

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0126.8
(749 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
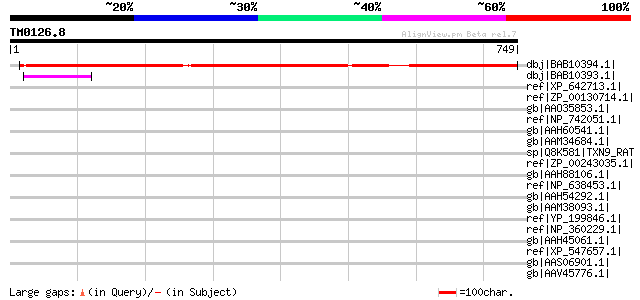
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB10394.1| unnamed protein product [Arabidopsis thaliana] g... 704 0.0
dbj|BAB10393.1| unnamed protein product [Arabidopsis thaliana] 73 4e-11
ref|XP_642713.1| hypothetical protein DDB0169096 [Dictyostelium ... 47 0.002
ref|ZP_00130714.1| COG0642: Signal transduction histidine kinase... 39 0.81
gb|AAO35853.1| Fe-S oxidoreductase [Clostridium tetani E88] gi|2... 37 2.4
ref|NP_742051.1| ATP binding protein associated with cell differ... 37 3.1
gb|AAH60541.1| Txndc9 protein [Rattus norvegicus] 37 3.1
gb|AAM34684.1| ES cell-related protein [Rattus norvegicus] 37 3.1
sp|Q8K581|TXN9_RAT Thioredoxin domain containing protein 9 (ES c... 37 3.1
ref|ZP_00243035.1| COG0642: Signal transduction histidine kinase... 37 3.1
gb|AAH88106.1| Txndc9 protein [Rattus norvegicus] 37 3.1
ref|NP_638453.1| two-component system sensor protein [Xanthomona... 36 4.0
gb|AAH54292.1| Apacd-prov protein [Xenopus laevis] 36 5.3
gb|AAM38093.1| two-component system sensor protein [Xanthomonas ... 36 5.3
ref|YP_199846.1| two-component system sensor protein [Xanthomona... 36 5.3
ref|NP_360229.1| osmolarity sensor protein envZ [EC:2.7.3.-] [Ri... 35 6.9
gb|AAH45061.1| Apacd-prov protein [Xenopus laevis] 35 9.0
ref|XP_547657.1| PREDICTED: similar to hypothetical protein [Can... 35 9.0
gb|AAS06901.1| CD45 [Aotus nigriceps] 35 9.0
gb|AAV45776.1| putative PAS/PAC sensing his kinase [Haloarcula m... 35 9.0
>dbj|BAB10394.1| unnamed protein product [Arabidopsis thaliana]
gi|15237930|ref|NP_197816.1| expressed protein
[Arabidopsis thaliana]
Length = 1634
Score = 704 bits (1816), Expect = 0.0
Identities = 371/740 (50%), Positives = 489/740 (65%), Gaps = 45/740 (6%)
Query: 15 NAKRKLVLRKDDDDTGKAYRFKILLPNGTSVELTLRDPQPQMPLPDFLQLVEDAYNGARE 74
+ KR LVL DDDD Y FK+LLPNGTSV+LTL++P+P++ + F+ LV+ Y+ AR+
Sbjct: 15 SVKRSLVL-DDDDDEDIFYNFKVLLPNGTSVKLTLKNPEPEISMQSFVNLVKKEYDNARK 73
Query: 75 HNQSMKRKKDINWIGAPLFLHDSTDAKIKNVIDFHNYKPHKCHILQLHGYFDSV*NAMQN 134
M ++ ++W F +S K+K ++ F +KP CHI++L +N
Sbjct: 74 DCLLMSKRMKVDWNSGGKFHLESNGGKMKGIVRFAAFKPDLCHIIRLDDGSGIASTMYEN 133
Query: 135 MWDLTPDIDLLLELPEEYSFQTALADLIDNSLQAVWSNGGNSRKLIRVNVGPDKISIFDN 194
+WDLTPD DLL ELPE YSF+TALADLIDNSLQAVW +RKLI V++ D I++FD
Sbjct: 134 LWDLTPDTDLLKELPENYSFETALADLIDNSLQAVWPYREGARKLISVDISGDHITVFDT 193
Query: 195 GPGMDDTDDNSLVKWGKLGASLHRNSKLQAIGGKPPYLMPYFGMFGYGGPIASMHLGR** 254
G GMD ++ NS+ KWGK+GASLHR+ K AIGG PPYL PYFGMFGYGGP ASM LGR
Sbjct: 194 GRGMDSSEGNSIDKWGKIGASLHRSQKTGAIGGNPPYLKPYFGMFGYGGPYASMFLGRCD 253
Query: 255 HSFPTSVQALPQNYIYLFIAPSHVLSHNRRASVSSKTKHVKKVYMLHLEREALLNTCSEV 314
SF LP + + L RR VSSKTK KKV+ L ++EAL++ S V
Sbjct: 254 FSF-----CLP--ILICIVLLRLTLFSVRRTLVSSKTKESKKVFTLQFKKEALIDNRSIV 306
Query: 315 --KWKTDGGIRDPSKDEIK-DCHGSFTMVEIFDPKVKDFDINRLQCHLKDIYFPYIQFLI 371
WKTDGG+RDPS++E+K HGSFT VEIF+ + I +LQC LKDIYFPYIQF +
Sbjct: 307 GKNWKTDGGMRDPSEEEMKLSPHGSFTKVEIFESEFDISKIYQLQCRLKDIYFPYIQFCL 366
Query: 372 VVFIQCDELSERGKTITPIKFQVNSVDLTEIQGGEVAITNLHSCNGPEFQFQLRLSFFSG 431
CDELS+ G+T P+ FQVN DL EI GGEVAITNLHS G F FQ+R + F G
Sbjct: 367 ATIFLCDELSKTGRTERPVAFQVNGEDLAEIAGGEVAITNLHS-KGQFFSFQIRFTLFGG 425
Query: 432 SRE--IQVANARLRVVYFPFTKGKENIERILEKLADDGCVIRENFLNFSRVSVRRLGRLL 489
R+ Q ANARL+ VYFP +GKE+IE+IL+ L ++GC + E+F F RVS+RRLGRLL
Sbjct: 426 KRKGTAQEANARLKFVYFPIVQGKESIEKILQSLEEEGCKVSESFQTFGRVSLRRLGRLL 485
Query: 490 PDARWTLLPFMDIRNKKGNRANILKRCSLRVKCFIETDAGFKPTQTKTDLAHHSPLTTAL 549
P+ RW +PFM ++GNRA+ L++ RVKCF++ DAGF PT +KTDLA +P + AL
Sbjct: 486 PEVRWDSIPFM----QRGNRASTLQKSCRRVKCFVDLDAGFSPTPSKTDLASQNPFSVAL 541
Query: 550 KNIGNKISDNQKGILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLE 609
+N G+K ++ +K DV I I ++ K ++ LE
Sbjct: 542 RNFGSKSTEKEK---------------------------DDDVNIVIHREGKSVSYAHLE 574
Query: 610 KEYVEWLLQMHGQYDEEADSGEDQAVIIVNPANKKELRISSDVIRVHKVLKRKEKLWKHG 669
++Y EW+L+MH +DEEA SG D+AV+IV +KK L I D +RVHK ++RKEK WK G
Sbjct: 575 EKYQEWVLEMHNTHDEEAASGLDEAVLIVGSLDKKALGILRDAVRVHKEVRRKEKTWKRG 634
Query: 670 ERIKVQKGACAGCHKNTVFATIEYFLLEGFEGDAGGDSRIICRSMDIPDKNGCLLSVTDE 729
+ IK+ +GA AG H N V+ATI+YFL+EGFE +AGGD+RI+CR +D P+ GC LS+ D
Sbjct: 635 QNIKILRGAYAGIHNNNVYATIDYFLIEGFEDEAGGDTRILCRPIDRPENEGCKLSIIDG 694
Query: 730 NASLEIRGSMSIPIGVIDSG 749
+ LE++ S+S+PI +IDSG
Sbjct: 695 ISKLEVQSSLSLPITIIDSG 714
>dbj|BAB10393.1| unnamed protein product [Arabidopsis thaliana]
Length = 1335
Score = 72.8 bits (177), Expect = 4e-11
Identities = 39/103 (37%), Positives = 62/103 (59%), Gaps = 2/103 (1%)
Query: 21 VLRKDDDDT-GKAYRFKILLPNGTSVELTLRDPQPQMPLPDFLQLVEDAY-NGAREHNQS 78
VL DDD+ K YR K+LLPN TSV L L +P+P+M + +F+ LV++ Y + S
Sbjct: 16 VLSLDDDEAEDKFYRLKVLLPNSTSVTLALTNPEPRMSMNNFVNLVKEEYEKSLNKCVLS 75
Query: 79 MKRKKDINWIGAPLFLHDSTDAKIKNVIDFHNYKPHKCHILQL 121
K++K ++W A + + KIK ++ F +KP C++L+L
Sbjct: 76 GKKRKRVDWNLAVKYYLEFNGEKIKEIVRFDKFKPDLCNVLRL 118
>ref|XP_642713.1| hypothetical protein DDB0169096 [Dictyostelium discoideum]
gi|28828603|gb|AAO51206.1| hypothetical protein
[Dictyostelium discoideum] gi|60470806|gb|EAL68778.1|
hypothetical protein DDB0169096 [Dictyostelium
discoideum]
Length = 2284
Score = 47.4 bits (111), Expect = 0.002
Identities = 31/86 (36%), Positives = 50/86 (58%), Gaps = 10/86 (11%)
Query: 137 DLTPDIDLLLEL-PEEYSFQTALADLIDNSLQAVWSNGGNSRKL-IRVN-----VGPDKI 189
D+TP ++LL+ EY+F A+A+ IDNS+QAV N R++ + +N + I
Sbjct: 173 DVTPSKNILLKAGTSEYNFPNAIAEFIDNSIQAVRLNPPGKREIKVTINRPNSDLRTTSI 232
Query: 190 SIFDNGPGMDDTDDNSLVKWGKLGAS 215
I+DNG GM + ++L +W +G S
Sbjct: 233 RIWDNGCGM---NKDALQRWATMGLS 255
>ref|ZP_00130714.1| COG0642: Signal transduction histidine kinase [Desulfovibrio
desulfuricans G20]
Length = 813
Score = 38.5 bits (88), Expect = 0.81
Identities = 30/118 (25%), Positives = 50/118 (41%), Gaps = 19/118 (16%)
Query: 102 IKNVIDFHNYKPHKCHILQLHGYFDSV*NAMQNMWDLTPDIDLL-LELPEEYS------- 153
+ N+++F + + +H D N +DL D +EL +Y+
Sbjct: 639 VSNMLEFSRRSESRRQLADMHALMDKTVELAANDYDLKKKYDFRHIELVRDYAPDLPGIH 698
Query: 154 -----FQTALADLIDNSLQAVWSNGGNS----RKLIRVNVGPDK--ISIFDNGPGMDD 200
+ L +L+ NS QA+ +S R +R + DK I + DNGPGMD+
Sbjct: 699 CTPTEIEQVLLNLLKNSAQAMAGRPKDSTEPPRITLRTRLQHDKVCIQVLDNGPGMDE 756
>gb|AAO35853.1| Fe-S oxidoreductase [Clostridium tetani E88]
gi|28210972|ref|NP_781916.1| Fe-S oxidoreductase
[Clostridium tetani E88]
Length = 444
Score = 37.0 bits (84), Expect = 2.4
Identities = 19/96 (19%), Positives = 46/96 (47%), Gaps = 2/96 (2%)
Query: 81 RKKDINWIGAPLFLHDSTDAKIKNVIDFHNYKPHKCHILQLHGYFDSV*NAMQNMWDLTP 140
++ D+ + F+ DS I +++ NYK + C +L + G + + +L P
Sbjct: 38 KEADVIIVNTCGFIEDSKKESIDTILEMSNYKNNNCKVLVVTGCLSQ--RYGEELQELLP 95
Query: 141 DIDLLLELPEEYSFQTALADLIDNSLQAVWSNGGNS 176
++D++L + + A+ I+ ++++ N N+
Sbjct: 96 EVDVMLGVNDYDKLSDAIKKSIEKGEKSLYCNYSNT 131
>ref|NP_742051.1| ATP binding protein associated with cell differentiation [Mus
musculus] gi|52789353|gb|AAH83077.1| ATP binding protein
associated with cell differentiation [Mus musculus]
gi|18605500|gb|AAH22947.1| Txndc9 protein [Mus musculus]
gi|50401676|sp|Q9CQ79|TXND9_MOUSE Thioredoxin domain
containing protein 9 (ATP binding protein associated
with cell differentiation) gi|26324474|dbj|BAC25991.1|
unnamed protein product [Mus musculus]
gi|12855662|dbj|BAB30412.1| unnamed protein product [Mus
musculus] gi|12847540|dbj|BAB27611.1| unnamed protein
product [Mus musculus] gi|12846349|dbj|BAB27134.1|
unnamed protein product [Mus musculus]
gi|12839124|dbj|BAB24440.1| unnamed protein product [Mus
musculus] gi|12833213|dbj|BAB22438.1| unnamed protein
product [Mus musculus]
Length = 226
Score = 36.6 bits (83), Expect = 3.1
Identities = 17/73 (23%), Positives = 37/73 (50%)
Query: 553 GNKISDNQKGILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLEKEY 612
GN D +L N + L+ LD+++ + + +E+ K++++ + +++
Sbjct: 3 GNGSVDMFSEVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQK 62
Query: 613 VEWLLQMHGQYDE 625
EWL + HG+Y E
Sbjct: 63 QEWLSKGHGEYRE 75
>gb|AAH60541.1| Txndc9 protein [Rattus norvegicus]
Length = 259
Score = 36.6 bits (83), Expect = 3.1
Identities = 17/73 (23%), Positives = 37/73 (50%)
Query: 553 GNKISDNQKGILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLEKEY 612
GN D +L N + L+ LD+++ + + +E+ K++++ + +++
Sbjct: 36 GNGSIDMFSEVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQK 95
Query: 613 VEWLLQMHGQYDE 625
EWL + HG+Y E
Sbjct: 96 QEWLSKGHGEYRE 108
>gb|AAM34684.1| ES cell-related protein [Rattus norvegicus]
Length = 252
Score = 36.6 bits (83), Expect = 3.1
Identities = 17/73 (23%), Positives = 37/73 (50%)
Query: 553 GNKISDNQKGILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLEKEY 612
GN D +L N + L+ LD+++ + + +E+ K++++ + +++
Sbjct: 29 GNGSIDMFSEVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQK 88
Query: 613 VEWLLQMHGQYDE 625
EWL + HG+Y E
Sbjct: 89 QEWLSKGHGEYRE 101
>sp|Q8K581|TXN9_RAT Thioredoxin domain containing protein 9 (ES cell-related protein)
gi|49248541|ref|NP_742029.2| thioredoxin domain
containing 9 [Rattus norvegicus]
Length = 226
Score = 36.6 bits (83), Expect = 3.1
Identities = 17/73 (23%), Positives = 37/73 (50%)
Query: 553 GNKISDNQKGILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLEKEY 612
GN D +L N + L+ LD+++ + + +E+ K++++ + +++
Sbjct: 3 GNGSIDMFSEVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQK 62
Query: 613 VEWLLQMHGQYDE 625
EWL + HG+Y E
Sbjct: 63 QEWLSKGHGEYRE 75
>ref|ZP_00243035.1| COG0642: Signal transduction histidine kinase [Rubrivivax
gelatinosus PM1]
Length = 440
Score = 36.6 bits (83), Expect = 3.1
Identities = 25/66 (37%), Positives = 37/66 (55%), Gaps = 5/66 (7%)
Query: 157 ALADLIDNSLQAVWSNGGNSRKLIRVNVGPDKISIFDNGPGMDDTDDNSLVKWGKLGASL 216
AL +LIDN+L+ +GG + + + V GP IS+ D+GPG+ LV+ + GAS
Sbjct: 343 ALRNLIDNALK----HGGPAAQ-VTVQAGPGSISVRDDGPGVPPDSLTRLVRPFERGASA 397
Query: 217 HRNSKL 222
S L
Sbjct: 398 AEGSGL 403
>gb|AAH88106.1| Txndc9 protein [Rattus norvegicus]
Length = 226
Score = 36.6 bits (83), Expect = 3.1
Identities = 17/73 (23%), Positives = 37/73 (50%)
Query: 553 GNKISDNQKGILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLEKEY 612
GN D +L N + L+ LD+++ + + +E+ K++++ + +++
Sbjct: 3 GNGSIDMFSEVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQK 62
Query: 613 VEWLLQMHGQYDE 625
EWL + HG+Y E
Sbjct: 63 QEWLSKGHGEYRE 75
>ref|NP_638453.1| two-component system sensor protein [Xanthomonas campestris pv.
campestris str. ATCC 33913] gi|21114329|gb|AAM42377.1|
two-component system sensor protein [Xanthomonas
campestris pv. campestris str. ATCC 33913]
gi|66767378|ref|YP_242140.1| two-component system sensor
protein [Xanthomonas campestris pv. campestris str.
8004] gi|66572710|gb|AAY48120.1| two-component system
sensor protein [Xanthomonas campestris pv. campestris
str. 8004]
Length = 385
Score = 36.2 bits (82), Expect = 4.0
Identities = 22/71 (30%), Positives = 39/71 (53%), Gaps = 7/71 (9%)
Query: 143 DLLLELPEEYSFQTALADLIDNSLQAVWSNGGNSRKLIRVNVGPDKISIFDNGPGMDDTD 202
DL+++ PE + AL +LI N+++ ++ G +RV V D + + D+GPG+ + D
Sbjct: 277 DLVIDAPES-ALSVALGNLIGNAVK--YTQDGQ----VRVRVLADAVEVIDSGPGLSEED 329
Query: 203 DNSLVKWGKLG 213
L + G G
Sbjct: 330 AAKLFQRGYRG 340
>gb|AAH54292.1| Apacd-prov protein [Xenopus laevis]
Length = 226
Score = 35.8 bits (81), Expect = 5.3
Identities = 16/63 (25%), Positives = 34/63 (53%)
Query: 563 ILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLEKEYVEWLLQMHGQ 622
++ N ++ LD +L K+ + +E+ K+R++ +++K+ EWL + HG+
Sbjct: 13 VMENQLLQTAKIMEEQLDAELEKLDKTDEDEMELLKERRLEALKKVQKQKQEWLSKGHGE 72
Query: 623 YDE 625
Y E
Sbjct: 73 YRE 75
>gb|AAM38093.1| two-component system sensor protein [Xanthomonas axonopodis pv.
citri str. 306] gi|21243975|ref|NP_643557.1|
two-component system sensor protein [Xanthomonas
axonopodis pv. citri str. 306]
Length = 385
Score = 35.8 bits (81), Expect = 5.3
Identities = 22/71 (30%), Positives = 39/71 (53%), Gaps = 7/71 (9%)
Query: 143 DLLLELPEEYSFQTALADLIDNSLQAVWSNGGNSRKLIRVNVGPDKISIFDNGPGMDDTD 202
DL+++ PE + AL +LI N+++ ++ G +RV V D + + D+GPG+ + D
Sbjct: 277 DLVIDAPES-ALSVALGNLIGNAVK--YTQDGQ----VRVRVLGDAVEVIDSGPGLSEED 329
Query: 203 DNSLVKWGKLG 213
L + G G
Sbjct: 330 AAKLFQRGYRG 340
>ref|YP_199846.1| two-component system sensor protein [Xanthomonas oryzae pv. oryzae
KACC10331] gi|58425424|gb|AAW74461.1| two-component
system sensor protein [Xanthomonas oryzae pv. oryzae
KACC10331]
Length = 450
Score = 35.8 bits (81), Expect = 5.3
Identities = 22/71 (30%), Positives = 39/71 (53%), Gaps = 7/71 (9%)
Query: 143 DLLLELPEEYSFQTALADLIDNSLQAVWSNGGNSRKLIRVNVGPDKISIFDNGPGMDDTD 202
DL+++ PE + AL +LI N+++ ++ G +RV V D + + D+GPG+ + D
Sbjct: 342 DLVIDAPES-ALSVALGNLIGNAVK--YTQDGQ----VRVRVRGDCVDVIDSGPGLSEED 394
Query: 203 DNSLVKWGKLG 213
L + G G
Sbjct: 395 AAKLFQRGYRG 405
>ref|NP_360229.1| osmolarity sensor protein envZ [EC:2.7.3.-] [Rickettsia conorii
str. Malish 7] gi|15619675|gb|AAL03130.1| osmolarity
sensor protein envZ [EC:2.7.3.-] [Rickettsia conorii
str. Malish 7] gi|25507972|pir||H97773 osmolarity sensor
protein envZ (EC 2.7.3.-) [imported] - Rickettsia
conorii (strain Malish 7)
Length = 479
Score = 35.4 bits (80), Expect = 6.9
Identities = 22/68 (32%), Positives = 39/68 (57%), Gaps = 4/68 (5%)
Query: 141 DIDLLLELPEEYSFQTALADLIDNSLQAVWSNGGNSRKLIRVNVGPDKISIFDNGPGMDD 200
++D L + +SF+ AL++LI N+++ +G + + N I I DNGPG+DD
Sbjct: 361 NLDKLEVCIKPHSFERALSNLIGNAIK----HGTKIKISSKDNSSTVAIIIEDNGPGIDD 416
Query: 201 TDDNSLVK 208
T+ + + K
Sbjct: 417 TETHLVFK 424
>gb|AAH45061.1| Apacd-prov protein [Xenopus laevis]
Length = 263
Score = 35.0 bits (79), Expect = 9.0
Identities = 16/63 (25%), Positives = 33/63 (51%)
Query: 563 ILINSYYTVTTLLSRFLDTQLTSYLKSADVAIEIFKDRKVLTPLQLEKEYVEWLLQMHGQ 622
++ N ++ LD +L K+ + +E+ K+R++ + +K+ EWL + HG+
Sbjct: 50 VMENQLLQTAKIMEEQLDAELEKLDKTDEDEMELLKERRLEALKKAQKQKQEWLSKGHGE 109
Query: 623 YDE 625
Y E
Sbjct: 110 YRE 112
>ref|XP_547657.1| PREDICTED: similar to hypothetical protein [Canis familiaris]
Length = 2290
Score = 35.0 bits (79), Expect = 9.0
Identities = 33/105 (31%), Positives = 45/105 (42%), Gaps = 9/105 (8%)
Query: 157 ALADLIDNSLQAVWSNGGNSRKLIRV----NVGPDKISIFDNGPGMDDTDDNS-----LV 207
ALA+LIDNSL A N G R I++ G +++ DNG GM N+ L
Sbjct: 374 ALAELIDNSLSATSRNTGIRRIQIKLLFDETQGKPAVAVIDNGRGMTSKQLNNWAVYRLS 433
Query: 208 KWGKLGASLHRNSKLQAIGGKPPYLMPYFGMFGYGGPIASMHLGR 252
K+ + G +S P L FG GG A +G+
Sbjct: 434 KFTRQGDFESDHSGYVRPVPVPRSLNSDISYFGVGGKQAVFFVGQ 478
>gb|AAS06901.1| CD45 [Aotus nigriceps]
Length = 1303
Score = 35.0 bits (79), Expect = 9.0
Identities = 21/73 (28%), Positives = 38/73 (51%), Gaps = 5/73 (6%)
Query: 351 DINRLQCHLKDIYFPYIQFLIVVFIQCDELSERGKTITPIKFQVNSVDLTEIQGGEVAIT 410
D N +C L+++ PY +++ ++ ++R T P F NS +E+Q V+IT
Sbjct: 435 DKNLTKCDLQNLK-PYTKYISSLYAYIIARAQRNGTAVPCHFTTNSARPSEVQNMTVSIT 493
Query: 411 NLHS----CNGPE 419
+ +S CN P+
Sbjct: 494 SENSMLVKCNAPK 506
>gb|AAV45776.1| putative PAS/PAC sensing his kinase [Haloarcula marismortui ATCC
43049] gi|55377632|ref|YP_135482.1| putative PAS/PAC
sensing his kinase [Haloarcula marismortui ATCC 43049]
Length = 576
Score = 35.0 bits (79), Expect = 9.0
Identities = 25/90 (27%), Positives = 45/90 (49%), Gaps = 13/90 (14%)
Query: 140 PDIDLLLELPEEYSFQTALADLIDNSLQAVWSNGG--NSRKLIRVNV--------GPDKI 189
PD++ +E P+ + + DLID++L+ V N N + + V+V G +
Sbjct: 446 PDVEFRVERPDRA--EVSAVDLIDSALRNVLENAAEHNDQSVPEVSVTVMGPTDDGDVTV 503
Query: 190 SIFDNGPGMDDTDDNSLVKWGKLGASLHRN 219
++ DNGPG+ D + +++ G A H N
Sbjct: 504 AVADNGPGLPDR-EQEVIETGTEMALKHSN 532
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.140 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,317,800,373
Number of Sequences: 2540612
Number of extensions: 57381841
Number of successful extensions: 120156
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 120133
Number of HSP's gapped (non-prelim): 26
length of query: 749
length of database: 863,360,394
effective HSP length: 136
effective length of query: 613
effective length of database: 517,837,162
effective search space: 317434180306
effective search space used: 317434180306
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0126.8


