

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
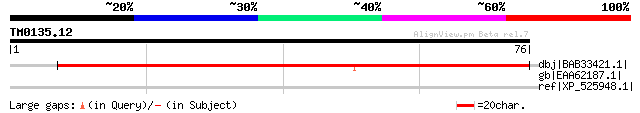
Query= TM0135.12
(76 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB33421.1| putative senescence-associated protein [Pisum sa... 115 3e-25
gb|EAA62187.1| predicted protein [Aspergillus nidulans FGSC A4] ... 42 0.004
ref|XP_525948.1| PREDICTED: similar to putative senescence-assoc... 38 0.059
>dbj|BAB33421.1| putative senescence-associated protein [Pisum sativum]
Length = 282
Score = 115 bits (288), Expect = 3e-25
Identities = 55/71 (77%), Positives = 58/71 (81%), Gaps = 2/71 (2%)
Query: 8 SPNSELEALSHNPTHGSFAPLTSQTSVMTNCANQRFLFN*VE--LRHFHQ*GKTNMSHDG 65
SP+S+LEA SHNPTHGSFAPL Q S MTNCANQRFL VE LRH HQ GKTN+SHDG
Sbjct: 9 SPDSDLEAFSHNPTHGSFAPLAFQPSAMTNCANQRFLSYYVELLLRHCHQWGKTNLSHDG 68
Query: 66 LIPTHVPYWWV 76
LIP HVPYWWV
Sbjct: 69 LIPAHVPYWWV 79
>gb|EAA62187.1| predicted protein [Aspergillus nidulans FGSC A4]
gi|67538150|ref|XP_662849.1| hypothetical protein
AN5245_2 [Aspergillus nidulans FGSC A4]
gi|49095842|ref|XP_409382.1| predicted protein
[Aspergillus nidulans FGSC A4]
Length = 198
Score = 42.0 bits (97), Expect = 0.004
Identities = 21/39 (53%), Positives = 24/39 (60%)
Query: 6 RYSPNSELEALSHNPTHGSFAPLTSQTSVMTNCANQRFL 44
R +S+LEA SH P GS A L QT+ TN NQRFL
Sbjct: 158 RSGTDSDLEAFSHYPADGSVAALPGQTAAKTNYLNQRFL 196
>ref|XP_525948.1| PREDICTED: similar to putative senescence-associated protein [Pan
troglodytes]
Length = 106
Score = 38.1 bits (87), Expect = 0.059
Identities = 17/27 (62%), Positives = 19/27 (69%)
Query: 10 NSELEALSHNPTHGSFAPLTSQTSVMT 36
+S+LE SHNPT GSFAPL Q S T
Sbjct: 65 DSDLEVFSHNPTDGSFAPLAPQPSTYT 91
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.332 0.139 0.486
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 121,307,216
Number of Sequences: 2540612
Number of extensions: 3077657
Number of successful extensions: 9591
Number of sequences better than 10.0: 3
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 9587
Number of HSP's gapped (non-prelim): 3
length of query: 76
length of database: 863,360,394
effective HSP length: 52
effective length of query: 24
effective length of database: 731,248,570
effective search space: 17549965680
effective search space used: 17549965680
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0135.12


