

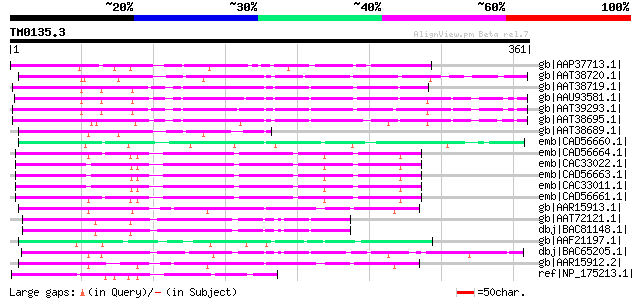
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.3
(361 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP37713.1| At3g23880 [Arabidopsis thaliana] gi|9294656|dbj|B... 124 4e-27
gb|AAT38720.1| putative F-Box protein [Solanum demissum] 114 5e-24
gb|AAT38719.1| putative F-Box protein [Solanum demissum] 107 7e-22
gb|AAU93581.1| putative F-box protein [Solanum demissum] 106 1e-21
gb|AAT39293.1| putative F-box-like protein [Solanum demissum] 103 6e-21
gb|AAT38695.1| putative F-box protein [Solanum demissum] 102 2e-20
gb|AAT38689.1| putative S haplotype-specific F-box protein [Sola... 83 1e-14
emb|CAD56660.1| S locus F-box (SLF)-S4D protein [Antirrhinum his... 81 4e-14
emb|CAD56664.1| S locus F-box (SLF)-S5 protein [Antirrhinum hisp... 81 6e-14
emb|CAC33022.1| SLF-S2 protein [Antirrhinum hispanicum] gi|13161... 80 7e-14
emb|CAD56663.1| S locus F-box (SLF)-S1 protein [Antirrhinum hisp... 79 2e-13
emb|CAC33011.1| S locus F-box (SLF)-S2-like protein [Antirrhinum... 79 2e-13
emb|CAD56661.1| S locus F-box (SLF)-S4 protein [Antirrhinum hisp... 77 1e-12
gb|AAR15913.1| S3 self-incompatibility locus-linked putative F-b... 75 2e-12
gb|AAT72121.1| SFB3 [Prunus avium] 74 5e-12
dbj|BAC81148.1| S-locus F-Box protein 3 [Prunus avium] 74 5e-12
gb|AAF21197.1| unknown protein [Arabidopsis thaliana] gi|1598347... 74 7e-12
dbj|BAC65205.1| S haplotype-specific F-box protein d [Prunus dul... 72 3e-11
gb|AAR15912.2| S2 self-incompatibility locus-linked putative F-b... 72 3e-11
ref|NP_175213.1| F-box family protein [Arabidopsis thaliana] gi|... 72 3e-11
>gb|AAP37713.1| At3g23880 [Arabidopsis thaliana] gi|9294656|dbj|BAB03005.1| unnamed
protein product [Arabidopsis thaliana]
gi|26449693|dbj|BAC41970.1| unknown protein [Arabidopsis
thaliana] gi|15229536|ref|NP_189030.1| F-box family
protein [Arabidopsis thaliana]
Length = 364
Score = 124 bits (311), Expect = 4e-27
Identities = 109/319 (34%), Positives = 147/319 (45%), Gaps = 49/319 (15%)
Query: 1 MEPPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLH--LHRSSSTTRNT 58
M P LP E+ EIL LPVKSL RF+CV SW+S+IS++ F H + +S T +T
Sbjct: 8 MFSPHNLPLEMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHALILETSKATTST 67
Query: 59 DFAYLQSLITSPR---------KSRSASTIAIP---------DDYDFCGTCNGLVCLHSS 100
Y +IT+ R +AST+ + D Y GTC+GLVC H
Sbjct: 68 KSPY--GVITTSRYHLKSCCIHSLYNASTVYVSEHDGELLGRDYYQVVGTCHGLVCFHVD 125
Query: 101 KYDRKDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDS---YFGFDYDSSTDTYKVVAVAV 157
Y + LWNP ++ + QR L D +GF YD S D YKVVA+
Sbjct: 126 -------YDKSLYLWNPTIK-LQQRLSSSDLETSDDECVVTYGFGYDESEDDYKVVALLQ 177
Query: 158 ALSWDTLETMVNVYNIGDDTCWRTIPVSHLPSMYL---KDDDAVYVNNTLNRLAILPNVE 214
+ET +Y+ WR+ + PS + K +Y+N TLN A
Sbjct: 178 QRHQVKIET--KIYST-RQKLWRS--NTSFPSGVVVADKSRSGIYINGTLNWAA----TS 228
Query: 215 DRDRCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVW 274
TI+S+D ++ +LP P C R F TLG LR CL + K + VW
Sbjct: 229 SSSSWTIISYDMSRDEFKELPGPVC-CGRGCFTM---TLGDLRGCLSMVCYCKGANADVW 284
Query: 275 QMKEFGLYKSWTLLFNIAG 293
MKEFG SW+ L +I G
Sbjct: 285 VMKEFGEVYSWSKLLSIPG 303
>gb|AAT38720.1| putative F-Box protein [Solanum demissum]
Length = 372
Score = 114 bits (285), Expect = 5e-24
Identities = 111/389 (28%), Positives = 174/389 (44%), Gaps = 64/389 (16%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHL-----------HR--SSS 53
LP E+ +EIL +P KSL++F CVSK+W +IS ++FIK HL HR
Sbjct: 9 LPHEIIIEILLKVPPKSLLKFMCVSKTWLELISSAKFIKTHLELIANDKEYSHHRIIFQE 68
Query: 54 TTRNTDFAYLQSLITSPRKSR---SASTIAIPDDYDF-CGTCNGLVCLHSSKYDRKDKYT 109
+ N L S++ R + S + P Y + G+ NGL+CL+S
Sbjct: 69 SACNFKVCCLPSMLNKERSTELFDIGSPMENPTIYTWIVGSVNGLICLYSK--------I 120
Query: 110 SHVRLWNPAMRSMSQRSPPLYLPI----VYDSYFGFDYDSSTDTYKVVAV-AVALSWDTL 164
LWNPA++ S++ P L + Y +GF YD + D YKVV + + +
Sbjct: 121 EETVLWNPAVKK-SKKLPTLGAKLRNGCSYYLKYGFGYDETRDDYKVVVIQCIYEDSGSC 179
Query: 165 ETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSF 224
+++VN+Y++ D+ WRTI + +L + +VN + A+ +V+ + C I+S
Sbjct: 180 DSVVNIYSLKADS-WRTI--NKFQGNFLVNSPGKFVNGKI-YWALSADVDTFNMCNIISL 235
Query: 225 DFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCI-CQKDKNMHFVVWQMKEFGLYK 283
D E+ +L LP + + +G LC+ C + N VW K+ G+
Sbjct: 236 DLADETWRRLELP-DSYGKGSYPLALGVVGSHLSVLCLNCIEGTNSD--VWIRKDCGVEV 292
Query: 284 SWTLLFNI-----------AGYAYDIPCRFFAMYMSENGDALLLSKLAVSQPQAVLYTQK 332
SWT +F + + +PC Y S + LLL P + Y
Sbjct: 293 SWTKIFTVDHPKDLGEFIFFTSIFSVPC-----YQSNKDEILLL-----LPPVILTYNGS 342
Query: 333 DNKLEVTDVANYIFNCYAMN-YIESLVPP 360
++EV D C A Y+ESLV P
Sbjct: 343 TRQVEVVD---RFEECAAAEIYVESLVDP 368
>gb|AAT38719.1| putative F-Box protein [Solanum demissum]
Length = 327
Score = 107 bits (266), Expect = 7e-22
Identities = 94/308 (30%), Positives = 138/308 (44%), Gaps = 33/308 (10%)
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHL-----------HRS 51
PP LP+EL EIL LP+KSL++F CVSKSW +IS F+K H+ HR
Sbjct: 8 PPLLLPDELITEILLKLPIKSLLKFMCVSKSWLQLISSPAFVKNHIKLTADDKGYIYHRL 67
Query: 52 SSTTRNTDFAY--LQSLITSPRKSRSASTIAIPDD-----YDFCGTCNGLVCLHSSKYDR 104
N DF + L L T + + I P + G+ NGL+C ++ +
Sbjct: 68 IFRNTNDDFKFCPLPPLFTQQQLIKELYHIDSPIERTTLSTHIVGSVNGLIC--AAHVRQ 125
Query: 105 KDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSY-FGFDYDSSTDTYKVVAVAVALSWDT 163
++ Y +WNP + + S+ P + D GF YD S D YKVV + +
Sbjct: 126 REAY-----IWNPTI-TKSKELPKSRSNLCSDGIKCGFGYDESRDDYKVVFIDYPIHRHN 179
Query: 164 LETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILS 223
T+VN+Y++ + W T+ L +L + +VN L + + + C I S
Sbjct: 180 HRTVVNIYSLRTKS-WTTLH-DQLQGFFLLNLHGRFVNGKLYWTS-SSCINNYKVCNITS 236
Query: 224 FDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYK 283
FD + +L LP C + + Y +G L CQ+ VW MK G+
Sbjct: 237 FDLADGTWERLELPSC--GKDNSYINVGVVGSDLSLLYTCQRGAATS-DVWIMKHSGVNV 293
Query: 284 SWTLLFNI 291
SWT LF I
Sbjct: 294 SWTKLFTI 301
>gb|AAU93581.1| putative F-box protein [Solanum demissum]
Length = 383
Score = 106 bits (265), Expect = 1e-21
Identities = 113/382 (29%), Positives = 171/382 (44%), Gaps = 50/382 (13%)
Query: 4 PQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHL-----------HRSS 52
P LP+EL EIL LP+KSL +F CVSKSW +IS F+K H+ HR
Sbjct: 8 PLLLPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKKHIKLTANDKGYIYHRLI 67
Query: 53 STTRNTDFAY--LQSLITSPRKSRSASTIAIPDD-----YDFCGTCNGLVCLHSSKYDRK 105
N DF + L L T+ + I P + G+ NGL+C+ + ++
Sbjct: 68 FRNTNNDFKFCPLPPLFTNQQLIEEILHIDSPIERTTLSTHIVGSVNGLICV--AHVRQR 125
Query: 106 DKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSY-FGFDYDSSTDTYKVVAVAVALSWDTL 164
+ Y +WNPA+ + S+ P + D GF YD S D YKVV + + +
Sbjct: 126 EAY-----IWNPAI-TKSKELPKSTSNLCSDGIKCGFGYDESRDDYKVVFIDYPIRHNH- 178
Query: 165 ETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSF 224
T+VN+Y++ ++ W T+ L ++L + +VN L + + + C I SF
Sbjct: 179 RTVVNIYSLRTNS-WTTLH-DQLQGIFLLNLHGRFVNGKLYWTS-STCINNYKVCNITSF 235
Query: 225 DFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYKS 284
D + L LP C + + Y +G L CQ VW MK G+ S
Sbjct: 236 DLADGTWGSLELPSC--GKDNSYINVGVVGSDLSLLYTCQLGAATS-DVWIMKHSGVNVS 292
Query: 285 WTLLF------NIAGYAYDIPCRFFAMYMSENGDALLLSKLAVSQPQAVLYTQKDNKLEV 338
WT LF NI + P F++++ +G+ LLL A+ ++Y +L+
Sbjct: 293 WTKLFTIKYPQNIKIHRCVAPAFTFSIHI-RHGEILLLLDSAI-----MIYDGSTRQLKH 346
Query: 339 TDVANYIFNCYAMNYIESLVPP 360
T N C + Y+ESLV P
Sbjct: 347 TFHVN---QCEEI-YVESLVNP 364
>gb|AAT39293.1| putative F-box-like protein [Solanum demissum]
Length = 384
Score = 103 bits (258), Expect = 6e-21
Identities = 111/383 (28%), Positives = 171/383 (43%), Gaps = 50/383 (13%)
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHL-----------HRS 51
P LP+EL EIL LP+KSL +F CVSKSW +IS F+K H+ HR
Sbjct: 8 PLLLLPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKNHIKLTANGKGYIYHRL 67
Query: 52 SSTTRNTDFAY--LQSLITSPRKSRSASTIAIPDD-----YDFCGTCNGLVCLHSSKYDR 104
N DF + L SL T + I P + G+ NGL+C ++ +
Sbjct: 68 IFRNTNDDFKFCPLPSLFTKQQLIEELFDIVSPIERTTLSTHIVGSVNGLIC--AAHVRQ 125
Query: 105 KDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSY-FGFDYDSSTDTYKVVAVAVALSWDT 163
++ Y +WNP + + S+ P + D GF YD S D YKVV + S
Sbjct: 126 REAY-----IWNPTI-TKSKELPKSRSNLCSDGIKCGFGYDESHDDYKVVFINYP-SHHN 178
Query: 164 LETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILS 223
++VN+Y++ ++ W T+ L ++L + +V L + + + C I S
Sbjct: 179 HRSVVNIYSLRTNS-WTTLH-DQLQGIFLLNLHCRFVKEKLYWTS-STCINNYKVCNITS 235
Query: 224 FDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYK 283
FD + L LP C + + Y +G L CQ+ + VW MK G+
Sbjct: 236 FDLADGTWESLELPSC--GKDNSYINVGVVGSDLSLLYTCQRGA-ANSDVWIMKHSGVNV 292
Query: 284 SWTLLF------NIAGYAYDIPCRFFAMYMSENGDALLLSKLAVSQPQAVLYTQKDNKLE 337
SWT LF NI + +P F++++ +G+ LL+ A+ ++Y +L+
Sbjct: 293 SWTKLFTIKYPQNIKIHRCVVPAFTFSIHI-RHGEILLVLDSAI-----MIYDGSTRQLK 346
Query: 338 VTDVANYIFNCYAMNYIESLVPP 360
T N C + Y+ESLV P
Sbjct: 347 HTFHVN---QCEEI-YVESLVNP 365
>gb|AAT38695.1| putative F-box protein [Solanum demissum]
Length = 427
Score = 102 bits (253), Expect = 2e-20
Identities = 111/389 (28%), Positives = 171/389 (43%), Gaps = 60/389 (15%)
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTT------- 55
P LP+EL EIL LPVKSL +F CVSKSW +IS F+K H+ ++
Sbjct: 49 PLLLLPDELITEILLKLPVKSLSKFMCVSKSWLQLISSPTFVKNHIKLTADDKGYIHHRL 108
Query: 56 --RNTD----FAYLQSLITSPRKSRSASTIAIPDDYD-----FCGTCNGLVCLHSSKYDR 104
RN D F L L T + + I P + G+ NGL+C+ + +
Sbjct: 109 IFRNIDGNFKFCSLPPLFTKQQHTEELFHIDSPIERSTLSTHIVGSVNGLICV---VHGQ 165
Query: 105 KDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSY-FGFDYDSSTDTYKVVAVAVAL---S 160
K+ Y +WNP + + S+ P + S +GF YD S D YKVV + S
Sbjct: 166 KEAY-----IWNPTI-TKSKELPKFTSNMCSSSIKYGFGYDESRDDYKVVFIHYPYNHSS 219
Query: 161 WDTLETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCT 220
+ T+V++Y++ +++ W T +L + +VN L + + C
Sbjct: 220 SSNMTTVVHIYSLRNNS-WTTF--RDQLQCFLVNHYGRFVNGKLYWTS-STCINKYKVCN 275
Query: 221 ILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCI---CQKDKNMHFVVWQMK 277
I SFD + L LP C + +D LGV+ L + CQ+ VW MK
Sbjct: 276 ITSFDLADGTWGSLDLPICGKDNFDI-----NLGVVGSDLSLLYTCQRGAATS-DVWIMK 329
Query: 278 EFGLYKSWTLLF------NIAGYAYDIPCRFFAMYMSENGDALLLSKLAVSQPQAVLYTQ 331
+ SWT LF NI + P F+++ + + LLL + A+ ++Y
Sbjct: 330 HSAVNVSWTKLFTIKYPQNIKTHRCFAPVFTFSIHF-RHSEILLLLRSAI-----MIYDG 383
Query: 332 KDNKLEVTDVANYIFNCYAMNYIESLVPP 360
+L+ T + + C + Y+ESLV P
Sbjct: 384 STRQLKHT---SDVMQCEEI-YVESLVNP 408
>gb|AAT38689.1| putative S haplotype-specific F-box protein [Solanum demissum]
Length = 240
Score = 83.2 bits (204), Expect = 1e-14
Identities = 62/197 (31%), Positives = 99/197 (49%), Gaps = 39/197 (19%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS------------- 53
LP E+ EIL LP KSL++FRCVSKSW +IS ++FIK HL ++++
Sbjct: 9 LPHEIIKEILLNLPPKSLLKFRCVSKSWLELISSAKFIKNHLKQTANDKEYSHHRIIFQE 68
Query: 54 TTRNTDFAYLQSLITSPRKSR---SASTIAIPDDYDF-CGTCNGLVCLHSSKYDRKDKYT 109
+ N L+S++ + + S + P Y + G+ NGL+CL+S
Sbjct: 69 SACNFKVCCLRSMLNKEQSTELFDIGSPMENPSIYTWIVGSVNGLICLYSK--------I 120
Query: 110 SHVRLWNPAMRSMSQRSPPLYLPI----VYDSYFGFDYDSSTDTYKVVAVAVALSWDTLE 165
LWNPA++ S++ P L + Y +GF YD + D YKV++ +
Sbjct: 121 EETVLWNPAVKK-SKKLPTLGAKLRNGCSYYLKYGFGYDETRDDYKVMS--------NMR 171
Query: 166 TMVNVYNIGDDTCWRTI 182
+VN+Y++ D+ W T+
Sbjct: 172 IVVNIYSLRTDS-WTTL 187
>emb|CAD56660.1| S locus F-box (SLF)-S4D protein [Antirrhinum hispanicum]
Length = 374
Score = 81.3 bits (199), Expect = 4e-14
Identities = 96/394 (24%), Positives = 157/394 (39%), Gaps = 74/394 (18%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRS-------------SS 53
+PE++ EIL WLPVKSL+R +C SK +I FI H+ +
Sbjct: 7 IPEDILKEILVWLPVKSLIRLKCASKHLDMLIKSQAFITSHMIKQRRNDGMLLVRRILPP 66
Query: 54 TTRNTDFAYLQSLITSPRKSRSASTIAI------------PDDYDFCGTCNGLVCLHSSK 101
+T N F++ + SP + I P+ D G CNG+VC+ +
Sbjct: 67 STYNDVFSFHD--VNSPELEEVLPKLPITLLSNPDEASFNPNIVDVLGPCNGIVCITGQE 124
Query: 102 YDRKDKYTSHVRLWNPAMRSMSQ--RSPPLYLPIVYDSYFGFDYDSS-TDTYKVVAV--- 155
+ L NPA+R + +P P Y G + S+ T+ +KV+ +
Sbjct: 125 ---------DIILCNPALREFRKLPSAPISCRPPCYSIRTGGGFGSTCTNNFKVILMNTL 175
Query: 156 -AVALSWDTLETMVNVYNIGDDTCWRTIP--VSHLPSMYLKDDDAVYVNNTLNRLAILPN 212
+ + +++YN +D+ WR I +P ++ ++ +
Sbjct: 176 YTARVDGRDAQHRIHLYNSNNDS-WREINDFAIVMPVVFSYQCSELFFKGACHWNGRTSG 234
Query: 213 VEDRDRCTILSFDFGKESCAQLPLP----YCPQSRYDFYRTWPTLGVLRDCLCICQKDKN 268
D IL+FD E Q P C +++F +L +C + +
Sbjct: 235 ETTPD--VILTFDVSTEVFGQFEHPSGFKLCTGLQHNFM-------ILNECFASVRSEVV 285
Query: 269 MHFV-VWQMKEFGLYKSWTLLFNIAGYAYDIPCRFF---AMYMSENGDALLLSKLAVSQP 324
+ VW MKE+G+ +SWT F I + P F+ + +NGD L S
Sbjct: 286 RCLIEVWVMKEYGIKQSWTKKFVIGPHEIGCPFLFWRNNEELLGDNGDGQLAS------- 338
Query: 325 QAVLYTQKDNKLEVTDVANYIFNCYAMNYIESLV 358
VL+T N+++ +V F A Y ESLV
Sbjct: 339 -FVLHT---NEIKTFEVYAKFFTLRACLYKESLV 368
>emb|CAD56664.1| S locus F-box (SLF)-S5 protein [Antirrhinum hispanicum]
Length = 376
Score = 80.9 bits (198), Expect = 6e-14
Identities = 89/320 (27%), Positives = 135/320 (41%), Gaps = 58/320 (18%)
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS----------- 53
+F +++ EIL + VKSL+RFRCVSKSW S+I + FI HL R +
Sbjct: 5 RFPRQDVISEILLFSSVKSLLRFRCVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYV 64
Query: 54 -TTRNTDFAYLQSLITSPRKSRSASTIAIP------DDYDF---------CGTCNGLVCL 97
T F++ I SP+ + P DYD+ G CNGL+CL
Sbjct: 65 RTPERDMFSFYN--INSPKLEELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLICL 122
Query: 98 HSSKYDRKDKYTSHVRLWNPAMRSMSQRSP-PLYLPIVY-DSYFGFDY-DSSTDTYKVVA 154
Y V L NPA+R + + P P P + G+ + ++ D YKVV
Sbjct: 123 ---------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKVVL 173
Query: 155 VAVALSWDTLETMVNVYNIGDDT-CWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNV 213
+ S + +N+Y DT W+ I P Y+ N + A N
Sbjct: 174 IE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPC---NELFFKGAFHWNA 227
Query: 214 EDRD---RCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMH 270
D IL+FD E ++ P+C F ++ +L L +CL + + + M
Sbjct: 228 NSTDIFYADFILTFDIITEVFKEMAYPHC---LAQFSNSFLSLMSLNECLAMVRYKEWME 284
Query: 271 ----FVVWQMKEFGLYKSWT 286
F +W M ++G+ +SWT
Sbjct: 285 EPELFDIWVMNQYGVRESWT 304
>emb|CAC33022.1| SLF-S2 protein [Antirrhinum hispanicum] gi|13161526|emb|CAC33010.1|
S locus F-box (SLF)-S2 protein [Antirrhinum hispanicum]
Length = 376
Score = 80.5 bits (197), Expect = 7e-14
Identities = 88/321 (27%), Positives = 137/321 (42%), Gaps = 60/321 (18%)
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQ 64
+F +++ EIL + VKSL+RFRCVSKSW S+I + FI HL R + N + ++
Sbjct: 5 RFPRQDVISEILLFSSVKSLLRFRCVSKSWCSLIKSNDFIDNHLLRRQT---NGNVMVVK 61
Query: 65 SLITSPRKSR-SASTIAIPD------------------DYDF---------CGTCNGLVC 96
+ +P + S I P+ DYD+ G CNGL+C
Sbjct: 62 RYVRTPERDMFSFYNINSPELDELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLIC 121
Query: 97 LHSSKYDRKDKYTSHVRLWNPAMRSMSQRSP-PLYLPIVY-DSYFGFDY-DSSTDTYKVV 153
L Y V L NPA+R + + P P P + G+ + ++ D YKVV
Sbjct: 122 L---------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKVV 172
Query: 154 AVAVALSWDTLETMVNVYNIGDDT-CWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPN 212
+ S + +N+Y DT W+ I P Y+ N + A N
Sbjct: 173 LIE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPC---NELFFKGAFHWN 226
Query: 213 VEDRD---RCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNM 269
D IL+FD E ++ P+C F ++ +L L +CL + + + M
Sbjct: 227 ANSTDIFYADFILTFDIITEVFKEMAYPHC---LAQFSNSFLSLMSLNECLAMVRYKEWM 283
Query: 270 H----FVVWQMKEFGLYKSWT 286
F +W M ++G+ +SWT
Sbjct: 284 EDPELFDIWVMNQYGVRESWT 304
>emb|CAD56663.1| S locus F-box (SLF)-S1 protein [Antirrhinum hispanicum]
Length = 376
Score = 79.3 bits (194), Expect = 2e-13
Identities = 88/321 (27%), Positives = 137/321 (42%), Gaps = 60/321 (18%)
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQ 64
+F +++ EIL + VKSL+RFR VSKSW S+I + FI HL R + N + ++
Sbjct: 5 RFPRQDVTSEILLFSSVKSLLRFRLVSKSWCSLIKSNDFIDNHLLRRQT---NGNVMVVK 61
Query: 65 SLITSPRKSR-SASTIAIPD------------------DYDF---------CGTCNGLVC 96
+ +P + S I P+ DYD+ G CNGL+C
Sbjct: 62 RYVRTPERDMFSFYNINSPELDELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLIC 121
Query: 97 LHSSKYDRKDKYTSHVRLWNPAMRSMSQRSP-PLYLPIVY-DSYFGFDY-DSSTDTYKVV 153
L Y V L NPA+R + + P P P + G+ + ++ D YKVV
Sbjct: 122 L---------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKVV 172
Query: 154 AVAVALSWDTLETMVNVYNIGDDT-CWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPN 212
+ S + +N+Y DT W+ I P Y+ N + A N
Sbjct: 173 LIE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPC---NELFFKGAFHWN 226
Query: 213 VEDRD---RCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNM 269
D IL+FD E ++ P+C F ++ +L L +CL + + + M
Sbjct: 227 ANSTDIFYADFILTFDIITEVFKEMAYPHC---LAQFSNSFLSLMSLNECLAMVRYKEWM 283
Query: 270 H----FVVWQMKEFGLYKSWT 286
F +W MK++G+ +SWT
Sbjct: 284 EEPELFDIWVMKQYGVRESWT 304
>emb|CAC33011.1| S locus F-box (SLF)-S2-like protein [Antirrhinum hispanicum]
Length = 376
Score = 79.3 bits (194), Expect = 2e-13
Identities = 88/321 (27%), Positives = 137/321 (42%), Gaps = 60/321 (18%)
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQ 64
+F +++ EIL + VKSL+RFR VSKSW S+I + FI HL R + N + ++
Sbjct: 5 RFPRQDVTSEILLFSSVKSLLRFRLVSKSWCSLIKSNDFIDNHLLRRQT---NGNVMVVK 61
Query: 65 SLITSPRKSR-SASTIAIPD------------------DYDF---------CGTCNGLVC 96
+ +P + S I P+ DYD+ G CNGL+C
Sbjct: 62 RYVRTPERDMFSFYNINSPELDELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLIC 121
Query: 97 LHSSKYDRKDKYTSHVRLWNPAMRSMSQRSP-PLYLPIVY-DSYFGFDY-DSSTDTYKVV 153
L Y V L NPA+R + + P P P + G+ + ++ D YKVV
Sbjct: 122 L---------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKVV 172
Query: 154 AVAVALSWDTLETMVNVYNIGDDT-CWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPN 212
+ S + +N+Y DT W+ I P Y+ N + A N
Sbjct: 173 LIE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPC---NELFFKGAFHWN 226
Query: 213 VEDRD---RCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNM 269
D IL+FD E ++ P+C F ++ +L L +CL + + + M
Sbjct: 227 ANSTDIFYADFILTFDIITEVFKEMAYPHC---LAQFSNSFLSLMSLNECLAMVRYKEWM 283
Query: 270 H----FVVWQMKEFGLYKSWT 286
F +W MK++G+ +SWT
Sbjct: 284 EEPELFDIWVMKQYGVRESWT 304
>emb|CAD56661.1| S locus F-box (SLF)-S4 protein [Antirrhinum hispanicum]
Length = 376
Score = 76.6 bits (187), Expect = 1e-12
Identities = 89/320 (27%), Positives = 132/320 (40%), Gaps = 58/320 (18%)
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS----------- 53
+F +++ EIL + VKSL+RFR VSKSW S+I FI HL R +
Sbjct: 5 RFPRQDVTSEILLFSSVKSLLRFRLVSKSWCSLIKSHDFIDNHLLRRQTNGNVMVVKRYV 64
Query: 54 -TTRNTDFAYLQSLITSPRKSRSASTIAIP------DDYDF---------CGTCNGLVCL 97
T F++ I SP + P DYD+ G CNGLVCL
Sbjct: 65 RTPERDMFSFYD--INSPELDELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLVCL 122
Query: 98 HSSKYDRKDKYTSHVRLWNPAMRSMSQRSP-PLYLPIVY-DSYFGFDY-DSSTDTYKVVA 154
Y V L NPA+R + + P P P + G+ + ++ D YKVV
Sbjct: 123 ---------AYGDCVLLSNPALREIKRLPPTPFANPEGHCTDIIGYGFGNTCNDCYKVVL 173
Query: 155 VAVALSWDTLETMVNVYNIGDDT-CWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNV 213
+ S + +N+Y DT W+ I P Y+ N + A N
Sbjct: 174 IE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPC---NELFFKGAFHWNA 227
Query: 214 EDRD---RCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMH 270
D IL+FD E ++ P+C F ++ +L L +CL + + + M
Sbjct: 228 NSTDIFYADFILTFDIITEVFKEMAYPHC---LAQFSNSFLSLMSLNECLAMVRYKEWME 284
Query: 271 ----FVVWQMKEFGLYKSWT 286
F +W M ++G+ +SWT
Sbjct: 285 EPELFDIWVMNQYGVRESWT 304
>gb|AAR15913.1| S3 self-incompatibility locus-linked putative F-box protein S3-A113
[Petunia integrifolia subsp. inflata]
Length = 376
Score = 75.5 bits (184), Expect = 2e-12
Identities = 80/314 (25%), Positives = 132/314 (41%), Gaps = 53/314 (16%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS----------TTR 56
LP+++ + IL LPVKSL+RF+C K++ +II S FI LHL+ +++ + +
Sbjct: 4 LPQDVVIYILVMLPVKSLLRFKCSCKTFCNIIKSSTFIYLHLNHTTNVKDELVLLKRSFK 63
Query: 57 NTDFAYLQSLITSPRKSRSASTIAIPDD--------------YDFCGTCNGLVCLHSSKY 102
D+ + +S+++ +I D + G CNGL+ L
Sbjct: 64 TDDYNFYKSILSFLSSKEGYDFKSISPDVEIPHLTTTSACVFHQLIGPCNGLIAL----- 118
Query: 103 DRKDKYTSHVRLWNPAMRSMSQRSP-PLYLPIVYD---SYFGFDYDSSTDTYKVVAVA-V 157
D T+ V +NPA R P P +P + S GF +DS + YKVV ++ V
Sbjct: 119 --TDSLTTIV--FNPATRKYRLIPPCPFGIPRGFRRSISGIGFGFDSDANDYKVVRLSEV 174
Query: 158 ALSWDTLETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRD 217
E V++Y+ D+ WR + +P +Y + + A +V
Sbjct: 175 YKEPCDKEMKVDIYDFSVDS-WRELLGQEVPIVYWLPCAEILYKRNFHWFAFADDV---- 229
Query: 218 RCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKD------KNMHF 271
IL FD E L +P +D + + + + + IC D
Sbjct: 230 --VILCFDMNTEKFHNLGMP--DACHFDDGKCYGLVILCKCMTLICYPDPMPSSPTEKLT 285
Query: 272 VVWQMKEFGLYKSW 285
+W MKE+G +SW
Sbjct: 286 DIWIMKEYGEKESW 299
>gb|AAT72121.1| SFB3 [Prunus avium]
Length = 376
Score = 74.3 bits (181), Expect = 5e-12
Identities = 66/253 (26%), Positives = 108/253 (42%), Gaps = 37/253 (14%)
Query: 10 ELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNT----------- 58
E+ ++IL LP KSL+RF C KSW +I S F++ HLHR+ + +
Sbjct: 9 EILIDILVRLPAKSLIRFLCTCKSWSDLIGSSSFVRTHLHRNVTKHAHVYLLCLHHPQFE 68
Query: 59 ---------DFAYLQSLITSPRKSRSASTIAIP----DDYDFCGTCNGLVCLHSSKYDRK 105
D LQ + S K S ++ P + + G+ NGLVC+ D
Sbjct: 69 RQNDNDDPYDIEELQWSLFSNEKFEQFSNLSHPLENTEHFRIYGSSNGLVCMS----DEI 124
Query: 106 DKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSY-FGFDYDSSTDTYKVVAVAVALSWDTL 164
+ S +++WNP++R + + + F + + YK V + + +
Sbjct: 125 LNFDSPIQIWNPSVRKFRTLPMSTNINMKFSHVSLQFGFHPGVNDYKAVRM---MHTNKG 181
Query: 165 ETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSF 224
V VY++ D CW+ I V +P +LK + N +A +E C+I+SF
Sbjct: 182 ALAVEVYSLKTD-CWKMIEV--IPP-WLKCTWKHHKGTFFNGVA-YHIIEKGPICSIMSF 236
Query: 225 DFGKESCAQLPLP 237
D G E + P
Sbjct: 237 DSGSEKFEEFITP 249
>dbj|BAC81148.1| S-locus F-Box protein 3 [Prunus avium]
Length = 376
Score = 74.3 bits (181), Expect = 5e-12
Identities = 66/253 (26%), Positives = 108/253 (42%), Gaps = 37/253 (14%)
Query: 10 ELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNT----------- 58
E+ ++IL LP KSL+RF C KSW +I S F++ HLHR+ + +
Sbjct: 9 EILIDILVRLPAKSLIRFLCTCKSWSDLIGSSSFVRTHLHRNVTKHAHVYLLCLHHPQFE 68
Query: 59 ---------DFAYLQSLITSPRKSRSASTIAIP----DDYDFCGTCNGLVCLHSSKYDRK 105
D LQ + S K S ++ P + + G+ NGLVC+ D
Sbjct: 69 RQNDNDDPYDIEELQWSLFSNEKFEQFSNLSHPLENTEHFRIYGSSNGLVCMS----DEI 124
Query: 106 DKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSY-FGFDYDSSTDTYKVVAVAVALSWDTL 164
+ S +++WNP++R + + + F + + YK V + + +
Sbjct: 125 LNFDSPIQIWNPSVRKFRTLPMSTNINMKFSHVSLQFGFHPGVNDYKAVRM---MHTNKG 181
Query: 165 ETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTILSF 224
V VY++ D CW+ I V +P +LK + N +A +E C+I+SF
Sbjct: 182 ALAVEVYSLKTD-CWKMIEV--IPP-WLKCTWKHHKGTFFNGVA-YHIIEKGPICSIMSF 236
Query: 225 DFGKESCAQLPLP 237
D G E + P
Sbjct: 237 DSGSEKFEEFITP 249
>gb|AAF21197.1| unknown protein [Arabidopsis thaliana] gi|15983479|gb|AAL11607.1|
AT3g07870/F17A17_21 [Arabidopsis thaliana]
gi|18398079|ref|NP_566322.1| F-box family protein
[Arabidopsis thaliana]
Length = 417
Score = 73.9 bits (180), Expect = 7e-12
Identities = 81/327 (24%), Positives = 128/327 (38%), Gaps = 62/327 (18%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIK----------LHLHRSSSTTR 56
LPE++ +I S LP+ S+ R V +SW+S+++ + L LH S
Sbjct: 28 LPEDIIADIFSRLPISSIARLMFVCRSWRSVLTQHGRLSSSSSSPTKPCLLLHCDSPIRN 87
Query: 57 NTDFAYL----QSLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHV 112
F L + + T R AS++ ++D G+CNGL+CL S Y+ +
Sbjct: 88 GLHFLDLSEEEKRIKTKKFTLRFASSM---PEFDVVGSCNGLLCLSDSLYN------DSL 138
Query: 113 RLWNPAMRSMSQRSPPLYLPIVYDSY------FGFDYDSSTDTYKVVAVAVALSWDT--- 163
L+NP + L LP + Y FGF + T YKV+ + +
Sbjct: 139 YLYNPFTTN------SLELPECSNKYHDQELVFGFGFHEMTKEYKVLKIVYFRGSSSNNN 192
Query: 164 -----------LETMVNVYNIGDDT-----CWRTIPVSHLPSMYLKDDDAVYVNNTLNRL 207
++ V + + T WR++ P ++K VN L+
Sbjct: 193 GIYRGRGRIQYKQSEVQILTLSSKTTDQSLSWRSL--GKAPYKFVKRSSEALVNGRLH-F 249
Query: 208 AILPNVEDRDRCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDK 267
P DR +SFD E ++P P C RT L L+ CLC
Sbjct: 250 VTRPRRHVPDR-KFVSFDLEDEEFKEIPKPDCG----GLNRTNHRLVNLKGCLCAVVYGN 304
Query: 268 NMHFVVWQMKEFGLYKSWTLLFNIAGY 294
+W MK +G+ +SW ++I Y
Sbjct: 305 YGKLDIWVMKTYGVKESWGKEYSIGTY 331
>dbj|BAC65205.1| S haplotype-specific F-box protein d [Prunus dulcis]
Length = 376
Score = 72.0 bits (175), Expect = 3e-11
Identities = 89/391 (22%), Positives = 164/391 (41%), Gaps = 69/391 (17%)
Query: 9 EELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS--------------- 53
+E+ ++IL LP KSLVRF C KSW +I S F+ HLHR+ +
Sbjct: 8 KEILIDILVRLPAKSLVRFLCTCKSWSDLIGSSSFVGTHLHRNVTGHAQAYLLCLHHPNF 67
Query: 54 -TTRNTDFAY--------LQSLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDR 104
R+ D Y L S +T S+ + + + Y G+ NGLVC+ D
Sbjct: 68 ECQRDDDDPYFKEELQWSLFSNVTFEESSKLSHPLGSTEHYVIYGSSNGLVCIS----DE 123
Query: 105 KDKYTSHVRLWNPAMRSMSQRSPPLYLPI-VYDSYFG--FDYDSSTDTYKVVAVAVALSW 161
+ S + +WNP+++ + R+ P+ I + S+ F + S + Y+ V + L
Sbjct: 124 IMNFDSPIHIWNPSVKKL--RTTPISTSINIKFSHVALQFGFHSGVNDYRAVRM---LRT 178
Query: 162 DTLETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRDRCTI 221
+ V +Y++ D+ W I +P +LK + N +A +E ++
Sbjct: 179 NQNALAVEIYSLRTDS-WTMIEA--IPP-WLKCTWQHHQGTFFNGVA-YHIIEKGPTFSV 233
Query: 222 LSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCI------CQKDKNMHFVVWQ 275
+SFD G E + P + +R + V ++ +C+ C+++ + +W
Sbjct: 234 ISFDSGSEEFEEF---IAPDAICSLWRL--CIHVYKEQICLLFGYYSCEEEGMENIDLWV 288
Query: 276 MKEFGLYKSWTLLFNIAGYAYDIPCRFFAMYMSENGDALLLSK---------LAVSQPQA 326
++E K W + + YD F+ + + L++++ L + ++
Sbjct: 289 LQE----KRWK---QLCPFIYDPLDDFYQIIGISTDNKLIMARKDFSKGGVELQLGNYES 341
Query: 327 VLYTQKDNKLEVTDVANYIFNCYAMNYIESL 357
+ KL + F +AM Y+ESL
Sbjct: 342 KQVLETGIKLAIMRYGESEF-LFAMTYVESL 371
>gb|AAR15912.2| S2 self-incompatibility locus-linked putative F-box protein S2-A113
[Petunia integrifolia subsp. inflata]
gi|38261532|gb|AAR15911.1| S1 self-incompatibility
locus-linked putative F-box protein S1-A113 [Petunia
integrifolia subsp. inflata]
Length = 376
Score = 72.0 bits (175), Expect = 3e-11
Identities = 80/323 (24%), Positives = 133/323 (40%), Gaps = 71/323 (21%)
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS----------TTR 56
LP+++ + IL LPVKSL+RF+C K++ +II S F+ LHL+ ++ + +
Sbjct: 4 LPQDVVIYILVMLPVKSLLRFKCSCKTFCNIIKSSTFVNLHLNHTTKVKDELVLLKRSFK 63
Query: 57 NTDFAYLQSLITSPRKSRSASTIAIPDDYDF-----------------------CGTCNG 93
++ + +S++ S + +DYDF G CNG
Sbjct: 64 TDEYNFYKSIL---------SFFSSKEDYDFMPMSPDVEIPHLTTTSARVFHQLIGPCNG 114
Query: 94 LVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQRSP-PLYLPIVYD---SYFGFDYDSSTDT 149
L+ L D T+ V +NPA R P P +P + S GF +DS +
Sbjct: 115 LIAL-------TDSLTTIV--FNPATRKYRLIPPCPFGIPRGFRRSISGIGFGFDSDVND 165
Query: 150 YKVVAVA-VALSWDTLETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLA 208
YKVV ++ V E V++Y+ D+ WR + +P +Y + + A
Sbjct: 166 YKVVRLSEVYKEPCDKEMKVDIYDFSVDS-WRELLGQEVPIVYWLPCADILFKRNFHWFA 224
Query: 209 ILPNVEDRDRCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKD-- 266
+V IL FD E + +P +D + + + + + IC D
Sbjct: 225 FADDV------VILCFDMNTEKFHNMGMP--DACHFDDGKCYGLVILCKCMSLICYPDPM 276
Query: 267 ----KNMHFVVWQMKEFGLYKSW 285
+W MKE+G +SW
Sbjct: 277 PSSPTEKLTDIWIMKEYGEKESW 299
>ref|NP_175213.1| F-box family protein [Arabidopsis thaliana]
gi|9802587|gb|AAF99789.1| T2E6.11 [Arabidopsis thaliana]
Length = 389
Score = 71.6 bits (174), Expect = 3e-11
Identities = 64/203 (31%), Positives = 92/203 (44%), Gaps = 34/203 (16%)
Query: 2 EPPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFA 61
+P P +L EIL LPVKS+VRFRCVSK W SII+D FIK + SST ++ F
Sbjct: 20 KPTSSFPLDLASEILLRLPVKSVVRFRCVSKLWSSIITDPYFIKTY-ETQSSTRQSLLFC 78
Query: 62 YLQS----LITSPR---KSRSASTIAI-------PDDYDF---CGTCNGLVCLHSSKYDR 104
+ QS + + P+ S S+S AI P ++ + + +GL+C H
Sbjct: 79 FKQSDKLFVFSIPKHHYDSNSSSQAAIDRFQVKLPQEFSYPSPTESVHGLICFH------ 132
Query: 105 KDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALSWDTL 164
+ V +WNP+MR P + G YD +KVV + + D
Sbjct: 133 ---VLATVIVWNPSMRQFLTLPKPRKSWKELTVFLG--YDPIEGKHKVVCLPRNRTCDEC 187
Query: 165 ETMVNVYNIGD-DTCWRTIPVSH 186
+ V +G WRT+ H
Sbjct: 188 Q----VLTLGSAQKSWRTVKTKH 206
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.137 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 645,578,154
Number of Sequences: 2540612
Number of extensions: 26257013
Number of successful extensions: 47248
Number of sequences better than 10.0: 473
Number of HSP's better than 10.0 without gapping: 401
Number of HSP's successfully gapped in prelim test: 72
Number of HSP's that attempted gapping in prelim test: 46610
Number of HSP's gapped (non-prelim): 580
length of query: 361
length of database: 863,360,394
effective HSP length: 129
effective length of query: 232
effective length of database: 535,621,446
effective search space: 124264175472
effective search space used: 124264175472
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0135.3


