaps = 56/380 (14%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC+K+GLKKGPWTPEEDQKLLAYIEEHGHGSWR+LP KAGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDKLGLKKGPWTPEEDQKLLAYIEEHGHGSWRSLPEKAGLHRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKF+LQEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRL+KMG
Sbjct: 61 LRPDIKRGKFNLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLVKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKL--RSHLGTS 178
IDPVTHKPKN+ SS+ G SK AA LSH AQWESARLEAEARL RESKL H T
Sbjct: 121 IDPVTHKPKNETPLSSL--GLSKNAAILSHTAQWESARLEAEARLARESKLLHLQHYQTK 178
Query: 179 SSILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNVAPIMIEF 238
+S P +K LESP ST+SFSE + P IEF
Sbjct: 179 TSSQPHHHHGFTHKSLLPNWTTKPHEDQQQLESPTSTVSFSEMKES-------IPAKIEF 231
Query: 239 VGTSLERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELTMTMEATWPPSDESLRTITTA 298
VG+S ++KE E +W ++T HE T
Sbjct: 232 VGSSTGVTLMKEP-EHDW-------------INSTMHEFETTQ----------------- 260
Query: 299 ATTAARAHVLGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGESNSGDGGDGCGSDFYEDNK 358
+G+ ++EG T LLL +S DRS S + + GGD ++YEDNK
Sbjct: 261 ---------MGEG-IEEGFTGLLLGGDS-IDRSFSGDKNETAGESSGGD---CNYYEDNK 306
Query: 359 NYWNNILNLVNSSPTNSPMF 378
NY ++I N V+ SP++SPMF
Sbjct: 307 NYLDSIFNFVDPSPSDSPMF 326
>gb|AAS10051.1| MYB transcription factor [Arabidopsis thaliana]
Length = 345
Score = 364 bits (934), Expect = 3e-99
Identities = 216/395 (54%), Positives = 249/395 (62%), Gaps = 74/395 (18%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSP C+K GLKKGPWTPEEDQKLLAYIEEHGHGSWR+LP KAGLQRCGKSCRLRW+NY
Sbjct: 1 MGRSPSCDKAGLKKGPWTPEEDQKLLAYIEEHGHGSWRSLPEKAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKF++QEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRLIKMG
Sbjct: 61 LRPDIKRGKFTVQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLIKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSS 180
IDPVTHK KN+ L SS G SK AA LSHMAQWESARLEAEARL RESKL HL +
Sbjct: 121 IDPVTHKHKNETLSSS--TGQSKNAATLSHMAQWESARLEAEARLARESKL-LHLQHYQN 177
Query: 181 ILPSPSSSSLNKH----DTVKAWNNGSSVVGD--LESPKSTLSFSEN-----------NN 223
S++ +H T W + GD LESP ST++FSEN +
Sbjct: 178 NNNLNKSAAPQQHCFTQKTSTNWTKPNQGNGDQQLESPTSTVTFSENLLMPLGIPTDSSR 237
Query: 224 NNHNDNNVAPIMIEFV---GTSLERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELTMT 280
N +N+NN + MIE TS + +VKE E +W
Sbjct: 238 NRNNNNNESSAMIELAVSSSTSSDVSLVKEH-EHDW------------------------ 272
Query: 281 MEATWPPSDESLRTITTAATTAARAHVLGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGES 340
+R I G + EG T LL+ + R L P G E+
Sbjct: 273 -----------IRQIN-----------CGSGGIGEGFTSLLIGDS--VGRGL-PTGKNEA 307
Query: 341 NSGDGGDGCGSDFYEDNKNYWNNILNLVNSSPTNS 375
+G G + ++YEDNKNYWN+ILNLV+SSP++S
Sbjct: 308 TAGVGNES-EYNYYEDNKNYWNSILNLVDSSPSDS 341
>gb|AAO49410.1| MYB1 [Dendrobium sp. XMW-2002-1]
Length = 367
Score = 363 bits (933), Expect = 4e-99
Identities = 221/410 (53%), Positives = 259/410 (62%), Gaps = 81/410 (19%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC+KVGLKKGPWTPEEDQKLLAYIE+HGHGSWRALP KAGLQRCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDKVGLKKGPWTPEEDQKLLAYIEKHGHGSWRALPNKAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFS+QEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRL KMG
Sbjct: 61 LRPDIKRGKFSMQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLAKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRS------- 173
IDP+THKPK+D L S+ +GHSK ANL+HMAQWESARLEAEARLVRESKLRS
Sbjct: 121 IDPITHKPKSDNLSSA--DGHSKCTANLNHMAQWESARLEAEARLVRESKLRSSSISXNT 178
Query: 174 ---HLGTSSSILP-----SPSSSSLNKHDTVKAW-----------NNGSSVVGDLESPKS 214
+ +LP +P++S D + AW + G DLESP S
Sbjct: 179 TTIFTSKNQPLLPPAVPMTPAASPC--LDVLSAWQGAWTKPAANNSQGRGYTIDLESPTS 236
Query: 215 TLSFSENNNNNHNDNNVAPIMIEFVGTSLERGIVKEEGEQEWKGYKD---GMENSMPFTS 271
TLSFS DN +AP + G L +E +QEWK + M + PF S
Sbjct: 237 TLSFS--------DNMMAPTI---SGMGLAEDSTRE--DQEWKCLRKTGFSMHTAAPFVS 283
Query: 272 TTFHELTMTMEATWPPSDESLRTITTAATTAARAHVLGDPVVQEGLTDLLLNSNSDEDRS 331
T ++W GD G T +L + ++++
Sbjct: 284 TE--------ASSWLSESSG-----------------GD--FAAGFTGMLSDKANEQN-- 314
Query: 332 LSPEGGGESNSGDGG---DGCGSDFYED-NKNYWNNILNLVN-SSPTNSP 376
S +G +S++ + G D S+ ED NKNYW++ILNLVN SSP NSP
Sbjct: 315 -SEDGCNDSDNAECGSCVDVEESEGEEDENKNYWSSILNLVNSSSPPNSP 363
>ref|XP_482547.1| MYB27 protein [Oryza sativa (japonica cultivar-group)]
gi|27125807|emb|CAD44619.1| MYB27 protein [Oryza sativa
(japonica cultivar-group)] gi|42409479|dbj|BAD09835.1|
MYB27 protein [Oryza sativa (japonica cultivar-group)]
Length = 357
Score = 357 bits (916), Expect = 3e-97
Identities = 202/385 (52%), Positives = 241/385 (62%), Gaps = 37/385 (9%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCCEK+GLKKGPWTPEEDQKLLAYIEEHGHGSWRALP+KAGLQRCGKSCRLRW+NY
Sbjct: 1 MGRSPCCEKIGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPSKAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRL KMG
Sbjct: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLAKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSS 180
IDPVTHK N L+++ + +KV A+LSHMAQWESARLEAEARL RESK+R T S
Sbjct: 121 IDPVTHKAINGTLNNTAGDKSAKVTASLSHMAQWESARLEAEARLARESKMRIAASTPSK 180
Query: 181 I----LPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNVAPIMI 236
+ P+S+ D + AW + DLESP STL+F+ +N + + + +
Sbjct: 181 LHAQSTNPPASTPSPCFDVLNAWQSAKI---DLESPTSTLTFAGSNASMLPFSTTTALEL 237
Query: 237 EFVGTSL--ERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELTMTMEATWPPSDESLRT 294
+++ +R E E EWK M T H + E +W P
Sbjct: 238 SESNSNVWQQRSDELEGEESEWKFVSKQQLQGMHGKETEEH--FIGCEESWFP------- 288
Query: 295 ITTAATTAARAHVLGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGESNSGDGGDGCGSDFY 354
G + G T +LL+ ++ D S E ES++G
Sbjct: 289 --------------GTANIGAGFTGMLLDGSNMHDTS---ECWDESSNGQDEQRSQVSED 331
Query: 355 EDNKNYWNNILNLVNSS--PTNSPM 377
+NKNYWN I ++VNS P P+
Sbjct: 332 AENKNYWNGIFSMVNSEQPPLQPPL 356
>gb|AAK19616.1| GHMYB25 [Gossypium hirsutum]
Length = 309
Score = 335 bits (858), Expect = 2e-90
Identities = 189/306 (61%), Positives = 215/306 (69%), Gaps = 28/306 (9%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCCEKVGLKKGPWTPEEDQKLLAYIE+HGHGSWRALP KAGLQRCGKSCRLRW NY
Sbjct: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEQHGHGSWRALPLKAGLQRCGKSCRLRWINY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRL KMG
Sbjct: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLTKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSS- 179
IDPVTHKPK DAL S+ G+ AANLSHMAQWESARLEAEARLVRESKL S
Sbjct: 121 IDPVTHKPKTDALGST--TGNPIDAANLSHMAQWESARLEAEARLVRESKLVPSNPPQSN 178
Query: 180 ---SILPSPSSSSLNK-HDTVKAWNNGSSVV-------GDLESPKSTLSFSENNNN--NH 226
++ PSP+ ++ + D +KAW + +L+SP STL+F EN
Sbjct: 179 HFTAVAPSPTPATRPQCLDVLKAWQGVVCGLFTFNMDNNNLQSPTSTLNFMENTTTLPMS 238
Query: 227 NDNNVAPIMIEFVG-----TSLERG-IVK-EEGEQEWKGYKDGMENSMPFTSTTFHELTM 279
+ ++V + E G E G I+K E G + K+ +++ M HE+
Sbjct: 239 SSSSVNGMFNENFGWNSSINPCESGDILKVEYGSDQIPELKERLDHPM-----ELHEMDC 293
Query: 280 TMEATW 285
+ E TW
Sbjct: 294 SSEGTW 299
>emb|CAB43399.1| Myb-related transcription factor mixta-like 1 [Antirrhinum majus]
Length = 359
Score = 334 bits (857), Expect = 2e-90
Identities = 192/390 (49%), Positives = 236/390 (60%), Gaps = 43/390 (11%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC+KV LK+GPWTPEEDQKLL+YI+EHGHGSWRALP+KAGLQRCGKSCRLRWSNY
Sbjct: 1 MGRSPCCDKVSLKRGPWTPEEDQKLLSYIQEHGHGSWRALPSKAGLQRCGKSCRLRWSNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFSLQEEQ IIQLHA LGNRWSAIATHL KRTDNEIKNYWNTHLKKRL KMG
Sbjct: 61 LRPDIKRGKFSLQEEQAIIQLHAFLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLTKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSH------ 174
IDP+THKPK+ H + G KV ANLSHMAQWESARL+AEARLVRES+L SH
Sbjct: 121 IDPMTHKPKS---HDVLGCGQPKVVANLSHMAQWESARLQAEARLVRESRLVSHHYHSQL 177
Query: 175 LGTSSSILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLS---FSENNNNNHNDNNV 231
L +++I L D +K W+ + + S + F+ NN N + ++
Sbjct: 178 LNRATAITHPTLPPCL---DVLKVWHGAWTTRPGKDIITSAMFNGFFASNNGNLESPTSI 234
Query: 232 APIMIEFVGTSLERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELTMTMEATWPPSDES 291
+ S G++ E + M + + ++ S+E
Sbjct: 235 LKSSDNMLNASTSVGLLHE----------NPFITDMSYVGKPSNVYEDWVKGIMDNSNEL 284
Query: 292 LRTITTAATTAARAH---VLGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGESNSGDGGDG 348
+ I +T AH +G P EG T+L+ ++ +G D
Sbjct: 285 NKIIEPIDSTHYVAHDDDSIGFPGFMEGSTNLVTST--------------VRTNGPDDDN 330
Query: 349 CGSDFYEDNKNYWNNILNLVNSSPTNSPMF 378
F E++ NYW N+LN+VN SP SP+F
Sbjct: 331 VVGVFEENDINYWRNVLNVVN-SPMGSPVF 359
>emb|CAD40986.2| OSJNBa0072F16.11 [Oryza sativa (japonica cultivar-group)]
gi|50924814|ref|XP_472754.1| OSJNBa0072F16.11 [Oryza
sativa (japonica cultivar-group)]
Length = 345
Score = 332 bits (852), Expect = 9e-90
Identities = 198/392 (50%), Positives = 235/392 (59%), Gaps = 80/392 (20%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCCEK GLKKGPWTPEEDQKLLAYIE+HGHG WR+LP+KAGLQRCGKSCRLRW+NY
Sbjct: 1 MGRSPCCEKEGLKKGPWTPEEDQKLLAYIEQHGHGCWRSLPSKAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRL KMG
Sbjct: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLAKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNG----------HSKVAANLSHMAQWESARLEAEARLVRESK 170
IDPVTHKP++D + G H+K AA+LSH AQWESARLEAEARL RE+K
Sbjct: 121 IDPVTHKPRSDVAGAGGGGGGAAGGAAGAQHAKAAAHLSHTAQWESARLEAEARLAREAK 180
Query: 171 LRSHLGTS---SSILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHN 227
LR+ ++ + LP+P +S+ ++ L+SP STLSFSE
Sbjct: 181 LRALAASATPGAPHLPAPPASAA-----------AAAAAHGLDSPTSTLSFSE------- 222
Query: 228 DNNVAPIMIEFVGTSLERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELTMTMEATWPP 287
+ V ++E G + Q + Y + + + W
Sbjct: 223 -SAVLATVLEAHGAA--AAAAARAAMQPMQAYDEACK-----------------DQHWGD 262
Query: 288 SDESLRTITTAATTAARAHVLGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGESNSGDGGD 347
D A +G P G T LLL + ++ P G DG
Sbjct: 263 VD---------------AADVGFPGAGAGFTGLLLEGSLNQ----IPRPAGRDAEADG-- 301
Query: 348 GCGSDFY--EDNKNYWNNILNLVNSSPTNSPM 377
+F E+ KNYWN+ILNLVNSS ++PM
Sbjct: 302 ----EFQETEEEKNYWNSILNLVNSS--SAPM 327
>emb|CAA55725.1| mixta [Antirrhinum majus] gi|629737|pir||S45338 myb-related protein
MIXTA - garden snapdragon gi|743793|prf||2013346A
myb-related protein
Length = 321
Score = 303 bits (777), Expect = 4e-81
Identities = 159/249 (63%), Positives = 186/249 (73%), Gaps = 10/249 (4%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M RSPCC+KVG+KKGPWT +EDQKLLAYIEEHGHGSWR+LP KAGLQRCGKSCRLRW+NY
Sbjct: 1 MVRSPCCDKVGVKKGPWTVDEDQKLLAYIEEHGHGSWRSLPLKAGLQRCGKSCRLRWANY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG FSLQEEQTIIQLHALLGNRWSAIA+HL KRTDNEIKNYWNTHLKKRL +MG
Sbjct: 61 LRPDIKRGPFSLQEEQTIIQLHALLGNRWSAIASHLPKRTDNEIKNYWNTHLKKRLTRMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKL---RSHLGT 177
IDPVTHKP H+ + +G K ANL+H+AQWESARL+AE RLVRES+L + +GT
Sbjct: 121 IDPVTHKPHT---HNILGHGQPKDVANLNHIAQWESARLQAERRLVRESRLAQNNNKIGT 177
Query: 178 SSSILPSP--SSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNVAPI- 234
L P + + H N+ S+V + ++ + S +NNN ++D V I
Sbjct: 178 IQRRLTWPLCLDNEQSNHYHSALLNSTSAVGLNQDNSFTNYSARPDNNNIYDDYEVNNIM 237
Query: 235 -MIEFVGTS 242
MIEF S
Sbjct: 238 GMIEFNNNS 246
>gb|AAU13905.1| MYB transcription factor MYBML3 [Antirrhinum majus]
Length = 219
Score = 302 bits (773), Expect = 1e-80
Identities = 143/175 (81%), Positives = 161/175 (91%), Gaps = 1/175 (0%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSP C+K GLK+GPWTPEEDQKLLA I+EHGHG+WRALPAKAGL+RCGKSCRLRW+NY
Sbjct: 1 MGRSPFCDKTGLKRGPWTPEEDQKLLACIQEHGHGNWRALPAKAGLERCGKSCRLRWNNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFS QEEQTIIQLHALLGNRWSAIATHLS+RTDNEIKNYWNTH+KK+L KMG
Sbjct: 61 LRPDIKRGKFSSQEEQTIIQLHALLGNRWSAIATHLSRRTDNEIKNYWNTHIKKKLAKMG 120
Query: 121 IDPVTHKPKND-ALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSH 174
IDPVTHKP+ D AL S+ + SK AAN+SH+AQWESARLEAEARL R+SKL+++
Sbjct: 121 IDPVTHKPQRDHALSSNNAHVQSKNAANMSHLAQWESARLEAEARLARQSKLQAN 175
Score = 55.1 bits (131), Expect = 3e-06
Identities = 25/43 (58%), Positives = 30/43 (69%), Gaps = 5/43 (11%)
Query: 336 GGGESNSGDGGDGCGSDFYEDNKNYWNNILNLVNSSPTNSPMF 378
GGG G G D CG DNKNYW+NIL+LVN SP++SP+F
Sbjct: 179 GGGSDTGGGGSDCCG-----DNKNYWDNILDLVNFSPSDSPIF 216
>gb|AAT66778.1| putative MYB related protein [Solanum demissum]
Length = 338
Score = 285 bits (730), Expect = 1e-75
Identities = 177/393 (45%), Positives = 218/393 (55%), Gaps = 70/393 (17%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRS CC++ GLKKGPWT EEDQKLL++I+++G GSWR LPAKAGLQRCGKSCRLRW NY
Sbjct: 1 MGRSKCCDEEGLKKGPWTHEEDQKLLSFIDKYGCGSWRGLPAKAGLQRCGKSCRLRWINY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFSLQEE+TII LHA LGNRWS IAT+L RTDNEIKNYWN+ LKKRL+KMG
Sbjct: 61 LRPDIKRGKFSLQEERTIIHLHAFLGNRWSTIATYLPSRTDNEIKNYWNSRLKKRLMKMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSS- 179
IDP+THKP N A G SK ANLSHMA+WESARLEAEARLVR+SK+ + +S
Sbjct: 121 IDPMTHKP-NAA-------GSSKYVANLSHMAEWESARLEAEARLVRKSKILFNKNNNSH 172
Query: 180 --SILPSPSSSSLNKH------DTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDNNV 231
+I P+ S L H D +KAW S+ + + + + + N N
Sbjct: 173 NYNINPTTISQQLPYHQQLPCLDILKAWQMTSTKLPTINDISHAILLNNSRNKN------ 226
Query: 232 APIMIEFVGTSLERGIVKEEGEQEWKGYKDGMENSMPFTSTTFHELTMTMEATWPPSDES 291
+E+S+P T + + +
Sbjct: 227 -------------------------------LESSIPSTLNSSENIFA----------NN 245
Query: 292 LRTITTAATTAARAHVLG--DPVVQEGLTDLLLNSNSDEDRSLSPEGGGESNSGDGGDGC 349
+ T T A H L + ++ L S E + PE S +G D
Sbjct: 246 VPTTTKVADDHQNLHNLSTINSCFEDDQLQTELPSFMQEFSGVFPEYTQNSTNGLQVDNI 305
Query: 350 GSDFY---EDNKNY-WNNILNLVNSSPTNSPMF 378
FY EDNK WNN N + +SP SP+F
Sbjct: 306 MGSFYGDFEDNKLINWNNFPNYLVNSPIGSPVF 338
>dbj|BAD29569.1| MYB27 protein-like [Oryza sativa (japonica cultivar-group)]
Length = 167
Score = 254 bits (650), Expect = 2e-66
Identities = 121/148 (81%), Positives = 132/148 (88%), Gaps = 2/148 (1%)
Query: 1 MGRSPCCEK-VGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSN 59
MGRSPCCEK GLKKGPWTPEEDQKLLAYIE+HGHG WR+LP KAGL+RCGKSCRLRW+N
Sbjct: 1 MGRSPCCEKEAGLKKGPWTPEEDQKLLAYIEQHGHGCWRSLPTKAGLRRCGKSCRLRWTN 60
Query: 60 YLRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKM 119
YLRPDIKRGKF+LQEEQTIIQLHALLGNRWSAIATHL KRTDNEIKNYWNTHLKKRL +M
Sbjct: 61 YLRPDIKRGKFTLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLARM 120
Query: 120 GIDPVTHKPKNDALHSSIDNGHSKVAAN 147
GIDPVTHK N ++ SS + S+V +
Sbjct: 121 GIDPVTHK-VNSSMSSSSSSLTSEVVTD 147
>emb|CAB71055.1| putative transcription factor (MYB17) [Arabidopsis thaliana]
gi|15233066|ref|NP_191684.1| myb family transcription
factor (MYB17) [Arabidopsis thaliana]
gi|41619302|gb|AAS10071.1| MYB transcription factor
[Arabidopsis thaliana] gi|11273969|pir||T47917 probable
transcription factor MYB17 - Arabidopsis thaliana
Length = 299
Score = 243 bits (620), Expect = 7e-63
Identities = 116/173 (67%), Positives = 137/173 (79%), Gaps = 4/173 (2%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGR+PCC+K+GLKKGPWTPEED+ L+A+I+++GHGSWR LP AGL RCGKSCRLRW+NY
Sbjct: 1 MGRTPCCDKIGLKKGPWTPEEDEVLVAHIKKNGHGSWRTLPKLAGLLRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG F+ EE+ +IQLHA+LGNRW+AIA L RTDNEIKN WNTHLKKRL+ MG
Sbjct: 61 LRPDIKRGPFTADEEKLVIQLHAILGNRWAAIAAQLPGRTDNEIKNLWNTHLKKRLLSMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRS 173
+DP TH+P L S + + HMAQWESAR+EAEARL RES L S
Sbjct: 121 LDPRTHEP----LPSYGLAKQAPSSPTTRHMAQWESARVEAEARLSRESMLFS 169
>ref|NP_174726.1| myb family transcription factor [Arabidopsis thaliana]
gi|5668784|gb|AAD46010.1| Strong similarity to M4
protein gb|X90381 from Arabidopsis thaliana and contains
2 PF|00249 Myb-like DNA-binding domains. EST gb|H36793
comes from this gene gi|41619130|gb|AAS10030.1| MYB
transcription factor [Arabidopsis thaliana]
gi|25294085|pir||D86470 F21H2.9 protein - Arabidopsis
thaliana
Length = 365
Score = 239 bits (611), Expect = 8e-62
Identities = 123/223 (55%), Positives = 158/223 (70%), Gaps = 17/223 (7%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC++ GLKKGPWTPEEDQKL+ YI +HGHGSWRALP A L RCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDENGLKKGPWTPEEDQKLIDYIHKHGHGSWRALPKLADLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFS +EEQTI+ LH++LGN+WSAIATHL RTDNEIKN+WNTHLKK+LI+MG
Sbjct: 61 LRPDIKRGKFSAEEEQTILHLHSILGNKWSAIATHLQGRTDNEIKNFWNTHLKKKLIQMG 120
Query: 121 IDPVTHKPKNDALHSSIDN--GHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTS 178
IDPVTH+P+ D L +S+ + + + +Q+ S + EA A+L L+ +S
Sbjct: 121 IDPVTHQPRTD-LFASLPQLIALANLKDLIEQTSQFSSMQGEA-AQLANLQYLQRMFNSS 178
Query: 179 SSILP------SPSS-------SSLNKHDTVKAWNNGSSVVGD 208
+S+ SPSS ++N +++ +WN + D
Sbjct: 179 ASLTNNNGNNFSPSSILDIDQHHAMNLLNSMVSWNKDQNPAFD 221
>gb|AAF26965.1| putative MYB family transcription factor [Arabidopsis thaliana]
gi|7644370|gb|AAF65560.1| putative transcription factor
[Arabidopsis thaliana] gi|15233021|ref|NP_186944.1| myb
family transcription factor (MYB107) [Arabidopsis
thaliana] gi|41619226|gb|AAS10053.1| MYB transcription
factor [Arabidopsis thaliana]
Length = 321
Score = 239 bits (610), Expect = 1e-61
Identities = 121/230 (52%), Positives = 161/230 (69%), Gaps = 7/230 (3%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC++ GLKKGPWTPEEDQKL+ +I +HGHGSWRALP +AGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDESGLKKGPWTPEEDQKLINHIRKHGHGSWRALPKQAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG F+ +EEQTII LH+LLGN+WS+IA HL RTDNEIKNYWNTH++K+LI+MG
Sbjct: 61 LRPDIKRGNFTAEEEQTIINLHSLLGNKWSSIAGHLPGRTDNEIKNYWNTHIRKKLIQMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQW-ESARLEAEARLVRESKLRSHLGTSS 179
IDPVTH+P+ D L+ AAN +++ ++ +L+A + + + L S + S
Sbjct: 121 IDPVTHRPRTDHLNVLAALPQLLAAANFNNLLNLNQNIQLDATS-VAKAQLLHSMIQVLS 179
Query: 180 SILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLSFSENNNNNHNDN 229
+ + +SSS + H T SS + +L P + + +H D+
Sbjct: 180 N---NNTSSSFDIHHTTNNLFGQSSFLENL--PNIENPYDQTQGLSHIDD 224
>dbj|BAC75672.1| transcription factor MYB102 [Lotus corniculatus var. japonicus]
Length = 336
Score = 237 bits (605), Expect = 4e-61
Identities = 121/239 (50%), Positives = 165/239 (68%), Gaps = 14/239 (5%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC++ GLKKGPWTPEEDQKL+ +I++HGHGSWRALP AGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDENGLKKGPWTPEEDQKLVEHIQKHGHGSWRALPKLAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFS +EEQTI+ LH++LGN+WS IATHL RTDNEIKN+WNTHLKK+LI+MG
Sbjct: 61 LRPDIKRGKFSPEEEQTILHLHSILGNKWSTIATHLPGRTDNEIKNFWNTHLKKKLIQMG 120
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEA-RLVRESKLRSHLGTSS 179
DP+TH+P+ D + + + AN++ + + +A +L + L L +S+
Sbjct: 121 FDPMTHQPRTDLVSTL---PYLLALANMTQLMDHNHQSFDEQAVQLAKLQYLNYLLQSSN 177
Query: 180 SILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTL--SFSENNNN----NHNDNNVA 232
+ L +S N ++A+ S++ + + K L FS+ + +H+ NN+A
Sbjct: 178 NPLCMTNSYDQNALTNMEAF----SLLNSISNVKENLGVDFSQMDTQTSLFSHDGNNMA 232
>ref|XP_483052.1| putative transcription factor Myb protein [Oryza sativa (japonica
cultivar-group)] gi|42408185|dbj|BAD09322.1| putative
transcription factor Myb protein [Oryza sativa (japonica
cultivar-group)]
Length = 357
Score = 235 bits (600), Expect = 1e-60
Identities = 102/131 (77%), Positives = 119/131 (89%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC++ GLKKGPWTPEED+KLL YI+++GHGSWR LP AGL RCGKSCRLRW+NY
Sbjct: 1 MGRSPCCDESGLKKGPWTPEEDEKLLHYIQKNGHGSWRTLPRLAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKFS +EEQTI+ LH++LGN+WSAIATHL RTDNEIKN+WNTHLKKRLI+MG
Sbjct: 61 LRPDIKRGKFSQEEEQTILHLHSILGNKWSAIATHLPGRTDNEIKNFWNTHLKKRLIQMG 120
Query: 121 IDPVTHKPKND 131
DP+TH+P+ D
Sbjct: 121 FDPMTHRPRTD 131
>emb|CAA72218.1| myb [Oryza sativa (japonica cultivar-group)] gi|7438328|pir||T03828
myb protein - rice
Length = 368
Score = 234 bits (596), Expect = 4e-60
Identities = 101/133 (75%), Positives = 118/133 (87%)
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGR+PCCEK GLKKGPWTPEED+KL+AYI+EHG G+WR LP AGL RCGKSCRLRW+NY
Sbjct: 1 MGRAPCCEKSGLKKGPWTPEEDEKLIAYIKEHGQGNWRTLPKNAGLSRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRG+FS +EE+ IIQLH++LGN+WSAIA L RTDNEIKNYWNTH++KRL++MG
Sbjct: 61 LRPDIKRGRFSFEEEEAIIQLHSILGNKWSAIAARLPGRTDNEIKNYWNTHIRKRLLRMG 120
Query: 121 IDPVTHKPKNDAL 133
IDPVTH P+ D L
Sbjct: 121 IDPVTHAPRLDLL 133
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.310 0.128 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 713,916,544
Number of Sequences: 2540612
Number of extensions: 32375205
Number of successful extensions: 312180
Number of sequences better than 10.0: 1939
Number of HSP's better than 10.0 without gapping: 1480
Number of HSP's successfully gapped in prelim test: 496
Number of HSP's that attempted gapping in prelim test: 277502
Number of HSP's gapped (non-prelim): 12944
length of query: 378
length of database: 863,360,394
effective HSP length: 130
effective length of query: 248
effective length of database: 533,080,834
effective search space: 132204046832
effective search space used: 132204046832
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0141.9


