

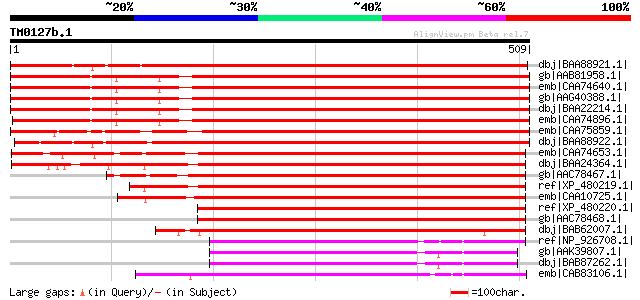
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0127b.1
(509 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA88921.1| sigma factor [Nicotiana tabacum] 700 0.0
gb|AAB81958.1| RNA polymerase sigma subunit 1 [Arabidopsis thali... 669 0.0
emb|CAA74640.1| plastid RNA polymerase sigma factor [Arabidopsis... 669 0.0
gb|AAG40388.1| At1g64860 [Arabidopsis thaliana] 667 0.0
dbj|BAA22214.1| plastid RNA polymerase sigma-subunit [Arabidopsi... 666 0.0
emb|CAA74896.1| sigma factorB [Arabidopsis thaliana] gi|25288494... 665 0.0
emb|CAA75859.1| Sig1 [Sinapis alba] gi|7488610|pir||T10470 trans... 664 0.0
dbj|BAA88922.1| sigma factor [Nicotiana tabacum] 657 0.0
emb|CAA74653.1| plastid RNA polymerase sigma factor [Sorghum bic... 498 e-139
dbj|BAA24364.1| plastid RNA polymerase sigma factor [Oryza sativ... 494 e-138
gb|AAC78467.1| RNA polymerase sigma factor 1 [Zea mays] 476 e-133
ref|XP_480219.1| putative plastid RNA polymerase sigma factor [O... 468 e-130
emb|CAA10725.1| chloroplast sigma factor [Triticum aestivum] 466 e-130
ref|XP_480220.1| putative plastid RNA polymerase sigma factor [O... 446 e-124
gb|AAC78468.1| RNA polymerase sigma factor 2 [Zea mays] 431 e-119
dbj|BAB62007.1| PpSIG1 [Physcomitrella patens] gi|14669828|dbj|B... 263 1e-68
ref|NP_926708.1| group2 RNA polymerase sigma factor [Gloeobacter... 190 1e-46
gb|AAK39807.1| RNA-polymerase sigma factor [Guillardia theta] gi... 187 5e-46
dbj|BAB87262.1| RNA polymerase sigma factor [Guillardia theta] 186 2e-45
emb|CAB83106.1| sigma factor 2 [Sinapis alba] 184 6e-45
>dbj|BAA88921.1| sigma factor [Nicotiana tabacum]
Length = 508
Score = 700 bits (1806), Expect = 0.0
Identities = 365/510 (71%), Positives = 429/510 (83%), Gaps = 6/510 (1%)
Query: 1 MATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQA 60
M+TAAVIGLS GKRLLSSS+YYSD+ EKL+C+SD + + K+V AKKSSDY+ +
Sbjct: 2 MSTAAVIGLSAGKRLLSSSFYYSDLNEKLTCTSDHSLPCHHVPSIKNVITAKKSSDYSPS 61
Query: 61 FPASDRPNQSIKALKEHVD--GVPAAAETWFQECDSNDLEVESSDLGYSVEALLLLQKSM 118
S R +QS K+LKEHVD P+ +WF+ + L+ ES D SVE LLLLQKSM
Sbjct: 62 C-VSSRKSQSAKSLKEHVDITADPSDVPSWFEMYEQ--LQNESDDEDESVEVLLLLQKSM 118
Query: 119 LEKQWSLSCEREVLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKIPGKTGSAMQACDA 178
LEKQW+LS E + LT P+ EK SKK+ +TCSG SAR+RR+++++++ + SA A
Sbjct: 119 LEKQWNLSLE-QTLTASPQHEKKSKKMHITCSGTSARRRRMDSRQRVLDRHSSATPTSAA 177
Query: 179 MQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGC 238
+RS+I PELLQNR KGYVKGVVSE+LL+HAEV++LS+KI+VGL L+E KSRLKE+LGC
Sbjct: 178 KPLRSIIGPELLQNRLKGYVKGVVSEELLTHAEVLQLSKKIQVGLYLEEQKSRLKERLGC 237
Query: 239 EPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQG 298
EPSDDQ+A SLK+SR++L++ IEC+LARERLAMSNVRLVMSIAQRYDN+GAEMADLVQG
Sbjct: 238 EPSDDQLAVSLKMSRTDLQSTLIECSLARERLAMSNVRLVMSIAQRYDNVGAEMADLVQG 297
Query: 299 GLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAK 358
GLIGLLRGIEKFD SKG+KISTYVYWWIRQGVSRALVENSRTLRLPT+LHERLSLIR AK
Sbjct: 298 GLIGLLRGIEKFDPSKGYKISTYVYWWIRQGVSRALVENSRTLRLPTYLHERLSLIRNAK 357
Query: 359 FRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIA 418
RLEE+GITP+++ IA SLNMSQKKVRNATEA SK++SLDREAFPSLNGLPG+T H YIA
Sbjct: 358 ARLEEKGITPSVNNIAASLNMSQKKVRNATEASSKMYSLDREAFPSLNGLPGETLHSYIA 417
Query: 419 DNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKRIGLS 478
DN LEN PWHGVD W LKDEVN LI+ TL +REREIIRLYYGLD ECLTWEDISKRIGLS
Sbjct: 418 DNHLENNPWHGVDVWALKDEVNNLISSTLRDREREIIRLYYGLDNECLTWEDISKRIGLS 477
Query: 479 RERVRQVGLVALEKLKHAARKREMEAMLLK 508
RERVRQVGLVALEKLKHAARKR+++AML+K
Sbjct: 478 RERVRQVGLVALEKLKHAARKRQLDAMLIK 507
>gb|AAB81958.1| RNA polymerase sigma subunit 1 [Arabidopsis thaliana]
Length = 502
Score = 669 bits (1727), Expect = 0.0
Identities = 354/515 (68%), Positives = 414/515 (79%), Gaps = 19/515 (3%)
Query: 1 MATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQA 60
MATAAVIGL+ GKRLLSSS+Y+SD+ EK +D S+ Y I KS AKK+S+Y +
Sbjct: 1 MATAAVIGLNTGKRLLSSSFYHSDVTEKFLSVNDHCSSQYHIASTKSGITAKKASNYRPS 60
Query: 61 FPASDRPNQSIKALKEHVDGVPAAAETWFQECDSNDLEVESSD----LGYSVEALLLLQK 116
FP+S+R QS KALKE VD V + + W +LE E D + +SVEA+LLLQK
Sbjct: 61 FPSSNRHTQSAKALKESVD-VASTEKPWLPNGTDKELEEECYDDDDLISHSVEAILLLQK 119
Query: 117 SMLEKQWSLSCEREVLTEHPRQEKSSKKV--AVTCSGVSARQRRINTKRKIPGKTGSAMQ 174
SMLEK W+LS E+ V +E+P + KK +TCSG+SARQRRI K+K
Sbjct: 120 SMLEKSWNLSFEKAVSSEYPGKGTIRKKKIPVITCSGISARQRRIGAKKKTN-------- 171
Query: 175 ACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKE 234
M +S + +GYVKGV+SE +LSHAEVV+LS+KIK GL LD+HKSRLK+
Sbjct: 172 ----MTHVKAVSDVSSGKQVRGYVKGVISEDVLSHAEVVRLSKKIKSGLRLDDHKSRLKD 227
Query: 235 KLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMAD 294
+LGCEPSD+Q+A SLKISR+EL+A +EC LARE+LAMSNVRLVMSIAQRYDN+GAEM+D
Sbjct: 228 RLGCEPSDEQLAVSLKISRAELQAWLMECHLAREKLAMSNVRLVMSIAQRYDNLGAEMSD 287
Query: 295 LVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLI 354
LVQGGLIGLLRGIEKFDSSKGF+ISTYVYWWIRQGVSRALV+NSRTLRLPTHLHERL LI
Sbjct: 288 LVQGGLIGLLRGIEKFDSSKGFRISTYVYWWIRQGVSRALVDNSRTLRLPTHLHERLGLI 347
Query: 355 RTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHH 414
R AK RL+E+GITP+IDRIA+SLNMSQKKVRNATEA+SKVFSLDR+AFPSLNGLPG+THH
Sbjct: 348 RNAKLRLQEKGITPSIDRIAESLNMSQKKVRNATEAVSKVFSLDRDAFPSLNGLPGETHH 407
Query: 415 CYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKR 474
YIAD RLEN PWHG D+ LK+EV+KLI+ TL ERE+EIIRLYYGLDKECLTWEDISKR
Sbjct: 408 SYIADTRLENNPWHGYDDLALKEEVSKLISATLGEREKEIIRLYYGLDKECLTWEDISKR 467
Query: 475 IGLSRERVRQVGLVALEKLKHAARKREMEAMLLKH 509
IGLSRERVRQVGLVALEKLKHAARKR+MEAM+LK+
Sbjct: 468 IGLSRERVRQVGLVALEKLKHAARKRKMEAMILKN 502
>emb|CAA74640.1| plastid RNA polymerase sigma factor [Arabidopsis thaliana]
gi|5478439|dbj|BAA82448.1| sigma factor SigA
[Arabidopsis thaliana] gi|2353171|gb|AAB69384.1| sigma
factor 1 [Arabidopsis thaliana]
gi|20259916|gb|AAM13305.1| RNA polymerase sigma subunit
1 [Arabidopsis thaliana] gi|5042421|gb|AAD38260.1| RNA
polymerase sigma subunit 1 [Arabidopsis thaliana]
gi|15217767|ref|NP_176666.1| RNA polymerase sigma
subunit SigA (sigA) / sigma factor 1 (SIG1) [Arabidopsis
thaliana] gi|17064900|gb|AAL32604.1| RNA polymerase
sigma subunit 1 [Arabidopsis thaliana]
gi|25288493|pir||H96671 RNA polymerase sigma subunit 1
[imported] - Arabidopsis thaliana
gi|2443408|dbj|BAA22421.1| SigA [Arabidopsis thaliana]
Length = 502
Score = 669 bits (1725), Expect = 0.0
Identities = 353/515 (68%), Positives = 414/515 (79%), Gaps = 19/515 (3%)
Query: 1 MATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQA 60
MATAAVIGL+ GKRLLSSS+Y+SD+ EK +D S+ Y I KS AKK+S+Y+ +
Sbjct: 1 MATAAVIGLNTGKRLLSSSFYHSDVTEKFLSVNDHCSSQYHIASTKSGITAKKASNYSPS 60
Query: 61 FPASDRPNQSIKALKEHVDGVPAAAETWFQECDSNDLEVESSD----LGYSVEALLLLQK 116
FP+S+R QS KALKE VD V + + W +LE E D + +SVEA+LLLQK
Sbjct: 61 FPSSNRHTQSAKALKESVD-VASTEKPWLPNGTDKELEEECYDDDDLISHSVEAILLLQK 119
Query: 117 SMLEKQWSLSCEREVLTEHPRQEKSSKKV--AVTCSGVSARQRRINTKRKIPGKTGSAMQ 174
SMLEK W+LS E+ V +E+P + KK +TCSG+SARQRRI K+K
Sbjct: 120 SMLEKSWNLSFEKAVSSEYPGKGTIRKKKIPVITCSGISARQRRIGAKKKTN-------- 171
Query: 175 ACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKE 234
M +S + +GYVKGV+SE +LSH EVV+LS+KIK GL LD+HKSRLK+
Sbjct: 172 ----MTHVKAVSDVSSGKQVRGYVKGVISEDVLSHVEVVRLSKKIKSGLRLDDHKSRLKD 227
Query: 235 KLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMAD 294
+LGCEPSD+Q+A SLKISR+EL+A +EC LARE+LAMSNVRLVMSIAQRYDN+GAEM+D
Sbjct: 228 RLGCEPSDEQLAVSLKISRAELQAWLMECHLAREKLAMSNVRLVMSIAQRYDNLGAEMSD 287
Query: 295 LVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLI 354
LVQGGLIGLLRGIEKFDSSKGF+ISTYVYWWIRQGVSRALV+NSRTLRLPTHLHERL LI
Sbjct: 288 LVQGGLIGLLRGIEKFDSSKGFRISTYVYWWIRQGVSRALVDNSRTLRLPTHLHERLGLI 347
Query: 355 RTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHH 414
R AK RL+E+GITP+IDRIA+SLNMSQKKVRNATEA+SKVFSLDR+AFPSLNGLPG+THH
Sbjct: 348 RNAKLRLQEKGITPSIDRIAESLNMSQKKVRNATEAVSKVFSLDRDAFPSLNGLPGETHH 407
Query: 415 CYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKR 474
YIAD RLEN PWHG D+ LK+EV+KLI+ TL ERE+EIIRLYYGLDKECLTWEDISKR
Sbjct: 408 SYIADTRLENNPWHGYDDLALKEEVSKLISATLGEREKEIIRLYYGLDKECLTWEDISKR 467
Query: 475 IGLSRERVRQVGLVALEKLKHAARKREMEAMLLKH 509
IGLSRERVRQVGLVALEKLKHAARKR+MEAM+LK+
Sbjct: 468 IGLSRERVRQVGLVALEKLKHAARKRKMEAMILKN 502
>gb|AAG40388.1| At1g64860 [Arabidopsis thaliana]
Length = 502
Score = 667 bits (1720), Expect = 0.0
Identities = 352/515 (68%), Positives = 414/515 (80%), Gaps = 19/515 (3%)
Query: 1 MATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQA 60
MATAAVIGL+ GKRLLSSS+Y+SD+ EK +D S+ Y I KS AKK+S+Y+ +
Sbjct: 1 MATAAVIGLNTGKRLLSSSFYHSDVTEKFLSVNDHCSSQYHIASTKSGITAKKASNYSPS 60
Query: 61 FPASDRPNQSIKALKEHVDGVPAAAETWFQECDSNDLEVESSD----LGYSVEALLLLQK 116
FP+S+R QS KALKE VD V + + W +LE E D + +SVEA+LLLQK
Sbjct: 61 FPSSNRHTQSAKALKESVD-VASTEKPWLPNGTDKELEEECYDDDDLISHSVEAILLLQK 119
Query: 117 SMLEKQWSLSCEREVLTEHPRQEKSSKKV--AVTCSGVSARQRRINTKRKIPGKTGSAMQ 174
SMLEK W+LS E+ V +E+P + KK +TCSG+SARQRRI K+K
Sbjct: 120 SMLEKSWNLSFEKAVSSEYPGKGTIRKKKIPVITCSGISARQRRIGAKKKTN-------- 171
Query: 175 ACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKE 234
M +S + +GYVKGV+SE +LSH EVV+LS+KIK GL LD+HKSRLK+
Sbjct: 172 ----MTHVKAVSDVSSGKQVRGYVKGVISEDVLSHVEVVRLSKKIKSGLRLDDHKSRLKD 227
Query: 235 KLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMAD 294
+LGCEPSD+Q+A SLKISR+EL+A +EC LARE+LAMSNVRLVMSIAQRY+N+GAEM+D
Sbjct: 228 RLGCEPSDEQLAVSLKISRAELQAWLMECHLAREKLAMSNVRLVMSIAQRYNNLGAEMSD 287
Query: 295 LVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLI 354
LVQGGLIGLLRGIEKFDSSKGF+ISTYVYWWIRQGVSRALV+NSRTLRLPTHLHERL LI
Sbjct: 288 LVQGGLIGLLRGIEKFDSSKGFRISTYVYWWIRQGVSRALVDNSRTLRLPTHLHERLGLI 347
Query: 355 RTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHH 414
R AK RL+E+GITP+IDRIA+SLNMSQKKVRNATEA+SKVFSLDR+AFPSLNGLPG+THH
Sbjct: 348 RNAKLRLQEKGITPSIDRIAESLNMSQKKVRNATEAVSKVFSLDRDAFPSLNGLPGETHH 407
Query: 415 CYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKR 474
YIAD RLEN PWHG D+ LK+EV+KLI+ TL ERE+EIIRLYYGLDKECLTWEDISKR
Sbjct: 408 SYIADTRLENNPWHGYDDLALKEEVSKLISATLGEREKEIIRLYYGLDKECLTWEDISKR 467
Query: 475 IGLSRERVRQVGLVALEKLKHAARKREMEAMLLKH 509
IGLSRERVRQVGLVALEKLKHAARKR+MEAM+LK+
Sbjct: 468 IGLSRERVRQVGLVALEKLKHAARKRKMEAMILKN 502
>dbj|BAA22214.1| plastid RNA polymerase sigma-subunit [Arabidopsis thaliana]
Length = 502
Score = 666 bits (1719), Expect = 0.0
Identities = 352/515 (68%), Positives = 413/515 (79%), Gaps = 19/515 (3%)
Query: 1 MATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQA 60
MATAAVIGL+ GKRLLSSS+Y+SD+ EK +D S+ Y I KS AKK+S+Y+ +
Sbjct: 1 MATAAVIGLNTGKRLLSSSFYHSDVTEKFLSVNDHCSSQYHIASTKSGITAKKASNYSPS 60
Query: 61 FPASDRPNQSIKALKEHVDGVPAAAETWFQECDSNDLEVESSD----LGYSVEALLLLQK 116
FP+S+R QS KALKE VD V + + W +LE E D + +SVEA+LLLQK
Sbjct: 61 FPSSNRHTQSAKALKESVD-VASTEKPWLPNGTDKELEEECYDDDDLISHSVEAILLLQK 119
Query: 117 SMLEKQWSLSCEREVLTEHPRQEKSSKKV--AVTCSGVSARQRRINTKRKIPGKTGSAMQ 174
SMLEK W+LS E+ V +E+P + KK +TCSG+SAR RRI K+K
Sbjct: 120 SMLEKSWNLSFEKAVSSEYPGKGTIRKKKIPVITCSGISARPRRIGAKKKTN-------- 171
Query: 175 ACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKE 234
M +S + +GYVKGV+SE +LSH EVV+LS+KIK GL LD+HKSRLK+
Sbjct: 172 ----MTHVKAVSDVSSGKQVRGYVKGVISEDVLSHVEVVRLSKKIKSGLRLDDHKSRLKD 227
Query: 235 KLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMAD 294
+LGCEPSD+Q+A SLKISR+EL+A +EC LARE+LAMSNVRLVMSIAQRYDN+GAEM+D
Sbjct: 228 RLGCEPSDEQLAVSLKISRAELQAWLMECHLAREKLAMSNVRLVMSIAQRYDNLGAEMSD 287
Query: 295 LVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLI 354
LVQGGLIGLLRGIEKFDSSKGF+ISTYVYWWIRQGVSRALV+NSRTLRLPTHLHERL LI
Sbjct: 288 LVQGGLIGLLRGIEKFDSSKGFRISTYVYWWIRQGVSRALVDNSRTLRLPTHLHERLGLI 347
Query: 355 RTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHH 414
R AK RL+E+GITP+IDRIA+SLNMSQKKVRNATEA+SKVFSLDR+AFPSLNGLPG+THH
Sbjct: 348 RNAKLRLQEKGITPSIDRIAESLNMSQKKVRNATEAVSKVFSLDRDAFPSLNGLPGETHH 407
Query: 415 CYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKR 474
YIAD RLEN PWHG D+ LK+EV+KLI+ TL ERE+EIIRLYYGLDKECLTWEDISKR
Sbjct: 408 SYIADTRLENNPWHGYDDLALKEEVSKLISATLGEREKEIIRLYYGLDKECLTWEDISKR 467
Query: 475 IGLSRERVRQVGLVALEKLKHAARKREMEAMLLKH 509
IGLSRERVRQVGLVALEKLKHAARKR+MEAM+LK+
Sbjct: 468 IGLSRERVRQVGLVALEKLKHAARKRKMEAMILKN 502
>emb|CAA74896.1| sigma factorB [Arabidopsis thaliana] gi|25288494|pir||T52642
transcription initiation factor sigma homolog B
[imported] - Arabidopsis thaliana (fragment)
Length = 500
Score = 665 bits (1716), Expect = 0.0
Identities = 351/513 (68%), Positives = 412/513 (79%), Gaps = 19/513 (3%)
Query: 3 TAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQAFP 62
TAAVIGL+ GKRLLSSS+Y+SD+ EK +D S+ Y I KS AKK+S+Y+ +FP
Sbjct: 1 TAAVIGLNTGKRLLSSSFYHSDVTEKFLSVNDHCSSQYHIASTKSGITAKKASNYSPSFP 60
Query: 63 ASDRPNQSIKALKEHVDGVPAAAETWFQECDSNDLEVESSD----LGYSVEALLLLQKSM 118
+S+R QS KALKE VD V + + W +LE E D + +SVEA+LLLQKSM
Sbjct: 61 SSNRHTQSAKALKESVD-VASTEKPWLPNGTDKELEEECYDDDDLISHSVEAILLLQKSM 119
Query: 119 LEKQWSLSCEREVLTEHPRQEKSSKKV--AVTCSGVSARQRRINTKRKIPGKTGSAMQAC 176
LEK W+LS E+ V +E+P + KK +TCSG+SARQRRI K+K
Sbjct: 120 LEKSWNLSFEKAVSSEYPGKGTIRKKKIPVITCSGISARQRRIGAKKKTN---------- 169
Query: 177 DAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKL 236
M +S + +GYVKGV+SE +LSH EVV+LS+KIK GL LD+HKSRLK++L
Sbjct: 170 --MTHVKAVSDVSSGKQVRGYVKGVISEDVLSHVEVVRLSKKIKSGLRLDDHKSRLKDRL 227
Query: 237 GCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLV 296
GCEPSD+Q+A SLKISR+EL+A +EC LARE+LAMSNVRLVMSIAQRYDN+GAEM+DLV
Sbjct: 228 GCEPSDEQLAVSLKISRAELQAWLMECHLAREKLAMSNVRLVMSIAQRYDNLGAEMSDLV 287
Query: 297 QGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRT 356
QGGLIGLLRGIEKFDSSKGF+ISTYVYWWIRQGVSRALV+NSRTLRLPTHLHERL LIR
Sbjct: 288 QGGLIGLLRGIEKFDSSKGFRISTYVYWWIRQGVSRALVDNSRTLRLPTHLHERLGLIRN 347
Query: 357 AKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCY 416
AK RL+E+GITP+IDRIA+SLNMSQKKVRNATEA+SKVFSLDR+AFPSLNGLPG+THH Y
Sbjct: 348 AKLRLQEKGITPSIDRIAESLNMSQKKVRNATEAVSKVFSLDRDAFPSLNGLPGETHHSY 407
Query: 417 IADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKRIG 476
IAD RLEN PWHG D+ LK+EV+KLI+ TL ERE+EIIRLYYGLDKECLTWEDISKRIG
Sbjct: 408 IADTRLENNPWHGYDDLALKEEVSKLISATLGEREKEIIRLYYGLDKECLTWEDISKRIG 467
Query: 477 LSRERVRQVGLVALEKLKHAARKREMEAMLLKH 509
LSRERVRQVGLVALEKLKHAARKR+MEAM+LK+
Sbjct: 468 LSRERVRQVGLVALEKLKHAARKRKMEAMILKN 500
>emb|CAA75859.1| Sig1 [Sinapis alba] gi|7488610|pir||T10470 transcription initiation
factor sigma 1 precursor, chloroplast - white mustard
Length = 481
Score = 664 bits (1712), Expect = 0.0
Identities = 355/512 (69%), Positives = 409/512 (79%), Gaps = 34/512 (6%)
Query: 1 MATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQI---VPAKSVTVAKKSSDY 57
MAT AVIGL+ GKRLLSSS+YYSD+ EK +D S+ Y + AK T + SS+Y
Sbjct: 1 MATTAVIGLNAGKRLLSSSFYYSDVTEKFLSVNDLCSSQYHTRSGITAKK-TSSSSSSNY 59
Query: 58 TQAFPASDRPNQSIKALKEHVDGVPAAAETWFQECDSNDLEVESSDLGYSVEALLLLQKS 117
+ +FP+S R QS KALKE V ++ + W +D E+E D+ +SVEALLLLQ+S
Sbjct: 60 SPSFPSSSRQTQSAKALKE---SVASSVQPWLPP--GSDQELEEDDIDHSVEALLLLQRS 114
Query: 118 MLEKQWSLSCEREVLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKIPGKTGSAMQACD 177
MLEKQW+LS E K+ KKV VTCSG+SARQRRI K+K K S +
Sbjct: 115 MLEKQWNLSFE-----------KTRKKVPVTCSGISARQRRIGAKKKTNVKAVSEVN--- 160
Query: 178 AMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLG 237
N KGYVKGV+S+ +LSHAEVV+LS+KIK GL LDEHKSRL ++LG
Sbjct: 161 -----------FQNNLVKGYVKGVISDHVLSHAEVVRLSKKIKSGLRLDEHKSRLTDRLG 209
Query: 238 CEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQ 297
CEPSD+Q+A SLKISR+EL+A +EC LARE+LAMSNVRLVMSIAQRYDNMGAEM+DLVQ
Sbjct: 210 CEPSDEQLAMSLKISRAELQAWLMECHLAREKLAMSNVRLVMSIAQRYDNMGAEMSDLVQ 269
Query: 298 GGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTA 357
GGLIGLLRGIEKFDSSKGF+ISTYVYWWIRQGVSRALV+NSRTLRLPTHLHERL LIR A
Sbjct: 270 GGLIGLLRGIEKFDSSKGFRISTYVYWWIRQGVSRALVDNSRTLRLPTHLHERLGLIRNA 329
Query: 358 KFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYI 417
K RL+E+GITP+IDRIA+SLNMSQKKVRNATEA+SK+FSLDR+AFPSLNGLPG+THH YI
Sbjct: 330 KLRLQEKGITPSIDRIAESLNMSQKKVRNATEAVSKIFSLDRDAFPSLNGLPGETHHSYI 389
Query: 418 ADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKRIGL 477
ADNRLEN PWHG D LKDEV+KLI+VTL EREREIIRLYYGLDKECLTWEDIS+RIGL
Sbjct: 390 ADNRLENNPWHGYDALALKDEVSKLISVTLGEREREIIRLYYGLDKECLTWEDISQRIGL 449
Query: 478 SRERVRQVGLVALEKLKHAARKREMEAMLLKH 509
SRERVRQVGLVALEKLKHAARKR+MEAM+LK+
Sbjct: 450 SRERVRQVGLVALEKLKHAARKRKMEAMILKN 481
>dbj|BAA88922.1| sigma factor [Nicotiana tabacum]
Length = 502
Score = 657 bits (1694), Expect = 0.0
Identities = 347/508 (68%), Positives = 406/508 (79%), Gaps = 11/508 (2%)
Query: 5 AVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAKSVTVAKKSSDYTQAFPAS 64
A+IGLS G R+LSSS+YYS + EK C SD + K+V AK SS+Y+ ++ S
Sbjct: 3 AIIGLSAGNRILSSSFYYSVLTEKF-CVSDHALPCHHFPSPKNVINAKNSSNYSPSY-VS 60
Query: 65 DRPNQSIKALKEHVD---GVPAAAETWFQECDSNDLEVESSDLGYSVEALLLLQKSMLEK 121
+R QS+KALKEHVD + E W Q L+ S SVE LLLLQKSMLEK
Sbjct: 61 NRKKQSVKALKEHVDIASNRDSDTEPWVQMF--KQLQNGSCSEEDSVEVLLLLQKSMLEK 118
Query: 122 QWSLSCEREVLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKIPGKTGSAMQACDAMQM 181
QW+LS E+ + E+ KK+ +TCSG SAR+RR++T+++ SA + ++
Sbjct: 119 QWNLSAEKAMTNEN----NCKKKMQITCSGTSARRRRMDTRKRDLLPKNSATSDSSSKRL 174
Query: 182 RSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPS 241
RS+I PELLQNRSKGY+ GVV ++LL+H EVV+LS+KIK GL L+E KSRLKE+LGCEPS
Sbjct: 175 RSVIGPELLQNRSKGYINGVVGQELLTHTEVVQLSKKIKFGLYLEEQKSRLKERLGCEPS 234
Query: 242 DDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLI 301
DDQ+A SLKISR++LR+ IEC+LARE+LAMSNVRLVMSIAQRYD G EMADL+QGGLI
Sbjct: 235 DDQLAISLKISRADLRSTLIECSLAREKLAMSNVRLVMSIAQRYDTNGTEMADLIQGGLI 294
Query: 302 GLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRL 361
GLLRGIEKFD S+GFKISTYVYWWIRQGVSRALVENSRTLRLP HLHERLSLIR AK +L
Sbjct: 295 GLLRGIEKFDHSRGFKISTYVYWWIRQGVSRALVENSRTLRLPIHLHERLSLIRNAKIKL 354
Query: 362 EERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNR 421
+E GITP+I++IA+ LNMSQKKVRNATEA SKVFSLDREAFPSLNGLPG+T H YIADNR
Sbjct: 355 KENGITPSIEKIAECLNMSQKKVRNATEASSKVFSLDREAFPSLNGLPGETLHSYIADNR 414
Query: 422 LENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKRIGLSRER 481
LENIPWHGVDEW LKDEVN LI+ TL EREREI+RLYYGLD ECLTWEDIS+RIGLSRER
Sbjct: 415 LENIPWHGVDEWMLKDEVNNLISSTLREREREIVRLYYGLDNECLTWEDISRRIGLSRER 474
Query: 482 VRQVGLVALEKLKHAARKREMEAMLLKH 509
VRQVGLVALEKLKHAARKR ++AML+KH
Sbjct: 475 VRQVGLVALEKLKHAARKRRLDAMLVKH 502
>emb|CAA74653.1| plastid RNA polymerase sigma factor [Sorghum bicolor]
gi|7489655|pir||T14826 transcription initiation factor
sigma 1 precursor, chloroplast - sorghum
Length = 500
Score = 498 bits (1282), Expect = e-139
Identities = 280/521 (53%), Positives = 363/521 (68%), Gaps = 42/521 (8%)
Query: 2 ATAAVIGLSGGKRLLSSSYYYSDIIEKLSCSSDFGSTHYQIVPAK-SVTV----AKKSSD 56
AT A+IGLS GKRLL++S+ +D+ + S Q PA +TV A SS
Sbjct: 3 ATPAMIGLSAGKRLLATSFGNTDLAPPDAHPS------LQFAPAAPKLTVVAHRASASSS 56
Query: 57 YTQAFPASDRPNQSIKALKEHVDGV----------PAAAETWFQECDSN-DLEVESSDLG 105
+ + PA +I+AL+ H P A E DS + E+ESS
Sbjct: 57 SSSSSPAGHARAHAIRALRNHSAPALAPPLPLPLPPPPAADPAPELDSEFEFELESS--- 113
Query: 106 YSVEALLLLQKSMLEKQWSLSCEREVLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKI 165
+EA++LLQ+SMLEKQW L E E + + + V V SGVSARQRR++ +R+
Sbjct: 114 --LEAIVLLQRSMLEKQWQLPFEDE---DEDDERLTKAAVVVARSGVSARQRRMSGRRRG 168
Query: 166 PGKTGSAMQACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSL 225
PG+ +SPEL+Q+R++ Y++G VS+ LL+H +V++LS KIK G+ L
Sbjct: 169 PGRKSVT------------VSPELMQSRNRIYLRGTVSKDLLTHKQVIQLSRKIKDGIWL 216
Query: 226 DEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRY 285
+S+LKEKLG EPS Q+A SL IS ELRA+ E LARE L MSN+RLV+SIAQ+Y
Sbjct: 217 QHQRSKLKEKLGNEPSYKQLAQSLHISPPELRARMRESFLAREMLTMSNIRLVISIAQKY 276
Query: 286 DNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPT 345
DN+G E+ADL+QGGLIGLLRGIEKFD+S+GF+ISTYVYWWIRQGVSRAL +NS+ RLPT
Sbjct: 277 DNLGVELADLIQGGLIGLLRGIEKFDASRGFRISTYVYWWIRQGVSRALADNSKAFRLPT 336
Query: 346 HLHERLSLIRTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSL 405
+LHERL IR AK+ LE++GI PT++ IA+SLN+S+KKV NATEA++KV SLD++AFPSL
Sbjct: 337 YLHERLIAIRGAKYALEDQGIAPTVENIAESLNISEKKVHNATEAVNKVLSLDQQAFPSL 396
Query: 406 NGLPGDTHHCYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKEC 465
NGLPGDT H YI D + N PWHG +E LK+EVN LIN L ERER+IIRLY+G+ K+C
Sbjct: 397 NGLPGDTLHSYIEDQNVANDPWHGFEERYLKEEVNNLINSNLNERERDIIRLYHGIGKQC 456
Query: 466 LTWEDISKRIGLSRERVRQVGLVALEKLKHAARKREMEAML 506
TWEDIS++ GLSRERVRQVGLVA+EKLKHAAR++ ++A+L
Sbjct: 457 HTWEDISRQFGLSRERVRQVGLVAMEKLKHAARRKHLDALL 497
>dbj|BAA24364.1| plastid RNA polymerase sigma factor [Oryza sativa (japonica
cultivar-group)]
Length = 519
Score = 494 bits (1271), Expect = e-138
Identities = 282/531 (53%), Positives = 366/531 (68%), Gaps = 43/531 (8%)
Query: 2 ATAAVIGLSGGKRLLSSSYYYSDII-EKLSCSSDFG-----------STHYQIVPA--KS 47
AT AVIGLS G RLLS+S+ +D++ +K+S G + + PA K
Sbjct: 3 ATPAVIGLSAGNRLLSASFGPTDLMPDKVSLGVGGGGGGGGGGGGGDAMSFAAAPATPKL 62
Query: 48 VTVAK---KSSDYTQAFPASDRPNQSIKALKEHVDGVPAAAETWFQECDSN----DLEVE 100
VA K S + +A Q ++AL+ H A A
Sbjct: 63 TAVAAHRLKLSPHGRA--------QVMRALRHHSSAAAALAPPPPPPPPPTPSPASRAAH 114
Query: 101 SSDLGYSVEALLLLQKSMLEKQWSLSCERE-----VLTEHPRQEKSSKKVAVTCSGVSAR 155
+ DL S+EA++LLQ+SMLEKQW L + + + + S V V S VSAR
Sbjct: 115 AHDLESSLEAIVLLQRSMLEKQWELPFDDDNHAMAIGLAEDDDDTSKATVVVARSSVSAR 174
Query: 156 QRRINTKRKIPGKTGSAMQACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKL 215
QRR++ +R+ K G+A A +SPEL+Q+R++ Y++G VS++LL+H +VV L
Sbjct: 175 QRRMSGRRRGRTKNGAAHFA---------VSPELIQSRNRIYLRGTVSKELLTHKQVVHL 225
Query: 216 SEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNV 275
S KIK G+ L + +S+LKEKLG EPS Q+A SLKIS ELR++ E LARE L MSN+
Sbjct: 226 SHKIKDGIWLQQQRSKLKEKLGNEPSYKQLAHSLKISPPELRSRMHESFLAREMLTMSNL 285
Query: 276 RLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALV 335
RLV+SIAQ+YDN+G E+ADL+QGGLIGLLRGIEKFD+S+GF+ISTYVYWWIRQGVSRAL
Sbjct: 286 RLVISIAQKYDNLGVELADLIQGGLIGLLRGIEKFDASRGFRISTYVYWWIRQGVSRALA 345
Query: 336 ENSRTLRLPTHLHERLSLIRTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVF 395
ENS+T RLPT+LHERL IR AK+ LE++GI PTI+ IA SLN+S+KKV NATEA++KV
Sbjct: 346 ENSKTFRLPTYLHERLIAIRGAKYELEDQGIAPTIENIAGSLNISEKKVLNATEAVNKVL 405
Query: 396 SLDREAFPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREII 455
SLD++AFPSLNGLPG+T H YI D + N PWHG +EW LK+EVNKL+N TL ERER+II
Sbjct: 406 SLDQQAFPSLNGLPGETLHSYIEDQNVANDPWHGFEEWYLKEEVNKLLNSTLNERERDII 465
Query: 456 RLYYGLDKECLTWEDISKRIGLSRERVRQVGLVALEKLKHAARKREMEAML 506
RLY+G+ K+C TWEDIS++ GLSRERVRQVGL+A+EKLKHAAR++ +EA+L
Sbjct: 466 RLYHGIGKQCHTWEDISRQFGLSRERVRQVGLIAMEKLKHAARRKNLEALL 516
>gb|AAC78467.1| RNA polymerase sigma factor 1 [Zea mays]
Length = 398
Score = 476 bits (1225), Expect = e-133
Identities = 249/411 (60%), Positives = 314/411 (75%), Gaps = 18/411 (4%)
Query: 96 DLEVESSDLGYSVEALLLLQKSMLEKQWSLSCEREVLTEHPRQEKSSKKVAVTCSGVSAR 155
+LE ESS +EA++LLQ+SMLEKQW L E ++ TE E V V SGVSAR
Sbjct: 3 ELEFESS-----LEAIVLLQRSMLEKQWELPFEDDISTE---DEDERSTVVVARSGVSAR 54
Query: 156 QRRINTKRKIPGKTGSAMQACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKL 215
QRR++ +R+ + +SPEL+Q+R++ Y++G VS+ LL+H +VV+L
Sbjct: 55 QRRMSGRRR----------GAFVGRKSVTVSPELMQSRNRIYLRGTVSKDLLTHKQVVQL 104
Query: 216 SEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNV 275
S KIK G+ L +S+LKEKLG EPS Q+A SL+I ELRA+ E LARE LAMSN
Sbjct: 105 SRKIKDGIWLQHQRSKLKEKLGNEPSYKQLAQSLRIPAPELRARMRESFLAREMLAMSNT 164
Query: 276 RLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALV 335
RLV+SIAQ+YDN+G E+ DL+QGGLIGLLRGIEKFD+S+GF+ISTYVYWWIRQGVSRAL
Sbjct: 165 RLVISIAQKYDNLGVELPDLIQGGLIGLLRGIEKFDASRGFRISTYVYWWIRQGVSRALA 224
Query: 336 ENSRTLRLPTHLHERLSLIRTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVF 395
+NSR+ RLPT+LHERL IR AK+ LE++GI PT++ IA+SLN+S+KKV NATEA+ KV
Sbjct: 225 DNSRSFRLPTYLHERLVAIRGAKYALEDQGIAPTVENIAESLNISEKKVHNATEAVHKVI 284
Query: 396 SLDREAFPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREII 455
SLD++AFPSLNGLPGDT H YI D + N PWHG +E LK+EVN LIN TL ERER+II
Sbjct: 285 SLDQQAFPSLNGLPGDTLHSYIEDQNVANDPWHGFEERYLKEEVNSLINSTLNERERDII 344
Query: 456 RLYYGLDKECLTWEDISKRIGLSRERVRQVGLVALEKLKHAARKREMEAML 506
RLY+G+ K+C TWEDIS++ GLSRERVRQVGLVA+EKLKHAAR+ ++A+L
Sbjct: 345 RLYHGIGKQCHTWEDISRQFGLSRERVRQVGLVAMEKLKHAARREHLDALL 395
>ref|XP_480219.1| putative plastid RNA polymerase sigma factor [Oryza sativa
(japonica cultivar-group)] gi|37806470|dbj|BAC99905.1|
putative plastid RNA polymerase sigma factor [Oryza
sativa (japonica cultivar-group)]
Length = 388
Score = 468 bits (1204), Expect = e-130
Identities = 241/394 (61%), Positives = 306/394 (77%), Gaps = 14/394 (3%)
Query: 118 MLEKQWSLSCERE-----VLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKIPGKTGSA 172
MLEKQW L + + + + S V V S VSARQRR++ +R+ K G+A
Sbjct: 1 MLEKQWELPFDDDNHAMAIGLAEDDDDTSKATVVVARSSVSARQRRMSGRRRGRTKNGAA 60
Query: 173 MQACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRL 232
A +SPEL+Q+R++ Y++G VS++LL+H +VV LS KIK G+ L + +S+L
Sbjct: 61 HFA---------VSPELIQSRNRIYLRGTVSKELLTHKQVVHLSHKIKDGIWLQQQRSKL 111
Query: 233 KEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEM 292
KEKLG EPS Q+A SLKIS ELR++ E LARE L MSN+RLV+SIAQ+YDN+G E+
Sbjct: 112 KEKLGNEPSYKQLAHSLKISPPELRSRMHESFLAREMLTMSNLRLVISIAQKYDNLGVEL 171
Query: 293 ADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLS 352
ADL+QGGLIGLLRGIEKFD+S+GF+ISTYVYWWIRQGVSRAL ENS+T RLPT+LHERL
Sbjct: 172 ADLIQGGLIGLLRGIEKFDASRGFRISTYVYWWIRQGVSRALAENSKTFRLPTYLHERLI 231
Query: 353 LIRTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDT 412
IR AK+ LE++GI PTI+ IA SLN+S+KKV NATEA++KV SLD++AFPSLNGLPG+T
Sbjct: 232 AIRGAKYELEDQGIAPTIENIAGSLNISEKKVLNATEAVNKVLSLDQQAFPSLNGLPGET 291
Query: 413 HHCYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDIS 472
H YI D + N PWHG +EW LK+EVNKL+N TL ERER+IIRLY+G+ K+C TWEDIS
Sbjct: 292 LHSYIEDQNVANDPWHGFEEWYLKEEVNKLLNSTLNERERDIIRLYHGIGKQCHTWEDIS 351
Query: 473 KRIGLSRERVRQVGLVALEKLKHAARKREMEAML 506
++ GLSRERVRQVGL+A+EKLKHAAR++ +EA+L
Sbjct: 352 RQFGLSRERVRQVGLIAMEKLKHAARRKNLEALL 385
>emb|CAA10725.1| chloroplast sigma factor [Triticum aestivum]
Length = 461
Score = 466 bits (1199), Expect = e-130
Identities = 240/416 (57%), Positives = 313/416 (74%), Gaps = 22/416 (5%)
Query: 106 YSVEALLLLQKSMLEKQWSLSCEREV---------------LTEHPRQEKSSKKVAVTCS 150
+S++A++LLQ+SMLEKQW L E E E + S V V S
Sbjct: 50 FSLDAIILLQRSMLEKQWQLPFEEEDDDDGGGDGHHVEAIGSAEDDDGKGRSSSVVVARS 109
Query: 151 GVSARQRRINTKRKIPGKTGSAMQACDAMQMRSLISPELLQNRSKGYVKGVVSEQLLSHA 210
GVSARQRR++ +R+ K GS + ISPELLQ+R++ Y++G VS++LL+
Sbjct: 110 GVSARQRRMSGRRRGRSKKGSGA-------VHLSISPELLQSRNRIYLRGTVSKELLTQK 162
Query: 211 EVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERL 270
+VV LS+KIK G+ L + +S+LKEKLG EPS Q+A SL+IS ELR++ E LARE L
Sbjct: 163 QVVHLSKKIKDGIWLQQQRSKLKEKLGNEPSYKQMAQSLRISTPELRSRMRESFLAREVL 222
Query: 271 AMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGV 330
MSN+RLV+SIAQ+YD +G E++DL+QGGLIGLLRGIEKFD+S+GF+ISTYVYWWIRQGV
Sbjct: 223 TMSNIRLVISIAQKYDKLGVELSDLIQGGLIGLLRGIEKFDASRGFRISTYVYWWIRQGV 282
Query: 331 SRALVENSRTLRLPTHLHERLSLIRTAKFRLEERGITPTIDRIAKSLNMSQKKVRNATEA 390
SRAL +NS+T RLPT+LHERL IR AK+ LE++G++PT IAK LN+S+KKV NATEA
Sbjct: 283 SRALADNSKTFRLPTYLHERLIAIRGAKYALEDQGVSPTTQNIAKLLNISEKKVHNATEA 342
Query: 391 ISKVFSLDREAFPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTLKDEVNKLINVTLVER 450
++K SLD++AFPSLNGLPGDT H YI D + N PWHG +EW LK+EVNKL+N L ER
Sbjct: 343 VNKALSLDQQAFPSLNGLPGDTLHSYIEDQNVANDPWHGFEEWYLKEEVNKLLNSNLTER 402
Query: 451 EREIIRLYYGLDKECLTWEDISKRIGLSRERVRQVGLVALEKLKHAARKREMEAML 506
ER+II+LY+G+ K+C TWEDIS++ GLSRERVRQVGL+A+EKLK AR+++++A+L
Sbjct: 403 ERDIIQLYHGIGKQCHTWEDISRQFGLSRERVRQVGLIAMEKLKQRARRKKLDALL 458
>ref|XP_480220.1| putative plastid RNA polymerase sigma factor [Oryza sativa
(japonica cultivar-group)] gi|37806471|dbj|BAC99906.1|
putative plastid RNA polymerase sigma factor [Oryza
sativa (japonica cultivar-group)]
Length = 342
Score = 446 bits (1148), Expect = e-124
Identities = 218/322 (67%), Positives = 274/322 (84%)
Query: 185 ISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQ 244
+SPEL+Q+R++ Y++G VS++LL+H +VV LS KIK G+ L + +S+LKEKLG EPS Q
Sbjct: 18 VSPELIQSRNRIYLRGTVSKELLTHKQVVHLSHKIKDGIWLQQQRSKLKEKLGNEPSYKQ 77
Query: 245 IAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLL 304
+A SLKIS ELR++ E LARE L MSN+RLV+SIAQ+YDN+G E+ADL+QGGLIGLL
Sbjct: 78 LAHSLKISPPELRSRMHESFLAREMLTMSNLRLVISIAQKYDNLGVELADLIQGGLIGLL 137
Query: 305 RGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRLEER 364
RGIEKFD+S+GF+ISTYVYWWIRQGVSRAL ENS+T RLPT+LHERL IR AK+ LE++
Sbjct: 138 RGIEKFDASRGFRISTYVYWWIRQGVSRALAENSKTFRLPTYLHERLIAIRGAKYELEDQ 197
Query: 365 GITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLEN 424
GI PTI+ IA SLN+S+KKV NATEA++KV SLD++AFPSLNGLPG+T H YI D + N
Sbjct: 198 GIAPTIENIAGSLNISEKKVLNATEAVNKVLSLDQQAFPSLNGLPGETLHSYIEDQNVAN 257
Query: 425 IPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKRIGLSRERVRQ 484
PWHG +EW LK+EVNKL+N TL ERER+IIRLY+G+ K+C TWEDIS++ GLSRERVRQ
Sbjct: 258 DPWHGFEEWYLKEEVNKLLNSTLNERERDIIRLYHGIGKQCHTWEDISRQFGLSRERVRQ 317
Query: 485 VGLVALEKLKHAARKREMEAML 506
VGL+A+EKLKHAAR++ +EA+L
Sbjct: 318 VGLIAMEKLKHAARRKNLEALL 339
>gb|AAC78468.1| RNA polymerase sigma factor 2 [Zea mays]
Length = 349
Score = 431 bits (1109), Expect = e-119
Identities = 213/322 (66%), Positives = 266/322 (82%)
Query: 185 ISPELLQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQ 244
ISPEL+Q+R++ Y++G VS+ LL+H +V++LS KIK G+ L +S+LKEKLG EPS Q
Sbjct: 25 ISPELMQSRNRIYLRGTVSKDLLTHKQVIQLSRKIKDGIWLQHQRSKLKEKLGNEPSYKQ 84
Query: 245 IAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLL 304
+A SL+IS ELRA+ E LARE L MSN+RLV+SIA +YDN+G ++ADL+QGGLIGLL
Sbjct: 85 LAQSLRISAPELRARMRESFLAREMLTMSNIRLVISIAHKYDNLGVDLADLIQGGLIGLL 144
Query: 305 RGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRLEER 364
RGIEKFD+S+GF+ISTYVYWWIRQGVSRAL +NS+T RLPT+LHERL IR AK+ LE++
Sbjct: 145 RGIEKFDASRGFRISTYVYWWIRQGVSRALADNSKTFRLPTYLHERLLAIRGAKYALEDQ 204
Query: 365 GITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLEN 424
GI PTI+ IA+SLN+S KKV NATEA+ KV SLD++AFPSLNGLPGDT H YI D + N
Sbjct: 205 GIAPTIENIAESLNISVKKVHNATEAVHKVLSLDQQAFPSLNGLPGDTLHSYIEDQNVAN 264
Query: 425 IPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKECLTWEDISKRIGLSRERVRQ 484
PWHG +E LK+EVN LIN L ERER+IIRLY+G+ K+C TWEDIS++ GLSRERVRQ
Sbjct: 265 DPWHGFEEMYLKEEVNNLINSNLNERERDIIRLYHGIGKQCHTWEDISRQFGLSRERVRQ 324
Query: 485 VGLVALEKLKHAARKREMEAML 506
V LVA+EKLKHAAR++ + A+L
Sbjct: 325 VRLVAMEKLKHAARRKHLNALL 346
>dbj|BAB62007.1| PpSIG1 [Physcomitrella patens] gi|14669828|dbj|BAB62006.1| PpSIG1
[Physcomitrella patens]
Length = 677
Score = 263 bits (671), Expect = 1e-68
Identities = 147/381 (38%), Positives = 239/381 (62%), Gaps = 22/381 (5%)
Query: 144 KVAVTCSGVSARQRRINTKRK---IPGKTGSAMQACDAMQMRSL-----------ISPEL 189
++ V SAR RR+ ++++ I GK S DA+ + L + +
Sbjct: 297 ELVVKSGSPSARTRRLESRQRRVAIAGKPTSP----DALTIHPLKKYQKGERVRRVKFDG 352
Query: 190 LQNRSKGYVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASL 249
+Q+ Y++ + LL+ E V LS+K+++GL+L E + +L +LG PS+++ A +
Sbjct: 353 VQDYLTTYLREITKINLLTKQEEVFLSKKLRIGLNLLEERKKLAAELGYVPSNEEWAPHM 412
Query: 250 KISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEK 309
+S +L K E AR+++ M+N+RLV+S+A++Y + G EMADL+Q G IGLLRG+EK
Sbjct: 413 NLSVGDLVRKLGEAEWARDKMVMANLRLVVSVAKQYHSFGLEMADLIQEGSIGLLRGVEK 472
Query: 310 FDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRLEERGITPT 369
FD ++G+K+STYV+WWIRQGV+RA+ ++SRT+RLP H+H+ L IR AK L G T +
Sbjct: 473 FDHTRGYKLSTYVHWWIRQGVTRAIADHSRTVRLPVHVHDTLGRIRKAKSTLLSEGCTAS 532
Query: 370 IDRIAKSLNMSQKKVRNATEAISKVFSLDRE-AFPSLNGLPGDTHHCYIADNRLENIPWH 428
+ I+++ N++ K++N + K+ SLD+E +L G+T H +AD +EN PW
Sbjct: 533 VQEISEATNLTGTKIKNTLQVTKKLVSLDKEIGRNTLQSGDGETLHSLVADRDIENQPWT 592
Query: 429 GVDEWTLKDEVNKLINVTLVEREREIIRLYYGLDKE---CLTWEDISKRIGLSRERVRQV 485
V+ +KD+V+KL+ L RER+I+RL+YG+ +E ++ E IS R G+SRERVRQ+
Sbjct: 593 IVERMLMKDDVDKLLTSQLGPRERDIVRLHYGVGREDGSVMSLESISYRYGVSRERVRQL 652
Query: 486 GLVALEKLKHAARKREMEAML 506
A+ KL+ +R++ ++ L
Sbjct: 653 ETAAMRKLQIKSREQGLDRYL 673
Score = 35.4 bits (80), Expect = 4.3
Identities = 73/333 (21%), Positives = 132/333 (38%), Gaps = 39/333 (11%)
Query: 69 QSIKALKEHVDGVPAAAETWFQECDSNDLEVESSDLGYSVEALLLLQKSMLEKQWSLSCE 128
+++ L EH+ GV A E ++L SS L +A LL++SMLE QW+L+ +
Sbjct: 101 RALTVLHEHISGV-ALQNIALSE---DELSRYSSILP---DASTLLRRSMLEAQWALTLK 153
Query: 129 REVLTEHPRQEKSSKKVAVTCSGVSARQRRINTKRKIPGKTGSA---MQACDAMQMRSLI 185
+ + ++K+S++ S T+ PG T +A + D +
Sbjct: 154 ELMHEQKMEEQKNSRRAGKGKKSFSLEV----TEDVSPGSTSTATGDLAVEDLTDEEKVT 209
Query: 186 SPELLQNRSKGYVKGVVSEQLLSHAEVVKL----SEKI----KVGLSLDE---------- 227
++ + R KG +K V +Q S E + SE++ + G L +
Sbjct: 210 ERKVTRRRGKGKLKQVERKQDTSLQETGNVHTEGSEEVVDYFRSGEQLKDLNTARRGGEG 269
Query: 228 HKSRLKEKLGCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDN 287
++ + KL +D S +EL K+ + RL R ++IA + +
Sbjct: 270 KLTKFERKLDSSLVEDACPESTSTMTTELVVKSGSPSARTRRLESRQRR--VAIAGKPTS 327
Query: 288 MGAEMADLVQGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHL 347
A ++ G KFD + Y+ ++R+ L+ + L L
Sbjct: 328 PDALTIHPLKKYQKGERVRRVKFDG-----VQDYLTTYLREITKINLLTKQEEVFLSKKL 382
Query: 348 HERLSLIRTAKFRLEERGITPTIDRIAKSLNMS 380
L+L+ K E G P+ + A +N+S
Sbjct: 383 RIGLNLLEERKKLAAELGYVPSNEEWAPHMNLS 415
>ref|NP_926708.1| group2 RNA polymerase sigma factor [Gloeobacter violaceus PCC 7421]
gi|35214335|dbj|BAC91703.1| group2 RNA polymerase sigma
factor [Gloeobacter violaceus PCC 7421]
Length = 362
Score = 190 bits (482), Expect = 1e-46
Identities = 112/312 (35%), Positives = 183/312 (57%), Gaps = 9/312 (2%)
Query: 197 YVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISRSEL 256
Y++ + LL E V+L+++++ + L K L+ LG E SD+Q A + K+ EL
Sbjct: 54 YLQEIGRVPLLGRDEEVELAQQVQKLMKLTALKESLQTSLGSEYSDEQWAEAAKVGLFEL 113
Query: 257 RAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKGF 316
+ + + + A++++ +N+RLV+S+A++Y N G E+ DLVQ G +GL R +EKFD +KG+
Sbjct: 114 KRQVLAGSRAKKKMIEANLRLVVSVAKKYQNRGLELLDLVQEGTLGLERAVEKFDPTKGY 173
Query: 317 KISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRLEER-GITPTIDRIAK 375
+ STY YWWIRQG++RA+ SRT+RLP H+ E+L+ I+ + +L + G TPTI IA+
Sbjct: 174 RFSTYAYWWIRQGITRAIATQSRTIRLPVHITEKLNRIKKTQRKLSQNLGRTPTISEIAE 233
Query: 376 SLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIADNRLENIPWHGVDEWTL 435
L M VR + + SL+ G DT + + E P + +L
Sbjct: 234 ELEMEPAHVRELMSRVPRSVSLETRV-----GKDKDTELGELLEAE-EVSPEESAVQESL 287
Query: 436 KDEVNKLINVTLVEREREIIRLYYG-LDKECLTWEDISKRIGLSRERVRQVGLVALEKLK 494
+ +V +L++ L RER++I + YG LD + +I + + LSRERVRQ+ AL+KL+
Sbjct: 288 RRDVKELLD-DLTLRERDVITMRYGLLDGRTYSLSEIGRALDLSRERVRQIESKALQKLR 346
Query: 495 HAARKREMEAML 506
R+ ++ L
Sbjct: 347 QPRRRAKVRDYL 358
>gb|AAK39807.1| RNA-polymerase sigma factor [Guillardia theta]
gi|25288491|pir||D90084 RNA-polymerase sigma factor
[imported] - Guillardia theta nucleomorph
gi|13812120|ref|NP_113247.1| RNA-polymerase sigma factor
[Guillardia theta]
Length = 460
Score = 187 bits (476), Expect = 5e-46
Identities = 114/307 (37%), Positives = 182/307 (59%), Gaps = 13/307 (4%)
Query: 197 YVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISR-SE 255
Y++ + +LL E ++L+ +I L + K L++KL EP++ + A+ K+S +
Sbjct: 118 YLQLIGRVRLLRPDEEIQLAREISQLLHWEHVKISLRDKLRREPTNSEWTAACKVSDVRK 177
Query: 256 LRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKG 315
R+ A+ER+ +N+RLV+SIA+RY N G + DL+Q G +GL+R EKFDS KG
Sbjct: 178 FRSILHNARRAKERMVAANLRLVVSIAKRYVNRGMSLQDLIQEGSLGLIRAAEKFDSEKG 237
Query: 316 FKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRLE-ERGITPTIDRIA 374
FK STY WW++Q V+RA+ ++SRT+RLP HL++ +S IR L E G +PT + IA
Sbjct: 238 FKFSTYATWWVKQAVTRAIADHSRTIRLPVHLYDTISAIRKTTRNLNIEIGRSPTEEEIA 297
Query: 375 KSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIAD--NRLENIPWHGVDE 432
K ++MS +K+RN ++ SL+R L D ++D + P V++
Sbjct: 298 KRMDMSVEKLRNVLQSAQPTLSLERP-------LKNDDDTSQLSDFIEADQESPEEHVEK 350
Query: 433 WTLKDEVNKLINVTLVEREREIIRLYYGLDK-ECLTWEDISKRIGLSRERVRQVGLVALE 491
L+D++ +IN +L RER+++R+ YGLD T E+I + ++RERVRQ+ A+
Sbjct: 351 SMLRDDLENVIN-SLSPRERDVVRMRYGLDDGRIKTLEEIGQIFSVTRERVRQIEAKAIR 409
Query: 492 KLKHAAR 498
KL+ R
Sbjct: 410 KLRQPYR 416
>dbj|BAB87262.1| RNA polymerase sigma factor [Guillardia theta]
Length = 490
Score = 186 bits (471), Expect = 2e-45
Identities = 113/307 (36%), Positives = 181/307 (58%), Gaps = 13/307 (4%)
Query: 197 YVKGVVSEQLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKLGCEPSDDQIAASLKISR-SE 255
Y++ + +LL E ++L+ +I L + K L+EKL EP+ + A+ K+S +
Sbjct: 135 YLQLIGRVRLLRPEEEIQLAREISQLLHWEHVKISLREKLQREPTSSEWTAACKVSDVKK 194
Query: 256 LRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLVQGGLIGLLRGIEKFDSSKG 315
R+ A+ER+ +N+RLV+SIA+RY N G + DL+Q G +GL+R EKFDS KG
Sbjct: 195 FRSILHNARRAKERMVAANLRLVVSIAKRYVNRGMSLQDLIQEGSLGLIRAAEKFDSEKG 254
Query: 316 FKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRTAKFRLE-ERGITPTIDRIA 374
FK STY WW++Q V+RA+ ++SRT+RLP HL++ +S IR L E G +PT + IA
Sbjct: 255 FKFSTYATWWVKQAVTRAIADHSRTIRLPVHLYDTISAIRKTTRTLNIEIGRSPTEEEIA 314
Query: 375 KSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHCYIAD--NRLENIPWHGVDE 432
+ ++MS +K+R+ ++ SL+R L D ++D + P V++
Sbjct: 315 EKMDMSVEKLRSVLQSAQPTLSLERP-------LKNDDDASQLSDFIEADQESPEEHVEK 367
Query: 433 WTLKDEVNKLINVTLVEREREIIRLYYGLDK-ECLTWEDISKRIGLSRERVRQVGLVALE 491
L+D++ +IN +L RER+++R+ YGLD T E+I + ++RERVRQ+ A+
Sbjct: 368 SMLRDDLENVIN-SLSPRERDVVRMRYGLDDGRIKTLEEIGQIFSVTRERVRQIEAKAIR 426
Query: 492 KLKHAAR 498
KL+ R
Sbjct: 427 KLRQPYR 433
>emb|CAB83106.1| sigma factor 2 [Sinapis alba]
Length = 575
Score = 184 bits (467), Expect = 6e-45
Identities = 126/393 (32%), Positives = 220/393 (55%), Gaps = 15/393 (3%)
Query: 124 SLSCEREVLTE-HPRQEKSSKKVAVTCSGVSARQRRINTKR-KIPGKTGSAMQAC----D 177
SL C +E + P +++ +K++V+ + S RQ +R K KT S MQ+
Sbjct: 183 SLECNKEASDDLTPAEKQEVEKLSVSSAVRSTRQTERKARRVKGLEKTASGMQSSVKTGS 242
Query: 178 AMQMRSLISPELLQNRSKGYVKGVVSE-QLLSHAEVVKLSEKIKVGLSLDEHKSRLKEKL 236
+ + + + S E+ N Y++ S +LL+ E +LSE I+ L L+ ++ L E+
Sbjct: 243 SSRKKRVASQEIDHNDPLRYLRMTTSSSKLLTAREEQQLSEGIQDLLKLERLQAELTERC 302
Query: 237 GCEPSDDQIAASLKISRSELRAKTIECTLARERLAMSNVRLVMSIAQRYDNMGAEMADLV 296
G +P+ Q A++ + + LR + T ++R+ SN+RLV+SIA+ Y G + DLV
Sbjct: 303 GHQPTFSQWASAAGVDKKTLRKRITHGTQCKDRMIKSNIRLVISIAKNYQGAGMDFQDLV 362
Query: 297 QGGLIGLLRGIEKFDSSKGFKISTYVYWWIRQGVSRALVENSRTLRLPTHLHERLSLIRT 356
Q G GL+RG EKFD++KGFK STY +WWI+Q V ++L + SR +RLP H+ E ++
Sbjct: 363 QEGCRGLVRGAEKFDATKGFKFSTYAHWWIKQAVRKSLSDQSRMIRLPFHMVEATYRVKE 422
Query: 357 AKFRL-EERGITPTIDRIAKSLNMSQKKVRNATEAISKVFSLDREAFPSLNGLPGDTHHC 415
A+ +L E G P + +A++ +S K++ + SLD++ +LN P +
Sbjct: 423 ARKQLYSETGKQPKNEEVAEATGLSMKRLMAVLLSPKPPRSLDQKIGINLNLKPSEV--- 479
Query: 416 YIADNRLENIPWHGVDEWTLKDEVNKLINVTLVEREREIIRLYYGL-DKECLTWEDISKR 474
IAD E + ++ ++ +++K+++ +L RE+++IR +G+ D T ++I +
Sbjct: 480 -IADPEAETSEDILMKQF-MRQDLDKVLD-SLSTREKQVIRWRFGMEDGRMKTLQEIGET 536
Query: 475 IGLSRERVRQVGLVALEKLKHAARKREMEAMLL 507
+G+SRERVRQ+ A KLK+ R ++ L+
Sbjct: 537 MGVSRERVRQIDTSAFRKLKNKKRNSHLQQYLV 569
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.131 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 770,910,416
Number of Sequences: 2540612
Number of extensions: 29748041
Number of successful extensions: 97812
Number of sequences better than 10.0: 1352
Number of HSP's better than 10.0 without gapping: 1241
Number of HSP's successfully gapped in prelim test: 112
Number of HSP's that attempted gapping in prelim test: 94595
Number of HSP's gapped (non-prelim): 1569
length of query: 509
length of database: 863,360,394
effective HSP length: 132
effective length of query: 377
effective length of database: 527,999,610
effective search space: 199055852970
effective search space used: 199055852970
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0127b.1


