

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.8
(935 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
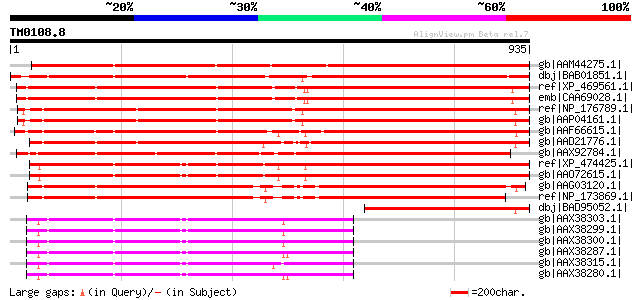
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM44275.1| receptor-like kinase RHG4 [Glycine max] gi|265185... 1368 0.0
dbj|BAB01851.1| unnamed protein product [Arabidopsis thaliana] g... 1123 0.0
ref|XP_469561.1| gibberellin-induced receptor-like kinase TMK [O... 892 0.0
emb|CAA69028.1| TMK [Oryza sativa (indica cultivar-group)] gi|74... 890 0.0
ref|NP_176789.1| leucine-rich repeat protein kinase, putative (T... 888 0.0
gb|AAP04161.1| putative receptor protein kinase (TMK1) [Arabidop... 887 0.0
gb|AAF66615.1| LRR receptor-like protein kinase [Nicotiana tabacum] 878 0.0
gb|AAD21776.1| putative receptor-like protein kinase [Arabidopsi... 872 0.0
gb|AAX92784.1| receptor-like kinase RHG4 [Oryza sativa (japonica... 785 0.0
ref|XP_474425.1| OSJNBa0088H09.21 [Oryza sativa (japonica cultiv... 785 0.0
gb|AAO72615.1| receptor-like protein kinase-like protein [Oryza ... 780 0.0
gb|AAG03120.1| F5A9.23 [Arabidopsis thaliana] 780 0.0
ref|NP_173869.1| leucine-rich repeat family protein / protein ki... 777 0.0
dbj|BAD95052.1| putative receptor-like protein kinase [Arabidops... 395 e-108
gb|AAX38303.1| receptor-like protein kinase [Solanum habrochaites] 374 e-102
gb|AAX38299.1| receptor-like protein kinase [Solanum habrochaite... 374 e-102
gb|AAX38300.1| receptor-like protein kinase [Solanum habrochaites] 374 e-102
gb|AAX38287.1| receptor-like protein kinase [Lycopersicon peruvi... 374 e-102
gb|AAX38315.1| receptor-like protein kinase [Lycopersicon chmiel... 374 e-101
gb|AAX38280.1| receptor-like protein kinase [Lycopersicon peruvi... 374 e-101
>gb|AAM44275.1| receptor-like kinase RHG4 [Glycine max] gi|26518502|gb|AAN80746.1|
receptor-like kinase RHG4 [Glycine max]
Length = 893
Score = 1368 bits (3542), Expect = 0.0
Identities = 694/899 (77%), Positives = 768/899 (85%), Gaps = 9/899 (1%)
Query: 40 MSKLLKSLSPPPSDWSSTTPFCQWDGIKCDSSNRVTTISLASRSLTGTLPSDLNSLSQLT 99
MS LKSL+PPPS WS TTPFCQW GI+CDSS+ VT+ISLAS+SLTGTLPSDLNSLSQL
Sbjct: 1 MSNFLKSLTPPPSGWSETTPFCQWKGIQCDSSSHVTSISLASQSLTGTLPSDLNSLSQLR 60
Query: 100 SLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGLTDLQTLSLSDNPNLSPW 159
+LSLQ+N+++G +PSL+NLS L+T + RNNF+SV +FA LT LQTLSL NP L PW
Sbjct: 61 TLSLQDNSLTGTLPSLSNLSFLQTVYFNRNNFSSVSPTAFASLTSLQTLSLGSNPALQPW 120
Query: 160 TLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVRLSYNNLTGALPNSL-AASA 218
+ PT+LT S+NLI L+L T LTG LP+ FDKFP LQ +RLSYNNLTG LP+S AA+
Sbjct: 121 SFPTDLTSSSNLIDLDLATVSLTGPLPD-IFDKFPSLQHLRLSYNNLTGNLPSSFSAANN 179
Query: 219 IENLWLNNQDNGLSGTIDVLSNMTQLAQVWLHKNQFTGPIPDLSQCSNLFDLQLRDNQLT 278
+E LWLNNQ GLSGT+ VLSNM+ L Q WL+KNQFTG IPDLSQC+ L DLQLRDNQLT
Sbjct: 180 LETLWLNNQAAGLSGTLLVLSNMSALNQSWLNKNQFTGSIPDLSQCTALSDLQLRDNQLT 239
Query: 279 GPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDGINSFCKDTPGPCDARVMVLL 338
G VP SL L SL+ VSLDNNELQGP P FGKGV VTLDGINSFC DTPG CD RVMVLL
Sbjct: 240 GVVPASLTSLPSLKKVSLDNNELQGPVPVFGKGVNVTLDGINSFCLDTPGNCDPRVMVLL 299
Query: 339 HIAGAFGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQGLQGTISPAFANLTDL 398
IA AFGYPI+ A SWKGNDPC GW++VVC +G KIITVN KQGLQGTISPAFANLTDL
Sbjct: 300 QIAEAFGYPIRLAESWKGNDPCDGWNYVVCAAG-KIITVNFEKQGLQGTISPAFANLTDL 358
Query: 399 RSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKVKLLTAGNVLLGQTTG 458
R+L+LNGNNL GSIP+SL TL QL+TL+VSDNNLSG VPKFPPKVKL+TAGN LLG+
Sbjct: 359 RTLFLNGNNLIGSIPDSLITLPQLQTLDVSDNNLSGLVPKFPPKVKLVTAGNALLGKPLS 418
Query: 459 SGGGAGGKTTPSAGSTPEGSHGESGNG-SSLTPGWIAGIVIIVLFFVAVVLFVSCKCYAK 517
GGG G TTPS GS+ GS GES G SS++PGWIAGIV+IVLFF+AVVLFVS KC+
Sbjct: 419 PGGGPSG-TTPS-GSSTGGSGGESSKGNSSVSPGWIAGIVVIVLFFIAVVLFVSWKCFVN 476
Query: 518 RRHGKFSRVANPVNGNGNVKLDVISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSI 577
+ GKFSRV NG G KLD + VSNGY G P ELQSQSSGD S+LH DG T SI
Sbjct: 477 KLQGKFSRVKGHENGKGGFKLDAVHVSNGYGGVPVELQSQSSGDRSDLHALDG--PTFSI 534
Query: 578 LVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVL 637
VL+QVT NFSE+NILGRGGFGVVYKG+L DGTKIAVKRMESVAMGNKGL EF+AEI VL
Sbjct: 535 QVLQQVTNNFSEENILGRGGFGVVYKGQLHDGTKIAVKRMESVAMGNKGLKEFEAEIAVL 594
Query: 638 SKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDV 697
SKVRHRHLVALLG+CING ERLLVYEYMPQGTLTQHLFEW+E GY PLTWKQRV +ALDV
Sbjct: 595 SKVRHRHLVALLGYCINGIERLLVYEYMPQGTLTQHLFEWQEQGYVPLTWKQRVVIALDV 654
Query: 698 ARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFG 757
ARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFG
Sbjct: 655 ARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFG 714
Query: 758 YLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENI 817
YLAPEYAATGRVTTKVD+YAFG+VLMELITGR+ALDD++PDERSHLVTWFRRVLINKENI
Sbjct: 715 YLAPEYAATGRVTTKVDIYAFGIVLMELITGRKALDDTVPDERSHLVTWFRRVLINKENI 774
Query: 818 PKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSR-H 876
PKAIDQTLNPDEETMESIYKV+ELAGHCTAREP QRPDMGHAVNVLVP+VEQWKP+S
Sbjct: 775 PKAIDQTLNPDEETMESIYKVAELAGHCTAREPYQRPDMGHAVNVLVPLVEQWKPSSHDE 834
Query: 877 EDDGHDSEPHMSLPQVLQRWQANEGTSTIFNDMSLSQTQSSINSKTYGFADSFDSLDCR 935
E+DG + MSLPQ L+RWQANEGTS+IFND+S+SQTQSSI+SK GFAD+FDS+DCR
Sbjct: 835 EEDGSGGDLQMSLPQALRRWQANEGTSSIFNDISISQTQSSISSKPVGFADTFDSMDCR 893
>dbj|BAB01851.1| unnamed protein product [Arabidopsis thaliana]
gi|15229508|ref|NP_189017.1| leucine-rich repeat family
protein / protein kinase family protein [Arabidopsis
thaliana]
Length = 928
Score = 1123 bits (2904), Expect = 0.0
Identities = 586/955 (61%), Positives = 706/955 (73%), Gaps = 51/955 (5%)
Query: 2 PHPKTLLSLSTLSLLFSITIIIAADDGPVFAATEDAAVMSKLLKSLSPPPSDWSSTTPFC 61
P P LL L T F+ ++ +D M L KS +PPPSDWSSTT FC
Sbjct: 4 PTPLLLLVLLTTITFFTTSV------------ADDQTAMLALAKSFNPPPSDWSSTTDFC 51
Query: 62 QWDGIKCDSSNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSAL 121
+W G++C + RVTTISLA +SLTG + ++++LS+L S+S+Q N +SG IPS A LS+L
Sbjct: 52 KWSGVRC-TGGRVTTISLADKSLTGFIAPEISTLSELKSVSIQRNKLSGTIPSFAKLSSL 110
Query: 122 KTAFLGRNNFTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARL 181
+ ++ NNF V + +FAGLT LQ LSLSDN N++ W+ P+EL ST+L T+ L +
Sbjct: 111 QEIYMDENNFVGVETGAFAGLTSLQILSLSDNNNITTWSFPSELVDSTSLTTIYLDNTNI 170
Query: 182 TGQLPESFFDKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQDNGLSGTIDVLSNM 241
G LP+ FD LQ++RLSYNN+TG LP SL S+I+NLW+NNQD G+SGTI+VLS+M
Sbjct: 171 AGVLPD-IFDSLASLQNLRLSYNNITGVLPPSLGKSSIQNLWINNQDLGMSGTIEVLSSM 229
Query: 242 TQLAQVWLHKNQFTGPIPDLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNEL 301
T L+Q WLHKN F GPIPDLS+ NLFDLQLRDN LTG VP +L+ L SL+N+SLDNN+
Sbjct: 230 TSLSQAWLHKNHFFGPIPDLSKSENLFDLQLRDNDLTGIVPPTLLTLASLKNISLDNNKF 289
Query: 302 QGPFPAFGKGVKVTLDGINSFCKDTPGP-CDARVMVLLHIAGAFGYPIKFAGSWKGNDPC 360
QGP P F VKVT+D N FC G C +VM LL +AG GYP A SW+G+D C
Sbjct: 290 QGPLPLFSPEVKVTIDH-NVFCTTKAGQSCSPQVMTLLAVAGGLGYPSMLAESWQGDDAC 348
Query: 361 QGWSFVVCDS-GRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTL 419
GW++V CDS G+ ++T+NL K G G ISPA ANLT L+SLYLNGN+LTG IP+ LT +
Sbjct: 349 SGWAYVSCDSAGKNVVTLNLGKHGFTGFISPAIANLTSLKSLYLNGNDLTGVIPKELTFM 408
Query: 420 TQLETLEVSDNNLSGEVPKFPPKVKL-LTAGNVLLGQTTGSGGGAGGKTTPSAGSTPEGS 478
T L+ ++VS+NNL GE+PKFP VK GN LLG G G ++P G G
Sbjct: 409 TSLQLIDVSNNNLRGEIPKFPATVKFSYKPGNALLGTNGGDG------SSPGTGGASGGP 462
Query: 479 HGESGNGSSLTPGWIAGIVIIVLFFVAVVLFVSCKCYAKRRHGKFSR------------- 525
G SG G S G I G+++ VL F+A++ FV K KR++G+F+R
Sbjct: 463 GGSSGGGGSKV-GVIVGVIVAVLVFLAILGFVVYKFVMKRKYGRFNRTDPEKVGKILVSD 521
Query: 526 -VANPVNGNGNVKLDVISVSNGYSGAP-SELQSQSSGDHSELHVFDGGNSTMSILVLRQV 583
V+N +GNG +NG+ + L S SSGD+S+ + +GG+ T+ + VLRQV
Sbjct: 522 AVSNGGSGNGGY-------ANGHGANNFNALNSPSSGDNSDRFLLEGGSVTIPMEVLRQV 574
Query: 584 TGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHR 643
T NFSEDNILGRGGFGVVY GEL DGTK AVKRME AMGNKG++EFQAEI VL+KVRHR
Sbjct: 575 TNNFSEDNILGRGGFGVVYAGELHDGTKTAVKRMECAAMGNKGMSEFQAEIAVLTKVRHR 634
Query: 644 HLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEY 703
HLVALLG+C+NGNERLLVYEYMPQG L QHLFEW ELGY+PLTWKQRV++ALDVARGVEY
Sbjct: 635 HLVALLGYCVNGNERLLVYEYMPQGNLGQHLFEWSELGYSPLTWKQRVSIALDVARGVEY 694
Query: 704 LHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEY 763
LHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEY
Sbjct: 695 LHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEY 754
Query: 764 AATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQ 823
AATGRVTTKVDVYAFGVVLME++TGR+ALDDSLPDERSHLVTWFRR+LINKENIPKA+DQ
Sbjct: 755 AATGRVTTKVDVYAFGVVLMEILTGRKALDDSLPDERSHLVTWFRRILINKENIPKALDQ 814
Query: 824 TLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDS 883
TL DEETMESIY+V+ELAGHCTAREP QRPDMGHAVNVL P+VE+WKP+ + E++
Sbjct: 815 TLEADEETMESIYRVAELAGHCTAREPQQRPDMGHAVNVLGPLVEKWKPSCQEEEESFGI 874
Query: 884 EPHMSLPQVLQRWQANEGT--STIFN-DMSLSQTQSSINSKTYGFADSFDSLDCR 935
+ +MSLPQ LQRWQ NEGT ST+F+ D S SQTQSSI K GF ++FDS D R
Sbjct: 875 DVNMSLPQALQRWQ-NEGTSSSTMFHGDFSYSQTQSSIPPKASGFPNTFDSADGR 928
>ref|XP_469561.1| gibberellin-induced receptor-like kinase TMK [Oryza sativa
(japonica cultivar-group)] gi|28301932|gb|AAO38825.1|
gibberellin-induced receptor-like kinase TMK [Oryza
sativa (japonica cultivar-group)]
Length = 962
Score = 892 bits (2304), Expect = 0.0
Identities = 482/952 (50%), Positives = 638/952 (66%), Gaps = 37/952 (3%)
Query: 13 LSLLFSITIIIAADDGPVFAATEDAAVMSKLLKSLSPPPS-DWSSTTPFCQ---WDGIKC 68
+++ ++ +++ A G A DAA M + ++L + WS+ P W G+ C
Sbjct: 19 VAVALAVMLVVGAAAGET--AASDAAAMRAVARALGADKALGWSTGDPCSSPRAWAGVTC 76
Query: 69 DSSNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGR 128
DS+ RVT + + +RSLTG L ++ +L+ L L L +N+ISG +PSLA LS+L+ +
Sbjct: 77 DSAGRVTAVQVGNRSLTGRLAPEVRNLTALARLELFDNSISGELPSLAGLSSLQYLLVHN 136
Query: 129 NNFTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPES 188
N FT +P F GLT L +SL +NP PW LP +L T+L TA +TG LP+
Sbjct: 137 NGFTRIPPDFFKGLTALAAVSLDNNP-FDPWPLPADLADCTSLTNFSANTANVTGALPDF 195
Query: 189 FFDKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQ--DNGLSGTIDVLSNMTQLAQ 246
F P LQ + L++N ++G +P SLA + ++ LWLNNQ +N +G+I +SNMT L +
Sbjct: 196 FGTALPSLQRLSLAFNKMSGPVPASLATAPLQALWLNNQIGENQFNGSISFISNMTSLQE 255
Query: 247 VWLHKNQFTGPIPDLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFP 306
+WLH N FTGP+PD S ++L DL+LRDNQLTGPVP+SL+ L SL V+L NN LQGP P
Sbjct: 256 LWLHSNDFTGPLPDFSGLASLSDLELRDNQLTGPVPDSLLKLGSLTKVTLTNNLLQGPTP 315
Query: 307 AFGKGVKV-TLDGINSFCKDTPG-PCDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWS 364
F VK + FC TPG PCD RV +LL +A F YP K A +WKGNDPC G+
Sbjct: 316 KFADKVKADVVPTTERFCLSTPGQPCDPRVNLLLEVAAEFQYPAKLADNWKGNDPCDGYI 375
Query: 365 FVVCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLET 424
V CD+G I +N A+ G G+ISPA +T L+ L L NN+TG++P+ + L L
Sbjct: 376 GVGCDAGN-ITVLNFARMGFSGSISPAIGKITTLQKLILADNNITGTVPKEVAALPALTE 434
Query: 425 LEVSDNNLSGEVPKFPPKVKLLTA-GNVLLGQTT---GSGGGAGGKTTPSAGSTPEGSHG 480
+++S+NNL G++P F K L+ A GN +G+ GG+GG P G+ G G
Sbjct: 435 VDLSNNNLYGKLPTFAAKNVLVKANGNPNIGKDAPAPSGSGGSGGSNAPDGGN---GGDG 491
Query: 481 ESGNGSSLTPGWIAGIVIIVLFFVAVVLFVSCKCYAKRRHGKFSRVANP------VNGNG 534
+G+ SS + G IAG V+ + V ++ + CY KR+ F RV +P +G
Sbjct: 492 SNGSPSSSSAGIIAGSVVGAIAGVGLLAALGFYCY-KRKQKPFGRVQSPHAMVVHPRHSG 550
Query: 535 N----VKLDVISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSED 590
+ VK+ V + A SE SQ+S ++HV + GN +SI VLR VT NFS++
Sbjct: 551 SDPDMVKITVAGGNVNGGAAASETYSQASSGPRDIHVVETGNMVISIQVLRNVTNNFSDE 610
Query: 591 NILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLG 650
N+LGRGGFG VYKGEL DGTKIAVKRME+ MGNKGLNEF++EI VL+KVRHR+LV+LLG
Sbjct: 611 NVLGRGGFGTVYKGELHDGTKIAVKRMEAGVMGNKGLNEFKSEIAVLTKVRHRNLVSLLG 670
Query: 651 HCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQ 710
+C++GNER+LVYEYMPQGTL+QHLFEW+E PL WK+R+++ALDVARGVEYLHSLAQQ
Sbjct: 671 YCLDGNERILVYEYMPQGTLSQHLFEWKEHNLRPLEWKKRLSIALDVARGVEYLHSLAQQ 730
Query: 711 SFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGK-YSVETRLAGTFGYLAPEYAATGR 768
+FIHRDLKPSNILLGDDM+AKVADFGLV+ AP DGK SVETRLAGTFGYLAPEYA TGR
Sbjct: 731 TFIHRDLKPSNILLGDDMKAKVADFGLVRLAPADGKCVSVETRLAGTFGYLAPEYAVTGR 790
Query: 769 VTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPD 828
VTTK DV++FGV+LMELITGR+ALD++ P++ HLVTWFRR+ ++K+ KAID T++
Sbjct: 791 VTTKADVFSFGVILMELITGRKALDETQPEDSMHLVTWFRRMQLSKDTFQKAIDPTIDLT 850
Query: 829 EETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMS 888
EET+ S+ V+ELAGHC AREP+QRPDMGHAVNVL + + WKP+ DD + + M+
Sbjct: 851 EETLASVSTVAELAGHCCAREPHQRPDMGHAVNVLSTLSDVWKPSDPDSDDSYGIDLDMT 910
Query: 889 LPQVLQRWQANEGTS-----TIFNDMSLSQTQSSINSKTYGFADSFDSLDCR 935
LPQ L++WQA E +S T SL TQ+SI ++ GFA+SF S D R
Sbjct: 911 LPQALKKWQAFEDSSHFDGATSSFLASLDNTQTSIPTRPPGFAESFTSADGR 962
>emb|CAA69028.1| TMK [Oryza sativa (indica cultivar-group)] gi|7434420|pir||T04124
receptor-like protein kinase (EC 2.7.1.-) - rice
Length = 962
Score = 890 bits (2301), Expect = 0.0
Identities = 481/952 (50%), Positives = 637/952 (66%), Gaps = 37/952 (3%)
Query: 13 LSLLFSITIIIAADDGPVFAATEDAAVMSKLLKSLSPPPS-DWSSTTPFCQ---WDGIKC 68
+++ ++ +++ A G A DAA M + ++L + WS+ P W G+ C
Sbjct: 19 VAVALAVMLVVGAAAGDT--AASDAAAMRAVARALGADKALGWSTGDPCSSPRAWAGVTC 76
Query: 69 DSSNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGR 128
DS+ RVT + + +RSLTG L ++ +L+ L L L +N+ISG +PSLA LS+L+ +
Sbjct: 77 DSAGRVTAVQVGNRSLTGRLAPEVRNLTALARLELFDNSISGELPSLAGLSSLQYLLVHN 136
Query: 129 NNFTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPES 188
N FT +P F GLT L +SL +NP PW LP +L T+L TA +TG LP+
Sbjct: 137 NGFTRIPPDFFKGLTALAAVSLDNNP-FDPWPLPADLADCTSLTNFSANTANVTGALPDF 195
Query: 189 FFDKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQ--DNGLSGTIDVLSNMTQLAQ 246
F P LQ + L++N ++G +P SLA + ++ LWLNNQ +N +G+I +SNMT L +
Sbjct: 196 FGTALPSLQRLSLAFNKMSGPVPASLATAPLQALWLNNQIGENQFNGSISFISNMTSLQE 255
Query: 247 VWLHKNQFTGPIPDLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFP 306
+WLH N FTGP+PD S ++L DL+LRDNQLTGPVP+SL+ L SL V+L NN LQGP P
Sbjct: 256 LWLHSNDFTGPLPDFSGLASLSDLELRDNQLTGPVPDSLLKLGSLTKVTLTNNLLQGPTP 315
Query: 307 AFGKGVKV-TLDGINSFCKDTPG-PCDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWS 364
F VK + FC TPG PCD RV +LL +A F YP K A +WKGNDPC G+
Sbjct: 316 KFADKVKADVVPTTERFCLSTPGQPCDPRVSLLLEVAAGFQYPAKLADNWKGNDPCDGYI 375
Query: 365 FVVCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLET 424
V CD+G I +N A+ G G+ISPA +T L+ L L NN+TG++P+ + L L
Sbjct: 376 GVGCDAGN-ITVLNFARMGFSGSISPAIGKITTLQKLILADNNITGTVPKEVAALPALTE 434
Query: 425 LEVSDNNLSGEVPKFPPKVKLLTA-GNVLLGQTT---GSGGGAGGKTTPSAGSTPEGSHG 480
+++S+NNL G++P F K L+ A GN +G+ GG+GG P G+ G G
Sbjct: 435 VDLSNNNLYGKLPTFAAKNVLVKANGNPNIGKDAPAPSGSGGSGGSNAPDGGN---GGDG 491
Query: 481 ESGNGSSLTPGWIAGIVIIVLFFVAVVLFVSCKCYAKRRHGKFSRVANP------VNGNG 534
+G+ S + G IAG V+ + V ++ + CY KR+ F RV +P +G
Sbjct: 492 SNGSPSPSSAGIIAGSVVGAIAGVGLLAALGFYCY-KRKQKPFGRVQSPHAMVVHPRHSG 550
Query: 535 N----VKLDVISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSED 590
+ VK+ V + A SE SQ+S ++HV + GN +SI VLR VT NFS++
Sbjct: 551 SDPDMVKITVAGGNVNGGAAASETYSQASSGPRDIHVVETGNMVISIQVLRNVTNNFSDE 610
Query: 591 NILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLG 650
N+LGRGGFG VYKGEL DGTKIAVKRME+ MGNKGLNEF++EI VL+KVRHR+LV+LLG
Sbjct: 611 NVLGRGGFGTVYKGELHDGTKIAVKRMEAGVMGNKGLNEFKSEIAVLTKVRHRNLVSLLG 670
Query: 651 HCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQ 710
+C++GNER+LVYEYMPQGTL+QHLFEW+E PL WK+R+++ALDVARGVEYLHSLAQQ
Sbjct: 671 YCLDGNERILVYEYMPQGTLSQHLFEWKEHNLRPLEWKKRLSIALDVARGVEYLHSLAQQ 730
Query: 711 SFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGK-YSVETRLAGTFGYLAPEYAATGR 768
+FIHRDLKPSNILLGDDM+AKVADFGLV+ AP DGK SVETRLAGTFGYLAPEYA TGR
Sbjct: 731 TFIHRDLKPSNILLGDDMKAKVADFGLVRLAPADGKCVSVETRLAGTFGYLAPEYAVTGR 790
Query: 769 VTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPD 828
VTTK DV++FGV+LMELITGR+ALD++ P++ HLVTWFRR+ ++K+ KAID T++
Sbjct: 791 VTTKADVFSFGVILMELITGRKALDETQPEDSMHLVTWFRRMQLSKDTFQKAIDPTIDLT 850
Query: 829 EETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMS 888
EET+ S+ V+ELAGHC AREP+QRPDMGHAVNVL + + WKP+ DD + + M+
Sbjct: 851 EETLASVSTVAELAGHCCAREPHQRPDMGHAVNVLSTLSDVWKPSDPDSDDSYGIDLDMT 910
Query: 889 LPQVLQRWQANEGTS-----TIFNDMSLSQTQSSINSKTYGFADSFDSLDCR 935
LPQ L++WQA E +S T SL TQ+SI ++ GFA+SF S D R
Sbjct: 911 LPQALKKWQAFEDSSHFDGATSSFLASLDNTQTSIPTRPPGFAESFTSADGR 962
>ref|NP_176789.1| leucine-rich repeat protein kinase, putative (TMK1) [Arabidopsis
thaliana] gi|1174718|sp|P43298|TMK1_ARATH Putative
receptor protein kinase TMK1 precursor
gi|12322608|gb|AAG51302.1| receptor protein kinase
(TMK1), putative [Arabidopsis thaliana]
gi|166888|gb|AAA32876.1| protein kinase
Length = 942
Score = 888 bits (2295), Expect = 0.0
Identities = 495/952 (51%), Positives = 624/952 (64%), Gaps = 48/952 (5%)
Query: 15 LLFSITIII------AADDGPVFAATEDAAVMSKLLKSLSPPPS-DWSSTTPFCQWDGIK 67
LLFS T ++ A DG D + M L KSL+PP S WS P C+W I
Sbjct: 8 LLFSFTFLLLLSLSKADSDG-------DLSAMLSLKKSLNPPSSFGWSDPDP-CKWTHIV 59
Query: 68 CDSSNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLG 127
C + RVT I + L GTL DL +LS+L L LQ N ISGP+PSL+ L++L+ L
Sbjct: 60 CTGTKRVTRIQIGHSGLQGTLSPDLRNLSELERLELQWNNISGPVPSLSGLASLQVLMLS 119
Query: 128 RNNFTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPE 187
NNF S+PS F GLT LQ++ + +NP S W +P L ++ L +A ++G LP
Sbjct: 120 NNNFDSIPSDVFQGLTSLQSVEIDNNPFKS-WEIPESLRNASALQNFSANSANVSGSLPG 178
Query: 188 SFF-DKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQDNGLSGTIDVLSNMTQLAQ 246
D+FPGL + L++NNL G LP SLA S +++LWLN Q L+G I VL NMT L +
Sbjct: 179 FLGPDEFPGLSILHLAFNNLEGELPMSLAGSQVQSLWLNGQK--LTGDITVLQNMTGLKE 236
Query: 247 VWLHKNQFTGPIPDLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFP 306
VWLH N+F+GP+PD S L L LRDN TGPVP SL+ L SL+ V+L NN LQGP P
Sbjct: 237 VWLHSNKFSGPLPDFSGLKELESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQGPVP 296
Query: 307 AFGKGVKVTLD-GINSFCKDTPGPCDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWSF 365
F V V LD NSFC +PG CD RV LL IA +F YP + A SWKGNDPC W
Sbjct: 297 VFKSSVSVDLDKDSNSFCLSSPGECDPRVKSLLLIASSFDYPPRLAESWKGNDPCTNWIG 356
Query: 366 VVCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETL 425
+ C +G I ++L K L GTISP F + L+ + L NNLTG IP+ LTTL L+TL
Sbjct: 357 IACSNGN-ITVISLEKMELTGTISPEFGAIKSLQRIILGINNLTGMIPQELTTLPNLKTL 415
Query: 426 EVSDNNLSGEVPKFPPKVKLLTAGNVLLGQTTGSGGGAGGKTTPSAGSTP--EGSHGESG 483
+VS N L G+VP F V + T GN +G+ S G ++PS GS G G
Sbjct: 416 DVSSNKLFGKVPGFRSNVVVNTNGNPDIGKDKSSLSSPGS-SSPSGGSGSGINGDKDRRG 474
Query: 484 NGSSLTPGWIAGIVI---IVLFFVAVVLFVSCKCYAKRRHGKFSR-------VANPVNG- 532
SS G I G V+ + +F + +++F C+ K+R +FS V +P +
Sbjct: 475 MKSSTFIGIIVGSVLGGLLSIFLIGLLVF----CWYKKRQKRFSGSESSNAVVVHPRHSG 530
Query: 533 --NGNVKLDVISVSNGYSGAPSELQSQSSGDHSE-LHVFDGGNSTMSILVLRQVTGNFSE 589
N +VK+ V S G + + + + + + GN +SI VLR VT NFS
Sbjct: 531 SDNESVKITVAGSSVSVGGISDTYTLPGTSEVGDNIQMVEAGNMLISIQVLRSVTNNFSS 590
Query: 590 DNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALL 649
DNILG GGFGVVYKGEL DGTKIAVKRME+ + KG EF++EI VL+KVRHRHLV LL
Sbjct: 591 DNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLL 650
Query: 650 GHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQ 709
G+C++GNE+LLVYEYMPQGTL++HLFEW E G PL WKQR+T+ALDVARGVEYLH LA
Sbjct: 651 GYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDVARGVEYLHGLAH 710
Query: 710 QSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRV 769
QSFIHRDLKPSNILLGDDMRAKVADFGLV+ AP+GK S+ETR+AGTFGYLAPEYA TGRV
Sbjct: 711 QSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIETRIAGTFGYLAPEYAVTGRV 770
Query: 770 TTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKE-NIPKAIDQTLNPD 828
TTKVDVY+FGV+LMELITGR++LD+S P+E HLV+WF+R+ INKE + KAID T++ D
Sbjct: 771 TTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYINKEASFKKAIDTTIDLD 830
Query: 829 EETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMS 888
EET+ S++ V+ELAGHC AREP QRPDMGHAVN+L +VE WKP+ ++ +D + + MS
Sbjct: 831 EETLASVHTVAELAGHCCAREPYQRPDMGHAVNILSSLVELWKPSDQNPEDIYGIDLDMS 890
Query: 889 LPQVLQRWQANEGTSTIFNDM-----SLSQTQSSINSKTYGFADSFDSLDCR 935
LPQ L++WQA EG S + + SL TQ SI ++ YGFA+SF S+D R
Sbjct: 891 LPQALKKWQAYEGRSDLESSTSSLLPSLDNTQMSIPTRPYGFAESFTSVDGR 942
>gb|AAP04161.1| putative receptor protein kinase (TMK1) [Arabidopsis thaliana]
Length = 942
Score = 887 bits (2292), Expect = 0.0
Identities = 494/952 (51%), Positives = 624/952 (64%), Gaps = 48/952 (5%)
Query: 15 LLFSITIII------AADDGPVFAATEDAAVMSKLLKSLSPPPS-DWSSTTPFCQWDGIK 67
LLFS T ++ A DG D + M L KSL+PP S WS P C+W I
Sbjct: 8 LLFSFTFLLLLSLSKADSDG-------DLSAMLSLKKSLNPPSSFGWSDPDP-CKWTHIV 59
Query: 68 CDSSNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLG 127
C + RVT I + L GTL DL +LS+L L LQ N ISGP+PSL+ L++L+ L
Sbjct: 60 CTGTKRVTRIQIGHSGLQGTLSPDLRNLSELERLELQWNNISGPVPSLSGLASLQVLMLS 119
Query: 128 RNNFTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPE 187
NNF S+PS F GLT LQ++ + +NP S W +P L ++ L +A ++G LP
Sbjct: 120 NNNFDSIPSDVFQGLTSLQSVEIDNNPFKS-WEIPESLRNASALQNFSANSANVSGSLPG 178
Query: 188 SFF-DKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQDNGLSGTIDVLSNMTQLAQ 246
D+FPGL + L++NNL G LP SLA S +++LWLN Q L+G I VL NMT L +
Sbjct: 179 FLGPDEFPGLSILHLAFNNLEGELPMSLAGSQVQSLWLNGQK--LTGDITVLQNMTGLKE 236
Query: 247 VWLHKNQFTGPIPDLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFP 306
VWLH N+F+GP+PD S L L LRDN TGPVP SL+ L SL+ V+L NN LQGP P
Sbjct: 237 VWLHSNKFSGPLPDFSGLKELESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQGPVP 296
Query: 307 AFGKGVKVTLD-GINSFCKDTPGPCDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWSF 365
F V V LD NSFC +PG CD RV LL IA +F YP + A SWKGNDPC W
Sbjct: 297 VFKSSVSVDLDKDSNSFCLSSPGECDPRVKSLLLIASSFDYPPRLAESWKGNDPCTNWIG 356
Query: 366 VVCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETL 425
+ C +G I ++L K L GTISP F + L+ + L NNLTG IP+ LTTL L+TL
Sbjct: 357 IACSNGN-ITVISLEKMELTGTISPEFGAIKSLQRIILGINNLTGMIPQELTTLPNLKTL 415
Query: 426 EVSDNNLSGEVPKFPPKVKLLTAGNVLLGQTTGSGGGAGGKTTPSAGSTP--EGSHGESG 483
+VS N L G+VP F V + T GN +G+ S G ++PS GS G G
Sbjct: 416 DVSSNKLFGKVPGFRSNVVVNTNGNPDIGKDKSSLSSPGS-SSPSGGSGSGINGDKDRRG 474
Query: 484 NGSSLTPGWIAGIVI---IVLFFVAVVLFVSCKCYAKRRHGKFSR-------VANPVNG- 532
SS G I G V+ + +F + +++F C+ K+R +FS V +P +
Sbjct: 475 MKSSTFIGIIVGSVLGGLLSIFLIGLLVF----CWYKKRQKRFSGSESSNAVVVHPRHSG 530
Query: 533 --NGNVKLDVISVSNGYSGAPSELQSQSSGDHSE-LHVFDGGNSTMSILVLRQVTGNFSE 589
N +VK+ V S G + + + + + + GN +SI VLR VT NFS
Sbjct: 531 SDNESVKITVAGSSVSVGGISDTYTLPGTSEVGDNIQMVEAGNMLISIQVLRSVTNNFSS 590
Query: 590 DNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALL 649
DNILG GGFGVVYKGEL DGTKIAVKRME+ + KG EF++EI VL+KVRHRHLV LL
Sbjct: 591 DNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLL 650
Query: 650 GHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQ 709
G+C++GNE+LLVYEYMPQGTL++HLFEW E G PL WKQR+T+ALDVARGVEYLH LA
Sbjct: 651 GYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDVARGVEYLHGLAH 710
Query: 710 QSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRV 769
QSFIHRDLKPSNILLGDDMRAKVADFGLV+ AP+GK S+ETR+AGTFGYLAPEYA TGRV
Sbjct: 711 QSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIETRIAGTFGYLAPEYAVTGRV 770
Query: 770 TTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKE-NIPKAIDQTLNPD 828
TTKVDVY+FGV+LMELITGR++LD+S P+E HLV+WF+R+ INKE + KAID T++ D
Sbjct: 771 TTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYINKEASFKKAIDTTIDLD 830
Query: 829 EETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMS 888
EET+ S++ V+ELAGHC +REP QRPDMGHAVN+L +VE WKP+ ++ +D + + MS
Sbjct: 831 EETLASVHTVAELAGHCCSREPYQRPDMGHAVNILSSLVELWKPSDQNPEDIYGIDLDMS 890
Query: 889 LPQVLQRWQANEGTSTIFNDM-----SLSQTQSSINSKTYGFADSFDSLDCR 935
LPQ L++WQA EG S + + SL TQ SI ++ YGFA+SF S+D R
Sbjct: 891 LPQALKKWQAYEGRSDLESSTSSLLPSLDNTQMSIPTRPYGFAESFTSVDGR 942
>gb|AAF66615.1| LRR receptor-like protein kinase [Nicotiana tabacum]
Length = 945
Score = 878 bits (2269), Expect = 0.0
Identities = 488/951 (51%), Positives = 622/951 (65%), Gaps = 40/951 (4%)
Query: 9 SLSTLSLLFSITIIIAADDGPVFAATEDAAVMSKLLKSLSPPPS-DWSSTTPFCQWDGIK 67
S+ L LL + + + +G +A DAAVM +L K ++PP S W+ P C+W ++
Sbjct: 11 SVRLLVLLLYVVSSVYSQEG---SAANDAAVMQELKKRINPPSSLGWNDPDP-CKWGKVQ 66
Query: 68 CDSSNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLG 127
C RVT I + ++ L G+LP +LN+L++L +QNN ++G +PS + L +L++ L
Sbjct: 67 CTKDGRVTRIQIGNQGLKGSLPPNLNNLTELLVFEVQNNGLTGSLPSFSGLDSLQSLLLN 126
Query: 128 RNNFTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPE 187
N FTS+P+ F GLT LQ++ L N SPW++P L +T++ T +A +TG +P+
Sbjct: 127 NNGFTSIPTDFFDGLTSLQSVYLDKN-QFSPWSIPESLKSATSIQTFSAVSANITGTIPD 185
Query: 188 SFFDKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQDNGLSGTIDVLSNMTQLAQV 247
FFD F L ++ LS+NNL G+LP+S + S I++LWLN L+G+I V+ NMTQL +
Sbjct: 186 -FFDAFASLTNLHLSFNNLGGSLPSSFSGSQIQSLWLNGLKGRLNGSIAVIQNMTQLTRT 244
Query: 248 WLHK-NQFTGPIPDLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFP 306
K N F+ P+PD S S L + LRDN LTGPVPNSL+ L SL+ V L NN LQGP P
Sbjct: 245 SGCKANAFSSPLPDFSGLSQLQNCSLRDNSLTGPVPNSLVNLPSLKVVVLTNNFLQGPTP 304
Query: 307 AFGKGVKVT-LDGINSFCKDTPG-PCDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWS 364
F V+V L NSFC PG PCD+RV LL +A GYP +FA +WKGNDPC W
Sbjct: 305 KFPSSVQVDMLADTNSFCLSQPGVPCDSRVNTLLAVAKDVGYPREFAENWKGNDPCSPWM 364
Query: 365 FVVCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLET 424
+ CD G I +N K GL GTISP ++++T L+ L L NNL G+IP L L L
Sbjct: 365 GITCDGGN-ITVLNFQKMGLTGTISPNYSSITSLQKLILANNNLIGTIPNELALLPNLRE 423
Query: 425 LEVSDNNLSGEVPKFPPKVKLLTAGNVLLGQTTGSGGGAGGKTTPSAGSTPEGSHGESG- 483
L+VS+N L G++P F V L T GNV +G+ G TPS GSTP S G G
Sbjct: 424 LDVSNNQLYGKIPPFKSNVLLKTQGNVNIGKDNPPPPAPG---TPS-GSTPGSSDGSGGG 479
Query: 484 -----NGSSLTPGWIAGIVI--IVLFFVAVVLFVSCKCYAKRRHGKFSRVANPV------ 530
+G + G + G VI + V LFV C KR+ RV +P
Sbjct: 480 QTHANSGKKSSTGVVVGSVIGGVCAAVVLAGLFVFCLYRTKRKRS--GRVQSPHTVVIHP 537
Query: 531 --NGNGNVKLDVISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFS 588
+G+ + + + +G S S + GD LH+ + GN +SI VLR VT NFS
Sbjct: 538 HHSGSDQDAVKITIAGSSVNGGDSCGSSSAPGD---LHIVEAGNMVISIQVLRDVTNNFS 594
Query: 589 EDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVAL 648
E NILGRGGFG VYKGEL DGTK+AVKRMES M KGL+EF++EI VL+KVRHRHLV L
Sbjct: 595 EVNILGRGGFGTVYKGELHDGTKMAVKRMESGVMSEKGLDEFKSEIAVLTKVRHRHLVTL 654
Query: 649 LGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLA 708
LG+C++GNERLLVYEYMPQGTL+++LF W+E G PL W +R+T+ALDVARGVEYLH LA
Sbjct: 655 LGYCLDGNERLLVYEYMPQGTLSRYLFNWKEEGLKPLEWTRRLTIALDVARGVEYLHGLA 714
Query: 709 QQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGR 768
QQSFIHRDLKPSNILLGDDMRAKVADFGLV+ APD K SV TRLAGTFGYLAPEYA TGR
Sbjct: 715 QQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPDPKASVVTRLAGTFGYLAPEYAVTGR 774
Query: 769 VTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPD 828
VTTK+DV++FGV+LMELITGR+ALD+S P+E HLV WFRR+ INKE KAID T++ D
Sbjct: 775 VTTKIDVFSFGVILMELITGRKALDESQPEESMHLVPWFRRMHINKETFRKAIDPTVDLD 834
Query: 829 EETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMS 888
EET+ S+ V+ELAGH AREP+QRPDMGHAVNVL + E WKP ED+ + + MS
Sbjct: 835 EETLSSVSTVAELAGHSCAREPHQRPDMGHAVNVLSSLAELWKPAEVDEDEIYGIDYDMS 894
Query: 889 LPQVLQRWQANEGTSTIFNDMSL----SQTQSSINSKTYGFADSFDSLDCR 935
LPQ +++WQA EG S I S TQ+SI ++ GFADSF S D R
Sbjct: 895 LPQAVKKWQALEGMSGIDGSSSYLASSDNTQTSIPTRPSGFADSFTSADGR 945
>gb|AAD21776.1| putative receptor-like protein kinase [Arabidopsis thaliana]
gi|15226361|ref|NP_178291.1| leucine-rich repeat protein
kinase, putative [Arabidopsis thaliana]
gi|25287709|pir||E84429 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana
Length = 943
Score = 872 bits (2252), Expect = 0.0
Identities = 483/928 (52%), Positives = 622/928 (66%), Gaps = 41/928 (4%)
Query: 36 DAAVMSKLLKSLSPPPS-DWSSTTPFCQWDGIKCDSSNRVTTISLASRSLTGTLPSDLNS 94
D + M L SL+ DWS+ P C+W ++CD SNRVT I L + + GTLP++L S
Sbjct: 29 DDSTMQSLKSSLNLTSDVDWSNPNP-CKWQSVQCDGSNRVTKIQLKQKGIRGTLPTNLQS 87
Query: 95 LSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGLTDLQTLSLSDNP 154
LS+L L L N ISGPIP L+ LS L+T L N FTSVP F+G++ LQ + L +NP
Sbjct: 88 LSELVILELFLNRISGPIPDLSGLSRLQTLNLHDNLFTSVPKNLFSGMSSLQEMYLENNP 147
Query: 155 NLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFDK-FPGLQSVRLSYNNLTGALPNS 213
PW +P + ++T+L L L + G++P+ F + P L +++LS N L G LP S
Sbjct: 148 -FDPWVIPDTVKEATSLQNLTLSNCSIIGKIPDFFGSQSLPSLTNLKLSQNGLEGELPMS 206
Query: 214 LAASAIENLWLNNQDNGLSGTIDVLSNMTQLAQVWLHKNQFTGPIPDLSQCSNLFDLQLR 273
A ++I++L+LN Q L+G+I VL NMT L +V L NQF+GPIPDLS +L +R
Sbjct: 207 FAGTSIQSLFLNGQK--LNGSISVLGNMTSLVEVSLQGNQFSGPIPDLSGLVSLRVFNVR 264
Query: 274 DNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTL-DGINSFCKDTPGP-CD 331
+NQLTG VP SL+ L+SL V+L NN LQGP P FGK V V + + +NSFC + G CD
Sbjct: 265 ENQLTGVVPQSLVSLSSLTTVNLTNNYLQGPTPLFGKSVGVDIVNNMNSFCTNVAGEACD 324
Query: 332 ARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQGLQGTISPA 391
RV L+ +A +FGYP+K A SWKGN+PC W + C SG I VN+ KQ L GTISP+
Sbjct: 325 PRVDTLVSVAESFGYPVKLAESWKGNNPCVNWVGITC-SGGNITVVNMRKQDLSGTISPS 383
Query: 392 FANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKVKLLTAGNV 451
A LT L ++ L N L+G IP+ LTTL++L L+VS+N+ G PKF V L+T GN
Sbjct: 384 LAKLTSLETINLADNKLSGHIPDELTTLSKLRLLDVSNNDFYGIPPKFRDTVTLVTEGNA 443
Query: 452 LLGQT----TGSGGGAGGKTTPSAGSTPEGSHGESGNGSSLTPGWIAGIVIIVLFFVAVV 507
+G+ T GA + PS GS + +S N + P + G V+ L V +
Sbjct: 444 NMGKNGPNKTSDAPGASPGSKPSGGSDGSETSKKSSNVKIIVP--VVGGVVGALCLVGLG 501
Query: 508 LFVSCKCYAKRRHGKFSRVANPVNG----------NGNVKLDVISVSNGYSGAPSELQSQ 557
+ + YAK+R + +RV +P + N ++KL V + S+ SG S+ S
Sbjct: 502 VCL----YAKKRK-RPARVQSPSSNMVIHPHHSGDNDDIKLTV-AASSLNSGGGSDSYSH 555
Query: 558 SSGDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRM 617
S S++HV + GN +SI VLR VT NFSE+NILGRGGFG VYKGEL DGTKIAVKRM
Sbjct: 556 SGSAASDIHVVEAGNLVISIQVLRNVTNNFSEENILGRGGFGTVYKGELHDGTKIAVKRM 615
Query: 618 ESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEW 677
ES + +KGL EF++EITVL+K+RHRHLVALLG+C++GNERLLVYEYMPQGTL+QHLF W
Sbjct: 616 ESSVVSDKGLTEFKSEITVLTKMRHRHLVALLGYCLDGNERLLVYEYMPQGTLSQHLFHW 675
Query: 678 RELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGL 737
+E G PL W +R+ +ALDVARGVEYLH+LA QSFIHRDLKPSNILLGDDMRAKV+DFGL
Sbjct: 676 KEEGRKPLDWTRRLAIALDVARGVEYLHTLAHQSFIHRDLKPSNILLGDDMRAKVSDFGL 735
Query: 738 VKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLP 797
V+ APDGKYS+ETR+AGTFGYLAPEYA TGRVTTKVD+++ GV+LMELITGR+ALD++ P
Sbjct: 736 VRLAPDGKYSIETRVAGTFGYLAPEYAVTGRVTTKVDIFSLGVILMELITGRKALDETQP 795
Query: 798 DERSHLVTWFRRVLINKEN--IPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPD 855
++ HLVTWFRRV +K+ AID ++ D++T+ SI KV ELAGHC AREP QRPD
Sbjct: 796 EDSVHLVTWFRRVAASKDENAFKNAIDPNISLDDDTVASIEKVWELAGHCCAREPYQRPD 855
Query: 856 MGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMSLPQVLQRWQANEGTSTIFNDM------ 909
M H VNVL + QWKPT DD + + M LPQVL++WQA EG S +D
Sbjct: 856 MAHIVNVLSSLTVQWKPTETDPDDVYGIDYDMPLPQVLKKWQAFEGLSQTADDSGSSSSA 915
Query: 910 --SLSQTQSSINSKTYGFADSFDSLDCR 935
S TQ+SI ++ GFADSF S+D R
Sbjct: 916 YGSKDNTQTSIPTRPSGFADSFTSVDGR 943
>gb|AAX92784.1| receptor-like kinase RHG4 [Oryza sativa (japonica cultivar-group)]
Length = 912
Score = 785 bits (2026), Expect = 0.0
Identities = 446/904 (49%), Positives = 584/904 (64%), Gaps = 31/904 (3%)
Query: 13 LSLLFSITIIIAADDGPVFAATEDAAVMSKLLKSLSPPPSDWSSTTPFCQWDGIKCD--S 70
L LL + +++AA G +AT DA + L +S+ P W C ++G+ C+
Sbjct: 8 LPLLPFLLLLLAAAAGVAESAT-DAEAIHDLARSV--PALGWDGDN-VCGFEGVTCERGG 63
Query: 71 SNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNN 130
+ +VT ++LA R L+GTLP L+SL+ LT+L LQ NA++G +PSLA + +L L N
Sbjct: 64 AGKVTELNLADRGLSGTLPDSLSSLTSLTALQLQGNALTGAVPSLARMGSLARLALDGNA 123
Query: 131 FTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPESFF 190
FTS+P GLT LQ L++ + P L PW +P + ++L T A ++G P +
Sbjct: 124 FTSLPPDFLHGLTSLQYLTMENLP-LPPWPVPDAIANCSSLDTFSASNASISGPFP-AVL 181
Query: 191 DKFPGLQSVRLSYNNLTGALPNSLAAS-AIENLWLNNQ--DNGLSGTIDVLSNMTQLAQV 247
L+++RLSYNNLTG LP L++ A+E+L LNNQ D+ LSG IDV+++M L +
Sbjct: 182 ATLVSLRNLRLSYNNLTGGLPPELSSLIAMESLQLNNQRSDDKLSGPIDVIASMKSLKLL 241
Query: 248 WLHKNQFTGPIPDLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPA 307
W+ N+FTGPIPDL+ + L +RDN LTG VP SL GL SL+NVSL NN QGP PA
Sbjct: 242 WIQSNKFTGPIPDLNG-TQLEAFNVRDNMLTGVVPPSLTGLMSLKNVSLSNNNFQGPKPA 300
Query: 308 FGKGVKVTLDGINSFCKDTPGPCDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQ-GWSFV 366
F D N FC +TPGPC LL +A FGYP + A +WKGNDPC W +
Sbjct: 301 FAAIPGQDEDSGNGFCLNTPGPCSPLTTTLLQVAEGFGYPYELAKTWKGNDPCSPAWVGI 360
Query: 367 VCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLE 426
VC S + +NL+++ L G ISPA ANLT L L L+ NNLTG IP+ LTTL L L
Sbjct: 361 VCTSS-DVSMINLSRKNLSGRISPALANLTRLARLDLSNNNLTGVIPDVLTTLPSLTVLN 419
Query: 427 VSDNNLSGEVPKFPPKVKLLTAGNVLLGQTTGSGGGAGGKTTPSAGSTPEGSHGESGNGS 486
V++N L+GEVPKF P V +L GN L GQ++GS GG GG S+ S G G S
Sbjct: 420 VANNRLTGEVPKFKPSVNVLAQGN-LFGQSSGSSGGGGGSDGDSSSSDSAG-----GGKS 473
Query: 487 SLTPGWIAGIVI-IVLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNGNGN--VKLDVIS- 542
G I GI++ +++ F + L V + K+ KF V+ + + +K+ V+
Sbjct: 474 KPNTGMIIGIIVAVIILFACIALLVHHR--KKKNVEKFRPVSTKTSPAESEMMKIQVVGA 531
Query: 543 --VSNGYSGAPSELQSQSSGDHSE--LHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGF 598
+SNG S P+EL S S +S +F+ +S+ VL + T NFSED ILGRGGF
Sbjct: 532 NGISNGSSAFPTELYSHVSAANSSNISELFESHGMQLSVEVLLKATNNFSEDCILGRGGF 591
Query: 599 GVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNER 658
GVV+KG L +G +AVKR +S MG KG EF AEI VL KVRHRHLVALLG+C +GNER
Sbjct: 592 GVVFKGNL-NGKLVAVKRCDSGTMGTKGQEEFLAEIDVLRKVRHRHLVALLGYCTHGNER 650
Query: 659 LLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLK 718
LLVYEYM GTL +HL + ++ G+ PLTW QR+T+ALDVARG+EYLH LAQ++FIHRDLK
Sbjct: 651 LLVYEYMSGGTLREHLCDLQQSGFIPLTWTQRMTIALDVARGIEYLHGLAQETFIHRDLK 710
Query: 719 PSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAF 778
PSNILL D+RAKV+DFGLVK A D S+ TR+AGTFGYLAPEYA TG+VTTKVDVYA+
Sbjct: 711 PSNILLDQDLRAKVSDFGLVKLAKDTDKSLMTRIAGTFGYLAPEYATTGKVTTKVDVYAY 770
Query: 779 GVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKV 838
GV+LME+ITGR+ LDDSLPD+ +HLVT FRR +++KE K +D TL E S+ +V
Sbjct: 771 GVILMEMITGRKVLDDSLPDDETHLVTIFRRNILDKEKFRKFVDPTLELSAEGWTSLLEV 830
Query: 839 SELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMSLPQVLQRWQA 898
++LA HCTAREP QRPDM H VN L +V+QWKPT+ EDD M L Q L++W+
Sbjct: 831 ADLARHCTAREPYQRPDMCHCVNRLSSLVDQWKPTNIDEDDYEGETSEMGLHQQLEKWRC 890
Query: 899 NEGT 902
++ T
Sbjct: 891 DDFT 894
>ref|XP_474425.1| OSJNBa0088H09.21 [Oryza sativa (japonica cultivar-group)]
gi|21741125|emb|CAD41925.1| OSJNBa0070M12.3 [Oryza
sativa (japonica cultivar-group)]
gi|32488720|emb|CAE03463.1| OSJNBa0088H09.21 [Oryza
sativa (japonica cultivar-group)]
Length = 938
Score = 785 bits (2026), Expect = 0.0
Identities = 427/924 (46%), Positives = 588/924 (63%), Gaps = 30/924 (3%)
Query: 36 DAAVMSKLLKSLSPPPS-----DWSSTTPFCQWDGIKCDSSNRVTTISLASRSLTGTLPS 90
D +V+ L +SL+ + D ++ P W I CD + RV I L + L GTLPS
Sbjct: 21 DLSVLHDLRRSLTNADAVLGWGDPNAADPCAAWPHISCDRAGRVNNIDLKNAGLAGTLPS 80
Query: 91 DLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGLTDLQTLSL 150
+L L LSLQNN +SG +PS +++L+ AFL N+F S+P+ F+GLT L +SL
Sbjct: 81 TFAALDALQDLSLQNNNLSGDLPSFRGMASLRHAFLNNNSFRSIPADFFSGLTSLLVISL 140
Query: 151 SDNP-NLSP--WTLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVRLSYNNLT 207
NP N+S WT+P ++ + L +L L LTG +P+ F LQ ++L+YN L+
Sbjct: 141 DQNPLNVSSGGWTIPADVAAAQQLQSLSLNGCNLTGAIPD-FLGAMNSLQELKLAYNALS 199
Query: 208 GALPNSLAASAIENLWLNNQDN--GLSGTIDVLSNMTQLAQVWLHKNQFTGPIPD-LSQC 264
G +P++ AS ++ LWLNNQ LSGT+D+++ M L Q WLH N F+GPIPD ++ C
Sbjct: 200 GPIPSTFNASGLQTLWLNNQHGVPKLSGTLDLIATMPNLEQAWLHGNDFSGPIPDSIADC 259
Query: 265 SNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDGINSFCK 324
L DL L NQL G VP +L + L++V LDNN L GP PA K K T N FC
Sbjct: 260 KRLSDLCLNSNQLVGLVPPALESMAGLKSVQLDNNNLLGPVPAI-KAPKYTYSQ-NGFCA 317
Query: 325 DTPG-PCDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQG 383
D PG C +VM LLH YP + SW GN+ C W + C +G + +NL + G
Sbjct: 318 DKPGVACSPQVMALLHFLAEVDYPKRLVASWSGNNSCVDWLGISCVAGN-VTMLNLPEYG 376
Query: 384 LQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKV 443
L GTIS + NL++L + L GNNLTG +P+SLT+L L+ L++S N+L+G +P F P V
Sbjct: 377 LNGTISDSLGNLSELSDINLIGNNLTGHVPDSLTSLRLLQKLDLSGNDLTGPLPTFSPSV 436
Query: 444 KLLTAGNVLLGQTTGSGGGAGGKTTPSAGSTPEGSHGESG----NGSSLTPGWIAGIVII 499
K+ GN+ T + G A K TP + S+ + G N + +A + +
Sbjct: 437 KVNVTGNLNFNGT--APGSAPSKDTPGSSSSRAPTLPGQGVLPENKKKRSAVVLATTIPV 494
Query: 500 VLFFVAVVLFVSCKCYAKRRHGKFSRVANPV-------NGNGNVKLDVISVSNGYSGAPS 552
+ VA+ + + K+R A+ V + + VK+ +++ S
Sbjct: 495 AVSVVALASVCAVLIFRKKRGSVPPNAASVVVHPRENSDPDNLVKIVMVNNDGNSSSTQG 554
Query: 553 ELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKI 612
S SS S++H+ D GN +++ VLR T NF++DN+LGRGGFGVVYKGEL DGT I
Sbjct: 555 NTLSGSSSRASDVHMIDTGNFVIAVQVLRGATKNFTQDNVLGRGGFGVVYKGELHDGTMI 614
Query: 613 AVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQ 672
AVKRME+ + NK L+EFQAEIT+L+KVRHR+LV++LG+ I GNERLLVYEYM G L++
Sbjct: 615 AVKRMEAAVISNKALDEFQAEITILTKVRHRNLVSILGYSIEGNERLLVYEYMSNGALSK 674
Query: 673 HLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKV 732
HLF+W++ PL+WK+R+ +ALDVARG+EYLH+LA Q +IHRDLK +NILLGDD RAKV
Sbjct: 675 HLFQWKQFELEPLSWKKRLNIALDVARGMEYLHNLAHQCYIHRDLKSANILLGDDFRAKV 734
Query: 733 ADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRAL 792
+DFGLVK+APDG +SV TRLAGTFGYLAPEYA TG++TTK DV++FGVVLMELITG A+
Sbjct: 735 SDFGLVKHAPDGNFSVATRLAGTFGYLAPEYAVTGKITTKADVFSFGVVLMELITGMTAI 794
Query: 793 DDS-LPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPN 851
D+S L +E +L +WF ++ +++ + AID TL+ +ET ESI ++ELAGHCT+REP
Sbjct: 795 DESRLEEETRYLASWFCQIRKDEDRLRAAIDPTLDQSDETFESISVIAELAGHCTSREPT 854
Query: 852 QRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMSLPQVLQRWQANEGTSTIFNDMSL 911
QRPDMGHAVNVLVPMVE+WKP + +D + H L Q+++ WQ E + T + +SL
Sbjct: 855 QRPDMGHAVNVLVPMVEKWKPVNDETEDYMGIDLHQPLLQMVKGWQDAEASMTDGSILSL 914
Query: 912 SQTQSSINSKTYGFADSFDSLDCR 935
++ SI ++ GFA+SF S D R
Sbjct: 915 EDSKGSIPARPAGFAESFTSADGR 938
>gb|AAO72615.1| receptor-like protein kinase-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 938
Score = 780 bits (2015), Expect = 0.0
Identities = 425/924 (45%), Positives = 587/924 (62%), Gaps = 30/924 (3%)
Query: 36 DAAVMSKLLKSLSPPPS-----DWSSTTPFCQWDGIKCDSSNRVTTISLASRSLTGTLPS 90
D +V+ L +SL+ + D ++ P W I CD + RV I L + L GTLP
Sbjct: 21 DLSVLHDLRRSLTXAEAVLGWGDPNAADPCAAWPHISCDRAGRVNNIDLKNAGLAGTLPF 80
Query: 91 DLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGLTDLQTLSL 150
+L L LSLQN+ +SG +PS +++L+ AFL N+F S+P+ F+GLT L +SL
Sbjct: 81 TFAALDALQDLSLQNHNLSGDLPSFRGMASLRHAFLNNNSFRSIPADFFSGLTSLLVISL 140
Query: 151 SDNP-NLSP--WTLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVRLSYNNLT 207
NP N+S WT+P ++ + L +L L LTG +P+ F LQ ++L+YN L+
Sbjct: 141 DQNPLNVSSGGWTIPADVAAAQQLQSLSLNGCNLTGAIPD-FLGAMNSLQELKLAYNALS 199
Query: 208 GALPNSLAASAIENLWLNNQDN--GLSGTIDVLSNMTQLAQVWLHKNQFTGPIPD-LSQC 264
G +P++ AS ++ LWLNNQ LSGT+D+++ M L Q WLH N F+GPIPD ++ C
Sbjct: 200 GPIPSTFNASGLQTLWLNNQHGVPKLSGTLDLIATMPNLEQAWLHGNDFSGPIPDSIADC 259
Query: 265 SNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDGINSFCK 324
L DL L NQL G VP +L + L++V LDNN L GP PA K K T N FC
Sbjct: 260 KRLSDLCLNSNQLVGLVPPALESMAGLKSVQLDNNNLLGPVPAI-KAPKYTYSQ-NGFCA 317
Query: 325 DTPG-PCDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQG 383
D PG C +VM LLH YP + SW GN+ C W + C +G + +NL + G
Sbjct: 318 DKPGVACSPQVMALLHFLAEVDYPKRLVASWSGNNSCVDWLGISCVAGN-VTMLNLPEYG 376
Query: 384 LQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKV 443
L GTIS + NL++L + L GNNLTG +P+SLT+L L+ L++S N+L+G +P F P V
Sbjct: 377 LNGTISDSLGNLSELSDINLIGNNLTGHVPDSLTSLRLLQKLDLSGNDLTGPLPTFSPSV 436
Query: 444 KLLTAGNVLLGQTTGSGGGAGGKTTPSAGSTPEGSHGESG----NGSSLTPGWIAGIVII 499
K+ GN+ T + G A K TP + S+ + G N + +A + +
Sbjct: 437 KVNVTGNLNFNGT--APGSAPSKDTPGSSSSRAPTLPGQGVLPENKKKRSAVVLATTIPV 494
Query: 500 VLFFVAVVLFVSCKCYAKRRHGKFSRVANPV-------NGNGNVKLDVISVSNGYSGAPS 552
+ VA+ + + K+R A+ V + + VK+ +++ S
Sbjct: 495 AVSVVALASVCAVLIFRKKRGSVPPNAASVVVHPRENSDPDNLVKIVMVNNDGNSSSTQG 554
Query: 553 ELQSQSSGDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKI 612
S SS S++H+ D GN +++ VLR T NF++DN+LGRGGFGVVYKGEL DGT I
Sbjct: 555 NTLSGSSSRASDVHMIDTGNFVIAVQVLRGATKNFTQDNVLGRGGFGVVYKGELHDGTMI 614
Query: 613 AVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQ 672
AVKRME+ + NK L+EFQAEIT+L+KVRHR+LV++LG+ I GNERLLVYEYM G L++
Sbjct: 615 AVKRMEAAVISNKALDEFQAEITILTKVRHRNLVSILGYSIEGNERLLVYEYMSNGALSK 674
Query: 673 HLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKV 732
HLF+W++ PL+WK+R+ +ALDVARG+EYLH+LA Q +IHRDLK +NILLGDD RAKV
Sbjct: 675 HLFQWKQFELEPLSWKKRLNIALDVARGMEYLHNLAHQCYIHRDLKSANILLGDDFRAKV 734
Query: 733 ADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRAL 792
+DFGLVK+APDG +SV TRLAGTFGYLAPEYA TG++TTK DV++FGVVLMELITG A+
Sbjct: 735 SDFGLVKHAPDGNFSVATRLAGTFGYLAPEYAVTGKITTKADVFSFGVVLMELITGMTAI 794
Query: 793 DDS-LPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPN 851
D+S L +E +L +WF ++ +++ + AID TL+ +ET ESI ++ELAGHCT+REP
Sbjct: 795 DESRLEEETRYLASWFCQIRKDEDRLRAAIDPTLDQSDETFESISVIAELAGHCTSREPT 854
Query: 852 QRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPHMSLPQVLQRWQANEGTSTIFNDMSL 911
QRPDMGHAVNVLVPMVE+WKP + +D + H L Q+++ WQ E + T + +SL
Sbjct: 855 QRPDMGHAVNVLVPMVEKWKPVNDETEDYMGIDLHQPLLQMVKGWQDAEASMTDGSILSL 914
Query: 912 SQTQSSINSKTYGFADSFDSLDCR 935
++ SI ++ GFA+SF S D R
Sbjct: 915 EDSKGSIPARPAGFAESFTSADGR 938
>gb|AAG03120.1| F5A9.23 [Arabidopsis thaliana]
Length = 924
Score = 780 bits (2013), Expect = 0.0
Identities = 439/918 (47%), Positives = 575/918 (61%), Gaps = 69/918 (7%)
Query: 33 ATEDAAVMSKLLKSLSPPPS-DWSSTTPFCQWDG-IKCDSSNRVTTISLASRSLTGTLPS 90
++ D AVM L SL + +WS + P C+W IKCD+SNRVT I + R ++G LP
Sbjct: 20 SSPDEAVMIALRDSLKLSGNPNWSGSDP-CKWSMFIKCDASNRVTAIQIGDRGISGKLPP 78
Query: 91 DLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGLTDLQTLSL 150
DL L+ LT + N ++GPIPSLA L +L T + N+FTSVP F+GL+ LQ +SL
Sbjct: 79 DLGKLTSLTKFEVMRNRLTGPIPSLAGLKSLVTVYANDNDFTSVPEDFFSGLSSLQHVSL 138
Query: 151 SDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFD--KFPGLQSVRLSYNNLTG 208
+NP W +P L +T+L+ L+G++P+ F+ F L +++LSYN+L
Sbjct: 139 DNNP-FDSWVIPPSLENATSLVDFSAVNCNLSGKIPDYLFEGKDFSSLTTLKLSYNSLVC 197
Query: 209 ALPNSLAASAIENLWLNNQDNG--LSGTIDVLSNMTQLAQVWLHKNQFTGPIPDLSQCSN 266
P + + S ++ L LN Q L G+I L MT L V L N F+GP+PD S +
Sbjct: 198 EFPMNFSDSRVQVLMLNGQKGREKLHGSISFLQKMTSLTNVTLQGNSFSGPLPDFSGLVS 257
Query: 267 LFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAF-GKGVKVTLDGINSFCKD 325
L +R+NQL+G VP+SL L SL +V+L NN LQGP P F +K L+G+NSFC D
Sbjct: 258 LKSFNVRENQLSGLVPSSLFELQSLSDVALGNNLLQGPTPNFTAPDIKPDLNGLNSFCLD 317
Query: 326 TPGP-CDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQGL 384
TPG CD RV LL I AFGYP+ FA WKGNDPC GW + C +G I +N GL
Sbjct: 318 TPGTSCDPRVNTLLSIVEAFGYPVNFAEKWKGNDPCSGWVGITC-TGTDITVINFKNLGL 376
Query: 385 QGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKVK 444
GTISP FA+ LR + L+ NNL G+IP+ L L+ L+TL+VS N L GEVP+F
Sbjct: 377 NGTISPRFADFASLRVINLSQNNLNGTIPQELAKLSNLKTLDVSKNRLCGEVPRF----- 431
Query: 445 LLTAGNVLLGQTTGS-----GGGAGGKTTPSAGSTPEGSHGESGNGSSLTPGWIAGIVII 499
N + TTG+ G AG K + +AG G + GI++
Sbjct: 432 -----NTTIVNTTGNFEDCPNGNAGKKASSNAGKIV---------------GSVIGILLA 471
Query: 500 VLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNGNGNVKLDVISVSNGYSGAPSELQSQSS 559
+L + F+ K K ++ K +P + + I++ N +G S+S
Sbjct: 472 LLLIGVAIFFLVKK---KMQYHKM----HPQQQSSDQDAFKITIENLCTGV-----SESG 519
Query: 560 GDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMES 619
++ H+ + GN +SI VLR T NF E NILGRGGFG+VYKGEL DGTKIAVKRMES
Sbjct: 520 FSGNDAHLGEAGNIVISIQVLRDATYNFDEKNILGRGGFGIVYKGELHDGTKIAVKRMES 579
Query: 620 VAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRE 679
+ KGL+EF++EI VL++VRHR+LV L G+C+ GNERLLVY+YMPQGTL++H+F W+E
Sbjct: 580 SIISGKGLDEFKSEIAVLTRVRHRNLVVLHGYCLEGNERLLVYQYMPQGTLSRHIFYWKE 639
Query: 680 LGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVK 739
G PL W +R+ +ALDVARGVEYLH+LA QSFIHRDLKPSNILLGDDM AKVADFGLV+
Sbjct: 640 EGLRPLEWTRRLIIALDVARGVEYLHTLAHQSFIHRDLKPSNILLGDDMHAKVADFGLVR 699
Query: 740 NAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDE 799
AP+G S+ET++AGTFGYLAPEYA TGRVTTKVDVY+FGV+LMEL+TGR+ALD + +E
Sbjct: 700 LAPEGTQSIETKIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELLTGRKALDVARSEE 759
Query: 800 RSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHA 859
HL TWFRR+ INK + PKAID+ + +EET+ SI V+ELA C++REP RPDM H
Sbjct: 760 EVHLATWFRRMFINKGSFPKAIDEAMEVNEETLRSINIVAELANQCSSREPRDRPDMNHV 819
Query: 860 VNVLVPMVEQWKPTSRHED--DGHDSEPHMSLPQVLQRWQANEGTSTIFNDMSL------ 911
VNVLV +V QWKPT R D D + + LPQ++ S F D +L
Sbjct: 820 VNVLVSLVVQWKPTERSSDSEDIYGIDYDTPLPQLIL-------DSCFFGDNTLTSIPSR 872
Query: 912 -SQTQSSINSKTYGFADS 928
S+ +S+ S FADS
Sbjct: 873 PSELESTFKSGQGRFADS 890
>ref|NP_173869.1| leucine-rich repeat family protein / protein kinase family protein
[Arabidopsis thaliana] gi|9743346|gb|AAF97970.1|
F21J9.31 [Arabidopsis thaliana]
Length = 886
Score = 777 bits (2006), Expect = 0.0
Identities = 428/876 (48%), Positives = 560/876 (63%), Gaps = 55/876 (6%)
Query: 33 ATEDAAVMSKLLKSLSPPPS-DWSSTTPFCQWDG-IKCDSSNRVTTISLASRSLTGTLPS 90
++ D AVM L SL + +WS + P C+W IKCD+SNRVT I + R ++G LP
Sbjct: 20 SSPDEAVMIALRDSLKLSGNPNWSGSDP-CKWSMFIKCDASNRVTAIQIGDRGISGKLPP 78
Query: 91 DLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGLTDLQTLSL 150
DL L+ LT + N ++GPIPSLA L +L T + N+FTSVP F+GL+ LQ +SL
Sbjct: 79 DLGKLTSLTKFEVMRNRLTGPIPSLAGLKSLVTVYANDNDFTSVPEDFFSGLSSLQHVSL 138
Query: 151 SDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFD--KFPGLQSVRLSYNNLTG 208
+NP W +P L +T+L+ L+G++P+ F+ F L +++LSYN+L
Sbjct: 139 DNNP-FDSWVIPPSLENATSLVDFSAVNCNLSGKIPDYLFEGKDFSSLTTLKLSYNSLVC 197
Query: 209 ALPNSLAASAIENLWLNNQDNG--LSGTIDVLSNMTQLAQVWLHKNQFTGPIPDLSQCSN 266
P + + S ++ L LN Q L G+I L MT L V L N F+GP+PD S +
Sbjct: 198 EFPMNFSDSRVQVLMLNGQKGREKLHGSISFLQKMTSLTNVTLQGNSFSGPLPDFSGLVS 257
Query: 267 LFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAF-GKGVKVTLDGINSFCKD 325
L +R+NQL+G VP+SL L SL +V+L NN LQGP P F +K L+G+NSFC D
Sbjct: 258 LKSFNVRENQLSGLVPSSLFELQSLSDVALGNNLLQGPTPNFTAPDIKPDLNGLNSFCLD 317
Query: 326 TPGP-CDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQGL 384
TPG CD RV LL I AFGYP+ FA WKGNDPC GW + C +G I +N GL
Sbjct: 318 TPGTSCDPRVNTLLSIVEAFGYPVNFAEKWKGNDPCSGWVGITC-TGTDITVINFKNLGL 376
Query: 385 QGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKVK 444
GTISP FA+ LR + L+ NNL G+IP+ L L+ L+TL+VS N L GEVP+F
Sbjct: 377 NGTISPRFADFASLRVINLSQNNLNGTIPQELAKLSNLKTLDVSKNRLCGEVPRF----- 431
Query: 445 LLTAGNVLLGQTTGS-----GGGAGGKTTPSAGSTPEGSHGESGNGSSLTPGWIAGIVII 499
N + TTG+ G AG K + +AG G + GI++
Sbjct: 432 -----NTTIVNTTGNFEDCPNGNAGKKASSNAGKIV---------------GSVIGILLA 471
Query: 500 VLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNGNGNVKLDVISVSNGYSGAPSELQSQSS 559
+L + F+ K K ++ K +P + + I++ N +G S+S
Sbjct: 472 LLLIGVAIFFLVKK---KMQYHKM----HPQQQSSDQDAFKITIENLCTGV-----SESG 519
Query: 560 GDHSELHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMES 619
++ H+ + GN +SI VLR T NF E NILGRGGFG+VYKGEL DGTKIAVKRMES
Sbjct: 520 FSGNDAHLGEAGNIVISIQVLRDATYNFDEKNILGRGGFGIVYKGELHDGTKIAVKRMES 579
Query: 620 VAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRE 679
+ KGL+EF++EI VL++VRHR+LV L G+C+ GNERLLVY+YMPQGTL++H+F W+E
Sbjct: 580 SIISGKGLDEFKSEIAVLTRVRHRNLVVLHGYCLEGNERLLVYQYMPQGTLSRHIFYWKE 639
Query: 680 LGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVK 739
G PL W +R+ +ALDVARGVEYLH+LA QSFIHRDLKPSNILLGDDM AKVADFGLV+
Sbjct: 640 EGLRPLEWTRRLIIALDVARGVEYLHTLAHQSFIHRDLKPSNILLGDDMHAKVADFGLVR 699
Query: 740 NAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDE 799
AP+G S+ET++AGTFGYLAPEYA TGRVTTKVDVY+FGV+LMEL+TGR+ALD + +E
Sbjct: 700 LAPEGTQSIETKIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELLTGRKALDVARSEE 759
Query: 800 RSHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHA 859
HL TWFRR+ INK + PKAID+ + +EET+ SI V+ELA C++REP RPDM H
Sbjct: 760 EVHLATWFRRMFINKGSFPKAIDEAMEVNEETLRSINIVAELANQCSSREPRDRPDMNHV 819
Query: 860 VNVLVPMVEQWKPTSRHED--DGHDSEPHMSLPQVL 893
VNVLV +V QWKPT R D D + + LPQ++
Sbjct: 820 VNVLVSLVVQWKPTERSSDSEDIYGIDYDTPLPQLI 855
>dbj|BAD95052.1| putative receptor-like protein kinase [Arabidopsis thaliana]
Length = 306
Score = 395 bits (1016), Expect = e-108
Identities = 197/306 (64%), Positives = 238/306 (77%), Gaps = 10/306 (3%)
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVAR 699
+RHRHLVALLG+C++GNERLLVYEYMP+GTL+QHLF W+E G PL W +R+ +ALDVAR
Sbjct: 1 MRHRHLVALLGYCLDGNERLLVYEYMPRGTLSQHLFHWKEEGRKPLDWTRRLAIALDVAR 60
Query: 700 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYL 759
GVEYLH+LA QSFIHRDLKPSNILLGDDMRAKV+DFGLV+ APDGKYS+ETR+AGTFGYL
Sbjct: 61 GVEYLHTLAHQSFIHRDLKPSNILLGDDMRAKVSDFGLVRLAPDGKYSIETRVAGTFGYL 120
Query: 760 APEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKEN--I 817
APEYA TGRVTTKVD+++ GV+LMELITGR+ALD++ P++ HLVTWFRRV +K+
Sbjct: 121 APEYAVTGRVTTKVDIFSLGVILMELITGRKALDETQPEDSVHLVTWFRRVAASKDENAF 180
Query: 818 PKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHE 877
AID ++ D++T+ SI KV ELAGHC AREP QRPDM H VNVL + QWKPT
Sbjct: 181 KNAIDPNISLDDDTVASIEKVWELAGHCCAREPYQRPDMAHIVNVLSSLTVQWKPTETDP 240
Query: 878 DDGHDSEPHMSLPQVLQRWQANEGTSTIFNDM--------SLSQTQSSINSKTYGFADSF 929
DD + + M LPQVL++WQA EG S +D S TQ+SI ++ GFADSF
Sbjct: 241 DDVYGIDYDMPLPQVLKKWQAFEGLSQTADDSGSSSSAYGSKDNTQTSIPTRPSGFADSF 300
Query: 930 DSLDCR 935
S+D R
Sbjct: 301 TSVDGR 306
>gb|AAX38303.1| receptor-like protein kinase [Solanum habrochaites]
Length = 628
Score = 374 bits (961), Expect = e-102
Identities = 235/626 (37%), Positives = 333/626 (52%), Gaps = 42/626 (6%)
Query: 30 VFAATE--DAAVMSKLLKSLSPP-----PSDWSSTTPFCQWDGIKCDSSNRVTTISLASR 82
VF T+ D +V+++ K L P P + W I C S +R+ I +
Sbjct: 6 VFTVTDPNDLSVINEFRKGLENPEVLKWPENGGDPCGSPVWPHIVC-SGSRIQQIQVMGL 64
Query: 83 SLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGL 142
L G LP +LN LS+LT L LQ N SG +PS + LS L A+L N F S+P F GL
Sbjct: 65 GLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELSFAYLDFNQFDSIPLDFFDGL 124
Query: 143 TDLQTLSLSDNP--NLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVR 200
+LQ L+L +NP S W+LP L S LI L + L G LPE F L+ +
Sbjct: 125 VNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCNLAGPLPE-FLGTMSSLEVLL 183
Query: 201 LSYNNLTGALPNSLAASAIENLWLNNQD-NGLSGTIDVLSNMTQLAQVWLHKNQFTGPIP 259
LS N L+G +P + + ++ LWLN+Q +G+SG+IDV++ M L +WLH NQF+G IP
Sbjct: 184 LSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVATMVSLTHLWLHGNQFSGKIP 243
Query: 260 -DLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDG 318
++ +NL DL + N L G +P SL + L N+ L+NN GP P F K V+
Sbjct: 244 VEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNNHFMGPVPKF-KATNVSFMS 301
Query: 319 INSFCKDTPGP-CDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQG-WSFVVCDSGRKIIT 376
NSFC+ G C VM LL YP + SW GN+PC G W + CD +K+
Sbjct: 302 -NSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSGNNPCDGRWWGISCDDNQKVSV 360
Query: 377 VNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEV 436
+NL K L GT+SP+ ANL + +YL NNL+G +P S T+L L L++S+NN+S +
Sbjct: 361 INLPKSNLSGTLSPSIANLESVTRIYLESNNLSGFVPSSWTSLKSLSVLDLSNNNISPPL 420
Query: 437 PKFPPKVKLLTAGN-VLLGQTTGSGGGAGGKTTPSAGSTPEGSHGESGNGSSL--TPGW- 492
PKF +KL+ GN L G+ TTP+ T + SS+ PG
Sbjct: 421 PKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPTSSVPPSRPNSSSSVIFKPGEQ 480
Query: 493 -----------------IAGIVIIVLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNGNGN 535
IAG +++V + + ++V CK + + V +P + + +
Sbjct: 481 SPEKKDSKSKIAIVVVPIAGFLLLVFLAIPLYIYV-CKKSKDKHQAPTALVVHPRDPSDS 539
Query: 536 VKLDVISVSNGYSGAPSELQSQSSG--DHSELHVFDGGNSTMSILVLRQVTGNFSEDNIL 593
+ I+++N +G+ S + + S E H+ + GN +S+ VLR VT NFS +N L
Sbjct: 540 DNVVKIAIANQTNGSLSTVNASGSASIQSGESHMIEAGNLLISVQVLRNVTKNFSPENEL 599
Query: 594 GRGGFGVVYKGELQDGTKIAVKRMES 619
GRGGFGVVYKGEL DGT+IAVKRME+
Sbjct: 600 GRGGFGVVYKGELDDGTQIAVKRMEA 625
>gb|AAX38299.1| receptor-like protein kinase [Solanum habrochaites]
gi|61105037|gb|AAX38298.1| receptor-like protein kinase
[Solanum habrochaites]
Length = 628
Score = 374 bits (961), Expect = e-102
Identities = 235/626 (37%), Positives = 333/626 (52%), Gaps = 42/626 (6%)
Query: 30 VFAATE--DAAVMSKLLKSLSPP-----PSDWSSTTPFCQWDGIKCDSSNRVTTISLASR 82
VF T+ D +V+++ K L P P + W I C S +R+ I +
Sbjct: 6 VFTVTDPNDLSVINEFRKGLENPEVLKWPENGGDPCGSPVWPHIVC-SGSRIQQIQVMGL 64
Query: 83 SLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGL 142
L G LP +LN LS+LT L LQ N SG +PS + LS L A+L N F S+P F GL
Sbjct: 65 GLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELSFAYLDFNQFDSIPLDFFDGL 124
Query: 143 TDLQTLSLSDNP--NLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVR 200
+LQ L+L +NP S W+LP L S LI L + L G LPE F L+ +
Sbjct: 125 VNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCNLAGPLPE-FLGTMSSLEVLL 183
Query: 201 LSYNNLTGALPNSLAASAIENLWLNNQD-NGLSGTIDVLSNMTQLAQVWLHKNQFTGPIP 259
LS N L+G +P + + ++ LWLN+Q +G+SG+IDV++ M L +WLH NQF+G IP
Sbjct: 184 LSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVATMVSLTHLWLHGNQFSGKIP 243
Query: 260 -DLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDG 318
++ +NL DL + N L G +P SL + L N+ L+NN GP P F K V+
Sbjct: 244 VEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNNHFMGPVPKF-KATNVSFMS 301
Query: 319 INSFCKDTPGP-CDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQG-WSFVVCDSGRKIIT 376
NSFC+ G C VM LL YP + SW GN+PC G W + CD +K+
Sbjct: 302 -NSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVQSWSGNNPCDGRWWGISCDDNQKVSV 360
Query: 377 VNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEV 436
+NL K L GT+SP+ ANL + +YL NNL+G +P S T+L L L++S+NN+S +
Sbjct: 361 INLPKSNLSGTLSPSIANLESVTRIYLESNNLSGFVPSSWTSLKSLSVLDLSNNNISPPL 420
Query: 437 PKFPPKVKLLTAGN-VLLGQTTGSGGGAGGKTTPSAGSTPEGSHGESGNGSSL--TPGW- 492
PKF +KL+ GN L G+ TTP+ T + SS+ PG
Sbjct: 421 PKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPTSSVPPSRPNSSSSVIFKPGEQ 480
Query: 493 -----------------IAGIVIIVLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNGNGN 535
IAG +++V + + ++V CK + + V +P + + +
Sbjct: 481 SPEKKDSKSKIAIVVVPIAGFLLLVFLAIPLYIYV-CKKSKDKHQAPTALVVHPRDPSDS 539
Query: 536 VKLDVISVSNGYSGAPSELQSQSSG--DHSELHVFDGGNSTMSILVLRQVTGNFSEDNIL 593
+ I+++N +G+ S + + S E H+ + GN +S+ VLR VT NFS +N L
Sbjct: 540 DNVVKIAIANQTNGSLSTVNASGSASIQSGESHMIEAGNLLISVQVLRNVTKNFSPENEL 599
Query: 594 GRGGFGVVYKGELQDGTKIAVKRMES 619
GRGGFGVVYKGEL DGT+IAVKRME+
Sbjct: 600 GRGGFGVVYKGELDDGTQIAVKRMEA 625
>gb|AAX38300.1| receptor-like protein kinase [Solanum habrochaites]
Length = 628
Score = 374 bits (960), Expect = e-102
Identities = 234/626 (37%), Positives = 333/626 (52%), Gaps = 42/626 (6%)
Query: 30 VFAATE--DAAVMSKLLKSLSPP-----PSDWSSTTPFCQWDGIKCDSSNRVTTISLASR 82
VF T+ D +V+++ K L P P + W I C S +R+ I +
Sbjct: 6 VFTVTDPNDLSVINEFRKGLENPEVLKWPENGGDPCGSPVWPHIVC-SGSRIQQIQVMGL 64
Query: 83 SLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGL 142
L G LP +LN LS+LT L LQ N SG +PS + LS L A+L N F ++P F GL
Sbjct: 65 GLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELSFAYLDFNQFDTIPLDFFDGL 124
Query: 143 TDLQTLSLSDNP--NLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVR 200
+LQ L+L +NP S W+LP L S LI L + L G LPE F L+ +
Sbjct: 125 VNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCNLAGPLPE-FLGTMSSLEVLL 183
Query: 201 LSYNNLTGALPNSLAASAIENLWLNNQD-NGLSGTIDVLSNMTQLAQVWLHKNQFTGPIP 259
LS N L+G +P + + ++ LWLN+Q +G+SG+IDV++ M L +WLH NQF+G IP
Sbjct: 184 LSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVATMVSLTHIWLHGNQFSGKIP 243
Query: 260 -DLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDG 318
++ +NL DL + N L G +P SL + L N+ L+NN GP P F K V+
Sbjct: 244 VEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNNHFMGPVPKF-KATNVSFMS 301
Query: 319 INSFCKDTPGP-CDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQG-WSFVVCDSGRKIIT 376
NSFC+ G C VM LL YP + SW GN+PC G W + CD +K+
Sbjct: 302 -NSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSGNNPCDGRWWGISCDDNQKVSV 360
Query: 377 VNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEV 436
+NL K L GT+SP+ ANL + +YL NNL+G +P S T+L L L++S+NN+S +
Sbjct: 361 INLPKSNLSGTLSPSIANLESVTRIYLESNNLSGFVPSSWTSLKSLSVLDLSNNNISPPL 420
Query: 437 PKFPPKVKLLTAGN-VLLGQTTGSGGGAGGKTTPSAGSTPEGSHGESGNGSSL--TPGW- 492
PKF +KL+ GN L G+ TTP+ T + SS+ PG
Sbjct: 421 PKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPTSSVPPSRPNSSSSVIFKPGEQ 480
Query: 493 -----------------IAGIVIIVLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNGNGN 535
IAG +++V + + ++V CK + + V +P + + +
Sbjct: 481 SPEKKDSKSKIAIVVVPIAGFLLLVFLAIPLYIYV-CKKSKDKHQAPTALVVHPRDPSDS 539
Query: 536 VKLDVISVSNGYSGAPSELQSQSSG--DHSELHVFDGGNSTMSILVLRQVTGNFSEDNIL 593
+ I+++N +G+ S + + S E H+ + GN +S+ VLR VT NFS +N L
Sbjct: 540 DNVVKIAIANQTNGSLSTVNASGSASIQSGESHMIEAGNLLISVQVLRNVTKNFSPENEL 599
Query: 594 GRGGFGVVYKGELQDGTKIAVKRMES 619
GRGGFGVVYKGEL DGT+IAVKRME+
Sbjct: 600 GRGGFGVVYKGELDDGTQIAVKRMEA 625
>gb|AAX38287.1| receptor-like protein kinase [Lycopersicon peruvianum]
Length = 629
Score = 374 bits (960), Expect = e-102
Identities = 239/626 (38%), Positives = 337/626 (53%), Gaps = 41/626 (6%)
Query: 30 VFAATE--DAAVMSKLLKSLSPP-----PSDWSSTTPFCQWDGIKCDSSNRVTTISLASR 82
VF T+ D +V+++ K L P P + W I C S +R+ I +
Sbjct: 6 VFTVTDPNDLSVINEFRKGLENPEALKWPENGGDPCGSPVWPHIVC-SGSRIQQIQVMGL 64
Query: 83 SLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGL 142
L G LP +LN LS+LT L LQ N SG +PS + LS L A+L N F ++P F GL
Sbjct: 65 GLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELSFAYLDFNQFDTIPLDFFDGL 124
Query: 143 TDLQTLSLSDNP--NLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVR 200
+LQ L+L +NP S W+LP L S LI L + L G LPE F L+ +
Sbjct: 125 VNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCNLAGPLPE-FLGTMSSLEVLL 183
Query: 201 LSYNNLTGALPNSLAASAIENLWLNNQD-NGLSGTIDVLSNMTQLAQVWLHKNQFTGPIP 259
LS N L+G +P + + ++ LWLN+Q +G+SG+IDV++ M L +WLH NQF+G IP
Sbjct: 184 LSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVATMVSLTHIWLHGNQFSGKIP 243
Query: 260 -DLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDG 318
++ +NL DL + N L G +P SL + L N+ L+NN GP P F K V+
Sbjct: 244 VEIGNITNLKDLSVNTNNLVGLIPESLANMR-LDNLDLNNNHFMGPVPKF-KATNVSFMS 301
Query: 319 INSFCKDTPGP-CDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQG-WSFVVCDSGRKIIT 376
NSFC+ G C VM LL YP + SW GN+PC G W + CD +K+
Sbjct: 302 -NSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSGNNPCDGRWWGISCDDNQKVSV 360
Query: 377 VNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEV 436
+NL K L GT+SP+ ANL + +YL NNL+G +P S T+L L L++S+NN+S +
Sbjct: 361 INLPKSNLSGTLSPSIANLVTVTRIYLESNNLSGFVPSSWTSLKSLSILDLSNNNISPPL 420
Query: 437 PKFPPKVKLLTAGN-VLLGQTTGSGGGAGGKTTPSAGSTPEGSHGESGNGSS---LTPG- 491
PKF +KL+ GN L G+ TTP+ T N SS PG
Sbjct: 421 PKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPTSSVVPSSRPNSSSSVIFKPGE 480
Query: 492 ----------WIAGIVI-----IVLFFVAVVLFV-SCKCYAKRRHGKFSRVANPVNGNGN 535
IA +V+ ++L F+A+ L++ CK + + V +P + + +
Sbjct: 481 QSPEKKDSKSKIAIVVVPIAGSLLLVFLAIPLYIYVCKKSKDKHQAPTALVVHPRDPSDS 540
Query: 536 VKLDVISVSNGYSGAPSELQSQSSGD-HS-ELHVFDGGNSTMSILVLRQVTGNFSEDNIL 593
+ I+++N +G+ S + + S HS E H+ + GN +S+ VLR VT NFS +N L
Sbjct: 541 DNVVKIAIANQTNGSLSTVNASGSASIHSGESHMIEAGNLLISVQVLRNVTKNFSPENEL 600
Query: 594 GRGGFGVVYKGELQDGTKIAVKRMES 619
GRGGFGVVYKGEL DGT+IAVKRME+
Sbjct: 601 GRGGFGVVYKGELDDGTQIAVKRMEA 626
>gb|AAX38315.1| receptor-like protein kinase [Lycopersicon chmielewskii]
gi|61105069|gb|AAX38314.1| receptor-like protein kinase
[Lycopersicon chmielewskii] gi|61105067|gb|AAX38313.1|
receptor-like protein kinase [Lycopersicon chmielewskii]
gi|61105065|gb|AAX38312.1| receptor-like protein kinase
[Lycopersicon chmielewskii] gi|61105063|gb|AAX38311.1|
receptor-like protein kinase [Lycopersicon chmielewskii]
gi|61105061|gb|AAX38310.1| receptor-like protein kinase
[Lycopersicon chmielewskii] gi|61105059|gb|AAX38309.1|
receptor-like protein kinase [Lycopersicon chmielewskii]
gi|61105057|gb|AAX38308.1| receptor-like protein kinase
[Lycopersicon chmielewskii] gi|61105055|gb|AAX38307.1|
receptor-like protein kinase [Lycopersicon chmielewskii]
gi|61105053|gb|AAX38306.1| receptor-like protein kinase
[Lycopersicon chmielewskii]
Length = 628
Score = 374 bits (959), Expect = e-101
Identities = 237/628 (37%), Positives = 336/628 (52%), Gaps = 46/628 (7%)
Query: 30 VFAATE--DAAVMSKLLKSLSPP-----PSDWSSTTPFCQWDGIKCDSSNRVTTISLASR 82
VF T+ D +V+++L K L P P + W I C S +R+ I +
Sbjct: 6 VFTVTDPNDLSVINELRKGLENPEVLKWPENGGDPCGSPVWPHIVC-SGSRIQQIQVMGL 64
Query: 83 SLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGL 142
L G LP +LN LS+LT L LQ N SG +PS + LS L A+L N F ++P F GL
Sbjct: 65 GLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELSFAYLDFNQFDTIPLDFFDGL 124
Query: 143 TDLQTLSLSDNP--NLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVR 200
+LQ L+L +NP S W+LP L S LI L + L G LPE F L+ +
Sbjct: 125 VNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCNLAGPLPE-FLGTMSSLEVLL 183
Query: 201 LSYNNLTGALPNSLAASAIENLWLNNQD-NGLSGTIDVLSNMTQLAQVWLHKNQFTGPIP 259
LS N L+G +P + + ++ LWLN+Q +G+SG+IDV++ M L +WLH NQF+G IP
Sbjct: 184 LSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVATMVSLTHLWLHGNQFSGKIP 243
Query: 260 -DLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDG 318
++ +NL DL + N L G +P SL + L N+ L+NN GP P F K V+
Sbjct: 244 VEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNNHFMGPVPKF-KAANVSFMS 301
Query: 319 INSFCKDTPGP-CDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQG-WSFVVCDSGRKIIT 376
NSFC+ G C VM LL YP + SW GN+PC G W + CD +K+
Sbjct: 302 -NSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSGNNPCDGRWWGISCDDNQKVSV 360
Query: 377 VNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEV 436
+NL K L GT+SP+ ANL + +YL NNL+G +P S T+L L L++S+NN+S +
Sbjct: 361 INLPKSNLSGTLSPSIANLETVTRIYLESNNLSGFVPSSWTSLKSLSILDLSNNNISPPL 420
Query: 437 PKFPPKVKLLTAGN-VLLGQTTGSGGGAGGKTTPSAGST--------------------- 474
PKF +KL+ GN L G+ TTP+ T
Sbjct: 421 PKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPTSSVPSSRPNSSSSVIFKPSEQ 480
Query: 475 -PEGSHGESGNGSSLTPGWIAGIVIIVLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNGN 533
PE +S + P IAG +++V + + ++V CK + + V +P + +
Sbjct: 481 SPEKKDSKSKIAIVVVP--IAGFLLLVCLAIPLYIYV-CKKSKDKHQAPTALVVHPRDPS 537
Query: 534 GNVKLDVISVSNGYSGAPSELQSQSSGD-HS-ELHVFDGGNSTMSILVLRQVTGNFSEDN 591
+ + I+++N +G+ S + + S HS E H+ + GN +S+ VLR VT NFS +N
Sbjct: 538 DSDNVVKIAIANQTNGSLSTVNASGSASIHSGESHMIEAGNLLISVQVLRNVTKNFSPEN 597
Query: 592 ILGRGGFGVVYKGELQDGTKIAVKRMES 619
LGRGGFGVVYKGEL DGT+IAVKRME+
Sbjct: 598 ELGRGGFGVVYKGELDDGTQIAVKRMEA 625
>gb|AAX38280.1| receptor-like protein kinase [Lycopersicon peruvianum]
Length = 628
Score = 374 bits (959), Expect = e-101
Identities = 239/625 (38%), Positives = 338/625 (53%), Gaps = 40/625 (6%)
Query: 30 VFAATE--DAAVMSKLLKSLSPP-----PSDWSSTTPFCQWDGIKCDSSNRVTTISLASR 82
VF T+ D +V+++ K L P P + W I C S +R+ I +
Sbjct: 6 VFTVTDPNDLSVINEFRKGLENPEVLKWPKNGGDPCGSPVWPHIVC-SGSRIQQIQVMGL 64
Query: 83 SLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTSVPSASFAGL 142
L G LP +LN LS+LT L LQ N SG +PS + LS L A+L N F ++P F GL
Sbjct: 65 GLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELSFAYLDFNQFDTIPLDFFDGL 124
Query: 143 TDLQTLSLSDNP--NLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFDKFPGLQSVR 200
+LQ L+L +NP S W+LP L S LI L + L G LPE F L+ +
Sbjct: 125 VNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCNLAGPLPE-FLGTMSSLEVLL 183
Query: 201 LSYNNLTGALPNSLAASAIENLWLNNQD-NGLSGTIDVLSNMTQLAQVWLHKNQFTGPIP 259
LS N L+G +P + + ++ LWLN+Q +G+SG+IDV++ M L +WLH NQF+G IP
Sbjct: 184 LSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVATMVSLTHLWLHGNQFSGKIP 243
Query: 260 -DLSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDG 318
++ +NL DL + N L G +P SL + L N+ L+NN GP P F K V+
Sbjct: 244 VEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNNHFMGPVPKF-KATNVSFMS 301
Query: 319 INSFCKDTPGP-CDARVMVLLHIAGAFGYPIKFAGSWKGNDPCQG-WSFVVCDSGRKIIT 376
NSFC+ G C VM LL YP + SW GN+PC G W + CD +K+
Sbjct: 302 -NSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSGNNPCDGRWWGISCDDNQKVSV 360
Query: 377 VNLAKQGLQGTISPAFANLTDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEV 436
+NL K L GT+SP+ ANL + +YL NNL+G +P S T+L L L++S+NN+S +
Sbjct: 361 INLPKSNLSGTLSPSIANLETVTRIYLESNNLSGFVPSSWTSLKSLSILDLSNNNISPPL 420
Query: 437 PKFPPKVKLLTAGN-VLLGQTTGSGGGAGGKTTP--SAGSTPEGSHGESGNGSSLTPG-- 491
PKF +KL+ GN L G+ TTP S S+ S S + PG
Sbjct: 421 PKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPTSSVPSSRPNSSSSVMFKPGEQ 480
Query: 492 ---------WIAGIVI-----IVLFFVAVVLFV-SCKCYAKRRHGKFSRVANPVNGNGNV 536
IA +V+ ++L F+A+ L++ CK + + V +P + + +
Sbjct: 481 SPEKKDSKSKIAIVVVPIAGSLLLVFLAIPLYIYVCKKSKDKHQAPTALVVHPRDPSDSD 540
Query: 537 KLDVISVSNGYSGAPSELQSQSSGD-HS-ELHVFDGGNSTMSILVLRQVTGNFSEDNILG 594
+ I+++N +G+ S + + S HS E H+ + GN +S+ VLR VT NFS +N LG
Sbjct: 541 NVVKIAIANQTNGSLSTVNASGSASIHSGESHMIEAGNLLISVQVLRNVTKNFSPENELG 600
Query: 595 RGGFGVVYKGELQDGTKIAVKRMES 619
RGGFGVVYKGEL DGT+IAVKRME+
Sbjct: 601 RGGFGVVYKGELDDGTQIAVKRMEA 625
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,647,505,060
Number of Sequences: 2540612
Number of extensions: 74129411
Number of successful extensions: 393318
Number of sequences better than 10.0: 23794
Number of HSP's better than 10.0 without gapping: 10466
Number of HSP's successfully gapped in prelim test: 13367
Number of HSP's that attempted gapping in prelim test: 278200
Number of HSP's gapped (non-prelim): 55753
length of query: 935
length of database: 863,360,394
effective HSP length: 138
effective length of query: 797
effective length of database: 512,755,938
effective search space: 408666482586
effective search space used: 408666482586
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0108.8


