

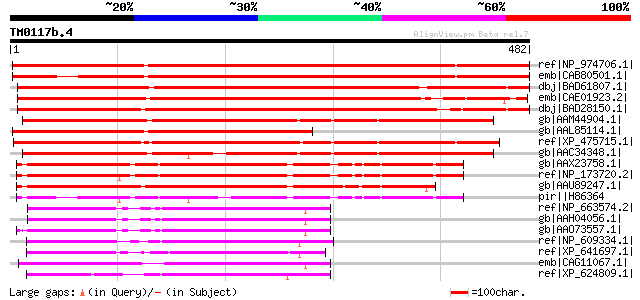
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0117b.4
(482 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_974706.1| expressed protein [Arabidopsis thaliana] 779 0.0
emb|CAB80501.1| putative protein [Arabidopsis thaliana] gi|45393... 731 0.0
dbj|BAD61807.1| MAP kinase activating protein-like [Oryza sativa... 683 0.0
emb|CAE01923.2| OSJNBb0078D11.6 [Oryza sativa (japonica cultivar... 589 e-167
dbj|BAD28150.1| putative MAP kinase activating protein [Oryza sa... 577 e-163
gb|AAM44904.1| unknown protein [Arabidopsis thaliana] gi|1433472... 476 e-133
gb|AAL85114.1| unknown protein [Arabidopsis thaliana] gi|1502811... 451 e-125
ref|XP_475715.1| unknown protein [Oryza sativa (japonica cultiva... 446 e-124
gb|AAC34348.1| Unknown protein [Arabidopsis thaliana] gi|7487004... 434 e-120
gb|AAX23758.1| hypothetical protein At1g23070 [Arabidopsis thali... 342 2e-92
ref|NP_173720.2| hypothetical protein [Arabidopsis thaliana] 333 5e-90
gb|AAU89247.1| hypothetical protein [Oryza sativa (japonica cult... 330 8e-89
pir||H86364 hypothetical protein T26J12.15 - Arabidopsis thalian... 269 1e-70
ref|NP_663574.2| transmembrane protein 34 [Mus musculus] gi|2632... 190 1e-46
gb|AAH04056.1| Similar to CG5850 gene product [Mus musculus] 190 1e-46
gb|AAO73557.1| hypothetical protein FLJ10846-like protein [Rattu... 189 2e-46
ref|NP_609334.1| CG5850-PA [Drosophila melanogaster] gi|22946075... 181 3e-44
ref|XP_641697.1| hypothetical protein DDB0205844 [Dictyostelium ... 177 7e-43
emb|CAG11067.1| unnamed protein product [Tetraodon nigroviridis] 176 1e-42
ref|XP_624809.1| PREDICTED: similar to CG5850-PA [Apis mellifera] 175 2e-42
>ref|NP_974706.1| expressed protein [Arabidopsis thaliana]
Length = 485
Score = 779 bits (2012), Expect = 0.0
Identities = 377/481 (78%), Positives = 427/481 (88%), Gaps = 5/481 (1%)
Query: 3 IIQLLHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIE 62
++ ++ P WA+ +A FL++TL+LS++L+F+HLS YKNPEEQKFLIGVILMVPCYSIE
Sbjct: 9 LLAAAYSAPAWASFMAGAFLVLTLSLSLFLVFDHLSTYKNPEEQKFLIGVILMVPCYSIE 68
Query: 63 SFVSLVNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLL 122
SF SLV PSISVDCGILRDCYESFAMYCFGRYLVAC+GGEERTIEFMERQGR + KTPLL
Sbjct: 69 SFASLVKPSISVDCGILRDCYESFAMYCFGRYLVACIGGEERTIEFMERQGRKSFKTPLL 128
Query: 123 LHHHSHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFG 182
H D KG + HPFP+ FLKPW L+ FYQ+VKFGIVQYMIIKS T++ A+ILEAFG
Sbjct: 129 DHK---DEKGIIKHPFPMNLFLKPWRLSPWFYQVVKFGIVQYMIIKSLTALTALILEAFG 185
Query: 183 VYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFL 242
VYCEGEFK GCGYPY+AVVLNFSQSWALYCLVQFY TKDELAHI+PLAKFLTFKSIVFL
Sbjct: 186 VYCEGEFKWGCGYPYLAVVLNFSQSWALYCLVQFYGATKDELAHIQPLAKFLTFKSIVFL 245
Query: 243 TWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMG 302
TWWQGVAIALL + GLFKS IAQ LQ K+SVQDFIICIEMGIAS+VHLYVFPAKPY LMG
Sbjct: 246 TWWQGVAIALLSSLGLFKSSIAQSLQLKTSVQDFIICIEMGIASVVHLYVFPAKPYGLMG 305
Query: 303 DRHPGSISVLGDY-SADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGG 361
DR GS+SVLGDY S DCP+DPDEIRDSERPTK+RLP DVD +SGMTI+ES+RDV +GG
Sbjct: 306 DRFTGSVSVLGDYASVDCPIDPDEIRDSERPTKVRLPHPDVDIRSGMTIKESMRDVFVGG 365
Query: 362 SGYIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPAR 421
YIVKDV+FTV QAVEP+EK IT+FNEKLH+ISQNIKKHDK++RR+KDDSC+ SSSP+R
Sbjct: 366 GEYIVKDVRFTVTQAVEPMEKSITKFNEKLHKISQNIKKHDKEKRRVKDDSCM-SSSPSR 424
Query: 422 RVIRGIDDPLLNGSISDSGISRAKKHRRKSGYTSAESGGESSSDQSYGAYQVRGHRWVTK 481
RVIRGIDDPLLNGS SDSG++R KKHRRKSGYTSAESGGESSSDQ+YG ++VRG RW+TK
Sbjct: 425 RVIRGIDDPLLNGSFSDSGVTRTKKHRRKSGYTSAESGGESSSDQAYGGFEVRGRRWITK 484
Query: 482 E 482
+
Sbjct: 485 D 485
>emb|CAB80501.1| putative protein [Arabidopsis thaliana] gi|4539344|emb|CAB37492.1|
putative protein [Arabidopsis thaliana]
gi|7485951|pir||T05664 hypothetical protein F22I13.130 -
Arabidopsis thaliana
Length = 466
Score = 731 bits (1886), Expect = 0.0
Identities = 358/481 (74%), Positives = 408/481 (84%), Gaps = 24/481 (4%)
Query: 3 IIQLLHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIE 62
++ ++ P WA+ +A FL++TL+LS++L+F+HLS YKNPE
Sbjct: 9 LLAAAYSAPAWASFMAGAFLVLTLSLSLFLVFDHLSTYKNPE------------------ 50
Query: 63 SFVSLVNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLL 122
F SLV PSISVDCGILRDCYESFAMYCFGRYLVAC+GGEERTIEFMERQGR + KTPLL
Sbjct: 51 -FASLVKPSISVDCGILRDCYESFAMYCFGRYLVACIGGEERTIEFMERQGRKSFKTPLL 109
Query: 123 LHHHSHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFG 182
H D KG + HPFP+ FLKPW L+ FYQ+VKFGIVQYMIIKS T++ A+ILEAFG
Sbjct: 110 DHK---DEKGIIKHPFPMNLFLKPWRLSPWFYQVVKFGIVQYMIIKSLTALTALILEAFG 166
Query: 183 VYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFL 242
VYCEGEFK GCGYPY+AVVLNFSQSWALYCLVQFY TKDELAHI+PLAKFLTFKSIVFL
Sbjct: 167 VYCEGEFKWGCGYPYLAVVLNFSQSWALYCLVQFYGATKDELAHIQPLAKFLTFKSIVFL 226
Query: 243 TWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMG 302
TWWQGVAIALL + GLFKS IAQ LQ K+SVQDFIICIEMGIAS+VHLYVFPAKPY LMG
Sbjct: 227 TWWQGVAIALLSSLGLFKSSIAQSLQLKTSVQDFIICIEMGIASVVHLYVFPAKPYGLMG 286
Query: 303 DRHPGSISVLGDY-SADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGG 361
DR GS+SVLGDY S DCP+DPDEIRDSERPTK+RLP DVD +SGMTI+ES+RDV +GG
Sbjct: 287 DRFTGSVSVLGDYASVDCPIDPDEIRDSERPTKVRLPHPDVDIRSGMTIKESMRDVFVGG 346
Query: 362 SGYIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPAR 421
YIVKDV+FTV QAVEP+EK IT+FNEKLH+ISQNIKKHDK++RR+KDDSC+ SSSP+R
Sbjct: 347 GEYIVKDVRFTVTQAVEPMEKSITKFNEKLHKISQNIKKHDKEKRRVKDDSCM-SSSPSR 405
Query: 422 RVIRGIDDPLLNGSISDSGISRAKKHRRKSGYTSAESGGESSSDQSYGAYQVRGHRWVTK 481
RVIRGIDDPLLNGS SDSG++R KKHRRKSGYTSAESGGESSSDQ+YG ++VRG RW+TK
Sbjct: 406 RVIRGIDDPLLNGSFSDSGVTRTKKHRRKSGYTSAESGGESSSDQAYGGFEVRGRRWITK 465
Query: 482 E 482
+
Sbjct: 466 D 466
>dbj|BAD61807.1| MAP kinase activating protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 479
Score = 683 bits (1763), Expect = 0.0
Identities = 333/477 (69%), Positives = 389/477 (80%), Gaps = 13/477 (2%)
Query: 8 HTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSL 67
+ PP+WA+I A +F++ +L+LS++LLF HLSAYKNPEEQKFL+GVILMVPCY++ES++SL
Sbjct: 14 YAPPIWASITAGIFVITSLSLSLFLLFNHLSAYKNPEEQKFLVGVILMVPCYAVESYISL 73
Query: 68 VNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHS 127
VNPSISVD ILRD YE+FAMYCFGRYLVACLGGE+RTIEF++R+G S S PLL H
Sbjct: 74 VNPSISVDIEILRDGYEAFAMYCFGRYLVACLGGEDRTIEFLKREGSSGSDVPLLDHETG 133
Query: 128 HDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEG 187
Y VNHPFP+ Y LKPW L FY ++KFG+VQY+IIK+ +I+AVILE+FGVYCEG
Sbjct: 134 QRY---VNHPFPMNYMLKPWPLGEWFYLVIKFGLVQYVIIKTICAILAVILESFGVYCEG 190
Query: 188 EFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQG 247
EFK CGY Y AVVLNFSQSWALYCLVQFY KDELAHIKPLAKFLTFKSIVFLTWWQG
Sbjct: 191 EFKWNCGYSYTAVVLNFSQSWALYCLVQFYAAIKDELAHIKPLAKFLTFKSIVFLTWWQG 250
Query: 248 VAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGDRHPG 307
V IALLY +GL + PIAQ LQ KSS+QDFIICIEMG+ASI HLYVFPAKPYE+MGDR G
Sbjct: 251 VVIALLYNWGLLRGPIAQELQFKSSIQDFIICIEMGVASIAHLYVFPAKPYEMMGDRFIG 310
Query: 308 SISVLGDY-SADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGGSGYIV 366
+SVLGDY S DCPLDPDE++DSERPTK RLP + I+ESVRDVV+GG YIV
Sbjct: 311 GVSVLGDYASVDCPLDPDEVKDSERPTKTRLPQPGDRVRCSTGIKESVRDVVLGGGEYIV 370
Query: 367 KDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRG 426
D+KFTV+ AVEP+ NEKLHRISQNIKKH+K++++ DDSCI S RVI G
Sbjct: 371 NDLKFTVNHAVEPI-------NEKLHRISQNIKKHEKEKKKTNDDSCINSQQSLSRVISG 423
Query: 427 IDDPLLNGSISD-SGISRAKKHRRKSGYTSAESGGESSSDQSYGAYQVRGHRWVTKE 482
IDDPLLNGS+SD SG +++KHRRKSGY SAESGGE SSDQ G Y++RGHRW+T+E
Sbjct: 424 IDDPLLNGSLSDNSGQKKSRKHRRKSGYGSAESGGE-SSDQGLGGYEIRGHRWITRE 479
>emb|CAE01923.2| OSJNBb0078D11.6 [Oryza sativa (japonica cultivar-group)]
gi|50927953|ref|XP_473504.1| OSJNBb0078D11.6 [Oryza
sativa (japonica cultivar-group)]
Length = 470
Score = 589 bits (1519), Expect = e-167
Identities = 291/477 (61%), Positives = 369/477 (77%), Gaps = 25/477 (5%)
Query: 8 HTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSL 67
+T P WAT+VA F+L++L+LS+YL+FEHLSAY NPEEQKF++GVILMVPCY+IES+VSL
Sbjct: 15 YTTPTWATLVAGFFVLLSLSLSIYLIFEHLSAYNNPEEQKFVLGVILMVPCYAIESYVSL 74
Query: 68 VNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHS 127
+NP+ SV CGILRD YE+FAMYCFGRY+ ACLGGE++TI F++R+G S S+ PLL H
Sbjct: 75 INPNTSVYCGILRDGYEAFAMYCFGRYITACLGGEDKTIAFLKREGGSGSRQPLLDHASE 134
Query: 128 HDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEG 187
KG ++H FP+ + LKPW L RFY I+KFGI QY+IIK+ T+ +++ LEAFGVYC+G
Sbjct: 135 ---KGIIHHHFPVNFILKPWRLGMRFYLIIKFGIFQYVIIKTVTASLSLFLEAFGVYCDG 191
Query: 188 EFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQG 247
EF L CGYPY A VLNFSQ WALYCLV++YT TKDELAHIKPLAKFL+FKSIVFLTWWQG
Sbjct: 192 EFNLRCGYPYFAAVLNFSQYWALYCLVEWYTATKDELAHIKPLAKFLSFKSIVFLTWWQG 251
Query: 248 VAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGDRHPG 307
V IA++Y+ GL +SP+AQ L+ KSS+QDFIICIEMGIASIVHLYVFPAKPYEL ++ PG
Sbjct: 252 VVIAIMYSLGLLRSPLAQSLELKSSIQDFIICIEMGIASIVHLYVFPAKPYELQANQSPG 311
Query: 308 SISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGGSGYIVK 367
++SVLGDY + P+DP EI++S RP KL+LP + D +S I+ESVRD V+G Y++K
Sbjct: 312 NVSVLGDYVSSDPVDPFEIKESNRPAKLKLPQLEPDERSTTNIKESVRDFVVGSGEYVIK 371
Query: 368 DVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRGI 427
D KFTV+QAV PVEK RF++ + K ++ +DD+ +++ SP R V RGI
Sbjct: 372 DFKFTVNQAVRPVEK---RFDKLM-----------KKNKKSQDDNWVSAVSPDRPV-RGI 416
Query: 428 DDPLLNGSISDSGISRAKKHRRK-SGYTSAES--GGESSSDQSYGAYQVRGHRWVTK 481
DDPLL GS SDSG ++ KKHRR S +A+S GG+ +SD Y++RG RW K
Sbjct: 417 DDPLLGGSTSDSGFTKGKKHRRAVSTVAAADSWGGGDLASD----GYEIRGRRWAVK 469
>dbj|BAD28150.1| putative MAP kinase activating protein [Oryza sativa (japonica
cultivar-group)] gi|50251340|dbj|BAD28316.1| putative
MAP kinase activating protein [Oryza sativa (japonica
cultivar-group)]
Length = 475
Score = 577 bits (1486), Expect = e-163
Identities = 281/476 (59%), Positives = 356/476 (74%), Gaps = 17/476 (3%)
Query: 8 HTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSL 67
+ P WA +++ F+L++++LSMYL+F+HLSAY NPEEQKF++GVILMVPCY++ES+VSL
Sbjct: 15 YAAPTWAILISGFFMLLSVSLSMYLIFQHLSAYNNPEEQKFVLGVILMVPCYAVESYVSL 74
Query: 68 VNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHS 127
VNP SV CGILRD YE+FAMYCFGRY+ ACLGGEERTI F++R+G S PLL H
Sbjct: 75 VNPDTSVYCGILRDAYEAFAMYCFGRYITACLGGEERTIAFLKREGGGDSGEPLL---HG 131
Query: 128 HDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEG 187
KG ++H FP+ Y LKPW + RFYQI+KFGI QY+IIK+ T+ +++IL+ FG YC+G
Sbjct: 132 ASEKGIIHHHFPVNYILKPWRMGVRFYQIIKFGIFQYVIIKTLTASLSLILQPFGAYCDG 191
Query: 188 EFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQG 247
EF L CGYPY A VLNFSQ WALYCLV++YT TKDELAHIKPLAKFL+FKSIVFLTWWQG
Sbjct: 192 EFNLRCGYPYFAAVLNFSQYWALYCLVEWYTATKDELAHIKPLAKFLSFKSIVFLTWWQG 251
Query: 248 VAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGD-RHP 306
+ IA++Y+ GL +SP+AQ L+ KSS+QDFIICIEMGIAS+VHLYVFPAKPY L+G+ R P
Sbjct: 252 IMIAIMYSLGLVRSPLAQSLELKSSIQDFIICIEMGIASVVHLYVFPAKPYSLLGNHRSP 311
Query: 307 GSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGGSGYIV 366
+ISVLGDY+A P+DPDEI+D RPTKLRLP + D ++ESVRD VIG Y++
Sbjct: 312 ENISVLGDYAATDPVDPDEIKDISRPTKLRLPQLEPDEIIVTNVKESVRDFVIGSGEYVI 371
Query: 367 KDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRG 426
KD+KFT+ QAV PV K + +K + Q+ +DD+ +++S+P +R I G
Sbjct: 372 KDLKFTMKQAVRPVGKRFEKLMKKKGKFGQS-----------RDDNWVSTSTP-QRAIHG 419
Query: 427 IDDPLLNGSISDSGISRAKKHRRKSGYTSAESGGESSSDQSYGAYQVRGHRWVTKE 482
IDDPL+ GS SDSGI R K+HRR E SDQ+ Y +RG RW K+
Sbjct: 420 IDDPLICGSSSDSGIGRGKRHRRDVSSAGVVDSWE-GSDQTSDGYVIRGRRWEIKK 474
>gb|AAM44904.1| unknown protein [Arabidopsis thaliana] gi|14334724|gb|AAK59540.1|
unknown protein [Arabidopsis thaliana]
gi|18411404|ref|NP_565152.1| expressed protein
[Arabidopsis thaliana]
Length = 484
Score = 476 bits (1224), Expect = e-133
Identities = 237/439 (53%), Positives = 317/439 (71%), Gaps = 9/439 (2%)
Query: 13 WATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSI 72
W + ASVF+++ + L MYL+FEHL++Y PEEQKFLIG+ILMVP Y++ESF+SLVN
Sbjct: 41 WPILSASVFVVIAILLPMYLIFEHLASYNQPEEQKFLIGLILMVPVYAVESFLSLVNSEA 100
Query: 73 SVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDYKG 132
+ +C ++RDCYE+FA+YCF RYL+ACL GEERTIEFME+Q TPLL S+ G
Sbjct: 101 AFNCEVIRDCYEAFALYCFERYLIACLDGEERTIEFMEQQTVITQSTPLLEGTCSY---G 157
Query: 133 TVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFKLG 192
V HPFP+ F+K W L +FY VK GIVQYMI+K +++A+ILEAFGVY EG+F
Sbjct: 158 VVEHPFPMNCFVKDWSLGPQFYHAVKIGIVQYMILKMICALLAMILEAFGVYGEGKFAWN 217
Query: 193 CGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIAL 252
GYPY+AVVLNFSQ+WALYCLVQFY V KD+LA IKPLAKFLTFKSIVFLTWWQG+ +A
Sbjct: 218 YGYPYLAVVLNFSQTWALYCLVQFYNVIKDKLAPIKPLAKFLTFKSIVFLTWWQGIIVAF 277
Query: 253 LYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGDRHPGSISVL 312
L++ GL K +A+ L K+ +QD+IICIEMGIA++VHLYVFPA PY+ G+R +++V+
Sbjct: 278 LFSMGLVKGSLAKEL--KTRIQDYIICIEMGIAAVVHLYVFPAAPYK-RGERCVRNVAVM 334
Query: 313 GDY-SADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGGSGYIVKDVKF 371
DY S D P DP+E++DSER T+ R D D + + +SVRDVV+G IV D++F
Sbjct: 335 SDYASIDVPPDPEEVKDSERTTRTRYGRHD-DREKRLNFPQSVRDVVLGSGEIIVDDMRF 393
Query: 372 TVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRVIRGIDDPL 431
TV VEPVE+GI + N H+IS+N+K+ ++ ++ KDDS + + + + + L
Sbjct: 394 TVSHVVEPVERGIAKINRTFHQISENVKRFEQQKKTTKDDSYVIPLNQWAKEFSDVHENL 453
Query: 432 LN-GSISDSGISRAKKHRR 449
+ GS+SDSG+ +H +
Sbjct: 454 YDGGSVSDSGLGSTNRHHQ 472
>gb|AAL85114.1| unknown protein [Arabidopsis thaliana] gi|15028111|gb|AAK76679.1|
unknown protein [Arabidopsis thaliana]
gi|18420224|ref|NP_568039.1| expressed protein
[Arabidopsis thaliana]
Length = 304
Score = 451 bits (1159), Expect = e-125
Identities = 217/279 (77%), Positives = 243/279 (86%), Gaps = 3/279 (1%)
Query: 3 IIQLLHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIE 62
++ ++ P WA+ +A FL++TL+LS++L+F+HLS YKNPEEQKFLIGVILMVPCYSIE
Sbjct: 9 LLAAAYSAPAWASFMAGAFLVLTLSLSLFLVFDHLSTYKNPEEQKFLIGVILMVPCYSIE 68
Query: 63 SFVSLVNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLL 122
SF SLV PSISVDCGILRDCYESFAMYCFGRYLVAC+GGEERTIEFMERQGR + KTPLL
Sbjct: 69 SFASLVKPSISVDCGILRDCYESFAMYCFGRYLVACIGGEERTIEFMERQGRKSFKTPLL 128
Query: 123 LHHHSHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFG 182
H D KG + HPFP+ FLKPW L+ FYQ+VKFGIVQYMIIKS T++ A+ILEAFG
Sbjct: 129 DHK---DEKGIIKHPFPMNLFLKPWRLSPWFYQVVKFGIVQYMIIKSLTALTALILEAFG 185
Query: 183 VYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFL 242
VYCEGEFK GCGYPY+AVVLNFSQSWALYCLVQFY TKDELAHI+PLAKFLTFKSIVFL
Sbjct: 186 VYCEGEFKWGCGYPYLAVVLNFSQSWALYCLVQFYGATKDELAHIQPLAKFLTFKSIVFL 245
Query: 243 TWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIE 281
TWWQGVAIALL + GLFKS IAQ LQ K+SVQDFIICIE
Sbjct: 246 TWWQGVAIALLSSLGLFKSSIAQSLQLKTSVQDFIICIE 284
>ref|XP_475715.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|46575956|gb|AAT01317.1| unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 488
Score = 446 bits (1147), Expect = e-124
Identities = 221/452 (48%), Positives = 312/452 (68%), Gaps = 8/452 (1%)
Query: 4 IQLLHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIES 63
+ L + P W + A + + +L LS++L+FEHL AY PEEQKFLIG+ILMVP Y+++S
Sbjct: 32 VSLSESLPSWPIVSAGISVTASLVLSLFLIFEHLCAYHQPEEQKFLIGLILMVPVYAVQS 91
Query: 64 FVSLVNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLL 123
F SL+N +++ C ++RDCYE+FAMYCF RYL+ACLGGEE TI FME + + + +PLL
Sbjct: 92 FFSLLNSNVAFICELMRDCYEAFAMYCFERYLIACLGGEESTIRFMEGRFQFSESSPLL- 150
Query: 124 HHHSHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGV 183
+DY G V HPFPL +F++ W L FY VK GIVQYMI+K +I+A+ ++ G+
Sbjct: 151 -DVDYDY-GIVKHPFPLNWFMRNWYLGPDFYHAVKVGIVQYMILKPICAILAIFMQLIGI 208
Query: 184 YCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLT 243
Y EG+F GYPY+A+VLNFSQ+WALYCL+QFYT TK++L IKPL+KFLTFKSIVFLT
Sbjct: 209 YGEGKFAWRYGYPYLAIVLNFSQTWALYCLIQFYTATKEKLEPIKPLSKFLTFKSIVFLT 268
Query: 244 WWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGD 303
WWQG+A+A L++ GLFK +AQ Q++ +QD+IIC+EMG+A++VHL VFPAKPY G+
Sbjct: 269 WWQGIAVAFLFSTGLFKGHLAQRFQTR--IQDYIICLEMGVAAVVHLKVFPAKPYR-RGE 325
Query: 304 RHPGSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGGSG 363
R +++V+ DY++ DP+E R+ + ++ D ++ +SVRDVV+G
Sbjct: 326 RSVSNVAVMSDYASLGASDPEEEREIDNVAIMQAARPD-SRDRRLSFPQSVRDVVLGSGE 384
Query: 364 YIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPARRV 423
+V DVK+TV VEPVE+ ++ N LH+IS+N+K+ +K +R+ KDDS + P
Sbjct: 385 IMVDDVKYTVSHVVEPVERSFSKINRTLHQISENVKQLEKQKRKAKDDSDV-PLEPFSEE 443
Query: 424 IRGIDDPLLNGSISDSGISRAKKHRRKSGYTS 455
D + GS+SDSG++R K K +S
Sbjct: 444 FAEAHDNVFGGSVSDSGLARKKYKNTKRAPSS 475
>gb|AAC34348.1| Unknown protein [Arabidopsis thaliana] gi|7487004|pir||T00451
hypothetical protein T14N5.8 - Arabidopsis thaliana
Length = 500
Score = 434 bits (1115), Expect = e-120
Identities = 227/466 (48%), Positives = 307/466 (65%), Gaps = 47/466 (10%)
Query: 13 WATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSI 72
W + ASVF+++ + L MYL+FEHL++Y PEEQKFLIG+ILMVP Y++ESF+SLVN
Sbjct: 41 WPILSASVFVVIAILLPMYLIFEHLASYNQPEEQKFLIGLILMVPVYAVESFLSLVNSEA 100
Query: 73 SVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDYKG 132
+ +C ++RDCYE+FA+YCF RYL+ACL GEERTIEFME+Q TPLL S+ G
Sbjct: 101 AFNCEVIRDCYEAFALYCFERYLIACLDGEERTIEFMEQQTVITQSTPLLEGTCSY---G 157
Query: 133 TVNHPFPLKYFLKPWILTRRFYQIVKFGIVQY---------------------------M 165
V HPFP+ F+K W L +FY VK GIVQY M
Sbjct: 158 VVEHPFPMNCFVKDWSLGPQFYHAVKIGIVQYVCVVNTFSGLWPMMYFCASTDINILLQM 217
Query: 166 IIKSFTSIMAVILEAFGVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELA 225
I+K +++A+ILEAFGVY EG+F N+ Q+WALYCLVQFY V KD+LA
Sbjct: 218 ILKMICALLAMILEAFGVYGEGKF-----------AWNYGQTWALYCLVQFYNVIKDKLA 266
Query: 226 HIKPLAKFLTFKSIVFLTWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIA 285
IKPLAKFLTFKSIVFLTWWQG+ +A L++ GL K +A+ L K+ +QD+IICIEMGIA
Sbjct: 267 PIKPLAKFLTFKSIVFLTWWQGIIVAFLFSMGLVKGSLAKEL--KTRIQDYIICIEMGIA 324
Query: 286 SIVHLYVFPAKPYELMGDRHPGSISVLGDY-SADCPLDPDEIRDSERPTKLRLPTTDVDA 344
++VHLYVFPA PY+ G+R +++V+ DY S D P DP+E++DSER T+ R D D
Sbjct: 325 AVVHLYVFPAAPYK-RGERCVRNVAVMSDYASIDVPPDPEEVKDSERTTRTRYGRHD-DR 382
Query: 345 KSGMTIRESVRDVVIGGSGYIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKD 404
+ + +SVRDVV+G IV D++FTV VEPVE+GI + N H+IS+N+K+ ++
Sbjct: 383 EKRLNFPQSVRDVVLGSGEIIVDDMRFTVSHVVEPVERGIAKINRTFHQISENVKRFEQQ 442
Query: 405 RRRIKDDSCIASSSPARRVIRGIDDPLLN-GSISDSGISRAKKHRR 449
++ KDDS + + + + + L + GS+SDSG+ +H +
Sbjct: 443 KKTTKDDSYVIPLNQWAKEFSDVHENLYDGGSVSDSGLGSTNRHHQ 488
>gb|AAX23758.1| hypothetical protein At1g23070 [Arabidopsis thaliana]
Length = 403
Score = 342 bits (877), Expect = 2e-92
Identities = 182/415 (43%), Positives = 264/415 (62%), Gaps = 24/415 (5%)
Query: 7 LHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVS 66
LH P + I+ F + + LS+Y + +HL Y NP EQK+++ V+ MVP Y+ ES +S
Sbjct: 12 LHLPSL---IIGGSFATVAICLSLYSILQHLRFYTNPAEQKWIVSVLFMVPVYATESIIS 68
Query: 67 LVNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHH 126
L N S+ C ILR+CYE+FA+Y FG YLVACLGGE R +E++E + SK PLL
Sbjct: 69 LSNSKFSLPCDILRNCYEAFALYSFGSYLVACLGGERRVVEYLENE----SKKPLLEEGA 124
Query: 127 SHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCE 186
+ K + F K+ P++L R + I KFG+VQYMI+K+F + + +LE GVY +
Sbjct: 125 NESKKKKKKNSF-WKFLCDPYVLGRELFVIEKFGLVQYMILKTFCAFLTFLLELLGVYGD 183
Query: 187 GEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQ 246
GEFK GYPYI VVLNFSQ WAL+CLVQFY VT + L IKPLAKF++FK+IVF TWWQ
Sbjct: 184 GEFKWYYGYPYIVVVLNFSQMWALFCLVQFYNVTHERLKEIKPLAKFISFKAIVFATWWQ 243
Query: 247 GVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGDRHP 306
G IALL +G+ + + + ++ +QDF+ICIEM IA++ HL+VFPA+PY H
Sbjct: 244 GFGIALLCYYGI----LPKEGRFQNGLQDFLICIEMAIAAVAHLFVFPAEPY------HY 293
Query: 307 GSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGGSGYIV 366
+S G +A+ E++ E + T V+A SG +I+ESV+D+VI G ++V
Sbjct: 294 IPVSECGKITAE--TSKTEVK-LEEGGLVETTETQVEA-SGTSIKESVQDIVIDGGQHVV 349
Query: 367 KDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPAR 421
KDV T++QA+ PVEKG+T+ + +H+ + + K+ + ++ + +S P +
Sbjct: 350 KDVVLTINQAIGPVEKGVTKIQDTIHQ--KLLDSDGKEETEVTEEVTVETSVPPK 402
>ref|NP_173720.2| hypothetical protein [Arabidopsis thaliana]
Length = 414
Score = 333 bits (855), Expect = 5e-90
Identities = 182/426 (42%), Positives = 264/426 (61%), Gaps = 35/426 (8%)
Query: 7 LHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVS 66
LH P + I+ F + + LS+Y + +HL Y NP EQK+++ V+ MVP Y+ ES +S
Sbjct: 12 LHLPSL---IIGGSFATVAICLSLYSILQHLRFYTNPAEQKWIVSVLFMVPVYATESIIS 68
Query: 67 LVNPSISVDCGILRDCYESFAMYCFGRYLVACLG-----------GEERTIEFMERQGRS 115
L N S+ C ILR+CYE+FA+Y FG YLVACLG GE R +E++E +
Sbjct: 69 LSNSKFSLPCDILRNCYEAFALYSFGSYLVACLGELSKIPNEWSGGERRVVEYLENE--- 125
Query: 116 ASKTPLLLHHHSHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMA 175
SK PLL + K + F K+ P++L R + I KFG+VQYMI+K+F + +
Sbjct: 126 -SKKPLLEEGANESKKKKKKNSF-WKFLCDPYVLGRELFVIEKFGLVQYMILKTFCAFLT 183
Query: 176 VILEAFGVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLT 235
+LE GVY +GEFK GYPYI VVLNFSQ WAL+CLVQFY VT + L IKPLAKF++
Sbjct: 184 FLLELLGVYGDGEFKWYYGYPYIVVVLNFSQMWALFCLVQFYNVTHERLKEIKPLAKFIS 243
Query: 236 FKSIVFLTWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPA 295
FK+IVF TWWQG IALL +G+ + + + ++ +QDF+ICIEM IA++ HL+VFPA
Sbjct: 244 FKAIVFATWWQGFGIALLCYYGI----LPKEGRFQNGLQDFLICIEMAIAAVAHLFVFPA 299
Query: 296 KPYELMGDRHPGSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVR 355
+PY H +S G +A+ E++ E + T V+A SG +I+ESV+
Sbjct: 300 EPY------HYIPVSECGKITAE--TSKTEVK-LEEGGLVETTETQVEA-SGTSIKESVQ 349
Query: 356 DVVIGGSGYIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIA 415
D+VI G ++VKDV T++QA+ PVEKG+T+ + +H+ + + K+ + ++ +
Sbjct: 350 DIVIDGGQHVVKDVVLTINQAIGPVEKGVTKIQDTIHQ--KLLDSDGKEETEVTEEVTVE 407
Query: 416 SSSPAR 421
+S P +
Sbjct: 408 TSVPPK 413
>gb|AAU89247.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 463
Score = 330 bits (845), Expect = 8e-89
Identities = 177/393 (45%), Positives = 253/393 (64%), Gaps = 17/393 (4%)
Query: 7 LHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVS 66
LHT V +V + F+L+ L +S++L+ +HL +Y NPEEQK++I V+ MVP Y+ ES +S
Sbjct: 17 LHTSVV---LVGAAFVLVALLVSLWLILQHLRSYSNPEEQKWIIAVLFMVPVYASESIIS 73
Query: 67 LVNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHH 126
L + S+ C ILR+CYE+FA+Y FGRYLVACLGGE + +E + R LL
Sbjct: 74 LWHSEFSLACDILRNCYEAFALYAFGRYLVACLGGERQVFRLLENKKREELTEQLL---E 130
Query: 127 SHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCE 186
S D N +F P L R Y I+KFG+VQYMI+KS + ++ ILE FG Y +
Sbjct: 131 SQDKAPVRNRSRVHIFFWDPNALGERLYTIIKFGLVQYMILKSLCAFLSSILELFGKYGD 190
Query: 187 GEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQ 246
GEFK GYPYIAVV+NFSQ+WALYCLV+FY T ++L I+PLAKF++FK+IVF TWWQ
Sbjct: 191 GEFKWYYGYPYIAVVINFSQTWALYCLVKFYNATHEKLQEIRPLAKFISFKAIVFATWWQ 250
Query: 247 GVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYELMGDRHP 306
G+ IA++ G+ + + + ++++QDF+ICIEM IA++ H +VF +PY+ +
Sbjct: 251 GLGIAIICHIGI----LPKEGKVQNAIQDFLICIEMAIAAVAHAFVFNVEPYQHIPVVEH 306
Query: 307 GSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIGGSGYIV 366
G I+ + + +D D+ + PT + T V+A G +I+ESV+DVVIGG ++V
Sbjct: 307 GEIT-SEESKLEVKVDSDD-DSNGTPTTIEEKETHVEA-PGTSIKESVQDVVIGGGHHVV 363
Query: 367 KDVKFTVHQAVEPVEKGIT----RFNEKLHRIS 395
KDV T+ QA+ PVEKG+ + + H IS
Sbjct: 364 KDVALTISQAIGPVEKGVEKGVGKIQDTFHHIS 396
>pir||H86364 hypothetical protein T26J12.15 - Arabidopsis thaliana
gi|2829904|gb|AAC00612.1| Hypothetical protein
[Arabidopsis thaliana]
Length = 379
Score = 269 bits (688), Expect = 1e-70
Identities = 162/421 (38%), Positives = 240/421 (56%), Gaps = 60/421 (14%)
Query: 7 LHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVS 66
LH P + I+ F + + LS+Y + +HL Y NP + +S
Sbjct: 12 LHLPSL---IIGGSFATVAICLSLYSILQHLRFYTNP-------------------AIIS 49
Query: 67 LVNPSISVDCGILRDCYESFAMYCFGRYLVACLG---GEERTIEFMERQGRSASKTPLLL 123
L N S+ C ILR+CYE+FA+Y FG YLVACLG GE R +E++E + SK PLL
Sbjct: 50 LSNSKFSLPCDILRNCYEAFALYSFGSYLVACLGELCGERRVVEYLENE----SKKPLLE 105
Query: 124 HHHSHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQY---MIIKSFTSIMAVILEA 180
+ K + F K+ P++L R + I KFG+VQY MI+K+F + + +LE
Sbjct: 106 EGANESKKKKKKNSF-WKFLCDPYVLGRELFVIEKFGLVQYVSQMILKTFCAFLTFLLEL 164
Query: 181 FGVYCEGEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIV 240
GVY +GEFK G Q WAL+CLVQFY VT + L IKPLAKF++FK+IV
Sbjct: 165 LGVYGDGEFKWYYG-----------QMWALFCLVQFYNVTHERLKEIKPLAKFISFKAIV 213
Query: 241 FLTWWQGVAIALLYTFGLFKSPIAQGLQSKSSVQDFIICIEMGIASIVHLYVFPAKPYEL 300
F TWWQG IALL +G+ + + + ++ +QDF+ICIEM IA++ HL+VFPA+PY
Sbjct: 214 FATWWQGFGIALLCYYGI----LPKEGRFQNGLQDFLICIEMAIAAVAHLFVFPAEPY-- 267
Query: 301 MGDRHPGSISVLGDYSADCPLDPDEIRDSERPTKLRLPTTDVDAKSGMTIRESVRDVVIG 360
H +S G +A+ E++ E + T V+A SG +I+ESV+D+VI
Sbjct: 268 ----HYIPVSECGKITAE--TSKTEVK-LEEGGLVETTETQVEA-SGTSIKESVQDIVID 319
Query: 361 GSGYIVKDVKFTVHQAVEPVEKGITRFNEKLHRISQNIKKHDKDRRRIKDDSCIASSSPA 420
G ++VKDV T++QA+ PVEKG+T+ + +H+ + + K+ + ++ + +S P
Sbjct: 320 GGQHVVKDVVLTINQAIGPVEKGVTKIQDTIHQ--KLLDSDGKEETEVTEEVTVETSVPP 377
Query: 421 R 421
+
Sbjct: 378 K 378
>ref|NP_663574.2| transmembrane protein 34 [Mus musculus] gi|26329069|dbj|BAC28273.1|
unnamed protein product [Mus musculus]
Length = 622
Score = 190 bits (482), Expect = 1e-46
Identities = 108/286 (37%), Positives = 162/286 (55%), Gaps = 23/286 (8%)
Query: 17 VASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSISVDC 76
+A +FLL+T+ +S++ + +HL Y PE QK +I ++ MVP YS++S+V+LV P I++
Sbjct: 51 IAGIFLLLTIPVSLWGILQHLVHYTQPELQKPIIRILWMVPIYSVDSWVALVYPKIAIYV 110
Query: 77 GILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDYKGTVNH 136
R+CYE++ +Y F +L L TI F L+LH + D + NH
Sbjct: 111 DTWRECYEAYVIYNFMIFLTNYL-----TIRFPN----------LILHLEAKDQQ---NH 152
Query: 137 PFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFKLGCGYP 196
PL PW + K G++QY +++ T++ A++ E VY EG F +
Sbjct: 153 ILPL-CCCPPWAMGEMLLFRCKLGVLQYTVVRPITTVTALVCEILDVYDEGNFGFSNAWT 211
Query: 197 YIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLYTF 256
Y+ ++ N SQ +A+YCL+ FY V K+EL+ I+P+ KFL K +VF+++WQ V IALL
Sbjct: 212 YLVILNNLSQLFAMYCLLLFYKVLKEELSPIQPVGKFLCVKLVVFVSFWQAVLIALLVKL 271
Query: 257 GLFKSPIAQGLQSKSSV----QDFIICIEMGIASIVHLYVFPAKPY 298
G+ QS +V QDFIICIEM A+I H Y F KPY
Sbjct: 272 GVISEKRTWEWQSAEAVATGLQDFIICIEMFFAAIAHHYTFSYKPY 317
>gb|AAH04056.1| Similar to CG5850 gene product [Mus musculus]
Length = 622
Score = 190 bits (482), Expect = 1e-46
Identities = 108/286 (37%), Positives = 162/286 (55%), Gaps = 23/286 (8%)
Query: 17 VASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSISVDC 76
+A +FLL+T+ +S++ + +HL Y PE QK +I ++ MVP YS++S+V+LV P I++
Sbjct: 51 IAGIFLLLTIPVSLWGILQHLVHYTQPELQKPIIRILWMVPIYSVDSWVALVYPKIAIYV 110
Query: 77 GILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDYKGTVNH 136
R+CYE++ +Y F +L L TI F L+LH + D + NH
Sbjct: 111 DTWRECYEAYVIYNFMIFLTNYL-----TIRFPN----------LILHLEAKDQQ---NH 152
Query: 137 PFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFKLGCGYP 196
PL PW + K G++QY +++ T++ A++ E VY EG F +
Sbjct: 153 ILPL-CCCPPWAMGEMLLFRCKLGVLQYTVVRPITTVTALVCEILDVYDEGNFGFSNAWT 211
Query: 197 YIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLYTF 256
Y+ ++ N SQ +A+YCL+ FY V K+EL+ I+P+ KFL K +VF+++WQ V IALL
Sbjct: 212 YLVILNNLSQLFAMYCLLLFYKVLKEELSPIQPVGKFLCVKLVVFVSFWQAVLIALLVKL 271
Query: 257 GLFKSPIAQGLQSKSSV----QDFIICIEMGIASIVHLYVFPAKPY 298
G+ QS +V QDFIICIEM A+I H Y F KPY
Sbjct: 272 GVISEKRTWEWQSAEAVATGLQDFIICIEMFFAAIAHHYTFSYKPY 317
>gb|AAO73557.1| hypothetical protein FLJ10846-like protein [Rattus norvegicus]
gi|31088932|ref|NP_847900.1| hypothetical protein
LOC291946 [Rattus norvegicus] gi|57920998|gb|AAH89112.1|
DNA sequence AY228474 [Rattus norvegicus]
Length = 503
Score = 189 bits (479), Expect = 2e-46
Identities = 112/296 (37%), Positives = 166/296 (55%), Gaps = 26/296 (8%)
Query: 7 LHTPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVS 66
+HT VW +A +FLL+T+ +SM + +HL Y PE QK +I ++ MVP YS++S+V+
Sbjct: 44 IHTK-VW--FIAGIFLLLTIPVSMCGILQHLVHYTQPELQKPIIRILWMVPIYSLDSWVA 100
Query: 67 LVNPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHH 126
L P I++ R+CYE++ +Y F +L L TI F L+LH
Sbjct: 101 LKYPKIAIYVDTWRECYEAYVIYNFMIFLTNYL-----TIRFPN----------LMLHLE 145
Query: 127 SHDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCE 186
+ D + NH PL PW + K G++QY +++ T++ +++ E GVY E
Sbjct: 146 AKDQQ---NHLPPL-CCCPPWAMGEMLLFRCKLGVLQYTVVRPITTVTSLVCEILGVYDE 201
Query: 187 GEFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQ 246
G F + Y+ ++ N SQ +A+YCL+ FY V K+EL+ I+P+ KFL K +VF+++WQ
Sbjct: 202 GNFSFSNAWTYLVILNNLSQLFAMYCLLLFYKVLKEELSPIQPVGKFLCVKLVVFVSFWQ 261
Query: 247 GVAIALLYTFGLFKSPIAQGLQSKSSV----QDFIICIEMGIASIVHLYVFPAKPY 298
V IALL G+ QS +V QDFIICIEM A+I H Y F KPY
Sbjct: 262 AVLIALLVKVGVISEKRTWEWQSAEAVATGLQDFIICIEMFFAAIAHHYTFSYKPY 317
>ref|NP_609334.1| CG5850-PA [Drosophila melanogaster] gi|22946075|gb|AAF52840.2|
CG5850-PA [Drosophila melanogaster]
gi|15291945|gb|AAK93241.1| LD32366p [Drosophila
melanogaster]
Length = 491
Score = 181 bits (460), Expect = 3e-44
Identities = 99/288 (34%), Positives = 161/288 (55%), Gaps = 23/288 (7%)
Query: 16 IVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSISVD 75
++ +F+L + +S++ + +H+ + P QK +I ++ MVP Y++ +++ L P S+
Sbjct: 53 LIGGLFVLSAVPVSIWHIIQHVIHFTKPILQKHIIRILWMVPIYALNAWIGLFFPKHSIY 112
Query: 76 CGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDYKGTVN 135
LR+CYE++ +Y F YL+ L G T +YK V
Sbjct: 113 VDSLRECYEAYVIYNFMVYLLNYLN-----------LGMDLEATM--------EYKPQVP 153
Query: 136 HPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFKLGCGY 195
H FPL ++PW++ R F K GI+QY +++ T+ ++VI E GVY EGEF +
Sbjct: 154 HFFPL-CCMRPWVMGREFIHNCKHGILQYTVVRPITTFISVICELCGVYGEGEFAGNVAF 212
Query: 196 PYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLYT 255
PYI VV N SQ A+YCLV FY K++L +KP+ KFL K++VF +++QGV + +L
Sbjct: 213 PYIVVVNNISQFVAMYCLVLFYRANKEDLKPMKPIPKFLCIKAVVFFSFFQGVLLNVLVY 272
Query: 256 FGLFKSPIAQGL---QSKSSVQDFIICIEMGIASIVHLYVFPAKPYEL 300
+ + K + S +Q+F+ICIEM IA++ H+Y FP P+ +
Sbjct: 273 YNIIKDIFGSDVGDTNLASLLQNFLICIEMFIAAVAHIYSFPHHPFHI 320
>ref|XP_641697.1| hypothetical protein DDB0205844 [Dictyostelium discoideum]
gi|60469728|gb|EAL67716.1| hypothetical protein
DDB0205844 [Dictyostelium discoideum]
Length = 351
Score = 177 bits (449), Expect = 7e-43
Identities = 107/280 (38%), Positives = 163/280 (58%), Gaps = 23/280 (8%)
Query: 16 IVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLVNPSISVD 75
IVA V + + LS YL+++HL Y NPE QK+++ +++MVP YS++S++SL +S+
Sbjct: 3 IVAGVCSGVAILLSFYLIYKHLRNYTNPELQKYIVRILIMVPIYSVDSWLSLRFVELSLY 62
Query: 76 CGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDYKGTVN 135
++RD YE++ +YCF +VA + ER + +E LLH K +
Sbjct: 63 FDVVRDTYEAYVLYCFFSLIVAYI---ERDFDLVE-----------LLHS-----KEPLP 103
Query: 136 HPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFKLGCGY 195
HPFPL K L R F K ++Q++ IK +I++++LE Y EG+F++G GY
Sbjct: 104 HPFPLTCLPKIK-LDRGFLTNCKRFVLQFVFIKPIVAIISLVLETQHKYGEGKFQVGTGY 162
Query: 196 PYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLYT 255
++ VV N S +LY LV +Y ++EL KPL KFL KSI+F ++WQ +AI+ L
Sbjct: 163 VWLTVVENISVGLSLYFLVLYYKAMEEELKPFKPLGKFLCIKSILFFSFWQSIAISFLVY 222
Query: 256 FGLFKSPIAQGL--QSKSSVQDFIICIEMGIASIVHLYVF 293
FG+ SPI S++QDFI C+EM I +I H + F
Sbjct: 223 FGVI-SPIGSWSVDNISSALQDFITCVEMVILAICHHFFF 261
>emb|CAG11067.1| unnamed protein product [Tetraodon nigroviridis]
Length = 411
Score = 176 bits (447), Expect = 1e-42
Identities = 99/295 (33%), Positives = 160/295 (53%), Gaps = 25/295 (8%)
Query: 9 TPPVWATIVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESFVSLV 68
+P A +A +F+ MT+ +S++ + +HL Y PE QK +I ++ MVP YS++S+++L
Sbjct: 41 SPHKKAWFIAGIFVFMTIPISLWGILQHLVHYTQPELQKPIIRILWMVPIYSLDSWIALK 100
Query: 69 NPSISVDCGILRDCYESFAMYCFGRYLVACLGGEERTIEFM-ERQGRSASKTPLLLHHHS 127
PSI++ R+CYE++ +Y F +L+ L + ++ M E Q + PL
Sbjct: 101 YPSIAIYVDTCRECYEAYVIYNFMTFLLNYLENQYPSLVLMLEVQEQQKHLPPLCC---- 156
Query: 128 HDYKGTVNHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEG 187
PW + K G++QY +++ T+++A+I + VY EG
Sbjct: 157 ----------------CPPWPMGEVLLWRCKLGVLQYTVVRPVTTVIALICQLCHVYDEG 200
Query: 188 EFKLGCGYPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQG 247
F + Y+ +V N SQ +A+YCLV FY ++EL IKP+ KFL K +VF+++WQ
Sbjct: 201 NFSSNNAWTYLVIVNNMSQLFAMYCLVLFYRTLREELGPIKPVGKFLCVKMVVFVSFWQA 260
Query: 248 VAIALLYTFGLFKSPIAQGLQSKSSV----QDFIICIEMGIASIVHLYVFPAKPY 298
V IALL G+ +S +V QDF+IC+EM +A+I H + F KPY
Sbjct: 261 VFIALLVKVGIISESHTWDWKSVEAVATGLQDFVICVEMFLAAIAHHFSFTYKPY 315
>ref|XP_624809.1| PREDICTED: similar to CG5850-PA [Apis mellifera]
Length = 421
Score = 175 bits (444), Expect = 2e-42
Identities = 102/289 (35%), Positives = 162/289 (55%), Gaps = 28/289 (9%)
Query: 16 IVASVFLLMTLALSMYLLFEHLSAYKNPEEQKFLIGVILMVPCYSIESF-VSLVNPSISV 74
+V F+L+ L ++ Y + +H+ Y P QK++I ++ MVP Y++ + + L + SI V
Sbjct: 48 LVGGAFVLLALPIAFYEIVQHMIYYTQPRLQKYIIRILWMVPIYAVNAVGIHLHSCSIYV 107
Query: 75 DCGILRDCYESFAMYCFGRYLVACLGGEERTIEFMERQGRSASKTPLLLHHHSHDYKGTV 134
D LR+CYE++ +Y F YL+A L + + H + V
Sbjct: 108 DS--LRECYEAYVIYNFMMYLLAYLDADRQL-------------------EHRLEISPQV 146
Query: 135 NHPFPLKYFLKPWILTRRFYQIVKFGIVQYMIIKSFTSIMAVILEAFGVYCEGEFKLGCG 194
+H FPL L W + R F + K GI+QY ++ T++++ I E GVY EGEF+
Sbjct: 147 HHMFPL-CCLPDWEMGREFVHMCKHGILQYTAVRPITTLISFICELNGVYGEGEFRTDVA 205
Query: 195 YPYIAVVLNFSQSWALYCLVQFYTVTKDELAHIKPLAKFLTFKSIVFLTWWQGVAIALLY 254
+PY+ + N SQ A+YCLV FY + L +KP+ KFL K++VF +++QGV IALL
Sbjct: 206 FPYMIALNNLSQFVAMYCLVLFYRANAEALKPMKPIGKFLCIKAVVFFSFFQGVIIALLV 265
Query: 255 TF----GLFKSPIAQGLQSKSS-VQDFIICIEMGIASIVHLYVFPAKPY 298
F +F + + +++ SS +QDF+ICIEM +A++ H Y F KP+
Sbjct: 266 YFNVISNIFNTNDIKDIRNISSKLQDFLICIEMFMAAVAHHYSFSYKPF 314
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 816,818,372
Number of Sequences: 2540612
Number of extensions: 34198346
Number of successful extensions: 80468
Number of sequences better than 10.0: 134
Number of HSP's better than 10.0 without gapping: 118
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 80026
Number of HSP's gapped (non-prelim): 206
length of query: 482
length of database: 863,360,394
effective HSP length: 132
effective length of query: 350
effective length of database: 527,999,610
effective search space: 184799863500
effective search space used: 184799863500
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0117b.4


