

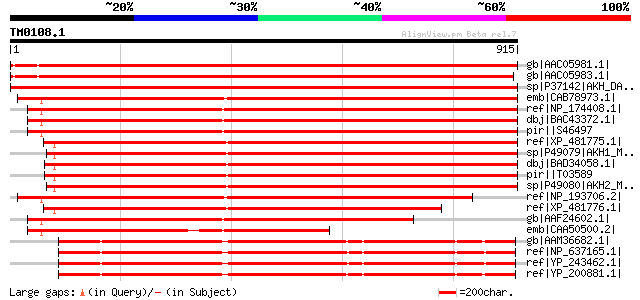
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.1
(915 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC05981.1| aspartokinase-homoserine dehydrogenase [Glycine m... 1620 0.0
gb|AAC05983.1| aspartokinase-homoserine dehydrogenase [Glycine m... 1600 0.0
sp|P37142|AKH_DAUCA Bifunctional aspartokinase/homoserine dehydr... 1387 0.0
emb|CAB78973.1| aspartate kinase-homoserine dehydrogenase-like p... 1386 0.0
ref|NP_174408.1| bifunctional aspartate kinase/homoserine dehydr... 1359 0.0
dbj|BAC43372.1| putative aspartate kinase-homoserine dehydrogena... 1357 0.0
pir||S46497 aspartate kinase (EC 2.7.2.4) / homoserine dehydroge... 1355 0.0
ref|XP_481775.1| putative aspartate kinase, homoserine dehydroge... 1320 0.0
sp|P49079|AKH1_MAIZE Bifunctional aspartokinase/homoserine dehyd... 1315 0.0
dbj|BAD34058.1| aspartate kinase-homoserine dehydrogenase [Oryza... 1289 0.0
pir||T03589 probable aspartate kinase (EC 2.7.2.4) / homoserine ... 1284 0.0
sp|P49080|AKH2_MAIZE Bifunctional aspartokinase/homoserine dehyd... 1270 0.0
ref|NP_193706.2| bifunctional aspartate kinase/homoserine dehydr... 1245 0.0
ref|XP_481776.1| putative aspartate kinase, homoserine dehydroge... 1101 0.0
gb|AAF24602.1| T19E23.1 [Arabidopsis thaliana] 1058 0.0
emb|CAA50500.2| beta-aspartyl phosphate homoserine [Arabidopsis ... 768 0.0
gb|AAM36682.1| aspartokinase; homoserine dehydrogenase I [Xantho... 681 0.0
ref|NP_637165.1| bifunctional aspartokinase/homoserine dehydroge... 669 0.0
ref|YP_243462.1| aspartokinase/homoserine dehydrogenase I [Xanth... 669 0.0
ref|YP_200881.1| aspartokinase; homoserine dehydrogenase I [Xant... 669 0.0
>gb|AAC05981.1| aspartokinase-homoserine dehydrogenase [Glycine max]
gi|7434562|pir||T06242 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean chloroplast
Length = 916
Score = 1620 bits (4196), Expect = 0.0
Identities = 821/918 (89%), Positives = 870/918 (94%), Gaps = 5/918 (0%)
Query: 1 MASLSASSLYHFSTISPSN---NTPDITKISQCQCLPFLPSHRSHSLRKALSLLPRGNQS 57
MAS SA+ + FS +SPS+ ++ + Q QC PF S SHSLRK L+L PRG ++
Sbjct: 1 MASFSAA-VAQFSRVSPSHTSLHSHSHGTLFQSQCRPFFLSRTSHSLRKGLTL-PRGREA 58
Query: 58 PSTKISASLTDVSPSVLVEEQQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERK 117
PST + AS TDVSP+V +EE+QL KGETWSVHKFGGTC+G+SQRIKNV DI+L+DDSERK
Sbjct: 59 PSTSVRASFTDVSPNVSLEEKQLPKGETWSVHKFGGTCVGTSQRIKNVADIILKDDSERK 118
Query: 118 LVVVSAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLH 177
LVVVSAMSKVTDMMY+LI+KAQSRDESY ++L+AVLEKHS TAHD+LDGD LATFLS+LH
Sbjct: 119 LVVVSAMSKVTDMMYDLIHKAQSRDESYTAALNAVLEKHSATAHDILDGDNLATFLSKLH 178
Query: 178 HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLI 237
HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNG DCKWMDTRDVLI
Sbjct: 179 HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGTDCKWMDTRDVLI 238
Query: 238 VNPTSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAI 297
VNPT SNQVDPDYLESE+RLE W+SLNPCKVIIATGFIASTPQ IPTTLKRDGSDFSAAI
Sbjct: 239 VNPTGSNQVDPDYLESEQRLEKWYSLNPCKVIIATGFIASTPQNIPTTLKRDGSDFSAAI 298
Query: 298 MGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV 357
MGALF+ARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV
Sbjct: 299 MGALFKARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV 358
Query: 358 MQYGIPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMA 417
M+YGIPI+IRNIFNLSAPGTKICHPSVND+ED N++NFVKGFATIDNLAL+NVEGTGMA
Sbjct: 359 MRYGIPIMIRNIFNLSAPGTKICHPSVNDHEDSQNLQNFVKGFATIDNLALVNVEGTGMA 418
Query: 418 GVPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGR 477
GVPGTASAIFGAVKDVGANVIMISQASSEHS+CFAVPEKEVKAVAEALQSRFR ALDNGR
Sbjct: 419 GVPGTASAIFGAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNGR 478
Query: 478 LSQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKRE 537
LSQVA+IPNCSILAAVGQKMASTPGVSA+LFNALAKANINVRAIAQGCSEYNITVV+KRE
Sbjct: 479 LSQVAVIPNCSILAAVGQKMASTPGVSASLFNALAKANINVRAIAQGCSEYNITVVVKRE 538
Query: 538 DCVKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILG 597
DC+KALRAVHSRFYLSRTTIAMGIIGPGLIGSTLL+QLRDQAS LKEEFNIDLRVMGILG
Sbjct: 539 DCIKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLEQLRDQASTLKEEFNIDLRVMGILG 598
Query: 598 SKSMLLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHY 657
SKSMLLSD GIDLARWRELR+E GEVAN+EKFVQHVHGNHFIPNTA+VDCTADSVIAG+Y
Sbjct: 599 SKSMLLSDVGIDLARWRELREERGEVANMEKFVQHVHGNHFIPNTALVDCTADSVIAGYY 658
Query: 658 YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 717
YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE
Sbjct: 659 YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 718
Query: 718 TGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVI 777
TGD+ILQIEGIFSGTLSYIFNNFKDGRAFSEVV EAKEAGYTEPDPRDDLSGTDVARKVI
Sbjct: 719 TGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVVSEAKEAGYTEPDPRDDLSGTDVARKVI 778
Query: 778 ILARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLR 837
ILARESGLKLELSNIPVES VPEPL+ACASAQEFMQ LP+FDQ F KK E+AENAGEVLR
Sbjct: 779 ILARESGLKLELSNIPVESPVPEPLRACASAQEFMQELPKFDQEFTKKQEDAENAGEVLR 838
Query: 838 YVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA 897
YVGVVDVT+KKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA
Sbjct: 839 YVGVVDVTNKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA 898
Query: 898 GGIFSDILRLASYLGAPS 915
GGIFSDILRLASYLGAPS
Sbjct: 899 GGIFSDILRLASYLGAPS 916
>gb|AAC05983.1| aspartokinase-homoserine dehydrogenase [Glycine max]
gi|7434563|pir||T06246 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) precursor -
soybean (fragment)
Length = 909
Score = 1600 bits (4144), Expect = 0.0
Identities = 811/911 (89%), Positives = 861/911 (94%), Gaps = 5/911 (0%)
Query: 1 MASLSASSLYHFSTISPSN---NTPDITKISQCQCLPFLPSHRSHSLRKALSLLPRGNQS 57
MAS SA+ + FS +SP+ ++ ++ QC PF S SHSLRK L+L PRG ++
Sbjct: 1 MASFSAA-VAQFSRVSPTLTLLHSHSHDRLFHSQCRPFFLSRPSHSLRKGLTL-PRGREA 58
Query: 58 PSTKISASLTDVSPSVLVEEQQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERK 117
PST + AS TDVSPSV +EE+QL KGETWSVHKFGGTC+G+SQRIKNV DI+L+DDSERK
Sbjct: 59 PSTTVRASFTDVSPSVSLEEKQLPKGETWSVHKFGGTCVGTSQRIKNVADIILKDDSERK 118
Query: 118 LVVVSAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLH 177
LVVVSAMSKVTDMMY+LI+KAQSRDESY+++LDAV EKHS TAHD+LDGD LA+FLS+LH
Sbjct: 119 LVVVSAMSKVTDMMYDLIHKAQSRDESYIAALDAVSEKHSATAHDILDGDNLASFLSKLH 178
Query: 178 HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLI 237
HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVI KNGADCKWMDTRDVLI
Sbjct: 179 HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVITKNGADCKWMDTRDVLI 238
Query: 238 VNPTSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAI 297
VNPT SNQVDPDYLESE+RLE W+SLNPCKVIIATGFIASTPQ IPTTLKRDGSDFSAAI
Sbjct: 239 VNPTGSNQVDPDYLESEQRLEKWYSLNPCKVIIATGFIASTPQNIPTTLKRDGSDFSAAI 298
Query: 298 MGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV 357
MGALF+ARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV
Sbjct: 299 MGALFKARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV 358
Query: 358 MQYGIPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMA 417
M+YGIPI+IRNIFNLSAPGTKICHPSVND+ED N++NFVKGFATIDNLAL+NVEGTGMA
Sbjct: 359 MRYGIPIMIRNIFNLSAPGTKICHPSVNDHEDRQNLQNFVKGFATIDNLALVNVEGTGMA 418
Query: 418 GVPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGR 477
GVPGTASAIFGAVKDVGANVIMISQASSEHS+CFAVPEKEVKAVAEALQSRFR ALDNGR
Sbjct: 419 GVPGTASAIFGAVKDVGANVIMISQASSEHSVCFAVPEKEVKAVAEALQSRFRQALDNGR 478
Query: 478 LSQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKRE 537
LSQVA+IPNCSILAAVGQKMASTPGVSA+LFNALAKANINVRAIAQGCSEYNITVV+KRE
Sbjct: 479 LSQVAVIPNCSILAAVGQKMASTPGVSASLFNALAKANINVRAIAQGCSEYNITVVVKRE 538
Query: 538 DCVKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILG 597
DC+KALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQAS LKEEFNIDLRVMGILG
Sbjct: 539 DCIKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASTLKEEFNIDLRVMGILG 598
Query: 598 SKSMLLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHY 657
SKSMLLSD GIDLARWRELR+E GEVAN+EKFVQHVHGNHFIPNTA+VDCTADS IAG+Y
Sbjct: 599 SKSMLLSDVGIDLARWRELREERGEVANVEKFVQHVHGNHFIPNTALVDCTADSAIAGYY 658
Query: 658 YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 717
YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE
Sbjct: 659 YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 718
Query: 718 TGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVI 777
TGD+ILQIEGIFSGTLSYIFNNFKDGRAFSEVV EAKEAGYTEPDPRDDLSGTDVARKVI
Sbjct: 719 TGDKILQIEGIFSGTLSYIFNNFKDGRAFSEVVSEAKEAGYTEPDPRDDLSGTDVARKVI 778
Query: 778 ILARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLR 837
ILARESGLKLELSNIPVESLVPEPL+ACASAQEFMQ P+FDQ F KK E+AENAGEVLR
Sbjct: 779 ILARESGLKLELSNIPVESLVPEPLRACASAQEFMQEPPKFDQEFTKKQEDAENAGEVLR 838
Query: 838 YVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA 897
YVGVVDVT++KGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA
Sbjct: 839 YVGVVDVTNEKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA 898
Query: 898 GGIFSDILRLA 908
GGIFSDILRLA
Sbjct: 899 GGIFSDILRLA 909
>sp|P37142|AKH_DAUCA Bifunctional aspartokinase/homoserine dehydrogenase, chloroplast
precursor (AK-HD) (AK-HSDH) [Includes: Aspartokinase ;
Homoserine dehydrogenase ] gi|464225|gb|AAA16972.1|
aspartokinase-homoserine dehydrogenase
Length = 921
Score = 1387 bits (3589), Expect = 0.0
Identities = 709/920 (77%), Positives = 794/920 (86%), Gaps = 5/920 (0%)
Query: 1 MASLSASSLYHFSTISPSNNTPDITKISQCQCL-PFLPSHRSHSLRKALSLLPRGNQSPS 59
++S + S Y + S TP K L P H+S SL K L RG + S
Sbjct: 2 LSSAISPSSYAAIAAAYSARTPIFNKKKTAAVLSPLSLFHQSPSLSKTGIFLHRGRKESS 61
Query: 60 TK--ISASLTDVSPSV--LVEEQQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSE 115
+K I+AS+T PS+ VE+ L +G WS+HKFGGTC+GSS+RI+NV +IV+EDDSE
Sbjct: 62 SKFYIAASVTTAVPSLDDSVEKVHLPRGAMWSIHKFGGTCVGSSERIRNVAEIVVEDDSE 121
Query: 116 RKLVVVSAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQ 175
RKLVVVSAMSKVTDMMY+LI KAQSRD+SY S+LDAV+EKH LTA DLLD D LA FL++
Sbjct: 122 RKLVVVSAMSKVTDMMYDLIYKAQSRDDSYESALDAVMEKHKLTAFDLLDEDDLARFLTR 181
Query: 176 LHHDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDV 235
L HD+ LKAMLRAIYIAGHATESF+DFVVGHGELWSAQ+LS VIRKNG DC WMDTRDV
Sbjct: 182 LQHDVITLKAMLRAIYIAGHATESFSDFVVGHGELWSAQLLSFVIRKNGGDCNWMDTRDV 241
Query: 236 LIVNPTSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSA 295
L+VNP SNQVDPDYLESEKRLE WFS N C+ I+ATGFIASTPQ IPTTLKRDGSDFSA
Sbjct: 242 LVVNPAGSNQVDPDYLESEKRLEKWFSSNQCQTIVATGFIASTPQNIPTTLKRDGSDFSA 301
Query: 296 AIMGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTII 355
AIMGAL RA QVTIWTDV+GVYSADPRKVSEAV+LKTLSYQEAWEMSYFGANVLHPRTI
Sbjct: 302 AIMGALLRAGQVTIWTDVNGVYSADPRKVSEAVVLKTLSYQEAWEMSYFGANVLHPRTIN 361
Query: 356 PVMQYGIPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTG 415
PVM+Y IPI+IRNIFNLSAPGT IC SV + ED + +++ VKGFATIDNLALINVEGTG
Sbjct: 362 PVMRYDIPIVIRNIFNLSAPGTMICRESVGETEDGLKLESHVKGFATIDNLALINVEGTG 421
Query: 416 MAGVPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDN 475
MAGVPGTA+AIFGAVKDVGANVIMISQASSEHSICFAVPE EVKAVA+AL++RFR ALD
Sbjct: 422 MAGVPGTATAIFGAVKDVGANVIMISQASSEHSICFAVPESEVKAVAKALEARFRQALDA 481
Query: 476 GRLSQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLK 535
GRLSQVA PNCSILA VGQKMASTPGVSATLFNALAKANINVRAIAQGC+EYNITVVL
Sbjct: 482 GRLSQVANNPNCSILATVGQKMASTPGVSATLFNALAKANINVRAIAQGCTEYNITVVLS 541
Query: 536 REDCVKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGI 595
REDCV+AL+AVHSRFYLSRTTIA+GI+GPGLIG+TLLDQLRDQA+ILKE IDLRVMGI
Sbjct: 542 REDCVRALKAVHSRFYLSRTTIAVGIVGPGLIGATLLDQLRDQAAILKENSKIDLRVMGI 601
Query: 596 LGSKSMLLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAG 655
GS++MLLS+ GIDL+RWRE++ E G+ A LEKFVQHV GNHFIP+T IVDCTADS +A
Sbjct: 602 TGSRTMLLSETGIDLSRWREVQKEKGQTAGLEKFVQHVRGNHFIPSTVIVDCTADSEVAS 661
Query: 656 HYYDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGL 715
HY+DWL +GIHV+TPNKKANSGPLDQYLKLRALQR+SYTHYFYEATV AGLPI++TL+GL
Sbjct: 662 HYHDWLCRGIHVITPNKKANSGPLDQYLKLRALQRRSYTHYFYEATVVAGLPIITTLQGL 721
Query: 716 LETGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARK 775
LETGD+IL+IEGIFSGTLSYIFNNFK FSEVV EAK AGYTEPDPRDDL+GTDVARK
Sbjct: 722 LETGDKILRIEGIFSGTLSYIFNNFKSTTPFSEVVSEAKAAGYTEPDPRDDLAGTDVARK 781
Query: 776 VIILARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEV 835
VIILAR SGLKLELS+IPV+SLVPEPL+ ASA+EF+ +LP+FD +K E+AENAGEV
Sbjct: 782 VIILARGSGLKLELSDIPVQSLVPEPLRGIASAEEFLLQLPQFDSDMTRKREDAENAGEV 841
Query: 836 LRYVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQV 895
LRYVGVVD ++KGVVEL+RYKK+HPFAQLSGSDNI AFTT RY QP I+RGPGAGA+V
Sbjct: 842 LRYVGVVDAVNQKGVVELKRYKKEHPFAQLSGSDNINAFTTERYNKQPPIIRGPGAGAEV 901
Query: 896 TAGGIFSDILRLASYLGAPS 915
TAGG+FSDILRLASYLGAPS
Sbjct: 902 TAGGVFSDILRLASYLGAPS 921
>emb|CAB78973.1| aspartate kinase-homoserine dehydrogenase-like protein [Arabidopsis
thaliana] gi|3250680|emb|CAA19688.1| aspartate
kinase-homoserine dehydrogenase-like protein
[Arabidopsis thaliana] gi|42572959|ref|NP_974576.1|
bifunctional aspartate kinase/homoserine dehydrogenase,
putative / AK-HSDH, putative [Arabidopsis thaliana]
gi|7434564|pir||T04752 aspartate kinase (EC 2.7.2.4) /
homoserine dehydrogenase (EC 1.1.1.3) T16H5.70 -
Arabidopsis thaliana
Length = 916
Score = 1386 bits (3588), Expect = 0.0
Identities = 702/910 (77%), Positives = 791/910 (86%), Gaps = 10/910 (1%)
Query: 14 TISPSNNTPDITKISQCQCLPFLPSHRSHSLRKALSLLPRGN-----QSPSTKISASLTD 68
T+SP N+ P QC +P R L + G + P +SA+ T
Sbjct: 9 TVSPPNSNPIRFGSFPPQCFLRVPKPRRLILPRFRKTTGGGGGLIRCELPDFHLSATATT 68
Query: 69 VSPSV---LVEEQQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMS 125
VS LV++ Q+ KGE WSVHKFGGTC+G+SQRI+NV ++++ D+SERKLVVVSAMS
Sbjct: 69 VSGVSTVNLVDQVQIPKGEMWSVHKFGGTCVGNSQRIRNVAEVIINDNSERKLVVVSAMS 128
Query: 126 KVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKA 185
KVTDMMY+LI KAQSRD+SY+S+L+AVLEKH LTA DLLDGD LA+FLS LH+DISNLKA
Sbjct: 129 KVTDMMYDLIRKAQSRDDSYLSALEAVLEKHRLTARDLLDGDDLASFLSHLHNDISNLKA 188
Query: 186 MLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQ 245
MLRAIYIAGHA+ESF+DFV GHGELWSAQMLS V+RK G +CKWMDTRDVLIVNPTSSNQ
Sbjct: 189 MLRAIYIAGHASESFSDFVAGHGELWSAQMLSYVVRKTGLECKWMDTRDVLIVNPTSSNQ 248
Query: 246 VDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRAR 305
VDPD+ ESEKRL+ WFSLNP K+IIATGFIASTPQ IPTTLKRDGSDFSAAIMGAL RAR
Sbjct: 249 VDPDFGESEKRLDKWFSLNPSKIIIATGFIASTPQNIPTTLKRDGSDFSAAIMGALLRAR 308
Query: 306 QVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPIL 365
QVTIWTDVDGVYSADPRKV+EAVIL+TLSYQEAWEMSYFGANVLHPRTIIPVM+Y IPI+
Sbjct: 309 QVTIWTDVDGVYSADPRKVNEAVILQTLSYQEAWEMSYFGANVLHPRTIIPVMRYNIPIV 368
Query: 366 IRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASA 425
IRNIFNLSAPGT IC P +D + + + VKGFATIDNLALINVEGTGMAGVPGTAS
Sbjct: 369 IRNIFNLSAPGTIICQPPEDDYD--LKLTTPVKGFATIDNLALINVEGTGMAGVPGTASD 426
Query: 426 IFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIP 485
IFG VKDVGANVIMISQASSEHS+CFAVPEKEV AV+EAL+SRF AL GRLSQ+ +IP
Sbjct: 427 IFGCVKDVGANVIMISQASSEHSVCFAVPEKEVNAVSEALRSRFSEALQAGRLSQIEVIP 486
Query: 486 NCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRA 545
NCSILAAVGQKMASTPGVS TLF+ALAKANINVRAI+QGCSEYN+TVV+KRED VKALRA
Sbjct: 487 NCSILAAVGQKMASTPGVSCTLFSALAKANINVRAISQGCSEYNVTVVIKREDSVKALRA 546
Query: 546 VHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSD 605
VHSRF+LSRTT+AMGI+GPGLIG+TLLDQLRDQA++LK+EFNIDLRV+GI GSK MLLSD
Sbjct: 547 VHSRFFLSRTTLAMGIVGPGLIGATLLDQLRDQAAVLKQEFNIDLRVLGITGSKKMLLSD 606
Query: 606 WGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGI 665
GIDL+RWREL +E G A+L+KF Q VHGNHFIPN+ +VDCTADS IA YYDWLRKGI
Sbjct: 607 IGIDLSRWRELLNEKGTEADLDKFTQQVHGNHFIPNSVVVDCTADSAIASRYYDWLRKGI 666
Query: 666 HVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQI 725
HV+TPNKKANSGPLDQYLKLR LQR+SYTHYFYEATVGAGLPI+STLRGLLETGD+IL+I
Sbjct: 667 HVITPNKKANSGPLDQYLKLRDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRI 726
Query: 726 EGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGL 785
EGI SGTLSY+FNNF R+FSEVV EAK AG+TEPDPRDDLSGTDVARKVIILARESGL
Sbjct: 727 EGICSGTLSYLFNNFVGDRSFSEVVTEAKNAGFTEPDPRDDLSGTDVARKVIILARESGL 786
Query: 786 KLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVT 845
KL+L+++P+ SLVPEPL+ C S +EFM++LP++D AK+ +AEN+GEVLRYVGVVD
Sbjct: 787 KLDLADLPIRSLVPEPLKGCTSVEEFMEKLPQYDGDLAKERLDAENSGEVLRYVGVVDAV 846
Query: 846 SKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDIL 905
++KG VELRRYKK+HPFAQL+GSDNIIAFTT RYKD PLIVRGPGAGAQVTAGGIFSDIL
Sbjct: 847 NQKGTVELRRYKKEHPFAQLAGSDNIIAFTTTRYKDHPLIVRGPGAGAQVTAGGIFSDIL 906
Query: 906 RLASYLGAPS 915
RLASYLGAPS
Sbjct: 907 RLASYLGAPS 916
>ref|NP_174408.1| bifunctional aspartate kinase/homoserine dehydrogenase / AK-HSDH
[Arabidopsis thaliana] gi|4512620|gb|AAD21689.1|
Identical to gb|X71364 gene for aspartate kinase
homoserine dehydrogenase from Arabidopsis thaliana
gi|25288158|pir||E86438 hypothetical protein F28K20.19 -
Arabidopsis thaliana
Length = 911
Score = 1359 bits (3518), Expect = 0.0
Identities = 683/891 (76%), Positives = 783/891 (87%), Gaps = 10/891 (1%)
Query: 33 LPFLPSHRSHSLRKALSLLPRGN--------QSPSTKISASLTDVSPSVLVEEQQLQKGE 84
+PF+ R S R + L R + + S ++ S+TD++ VE L KG+
Sbjct: 23 VPFIYGKRLVSNRVSFGKLRRRSCIGQCVRSELQSPRVLGSVTDLALDNSVENGHLPKGD 82
Query: 85 TWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDES 144
+W+VHKFGGTC+G+S+RIK+V +V++DDSERKLVVVSAMSKVTDMMY+LI++A+SRD+S
Sbjct: 83 SWAVHKFGGTCVGNSERIKDVAAVVVKDDSERKLVVVSAMSKVTDMMYDLIHRAESRDDS 142
Query: 145 YVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFV 204
Y+S+L VLEKH TA DLLDGD L++FL++L+ DI+NLKAMLRAIYIAGHATESF+DFV
Sbjct: 143 YLSALSGVLEKHRATAVDLLDGDELSSFLARLNDDINNLKAMLRAIYIAGHATESFSDFV 202
Query: 205 VGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLN 264
VGHGELWSAQML+ V+RK+G DC WMD RDVL+V PTSSNQVDPD++ESEKRLE WF+ N
Sbjct: 203 VGHGELWSAQMLAAVVRKSGLDCTWMDARDVLVVIPTSSNQVDPDFVESEKRLEKWFTQN 262
Query: 265 PCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKV 324
K+IIATGFIASTPQ IPTTLKRDGSDFSAAIM ALFR+ Q+TIWTDVDGVYSADPRKV
Sbjct: 263 SAKIIIATGFIASTPQNIPTTLKRDGSDFSAAIMSALFRSHQLTIWTDVDGVYSADPRKV 322
Query: 325 SEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSV 384
SEAV+LKTLSYQEAWEMSYFGANVLHPRTIIPVM+Y IPI+IRNIFNLSAPGT IC
Sbjct: 323 SEAVVLKTLSYQEAWEMSYFGANVLHPRTIIPVMKYDIPIVIRNIFNLSAPGTMICRQI- 381
Query: 385 NDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQAS 444
D+ED + VKGFATIDNLAL+NVEGTGMAGVPGTASAIF AVK+VGANVIMISQAS
Sbjct: 382 -DDEDGFKLDAPVKGFATIDNLALVNVEGTGMAGVPGTASAIFSAVKEVGANVIMISQAS 440
Query: 445 SEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVS 504
SEHS+CFAVPEKEVKAV+EAL SRFR AL GRLSQ+ IIPNCSILAAVGQKMASTPGVS
Sbjct: 441 SEHSVCFAVPEKEVKAVSEALNSRFRQALAGGRLSQIEIIPNCSILAAVGQKMASTPGVS 500
Query: 505 ATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGP 564
AT FNALAKANIN+RAIAQGCSE+NITVV+KREDC++ALRAVHSRFYLSRTT+A+GIIGP
Sbjct: 501 ATFFNALAKANINIRAIAQGCSEFNITVVVKREDCIRALRAVHSRFYLSRTTLAVGIIGP 560
Query: 565 GLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVA 624
GLIG TLLDQ+RDQA++LKEEF IDLRV+GI GS ML+S+ GIDL+RWREL E GE A
Sbjct: 561 GLIGGTLLDQIRDQAAVLKEEFKIDLRVIGITGSSKMLMSESGIDLSRWRELMKEEGEKA 620
Query: 625 NLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLK 684
++EKF Q+V GNHFIPN+ +VDCTAD+ IA YYDWL +GIHVVTPNKKANSGPLDQYLK
Sbjct: 621 DMEKFTQYVKGNHFIPNSVMVDCTADADIASCYYDWLLRGIHVVTPNKKANSGPLDQYLK 680
Query: 685 LRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGR 744
+R LQR+SYTHYFYEATVGAGLPI+STLRGLLETGD+IL+IEGIFSGTLSY+FNNF R
Sbjct: 681 IRDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRIEGIFSGTLSYLFNNFVGTR 740
Query: 745 AFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQA 804
+FSEVV EAK+AG+TEPDPRDDLSGTDVARKV ILARESGLKL+L +PV++LVP+PLQA
Sbjct: 741 SFSEVVAEAKQAGFTEPDPRDDLSGTDVARKVTILARESGLKLDLEGLPVQNLVPKPLQA 800
Query: 805 CASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQ 864
CASA+EFM++LP+FD+ +K+ EEAE AGEVLRYVGVVD KKG VEL+RYKKDHPFAQ
Sbjct: 801 CASAEEFMEKLPQFDEELSKQREEAEAAGEVLRYVGVVDAVEKKGTVELKRYKKDHPFAQ 860
Query: 865 LSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAPS 915
LSG+DNIIAFTT+RYK+QPLIVRGPGAGAQVTAGGIFSDILRLA YLGAPS
Sbjct: 861 LSGADNIIAFTTKRYKEQPLIVRGPGAGAQVTAGGIFSDILRLAFYLGAPS 911
>dbj|BAC43372.1| putative aspartate kinase-homoserine dehydrogenase [Arabidopsis
thaliana]
Length = 911
Score = 1357 bits (3511), Expect = 0.0
Identities = 681/891 (76%), Positives = 782/891 (87%), Gaps = 10/891 (1%)
Query: 33 LPFLPSHRSHSLRKALSLLPRGN--------QSPSTKISASLTDVSPSVLVEEQQLQKGE 84
+PF+ R S R + L R + + S ++ S+TD++ VE L KG+
Sbjct: 23 VPFIYGKRLVSNRVSFGKLRRRSCIGQCVRSELQSPRVLGSVTDLALDNSVENGHLPKGD 82
Query: 85 TWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDES 144
+W+VHKFGGTC+G+S+RIK+V +V++DDSERKLVVVSAMSKVTDMMY+LI++A+SRD+S
Sbjct: 83 SWAVHKFGGTCVGNSERIKDVAAVVVKDDSERKLVVVSAMSKVTDMMYDLIHRAESRDDS 142
Query: 145 YVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFV 204
Y+S+L VLEKH TA DLLDGD L++FL++L+ DI+NLKAMLRAIYIAGHATESF+DFV
Sbjct: 143 YLSALSGVLEKHRATAVDLLDGDELSSFLARLNDDINNLKAMLRAIYIAGHATESFSDFV 202
Query: 205 VGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLN 264
VGHGELWSAQML+ V+RK+G DC WMD RDVL+V PTSSNQVDPD++ESEKRLE WF+ N
Sbjct: 203 VGHGELWSAQMLAAVVRKSGLDCTWMDARDVLVVIPTSSNQVDPDFVESEKRLEKWFTQN 262
Query: 265 PCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKV 324
K+IIATGFIASTPQ IPTTLKRDGSDFSAAIM ALFR+ Q+TIWTDVDGVYSADPRKV
Sbjct: 263 SAKIIIATGFIASTPQNIPTTLKRDGSDFSAAIMSALFRSHQLTIWTDVDGVYSADPRKV 322
Query: 325 SEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSV 384
SEAV+LKTLSYQEAWEMSYFGANVLHPRTIIPVM+Y IPI+IRNIFNLSAPGT IC
Sbjct: 323 SEAVVLKTLSYQEAWEMSYFGANVLHPRTIIPVMKYDIPIVIRNIFNLSAPGTMICRQI- 381
Query: 385 NDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQAS 444
D+ED + VKGFATIDNLAL+NVEGTGMAGVPGTASAIF AVK+VGANVIMISQAS
Sbjct: 382 -DDEDGFKLDAPVKGFATIDNLALVNVEGTGMAGVPGTASAIFSAVKEVGANVIMISQAS 440
Query: 445 SEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVS 504
SEHS+CFAVPEKEVKAV+EAL SRFR AL GRLSQ+ IIPNCSILAAVGQKMASTPGVS
Sbjct: 441 SEHSVCFAVPEKEVKAVSEALNSRFRQALAGGRLSQIEIIPNCSILAAVGQKMASTPGVS 500
Query: 505 ATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGP 564
AT FNALAKANIN+RA+AQGCSE+NITVV+KREDC++ALRAVHSRFYLSRTT+A+GIIGP
Sbjct: 501 ATFFNALAKANINIRAMAQGCSEFNITVVVKREDCIRALRAVHSRFYLSRTTLAVGIIGP 560
Query: 565 GLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVA 624
GLIG TLLDQ+RDQA++LKEEF IDLRV+GI GS ML+S+ GIDL+RWREL E GE A
Sbjct: 561 GLIGGTLLDQIRDQAAVLKEEFKIDLRVIGITGSSKMLMSESGIDLSRWRELMKEEGEKA 620
Query: 625 NLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLK 684
++EKF Q+V GNHFIPN+ +VDCTAD+ IA YYDWL +GIHVVTPNKKANSGPLDQYLK
Sbjct: 621 DMEKFTQYVKGNHFIPNSVMVDCTADADIASCYYDWLLRGIHVVTPNKKANSGPLDQYLK 680
Query: 685 LRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGR 744
+R LQR+SYTHYFYEATVGAGLPI+STLRGLLETGD+IL+IEGIFSGTLSY+FNNF R
Sbjct: 681 IRDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRIEGIFSGTLSYLFNNFVGTR 740
Query: 745 AFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQA 804
+FSEVV EAK+AG+TEPDPRDDLSGTDVARKV ILARESGLKL+L +PV++LVP+PLQA
Sbjct: 741 SFSEVVAEAKQAGFTEPDPRDDLSGTDVARKVTILARESGLKLDLEGLPVQNLVPKPLQA 800
Query: 805 CASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQ 864
CASA+EFM++LP+FD+ +K+ EEAE AGEVLRYVGVVD KKG VEL+RYKKDHPFAQ
Sbjct: 801 CASAEEFMEKLPQFDEELSKQREEAEAAGEVLRYVGVVDAVEKKGTVELKRYKKDHPFAQ 860
Query: 865 LSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAPS 915
LSG+DNIIAFTT+RYK+QPLIVRGPGAGAQVTAGGIFSD LRLA YLGAPS
Sbjct: 861 LSGADNIIAFTTKRYKEQPLIVRGPGAGAQVTAGGIFSDFLRLAFYLGAPS 911
>pir||S46497 aspartate kinase (EC 2.7.2.4) / homoserine dehydrogenase (EC
1.1.1.3) precursor - Arabidopsis thaliana
Length = 911
Score = 1355 bits (3508), Expect = 0.0
Identities = 680/891 (76%), Positives = 781/891 (87%), Gaps = 10/891 (1%)
Query: 33 LPFLPSHRSHSLRKALSLLPRGN--------QSPSTKISASLTDVSPSVLVEEQQLQKGE 84
+PF+ R S R + L R + + S ++ S+TD++ VE L KG+
Sbjct: 23 VPFIYGKRLVSNRVSFGKLRRRSCIGQCVRSELQSPRVLGSVTDLALDNSVENGHLPKGD 82
Query: 85 TWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDES 144
+W+VHKFGGTC+G+S+RIK+V +V++DDSERKLVVVSAMSKVTDMMY+LI++A+SRD+S
Sbjct: 83 SWAVHKFGGTCVGNSERIKDVAAVVVKDDSERKLVVVSAMSKVTDMMYDLIHRAESRDDS 142
Query: 145 YVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFV 204
Y+S+L VLEKH TA DLLDGD L++FL++L+ DI+NLKAMLRAIYIAGHATESF+DFV
Sbjct: 143 YLSALSGVLEKHRATAVDLLDGDELSSFLARLNDDINNLKAMLRAIYIAGHATESFSDFV 202
Query: 205 VGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLN 264
VGHGELWSAQML+ V+RK+G DC WMD RDVL+V PTSSNQVDPD++ESEKRLE WF+ N
Sbjct: 203 VGHGELWSAQMLAAVVRKSGLDCTWMDARDVLVVIPTSSNQVDPDFVESEKRLEKWFTQN 262
Query: 265 PCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKV 324
K+IIATGFIASTPQ IPTTLKRDGSDFSAAIM ALFR+ Q+TIWTDVDGVYSADPRKV
Sbjct: 263 SAKIIIATGFIASTPQNIPTTLKRDGSDFSAAIMSALFRSHQLTIWTDVDGVYSADPRKV 322
Query: 325 SEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSV 384
SEAV+LKTLSYQEAWEMSYFGANVLHPRTIIPVM+Y IPI+IRNIFNLSAPGT IC
Sbjct: 323 SEAVVLKTLSYQEAWEMSYFGANVLHPRTIIPVMKYDIPIVIRNIFNLSAPGTMICRQI- 381
Query: 385 NDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQAS 444
D+ED + VKGFATIDNLAL+NVEG GMAGVPGTASAIF AVK+VGANVIMISQAS
Sbjct: 382 -DDEDGFKLDAPVKGFATIDNLALVNVEGAGMAGVPGTASAIFSAVKEVGANVIMISQAS 440
Query: 445 SEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVS 504
SEHS+CFAVPEKEVKAV+E L SRFR AL GRLSQ+ IIPNCSILAAVGQKMASTPGVS
Sbjct: 441 SEHSVCFAVPEKEVKAVSEELNSRFRQALAGGRLSQIEIIPNCSILAAVGQKMASTPGVS 500
Query: 505 ATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGP 564
AT FNALAKANIN+RAIAQGCSE+NITVV+KREDC++ALRAVHSRFYLSRTT+A+GIIGP
Sbjct: 501 ATFFNALAKANINIRAIAQGCSEFNITVVVKREDCIRALRAVHSRFYLSRTTLAVGIIGP 560
Query: 565 GLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVA 624
GLIG TLLDQ+RDQA++LKEEF IDLRV+GI GS ML+S+ GIDL+RWREL E GE A
Sbjct: 561 GLIGGTLLDQIRDQAAVLKEEFKIDLRVIGITGSSKMLMSESGIDLSRWRELMKEEGEKA 620
Query: 625 NLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLK 684
++EKF Q+V GNHFIPN+ +VDCTAD+ IA YYDWL +GIHVVTPNKKANSGPLDQYLK
Sbjct: 621 DMEKFTQYVKGNHFIPNSVMVDCTADADIASCYYDWLLRGIHVVTPNKKANSGPLDQYLK 680
Query: 685 LRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGR 744
+R LQR+SYTHYFYEATVGAGLPI+STLRGLLETGD+IL+IEGIFSGTLSY+FNNF R
Sbjct: 681 IRDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRIEGIFSGTLSYLFNNFAGTR 740
Query: 745 AFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQA 804
+FSEVV EAK+AG+TEPDPRDDLSGTDVARKV ILARESGLKL+L +P+++LVP+PLQA
Sbjct: 741 SFSEVVAEAKQAGFTEPDPRDDLSGTDVARKVTILARESGLKLDLEGLPIQNLVPKPLQA 800
Query: 805 CASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVVDVTSKKGVVELRRYKKDHPFAQ 864
CASA+EFM++LP+FD+ +K+ EEAE AGEVLRYVGVVD KKG VEL+RYKKDHPFAQ
Sbjct: 801 CASAEEFMEKLPQFDEELSKQREEAEAAGEVLRYVGVVDAVEKKGTVELKRYKKDHPFAQ 860
Query: 865 LSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGAPS 915
LSG+DNIIAFTT+RYK+QPLIVRGPGAGAQVTAGGIFSDILRLA YLGAPS
Sbjct: 861 LSGADNIIAFTTKRYKEQPLIVRGPGAGAQVTAGGIFSDILRLAFYLGAPS 911
>ref|XP_481775.1| putative aspartate kinase, homoserine dehydrogenase [Oryza sativa
(japonica cultivar-group)] gi|38423990|dbj|BAD01718.1|
putative aspartate kinase, homoserine dehydrogenase
[Oryza sativa (japonica cultivar-group)]
Length = 921
Score = 1320 bits (3415), Expect = 0.0
Identities = 660/858 (76%), Positives = 752/858 (86%), Gaps = 7/858 (0%)
Query: 62 ISASLTDVSPSVLVEEQQ----LQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERK 117
+S ++ + +V VE+ + L KG+ WSVHKFGGTCMG+SQRI+NV DI+L D SERK
Sbjct: 67 VSKNMLKPAAAVSVEQAEASAHLPKGDMWSVHKFGGTCMGTSQRIQNVADIILRDPSERK 126
Query: 118 LVVVSAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLH 177
LVVVSAMSKVTDMMY L+NKAQSRD+SY+++LD V EKH A DLL G+ LA FLSQLH
Sbjct: 127 LVVVSAMSKVTDMMYNLVNKAQSRDDSYITALDEVFEKHMAAAKDLLGGEDLARFLSQLH 186
Query: 178 HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLI 237
D+SNLKAMLRAI IAGHATESF+DFVVGHGE+WSAQ+LS I+K+G C WMDTR+VL+
Sbjct: 187 ADVSNLKAMLRAICIAGHATESFSDFVVGHGEIWSAQLLSFAIKKSGTPCSWMDTREVLV 246
Query: 238 VNPTSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAI 297
VNPT SNQVDPDYLESEKRLE WF+ P + IIATGFIASTP+ IPTTLKRDGSDFSAAI
Sbjct: 247 VNPTGSNQVDPDYLESEKRLEKWFARQPAETIIATGFIASTPENIPTTLKRDGSDFSAAI 306
Query: 298 MGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV 357
+G+L +A QVTIWTDVDGV+SADPRKVSEAVIL TLSYQEAWEMSYFGANVLHPRTIIPV
Sbjct: 307 IGSLVKAGQVTIWTDVDGVFSADPRKVSEAVILSTLSYQEAWEMSYFGANVLHPRTIIPV 366
Query: 358 MQYGIPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMA 417
M+Y IPI+IRN+FN+SAPGT IC N++ D+ + VK FATID L+L+NVEGTGMA
Sbjct: 367 MKYNIPIVIRNMFNISAPGTMICQQPANESGDL---EACVKAFATIDKLSLVNVEGTGMA 423
Query: 418 GVPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGR 477
GVPGTASAIFGAVKDVGANVIMISQASSEHS+CFAVPEKEV AV+ AL RFR AL GR
Sbjct: 424 GVPGTASAIFGAVKDVGANVIMISQASSEHSVCFAVPEKEVAAVSAALHVRFREALSAGR 483
Query: 478 LSQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKRE 537
LS+V +I NCSILAAVG KMASTPGVSATLF+ALAKANINVRAIAQGCSEYNITVVLK+E
Sbjct: 484 LSKVEVIHNCSILAAVGLKMASTPGVSATLFDALAKANINVRAIAQGCSEYNITVVLKQE 543
Query: 538 DCVKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILG 597
DCV+ALRA HSRF+LS+TT+A+GIIGPGLIG TLL+QL+DQA++LKE NIDLRVMGI G
Sbjct: 544 DCVRALRAAHSRFFLSKTTLAVGIIGPGLIGRTLLNQLKDQAAVLKENMNIDLRVMGITG 603
Query: 598 SKSMLLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHY 657
S++M+LSD GIDLA W+E E ANL+KFV H+ N PN +VDCTAD+ +A HY
Sbjct: 604 SRTMVLSDTGIDLAHWKEQLQTEAEPANLDKFVDHLSENQLFPNRVLVDCTADTSVASHY 663
Query: 658 YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 717
YDWL+KGIHV+TPNKKANSGPLD+YLKLR LQR SYTHYFYEATVGAGLPI+STLRGLLE
Sbjct: 664 YDWLKKGIHVITPNKKANSGPLDKYLKLRTLQRASYTHYFYEATVGAGLPIISTLRGLLE 723
Query: 718 TGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVI 777
TGD+IL+IEGIFSGTLSYIFNNF+ RAFS+VV EAKEAGYTEPDPRDDLSGTDVARKVI
Sbjct: 724 TGDKILRIEGIFSGTLSYIFNNFEGTRAFSDVVSEAKEAGYTEPDPRDDLSGTDVARKVI 783
Query: 778 ILARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLR 837
ILARESGLKLELS+IPV SLVPE L++C++A E+MQ+LP FDQ +A++ ++AE AGEVLR
Sbjct: 784 ILARESGLKLELSDIPVRSLVPEALRSCSTADEYMQKLPSFDQDWARESKDAEAAGEVLR 843
Query: 838 YVGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTA 897
YVGVVD+ +K+G VELRRYKKDHPFAQLSGSDNIIAFTT RYK+QPLIVRGPGAGA+VTA
Sbjct: 844 YVGVVDLVNKEGQVELRRYKKDHPFAQLSGSDNIIAFTTSRYKEQPLIVRGPGAGAEVTA 903
Query: 898 GGIFSDILRLASYLGAPS 915
GG+FSDILRLASYLGAPS
Sbjct: 904 GGVFSDILRLASYLGAPS 921
>sp|P49079|AKH1_MAIZE Bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplast
precursor (AK-HD 1) (AK-HSDH 1) [Includes: Aspartokinase
; Homoserine dehydrogenase ] gi|500851|gb|AAA74360.1|
aspartate kinase-homoserine dehydrogenase
Length = 920
Score = 1315 bits (3404), Expect = 0.0
Identities = 660/854 (77%), Positives = 744/854 (86%), Gaps = 7/854 (0%)
Query: 66 LTDVSPSVLVEEQQ----LQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVV 121
L + +V VE+ + L KG+ WSVHKFGGTCMG+S+RI NV DIVL D SERKLVVV
Sbjct: 70 LAPTAGAVSVEQAEAIADLPKGDMWSVHKFGGTCMGTSERIHNVADIVLRDPSERKLVVV 129
Query: 122 SAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDIS 181
SAMSKVTDMMY L+NKAQSRD+SY++ LD V +KH TA DLL G+ LA FLSQLH DIS
Sbjct: 130 SAMSKVTDMMYNLVNKAQSRDDSYIAVLDEVFDKHMTTAKDLLAGEDLARFLSQLHADIS 189
Query: 182 NLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPT 241
NLKAMLRAIYIAGHATESF+DFVVGHGELWSAQMLS I+K+G C WMDTR+VL+VNP+
Sbjct: 190 NLKAMLRAIYIAGHATESFSDFVVGHGELWSAQMLSYAIQKSGTPCSWMDTREVLVVNPS 249
Query: 242 SSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGAL 301
+NQVDPDYLESEKRLE WFS P + IIATGFIASTP+ IPTTLKRDGSDFSAAI+G+L
Sbjct: 250 GANQVDPDYLESEKRLEKWFSRCPAETIIATGFIASTPENIPTTLKRDGSDFSAAIIGSL 309
Query: 302 FRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYG 361
+ARQVTIWTDVDGV+SADPRKVSEAVIL TLSYQEAWEMSYFGANVLHPRTIIPVM+Y
Sbjct: 310 VKARQVTIWTDVDGVFSADPRKVSEAVILSTLSYQEAWEMSYFGANVLHPRTIIPVMKYN 369
Query: 362 IPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPG 421
IPI+IRNIFN SAPGT IC N+N D+ + VK FATID LAL+NVEGTGMAGVPG
Sbjct: 370 IPIVIRNIFNTSAPGTMICQQPANENGDL---EACVKAFATIDKLALVNVEGTGMAGVPG 426
Query: 422 TASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQV 481
TA+AIFGAVKDVGANVIMISQASSEHS+CFAVPEKEV V+ AL +RFR AL GRLS+V
Sbjct: 427 TANAIFGAVKDVGANVIMISQASSEHSVCFAVPEKEVALVSAALHARFREALAAGRLSKV 486
Query: 482 AIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVK 541
+I NCSILA VG +MASTPGVSATLF+ALAKANINVRAIAQGCSEYNIT+VLK+EDCV+
Sbjct: 487 EVIHNCSILATVGLRMASTPGVSATLFDALAKANINVRAIAQGCSEYNITIVLKQEDCVR 546
Query: 542 ALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSM 601
ALRA HSRF+LS+TT+A+GIIGPGLIG TLL+QL+DQA++LKE NIDLRVMGI GS++M
Sbjct: 547 ALRAAHSRFFLSKTTLAVGIIGPGLIGRTLLNQLKDQAAVLKENMNIDLRVMGIAGSRTM 606
Query: 602 LLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWL 661
LLSD G+DL +W+E E ANL+KFV H+ NHF PN +VDCTAD+ +A HYYDWL
Sbjct: 607 LLSDIGVDLTQWKEKLQTEAEPANLDKFVHHLSENHFFPNRVLVDCTADTSVASHYYDWL 666
Query: 662 RKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDR 721
+KGIHV+TPNKKANSGPLD+YLKLR LQR SYTHYFYEATVGAGLPI+STLRGLLETGD+
Sbjct: 667 KKGIHVITPNKKANSGPLDRYLKLRTLQRASYTHYFYEATVGAGLPIISTLRGLLETGDK 726
Query: 722 ILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILAR 781
IL+IEGIFSGTLSYIFNNF+ R FS+VV EAK+AGYTEPDPRDDLSGTDVARKVIILAR
Sbjct: 727 ILRIEGIFSGTLSYIFNNFEGARTFSDVVAEAKKAGYTEPDPRDDLSGTDVARKVIILAR 786
Query: 782 ESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGV 841
ESGL LELS+IPV SLVPE L++C SA E+MQ+LP FD+ +A++ + AE AGEVLRYVGV
Sbjct: 787 ESGLGLELSDIPVRSLVPEALKSCTSADEYMQKLPSFDEDWARERKNAEAAGEVLRYVGV 846
Query: 842 VDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIF 901
VDV SKKG VELR YK+DHPFAQLSGSDNIIAFTT RYKDQPLIVRGPGAGA+VTAGG+F
Sbjct: 847 VDVVSKKGQVELRAYKRDHPFAQLSGSDNIIAFTTSRYKDQPLIVRGPGAGAEVTAGGVF 906
Query: 902 SDILRLASYLGAPS 915
DILRL+SYLGAPS
Sbjct: 907 CDILRLSSYLGAPS 920
>dbj|BAD34058.1| aspartate kinase-homoserine dehydrogenase [Oryza sativa (japonica
cultivar-group)]
Length = 915
Score = 1289 bits (3336), Expect = 0.0
Identities = 647/857 (75%), Positives = 743/857 (86%), Gaps = 7/857 (0%)
Query: 63 SASLTDVSPSVLVEEQQ----LQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKL 118
S +L + ++ +E+ + L KG+ WSVHKFGGTCMG+ QRI+NV DIVL D SERKL
Sbjct: 62 SRNLLAAAAAISIEQAEVSTYLPKGDMWSVHKFGGTCMGTPQRIQNVADIVLGDSSERKL 121
Query: 119 VVVSAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHH 178
++VSAMSKVTDMM+ L++KAQSRD SYV++LD V KH A +LLDG+ LA FL+QLH
Sbjct: 122 IIVSAMSKVTDMMFNLVHKAQSRDNSYVTALDEVFNKHMAAAKELLDGEDLARFLAQLHS 181
Query: 179 DISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIV 238
DISNL+AMLRAI+IAGHATESF+DFVVGHGELWSAQMLS I+K+G C WMDTR+VL+V
Sbjct: 182 DISNLRAMLRAIFIAGHATESFSDFVVGHGELWSAQMLSYAIKKSGVPCSWMDTREVLVV 241
Query: 239 NPTSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIM 298
P+ SNQVDPDYLESEKRL+ WFS P ++IIATGFIAST + IPTTLKRDGSDFSA+I+
Sbjct: 242 KPSGSNQVDPDYLESEKRLQKWFSRQPAEIIIATGFIASTAENIPTTLKRDGSDFSASII 301
Query: 299 GALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVM 358
G+L RA QVTIWTDVDGV+SADPRKVSEAVIL TLSYQEAWEMSYFGANVLHPRTIIPVM
Sbjct: 302 GSLVRACQVTIWTDVDGVFSADPRKVSEAVILSTLSYQEAWEMSYFGANVLHPRTIIPVM 361
Query: 359 QYGIPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAG 418
+ IPI+IRN+FNLSAPGT IC N+N D+ VK FATID LAL+NVEGTGMAG
Sbjct: 362 KDNIPIVIRNMFNLSAPGTTICKQPANENADL---DACVKSFATIDKLALVNVEGTGMAG 418
Query: 419 VPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRL 478
VPGTASAIF A KDVGANVIMISQASSEHS+CFAVPEKEV AV+ AL RFR AL GRL
Sbjct: 419 VPGTASAIFSAAKDVGANVIMISQASSEHSVCFAVPEKEVAAVSTALHVRFREALAAGRL 478
Query: 479 SQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKRED 538
S+V +I CSILAAVG +MASTPGVSA LF+ALAKANINVRAIAQGCSEYNITVVLK+ED
Sbjct: 479 SKVEVIRGCSILAAVGLRMASTPGVSAILFDALAKANINVRAIAQGCSEYNITVVLKQED 538
Query: 539 CVKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGS 598
CV+ALRAVHSRF+LS+TT+A+GIIGPGLIG TLLDQL+DQA++LKE NIDLRV+GI GS
Sbjct: 539 CVRALRAVHSRFFLSKTTLAVGIIGPGLIGGTLLDQLKDQAAVLKENMNIDLRVIGISGS 598
Query: 599 KSMLLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYY 658
++M LSD G+DL +W+EL + E A+L+ FV+H+ NH PN +VDCTAD+ +A HYY
Sbjct: 599 RTMHLSDIGVDLNQWKELLRKEAEPADLDSFVRHLSENHVFPNKVLVDCTADTYVACHYY 658
Query: 659 DWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLET 718
DWL+KGIHV+TPNKKANSGPLD+YLKLR LQR SYTHYFYEATVGAGLPI+STLRGLLET
Sbjct: 659 DWLKKGIHVITPNKKANSGPLDRYLKLRTLQRASYTHYFYEATVGAGLPIISTLRGLLET 718
Query: 719 GDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVII 778
GD+IL+IEGIFSGTLSYIFNNF+ R FS VV EAKEAGYTEPDPRDDLSGTDVARKVII
Sbjct: 719 GDKILRIEGIFSGTLSYIFNNFEGTRTFSNVVAEAKEAGYTEPDPRDDLSGTDVARKVII 778
Query: 779 LARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRY 838
LARESGL+LELS+IPV+SLVPE L++C+SA EFMQ+LP FDQ + ++ +EAE AGEVLRY
Sbjct: 779 LARESGLRLELSDIPVKSLVPEALRSCSSADEFMQKLPSFDQDWDRQRDEAEAAGEVLRY 838
Query: 839 VGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAG 898
VGVVDV ++KG VEL+RYK+DHPFAQLSGSDNIIAFTT RYK+QPLIVRGPGAGA+VTAG
Sbjct: 839 VGVVDVANRKGRVELQRYKRDHPFAQLSGSDNIIAFTTSRYKEQPLIVRGPGAGAEVTAG 898
Query: 899 GIFSDILRLASYLGAPS 915
G+F DILRLASYLGAPS
Sbjct: 899 GVFCDILRLASYLGAPS 915
>pir||T03589 probable aspartate kinase (EC 2.7.2.4) / homoserine dehydrogenase
(EC 1.1.1.3) precursor - rice gi|1777375|dbj|BAA11417.1|
aspartate kinase-homoserine dehydrogenase [Oryza sativa
(japonica cultivar-group)]
Length = 915
Score = 1284 bits (3323), Expect = 0.0
Identities = 644/857 (75%), Positives = 742/857 (86%), Gaps = 7/857 (0%)
Query: 63 SASLTDVSPSVLVEEQQ----LQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKL 118
S +L + ++ +E+ + L KG+ WSVHKFGGTCMG+ QRI+NV DIVL D SERKL
Sbjct: 62 SRNLLAAAAAISIEQAEVSTYLPKGDMWSVHKFGGTCMGTPQRIQNVADIVLGDSSERKL 121
Query: 119 VVVSAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHH 178
++VSAMSKVTDMM+ L++KAQSRD SYV++LD V KH A +LLDG+ LA FL+QLH
Sbjct: 122 IIVSAMSKVTDMMFNLVHKAQSRDNSYVTALDEVFNKHMAAAKELLDGEDLARFLAQLHS 181
Query: 179 DISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIV 238
DISNL+AMLRAI+IAGHATESF+DFVVGHGELWSAQMLS I+K+G C WMDTR+VL+V
Sbjct: 182 DISNLRAMLRAIFIAGHATESFSDFVVGHGELWSAQMLSYAIKKSGVPCSWMDTREVLVV 241
Query: 239 NPTSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIM 298
P+ SNQVDPDYLESEKRL+ WFS P ++IIATGFIAST + IPTTLKRDGSDFSA+I+
Sbjct: 242 KPSGSNQVDPDYLESEKRLQKWFSRQPAEIIIATGFIASTAENIPTTLKRDGSDFSASII 301
Query: 299 GALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVM 358
G+L RA QVTIWTDVDGV+SADPRKVSEAVIL TLSYQEAWEMSYFGANVLHPRTIIPVM
Sbjct: 302 GSLVRACQVTIWTDVDGVFSADPRKVSEAVILSTLSYQEAWEMSYFGANVLHPRTIIPVM 361
Query: 359 QYGIPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAG 418
+ IPI+IRN+FNLSAPGT IC N+N D+ VK FATID LAL+NVEGTGMAG
Sbjct: 362 KDNIPIVIRNMFNLSAPGTTICKQPANENADL---DACVKSFATIDKLALVNVEGTGMAG 418
Query: 419 VPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRL 478
VPGTASAIF A KDVGAN IMISQASSEHS+CFAVPEKEV AV+ AL RFR AL GRL
Sbjct: 419 VPGTASAIFSAAKDVGANGIMISQASSEHSVCFAVPEKEVAAVSTALHVRFREALAAGRL 478
Query: 479 SQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKRED 538
S+V +I CSILAAVG +MASTPGVSA LF+ALAKANINVRAIAQGCSEYNITVVLK+ED
Sbjct: 479 SKVEVIRGCSILAAVGLRMASTPGVSAILFDALAKANINVRAIAQGCSEYNITVVLKQED 538
Query: 539 CVKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGS 598
CV+ALRAVHSRF+LS+TT+A+GIIGPGLIG TLLDQL+DQA++LKE NIDLRV+GI GS
Sbjct: 539 CVRALRAVHSRFFLSKTTLAVGIIGPGLIGGTLLDQLKDQAAVLKENMNIDLRVIGISGS 598
Query: 599 KSMLLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYY 658
++M LSD G+DL +W++L + E A+L+ FV+H+ NH PN +VDCTAD+ +A HYY
Sbjct: 599 RTMHLSDIGVDLNQWKDLLRKEAEPADLDSFVRHLSENHVFPNKVLVDCTADTYVACHYY 658
Query: 659 DWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLET 718
DWL+KGIHV+TPNKKANSGPLD+YLKLR LQR SYTHYFYEATVGAGLPI+STLRGLLET
Sbjct: 659 DWLKKGIHVITPNKKANSGPLDRYLKLRTLQRASYTHYFYEATVGAGLPIISTLRGLLET 718
Query: 719 GDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVII 778
GD+IL+IEGIFSGTLSYIFNNF+ R FS VV EAKEAGYTEPDPRDDLSGTDVARKVII
Sbjct: 719 GDKILRIEGIFSGTLSYIFNNFEGTRTFSNVVAEAKEAGYTEPDPRDDLSGTDVARKVII 778
Query: 779 LARESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRY 838
LARESGL+LELS+IPV+SL+PE L++C+SA EFMQ+LP FDQ + ++ +EAE AGEVLRY
Sbjct: 779 LARESGLRLELSDIPVKSLLPEALRSCSSADEFMQKLPSFDQDWDRQRDEAEAAGEVLRY 838
Query: 839 VGVVDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAG 898
VGVVDV ++KG VEL+RYK+DHPFAQLSGSDNIIAFTT RYK+QPLIVRGPGAGA+VTAG
Sbjct: 839 VGVVDVANRKGRVELQRYKRDHPFAQLSGSDNIIAFTTSRYKEQPLIVRGPGAGAEVTAG 898
Query: 899 GIFSDILRLASYLGAPS 915
G+F DILRLASYLGAPS
Sbjct: 899 GVFCDILRLASYLGAPS 915
>sp|P49080|AKH2_MAIZE Bifunctional aspartokinase/homoserine dehydrogenase 2, chloroplast
precursor (AK-HD 2) (AK-HSDH 2) [Includes: Aspartokinase
; Homoserine dehydrogenase ] gi|500853|gb|AAA74361.1|
aspartate kinase-homoserine dehydrogenase
Length = 917
Score = 1270 bits (3287), Expect = 0.0
Identities = 632/854 (74%), Positives = 730/854 (85%), Gaps = 7/854 (0%)
Query: 66 LTDVSPSVLVEEQQ----LQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVV 121
L + ++ VE+ + L KG+ WSVHKFGGTCMG+ +RI+ V +IVL D SERKL++V
Sbjct: 67 LRPAAAAISVEQDEVNTYLPKGDMWSVHKFGGTCMGTPKRIQCVANIVLGDSSERKLIIV 126
Query: 122 SAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDIS 181
SAMSKVTDMMY L+ KAQSRD+SY +L V EKH A DLLDG+ LA FLSQLH D+S
Sbjct: 127 SAMSKVTDMMYNLVQKAQSRDDSYAIALAEVFEKHMTAAKDLLDGEDLARFLSQLHSDVS 186
Query: 182 NLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPT 241
NL+AMLRAIYIAGHATESF+DFVVGHGELWSAQMLS I+K+GA C WMDTR+VL+V P+
Sbjct: 187 NLRAMLRAIYIAGHATESFSDFVVGHGELWSAQMLSYAIKKSGAPCSWMDTREVLVVTPS 246
Query: 242 SSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGAL 301
NQVDPDYLE EKRL+ WFS P ++I+ATGFIAST IPTTLKRDGSDFSAAI+G+L
Sbjct: 247 GCNQVDPDYLECEKRLQKWFSRQPAEIIVATGFIASTAGNIPTTLKRDGSDFSAAIVGSL 306
Query: 302 FRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYG 361
RARQVTIWTDVDGV+SADPRKVSEAVIL TLSYQEAWEMSYFGANVLHPRTIIPVM+
Sbjct: 307 VRARQVTIWTDVDGVFSADPRKVSEAVILSTLSYQEAWEMSYFGANVLHPRTIIPVMKDN 366
Query: 362 IPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPG 421
IPI+IRN+FNLSAPGT IC N+N D+ VK FAT+DNLAL+NVEGTGMAGVPG
Sbjct: 367 IPIVIRNMFNLSAPGTMICKQPANENGDL---DACVKSFATVDNLALVNVEGTGMAGVPG 423
Query: 422 TASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQV 481
TASAIF AVKDVGANVIMISQASSEHS+CFAVPEKEV V+ L RFR AL GRLS+V
Sbjct: 424 TASAIFSAVKDVGANVIMISQASSEHSVCFAVPEKEVAVVSAELHDRFREALAAGRLSKV 483
Query: 482 AIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVK 541
+I CSILAAVG +MASTPGVSA LF+ALAKANINVRAIAQGCSEYNITVVLK++DCV+
Sbjct: 484 EVINGCSILAAVGLRMASTPGVSAILFDALAKANINVRAIAQGCSEYNITVVLKQQDCVR 543
Query: 542 ALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSM 601
ALRA HSRF+LS+TT+A+GIIGPGLIG LL+QL++Q ++LKE NIDLRV+GI GS +M
Sbjct: 544 ALRAAHSRFFLSKTTLAVGIIGPGLIGGALLNQLKNQTAVLKENMNIDLRVIGITGSSTM 603
Query: 602 LLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWL 661
LLSD GIDL +W++L + E A++ FV H+ NH PN +VDCTAD+ +A HYYDWL
Sbjct: 604 LLSDTGIDLTQWKQLLQKEAEPADIGSFVHHLSDNHVFPNKVLVDCTADTSVASHYYDWL 663
Query: 662 RKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDR 721
+KGIHV+TPNKKANSGPLDQYLKLR +QR SYTHYFYEATVGAGLPI+STLRGLLETGD+
Sbjct: 664 KKGIHVITPNKKANSGPLDQYLKLRTMQRASYTHYFYEATVGAGLPIISTLRGLLETGDK 723
Query: 722 ILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILAR 781
IL+IEGIFSGTLSYIFNNF+ RAFS+VV EA+EAGYTEPDPRDDLSGTDVARKV++LAR
Sbjct: 724 ILRIEGIFSGTLSYIFNNFEGTRAFSDVVAEAREAGYTEPDPRDDLSGTDVARKVVVLAR 783
Query: 782 ESGLKLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGV 841
ESGL+LELS+IPV+SLVPE L +C+SA EFMQ+LP FD+ +A++ +AE AGEVLRYVG
Sbjct: 784 ESGLRLELSDIPVKSLVPETLASCSSADEFMQKLPSFDEDWARQRSDAEAAGEVLRYVGA 843
Query: 842 VDVTSKKGVVELRRYKKDHPFAQLSGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIF 901
+D ++ G VELRRY++DHPFAQLSGSDNIIAFTT RYK+QPLIVRGPGAGA+VTAGG+F
Sbjct: 844 LDAVNRSGQVELRRYRRDHPFAQLSGSDNIIAFTTSRYKEQPLIVRGPGAGAEVTAGGVF 903
Query: 902 SDILRLASYLGAPS 915
DILRLASYLGAPS
Sbjct: 904 CDILRLASYLGAPS 917
>ref|NP_193706.2| bifunctional aspartate kinase/homoserine dehydrogenase, putative /
AK-HSDH, putative [Arabidopsis thaliana]
Length = 859
Score = 1245 bits (3222), Expect = 0.0
Identities = 631/830 (76%), Positives = 716/830 (86%), Gaps = 10/830 (1%)
Query: 14 TISPSNNTPDITKISQCQCLPFLPSHRSHSLRKALSLLPRGN-----QSPSTKISASLTD 68
T+SP N+ P QC +P R L + G + P +SA+ T
Sbjct: 9 TVSPPNSNPIRFGSFPPQCFLRVPKPRRLILPRFRKTTGGGGGLIRCELPDFHLSATATT 68
Query: 69 VSPSV---LVEEQQLQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMS 125
VS LV++ Q+ KGE WSVHKFGGTC+G+SQRI+NV ++++ D+SERKLVVVSAMS
Sbjct: 69 VSGVSTVNLVDQVQIPKGEMWSVHKFGGTCVGNSQRIRNVAEVIINDNSERKLVVVSAMS 128
Query: 126 KVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKA 185
KVTDMMY+LI KAQSRD+SY+S+L+AVLEKH LTA DLLDGD LA+FLS LH+DISNLKA
Sbjct: 129 KVTDMMYDLIRKAQSRDDSYLSALEAVLEKHRLTARDLLDGDDLASFLSHLHNDISNLKA 188
Query: 186 MLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQ 245
MLRAIYIAGHA+ESF+DFV GHGELWSAQMLS V+RK G +CKWMDTRDVLIVNPTSSNQ
Sbjct: 189 MLRAIYIAGHASESFSDFVAGHGELWSAQMLSYVVRKTGLECKWMDTRDVLIVNPTSSNQ 248
Query: 246 VDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRAR 305
VDPD+ ESEKRL+ WFSLNP K+IIATGFIASTPQ IPTTLKRDGSDFSAAIMGAL RAR
Sbjct: 249 VDPDFGESEKRLDKWFSLNPSKIIIATGFIASTPQNIPTTLKRDGSDFSAAIMGALLRAR 308
Query: 306 QVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPIL 365
QVTIWTDVDGVYSADPRKV+EAVIL+TLSYQEAWEMSYFGANVLHPRTIIPVM+Y IPI+
Sbjct: 309 QVTIWTDVDGVYSADPRKVNEAVILQTLSYQEAWEMSYFGANVLHPRTIIPVMRYNIPIV 368
Query: 366 IRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASA 425
IRNIFNLSAPGT IC P +D + + + VKGFATIDNLALINVEGTGMAGVPGTAS
Sbjct: 369 IRNIFNLSAPGTIICQPPEDDYD--LKLTTPVKGFATIDNLALINVEGTGMAGVPGTASD 426
Query: 426 IFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIP 485
IFG VKDVGANVIMISQASSEHS+CFAVPEKEV AV+EAL+SRF AL GRLSQ+ +IP
Sbjct: 427 IFGCVKDVGANVIMISQASSEHSVCFAVPEKEVNAVSEALRSRFSEALQAGRLSQIEVIP 486
Query: 486 NCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRA 545
NCSILAAVGQKMASTPGVS TLF+ALAKANINVRAI+QGCSEYN+TVV+KRED VKALRA
Sbjct: 487 NCSILAAVGQKMASTPGVSCTLFSALAKANINVRAISQGCSEYNVTVVIKREDSVKALRA 546
Query: 546 VHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSD 605
VHSRF+LSRTT+AMGI+GPGLIG+TLLDQLRDQA++LK+EFNIDLRV+GI GSK MLLSD
Sbjct: 547 VHSRFFLSRTTLAMGIVGPGLIGATLLDQLRDQAAVLKQEFNIDLRVLGITGSKKMLLSD 606
Query: 606 WGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGI 665
GIDL+RWREL +E G A+L+KF Q VHGNHFIPN+ +VDCTADS IA YYDWLRKGI
Sbjct: 607 IGIDLSRWRELLNEKGTEADLDKFTQQVHGNHFIPNSVVVDCTADSAIASRYYDWLRKGI 666
Query: 666 HVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQI 725
HV+TPNKKANSGPLDQYLKLR LQR+SYTHYFYEATVGAGLPI+STLRGLLETGD+IL+I
Sbjct: 667 HVITPNKKANSGPLDQYLKLRDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRI 726
Query: 726 EGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGL 785
EGI SGTLSY+FNNF R+FSEVV EAK AG+TEPDPRDDLSGTDVARKVIILARESGL
Sbjct: 727 EGICSGTLSYLFNNFVGDRSFSEVVTEAKNAGFTEPDPRDDLSGTDVARKVIILARESGL 786
Query: 786 KLELSNIPVESLVPEPLQACASAQEFMQRLPEFDQMFAKKLEEAENAGEV 835
KL+L+++P+ SLVPEPL+ C S +EFM++LP++D AK+ +AEN+GEV
Sbjct: 787 KLDLADLPIRSLVPEPLKGCTSVEEFMEKLPQYDGDLAKERLDAENSGEV 836
>ref|XP_481776.1| putative aspartate kinase, homoserine dehydrogenase [Oryza sativa
(japonica cultivar-group)] gi|38423991|dbj|BAD01719.1|
putative aspartate kinase, homoserine dehydrogenase
[Oryza sativa (japonica cultivar-group)]
Length = 786
Score = 1101 bits (2848), Expect = 0.0
Identities = 549/721 (76%), Positives = 625/721 (86%), Gaps = 7/721 (0%)
Query: 62 ISASLTDVSPSVLVEEQQ----LQKGETWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERK 117
+S ++ + +V VE+ + L KG+ WSVHKFGGTCMG+SQRI+NV DI+L D SERK
Sbjct: 67 VSKNMLKPAAAVSVEQAEASAHLPKGDMWSVHKFGGTCMGTSQRIQNVADIILRDPSERK 126
Query: 118 LVVVSAMSKVTDMMYELINKAQSRDESYVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLH 177
LVVVSAMSKVTDMMY L+NKAQSRD+SY+++LD V EKH A DLL G+ LA FLSQLH
Sbjct: 127 LVVVSAMSKVTDMMYNLVNKAQSRDDSYITALDEVFEKHMAAAKDLLGGEDLARFLSQLH 186
Query: 178 HDISNLKAMLRAIYIAGHATESFTDFVVGHGELWSAQMLSLVIRKNGADCKWMDTRDVLI 237
D+SNLKAMLRAI IAGHATESF+DFVVGHGE+WSAQ+LS I+K+G C WMDTR+VL+
Sbjct: 187 ADVSNLKAMLRAICIAGHATESFSDFVVGHGEIWSAQLLSFAIKKSGTPCSWMDTREVLV 246
Query: 238 VNPTSSNQVDPDYLESEKRLETWFSLNPCKVIIATGFIASTPQKIPTTLKRDGSDFSAAI 297
VNPT SNQVDPDYLESEKRLE WF+ P + IIATGFIASTP+ IPTTLKRDGSDFSAAI
Sbjct: 247 VNPTGSNQVDPDYLESEKRLEKWFARQPAETIIATGFIASTPENIPTTLKRDGSDFSAAI 306
Query: 298 MGALFRARQVTIWTDVDGVYSADPRKVSEAVILKTLSYQEAWEMSYFGANVLHPRTIIPV 357
+G+L +A QVTIWTDVDGV+SADPRKVSEAVIL TLSYQEAWEMSYFGANVLHPRTIIPV
Sbjct: 307 IGSLVKAGQVTIWTDVDGVFSADPRKVSEAVILSTLSYQEAWEMSYFGANVLHPRTIIPV 366
Query: 358 MQYGIPILIRNIFNLSAPGTKICHPSVNDNEDMMNVKNFVKGFATIDNLALINVEGTGMA 417
M+Y IPI+IRN+FN+SAPGT IC N++ D+ + VK FATID L+L+NVEGTGMA
Sbjct: 367 MKYNIPIVIRNMFNISAPGTMICQQPANESGDL---EACVKAFATIDKLSLVNVEGTGMA 423
Query: 418 GVPGTASAIFGAVKDVGANVIMISQASSEHSICFAVPEKEVKAVAEALQSRFRHALDNGR 477
GVPGTASAIFGAVKDVGANVIMISQASSEHS+CFAVPEKEV AV+ AL RFR AL GR
Sbjct: 424 GVPGTASAIFGAVKDVGANVIMISQASSEHSVCFAVPEKEVAAVSAALHVRFREALSAGR 483
Query: 478 LSQVAIIPNCSILAAVGQKMASTPGVSATLFNALAKANINVRAIAQGCSEYNITVVLKRE 537
LS+V +I NCSILAAVG KMASTPGVSATLF+ALAKANINVRAIAQGCSEYNITVVLK+E
Sbjct: 484 LSKVEVIHNCSILAAVGLKMASTPGVSATLFDALAKANINVRAIAQGCSEYNITVVLKQE 543
Query: 538 DCVKALRAVHSRFYLSRTTIAMGIIGPGLIGSTLLDQLRDQASILKEEFNIDLRVMGILG 597
DCV+ALRA HSRF+LS+TT+A+GIIGPGLIG TLL+QL+DQA++LKE NIDLRVMGI G
Sbjct: 544 DCVRALRAAHSRFFLSKTTLAVGIIGPGLIGRTLLNQLKDQAAVLKENMNIDLRVMGITG 603
Query: 598 SKSMLLSDWGIDLARWRELRDESGEVANLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHY 657
S++M+LSD GIDLA W+E E ANL+KFV H+ N PN +VDCTAD+ +A HY
Sbjct: 604 SRTMVLSDTGIDLAHWKEQLQTEAEPANLDKFVDHLSENQLFPNRVLVDCTADTSVASHY 663
Query: 658 YDWLRKGIHVVTPNKKANSGPLDQYLKLRALQRQSYTHYFYEATVGAGLPIVSTLRGLLE 717
YDWL+KGIHV+TPNKKANSGPLD+YLKLR LQR SYTHYFYEATVGAGLPI+STLRGLLE
Sbjct: 664 YDWLKKGIHVITPNKKANSGPLDKYLKLRTLQRASYTHYFYEATVGAGLPIISTLRGLLE 723
Query: 718 TGDRILQIEGIFSGTLSYIFNNFKDGRAFSEVVQEAKEAGYTEPDPRDDLSGTDVARKVI 777
TGD+IL+IEGIFSGTLSYIFNNF+ RAFS+VV EAKEAGYTEPDPRDDLSGTDVARKV+
Sbjct: 724 TGDKILRIEGIFSGTLSYIFNNFEGTRAFSDVVSEAKEAGYTEPDPRDDLSGTDVARKVL 783
Query: 778 I 778
+
Sbjct: 784 L 784
>gb|AAF24602.1| T19E23.1 [Arabidopsis thaliana]
Length = 735
Score = 1058 bits (2737), Expect = 0.0
Identities = 533/705 (75%), Positives = 613/705 (86%), Gaps = 10/705 (1%)
Query: 33 LPFLPSHRSHSLRKALSLLPRGN--------QSPSTKISASLTDVSPSVLVEEQQLQKGE 84
+PF+ R S R + L R + + S ++ S+TD++ VE L KG+
Sbjct: 23 VPFIYGKRLVSNRVSFGKLRRRSCIGQCVRSELQSPRVLGSVTDLALDNSVENGHLPKGD 82
Query: 85 TWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDES 144
+W+VHKFGGTC+G+S+RIK+V +V++DDSERKLVVVSAMSKVTDMMY+LI++A+SRD+S
Sbjct: 83 SWAVHKFGGTCVGNSERIKDVAAVVVKDDSERKLVVVSAMSKVTDMMYDLIHRAESRDDS 142
Query: 145 YVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFV 204
Y+S+L VLEKH TA DLLDGD L++FL++L+ DI+NLKAMLRAIYIAGHATESF+DFV
Sbjct: 143 YLSALSGVLEKHRATAVDLLDGDELSSFLARLNDDINNLKAMLRAIYIAGHATESFSDFV 202
Query: 205 VGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLN 264
VGHGELWSAQML+ V+RK+G DC WMD RDVL+V PTSSNQVDPD++ESEKRLE WF+ N
Sbjct: 203 VGHGELWSAQMLAAVVRKSGLDCTWMDARDVLVVIPTSSNQVDPDFVESEKRLEKWFTQN 262
Query: 265 PCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKV 324
K+IIATGFIASTPQ IPTTLKRDGSDFSAAIM ALFR+ Q+TIWTDVDGVYSADPRKV
Sbjct: 263 SAKIIIATGFIASTPQNIPTTLKRDGSDFSAAIMSALFRSHQLTIWTDVDGVYSADPRKV 322
Query: 325 SEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSV 384
SEAV+LKTLSYQEAWEMSYFGANVLHPRTIIPVM+Y IPI+IRNIFNLSAPGT IC
Sbjct: 323 SEAVVLKTLSYQEAWEMSYFGANVLHPRTIIPVMKYDIPIVIRNIFNLSAPGTMICRQI- 381
Query: 385 NDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQAS 444
D+ED + VKGFATIDNLAL+NVEGTGMAGVPGTASAIF AVK+VGANVIMISQAS
Sbjct: 382 -DDEDGFKLDAPVKGFATIDNLALVNVEGTGMAGVPGTASAIFSAVKEVGANVIMISQAS 440
Query: 445 SEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVS 504
SEHS+CFAVPEKEVKAV+EAL SRFR AL GRLSQ+ IIPNCSILAAVGQKMASTPGVS
Sbjct: 441 SEHSVCFAVPEKEVKAVSEALNSRFRQALAGGRLSQIEIIPNCSILAAVGQKMASTPGVS 500
Query: 505 ATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGP 564
AT FNALAKANIN+RAIAQGCSE+NITVV+KREDC++ALRAVHSRFYLSRTT+A+GIIGP
Sbjct: 501 ATFFNALAKANINIRAIAQGCSEFNITVVVKREDCIRALRAVHSRFYLSRTTLAVGIIGP 560
Query: 565 GLIGSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVA 624
GLIG TLLDQ+RDQA++LKEEF IDLRV+GI GS ML+S+ GIDL+RWREL E GE A
Sbjct: 561 GLIGGTLLDQIRDQAAVLKEEFKIDLRVIGITGSSKMLMSESGIDLSRWRELMKEEGEKA 620
Query: 625 NLEKFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLK 684
++EKF Q+V GNHFIPN+ +VDCTAD+ IA YYDWL +GIHVVTPNKKANSGPLDQYLK
Sbjct: 621 DMEKFTQYVKGNHFIPNSVMVDCTADADIASCYYDWLLRGIHVVTPNKKANSGPLDQYLK 680
Query: 685 LRALQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIF 729
+R LQR+SYTHYFYEATVGAGLPI+STLRGLLETGD+IL+IEGIF
Sbjct: 681 IRDLQRKSYTHYFYEATVGAGLPIISTLRGLLETGDKILRIEGIF 725
>emb|CAA50500.2| beta-aspartyl phosphate homoserine [Arabidopsis thaliana]
Length = 555
Score = 768 bits (1984), Expect = 0.0
Identities = 397/553 (71%), Positives = 458/553 (82%), Gaps = 28/553 (5%)
Query: 33 LPFLPSHRSHSLRKALSLLPRGN--------QSPSTKISASLTDVSPSVLVEEQQLQKGE 84
+PF+ R S R + L R + + S ++ S+TD++ VE L KG+
Sbjct: 23 VPFIYGKRLVSNRVSFGKLRRRSCIGQCVRSELQSPRVLGSVTDLALDNSVENGHLPKGD 82
Query: 85 TWSVHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDES 144
+W+VHKFGGTC+G+S+RIK+V +V++DDSERKLVVVSAMSKVTDMMY+LI++A+SRD+S
Sbjct: 83 SWAVHKFGGTCVGNSERIKDVAAVVVKDDSERKLVVVSAMSKVTDMMYDLIHRAESRDDS 142
Query: 145 YVSSLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFV 204
Y+S+L VLEKH TA DLLDGD L++FL++L+ DI+NLKAMLRAIYIAGHATESF+DFV
Sbjct: 143 YLSALSGVLEKHRATAVDLLDGDELSSFLARLNDDINNLKAMLRAIYIAGHATESFSDFV 202
Query: 205 VGHGELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLN 264
VGHGELWSAQML+ V+RK+G DC WMD RDVL+V PTSSNQVDPD++ESEKRLE WF+ N
Sbjct: 203 VGHGELWSAQMLAAVVRKSGLDCTWMDARDVLVVIPTSSNQVDPDFVESEKRLEKWFTQN 262
Query: 265 PCKVIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKV 324
K+IIATGFIASTPQ IPTTLKRDGSDFSAAIM ALFR+ Q+TIWTDVDGVYSADPRK
Sbjct: 263 SAKIIIATGFIASTPQNIPTTLKRDGSDFSAAIMSALFRSHQLTIWTDVDGVYSADPRK- 321
Query: 325 SEAVILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSV 384
SYFGANVLHPRTIIPVM+Y IPI+IRNIFNLSAPGT IC
Sbjct: 322 -----------------SYFGANVLHPRTIIPVMKYDIPIVIRNIFNLSAPGTMICRQI- 363
Query: 385 NDNEDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQAS 444
D+ED + VKGFATIDNLAL+NVEG GMAGVPGTASAIF AVK+VGANVIMISQAS
Sbjct: 364 -DDEDGFKLDAPVKGFATIDNLALVNVEGAGMAGVPGTASAIFSAVKEVGANVIMISQAS 422
Query: 445 SEHSICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVS 504
SEHS+CFAVPEKEVKAV+E L SRFR AL GRLSQ+ IIPNCSILAAVGQKMASTPGVS
Sbjct: 423 SEHSVCFAVPEKEVKAVSEELNSRFRQALAGGRLSQIEIIPNCSILAAVGQKMASTPGVS 482
Query: 505 ATLFNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGP 564
AT FNALAKANIN+RAIAQGCSE+NITVV+KREDC++ALRAVHSRFYLSRTT+A+GIIGP
Sbjct: 483 ATFFNALAKANINIRAIAQGCSEFNITVVVKREDCIRALRAVHSRFYLSRTTLAVGIIGP 542
Query: 565 GLIGSTLLDQLRD 577
GLIG TLLDQ+RD
Sbjct: 543 GLIGGTLLDQIRD 555
>gb|AAM36682.1| aspartokinase; homoserine dehydrogenase I [Xanthomonas axonopodis
pv. citri str. 306] gi|21242564|ref|NP_642146.1|
aspartokinase [Xanthomonas axonopodis pv. citri str.
306]
Length = 835
Score = 681 bits (1757), Expect = 0.0
Identities = 378/828 (45%), Positives = 518/828 (61%), Gaps = 18/828 (2%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESYVS 147
VHKFGGT + ++R ++V ++L D ++ VVSAM VTD + EL A +
Sbjct: 22 VHKFGGTSVADAERYRHVAQLLLARDETVQVTVVSAMKGVTDALIELAELAAKNRPEWRE 81
Query: 148 SLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFVVGH 207
+H A LL G+ + L ++L +L A+ + G D V G
Sbjct: 82 RWHETRARHRGAAVALL-GEHSGPTVEWLDERFAHLSQILGALAVIGELPREVLDRVQGL 140
Query: 208 GELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLNPCK 267
GE++SAQ+L R G DC +D RDVL+VN VD D+ S +RL TW +P
Sbjct: 141 GEVYSAQLLGDHFRAIGEDCAVLDARDVLVVNRGELG-VDVDWEASAQRLATWRQAHPQT 199
Query: 268 VIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVSEA 327
++ TGF+A TTL R+GSD+S AI ALF A ++ IWTDVDGV SADPR V EA
Sbjct: 200 RVVVTGFVARDRADRVTTLGRNGSDYSGAIFAALFDADELHIWTDVDGVLSADPRVVPEA 259
Query: 328 VILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSVNDN 387
V L+TLSY EA E++YFGA V+HP+T+ P ++ G+PI+IRN F PGT+I S
Sbjct: 260 VQLETLSYDEACELAYFGAKVVHPQTMSPAIERGLPIIIRNTFQPEHPGTRITASSA--- 316
Query: 388 EDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQASSEH 447
V +KG +LA++N+EGTG+ GVPGTA +F A++ +V+MISQ SSEH
Sbjct: 317 -----VSGPIKGLTLSPDLAVLNLEGTGLIGVPGTAERVFAALRTAQVSVVMISQGSSEH 371
Query: 448 SICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVSATL 507
SIC V + E + AL F H L G++ +V + S+LAAVG MA PGV+A L
Sbjct: 372 SICCVVKQHESERARNALLQAFAHELTVGQVQRVQLTTGISVLAAVGDGMAGQPGVAARL 431
Query: 508 FNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGPGLI 567
F +L +A +N+ AIAQG SE NI+V + KALRA H+ F+LS T ++G+IGPG +
Sbjct: 432 FESLGRAQVNILAIAQGSSERNISVAIDAAHATKALRAAHAGFWLSPQTFSVGVIGPGNV 491
Query: 568 GSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVANLE 627
G+ LLDQLR+ L + N+DLR+ ++ MLL + G+ + WR+ S +LE
Sbjct: 492 GAALLDQLRNAQPQLLGKANLDLRLRAVVSRGRMLLDERGL-VGDWRDAFASSATPTDLE 550
Query: 628 KFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLKLRA 687
+F H+ H +P+T I+DC+ + +A Y WL GIHVVTPNK+A SGPL +Y +RA
Sbjct: 551 RFTTHLLSAH-LPHTVIIDCSGSAEVADRYAGWLAAGIHVVTPNKQAGSGPLARYEAIRA 609
Query: 688 LQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAFS 747
S + YEATVGAGLP+++TLR L++TGD + IEGIFSGTL+++FN ++ F+
Sbjct: 610 AADASGARFRYEATVGAGLPVITTLRDLVDTGDAVTSIEGIFSGTLAWLFNKYEGSVPFA 669
Query: 748 EVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQACAS 807
E+V +A+ GYTEPDPRDDLSG DVARK++ILARE+G + L ++ VESLVPE L+ AS
Sbjct: 670 ELVTQARGMGYTEPDPRDDLSGVDVARKLVILAREAGRSISLEDVQVESLVPEALRQ-AS 728
Query: 808 AQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVV--DVTSKKGVVELRRYKKDHPFAQL 865
+FM RLPE D FA++L +A G VLRYV + D G+VEL DH FA L
Sbjct: 729 VDDFMARLPEVDASFAQRLAQAHARGNVLRYVAQLPPDRAPSVGLVEL---PADHAFANL 785
Query: 866 SGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGA 913
+DN++ FTTRRY + PL+V+GPGAG +VTA G+F+D+LR+A+ GA
Sbjct: 786 RLTDNVVQFTTRRYCENPLVVQGPGAGPEVTAAGVFADLLRVAAGEGA 833
>ref|NP_637165.1| bifunctional aspartokinase/homoserine dehydrogenase I [Xanthomonas
campestris pv. campestris str. ATCC 33913]
gi|21112897|gb|AAM41089.1| bifunctional
aspartokinase/homoserine dehydrogenase I [Xanthomonas
campestris pv. campestris str. ATCC 33913]
Length = 835
Score = 669 bits (1727), Expect = 0.0
Identities = 373/828 (45%), Positives = 513/828 (61%), Gaps = 18/828 (2%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESYVS 147
VHKFGGT + ++R ++V ++L D ++ VVSAM VTD + EL A +
Sbjct: 22 VHKFGGTSVADAERYRHVAQLLLARDETVQVTVVSAMKGVTDALIELAELAAQNRPEWRE 81
Query: 148 SLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFVVGH 207
+H A LL G+ + L +L +L A+ + G D V G
Sbjct: 82 RWHETRARHRGAAVALL-GEHSGPTVEWLDERFEHLSQILAALAVIGELPREVLDRVQGL 140
Query: 208 GELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLNPCK 267
GE++SAQ+L R G DC +D RDVL+VN VD D+ S +RL+TW +P
Sbjct: 141 GEVYSAQLLGDHFRALGEDCAVLDARDVLVVNRGELG-VDVDWAVSAQRLDTWRQAHPQT 199
Query: 268 VIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVSEA 327
++ TGF+A TTL R+GSD+S AI ALF A ++ IWTDVDGV SADPR V EA
Sbjct: 200 RVVVTGFVARDRGDRITTLGRNGSDYSGAIFAALFDAHELHIWTDVDGVLSADPRVVPEA 259
Query: 328 VILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSVNDN 387
V L+TLSY EA E++YFGA V+HP+T+ P ++ G+PI+IRN F PGT+I S
Sbjct: 260 VQLETLSYDEACELAYFGAKVVHPQTMSPAIERGLPIIIRNTFQPEHPGTRITASSA--- 316
Query: 388 EDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQASSEH 447
V +KG +LA++N+EGTG+ GVPGTA +F A++ +V+MISQ SSEH
Sbjct: 317 -----VSGPIKGLTLSPDLAVLNLEGTGLIGVPGTAERVFAALRTAQVSVVMISQGSSEH 371
Query: 448 SICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVSATL 507
SIC V + E + AL F H + G++ +V + S+LAAVG MA PGV+A L
Sbjct: 372 SICCVVKQHESERARNALLQAFAHEITVGQVQRVQLTTGISVLAAVGDGMAGQPGVAARL 431
Query: 508 FNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGPGLI 567
F +L +A +N+ AIAQG SE NI+V + KALRA H+ F+LS T ++G+IGPG +
Sbjct: 432 FESLGRAQVNILAIAQGSSERNISVAIDAAHATKALRAAHAGFWLSPQTFSVGVIGPGNV 491
Query: 568 GSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVANLE 627
G+ LLDQLR L + N+DLR+ G++ MLL + + WR+ + +LE
Sbjct: 492 GAALLDQLRTAQPQLLGKANLDLRLRGVVSRGRMLLEERSVT-GDWRDAFAAASTPTDLE 550
Query: 628 KFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLKLRA 687
+F H+ H +P+T I+DC+ + +A Y WL GIHVVTPNK+A SGPL +Y +RA
Sbjct: 551 RFTAHLLSAH-LPHTVIIDCSGSAEVADRYAGWLAAGIHVVTPNKQAGSGPLARYEAIRA 609
Query: 688 LQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAFS 747
S + YEATVGAGLP+++TLR L++TGD + IEGIFSGTL+++FN + FS
Sbjct: 610 AADASGARFRYEATVGAGLPVITTLRDLVDTGDAVTSIEGIFSGTLAWLFNKYDGSVPFS 669
Query: 748 EVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQACAS 807
E+V +A+ GYTEPDPRDDLSG DVARK++ILARE+G + L ++ VESLVPE L+ AS
Sbjct: 670 ELVTQARGMGYTEPDPRDDLSGVDVARKLVILAREAGRAISLDDVSVESLVPEALRQ-AS 728
Query: 808 AQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVV--DVTSKKGVVELRRYKKDHPFAQL 865
+FM +L D FA++L +A G VLRYV + D G+VEL + H FA L
Sbjct: 729 VDDFMAQLQTVDAGFAQRLADAHARGNVLRYVAQLPPDRAPSVGLVELPAH---HAFANL 785
Query: 866 SGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGA 913
+DN++ FTTRRY + PL+V+GPGAG +VTA G+F+D+LR+A+ GA
Sbjct: 786 RLTDNVVQFTTRRYCENPLVVQGPGAGPEVTAAGVFADLLRVAAGEGA 833
>ref|YP_243462.1| aspartokinase/homoserine dehydrogenase I [Xanthomonas campestris
pv. campestris str. 8004] gi|66574032|gb|AAY49442.1|
aspartokinase/homoserine dehydrogenase I [Xanthomonas
campestris pv. campestris str. 8004]
Length = 835
Score = 669 bits (1726), Expect = 0.0
Identities = 374/828 (45%), Positives = 512/828 (61%), Gaps = 18/828 (2%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESYVS 147
VHKFGGT + ++R ++V ++L D ++ VVSAM VTD + EL A +
Sbjct: 22 VHKFGGTSVADAERYRHVAQLLLARDETVQVTVVSAMKGVTDALIELAELAAQNRPEWRE 81
Query: 148 SLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFVVGH 207
+H A LL G+ + L +L +L A+ + G D V G
Sbjct: 82 RWHETRARHRGAAVALL-GEHSGPTVEWLDERFEHLSQILAALAVIGELPREVLDRVQGL 140
Query: 208 GELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLNPCK 267
GE++SAQ+L R G DC +D RDVL+VN VD D+ S +RL+TW +P
Sbjct: 141 GEVYSAQLLGDHFRALGEDCAVLDARDVLVVNRGELG-VDVDWAVSAQRLDTWRQAHPQT 199
Query: 268 VIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVSEA 327
++ TGF+A TTL R+GSD+S AI ALF A ++ IWTDVDGV SADPR V EA
Sbjct: 200 RVVVTGFVARDRGDRITTLGRNGSDYSGAIFAALFDAHELHIWTDVDGVLSADPRVVPEA 259
Query: 328 VILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSVNDN 387
V L+TLSY EA E++YFGA V+HP+T+ P ++ G+PI+IRN F PGT+I S
Sbjct: 260 VQLETLSYDEACELAYFGAKVVHPQTMSPAIERGLPIIIRNTFQPEHPGTRISASSA--- 316
Query: 388 EDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQASSEH 447
V +KG +LA++N+EGTG+ GVPGTA +F A++ +V+MISQ SSEH
Sbjct: 317 -----VSGPIKGLTLSPDLAVLNLEGTGLIGVPGTAERVFAALRTAQVSVVMISQGSSEH 371
Query: 448 SICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVSATL 507
SIC V + E + AL F H L G++ +V + S+LAAVG MA PGV+A L
Sbjct: 372 SICCVVKQHESERARNALLQAFAHELTVGQVQRVQLTTGISVLAAVGDGMAGQPGVAARL 431
Query: 508 FNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGPGLI 567
F +L +A +N+ AIAQG SE NI+V + KALRA H+ F+LS T ++G+IGPG +
Sbjct: 432 FESLGRAQVNILAIAQGSSERNISVAIDAAHATKALRAAHAGFWLSPQTFSVGVIGPGNV 491
Query: 568 GSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVANLE 627
G+ LLDQLR L + N+DLR+ G++ MLL + WR+ + +LE
Sbjct: 492 GAALLDQLRTAQPQLLGKANLDLRLRGVVSRGRMLLEARSVT-GDWRDAFAAASTPTDLE 550
Query: 628 KFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLKLRA 687
+F H+ H +P+T I+DC+ + +A Y WL GIHVVTPNK+A SGPL +Y +RA
Sbjct: 551 RFTAHLLSAH-LPHTVIIDCSGSAEVADRYAGWLAAGIHVVTPNKQAGSGPLARYEAIRA 609
Query: 688 LQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAFS 747
S + YEATVGAGLP+++TLR L++TGD + IEGIFSGTL+++FN + FS
Sbjct: 610 AADASGARFRYEATVGAGLPVITTLRDLVDTGDAVTSIEGIFSGTLAWLFNKYDGSVPFS 669
Query: 748 EVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQACAS 807
E+V +A+ GYTEPDPRDDLSG DVARK++ILARE+G + L ++ VESLVPE L+ AS
Sbjct: 670 ELVTQARGMGYTEPDPRDDLSGVDVARKLVILAREAGRAISLDDVSVESLVPEALRQ-AS 728
Query: 808 AQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVV--DVTSKKGVVELRRYKKDHPFAQL 865
+FM +L D FA++L +A G VLRYV + D G+VEL + H FA L
Sbjct: 729 VDDFMAQLQTVDDGFAQRLADAHARGNVLRYVAQLPPDRAPSVGLVELPAH---HAFANL 785
Query: 866 SGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGA 913
+DN++ FTTRRY + PL+V+GPGAG +VTA G+F+D+LR+A+ GA
Sbjct: 786 RLTDNVVQFTTRRYCENPLVVQGPGAGPEVTAAGVFADLLRVAAGEGA 833
>ref|YP_200881.1| aspartokinase; homoserine dehydrogenase I [Xanthomonas oryzae pv.
oryzae KACC10331] gi|58426459|gb|AAW75496.1|
aspartokinase; homoserine dehydrogenase I [Xanthomonas
oryzae pv. oryzae KACC10331]
Length = 846
Score = 669 bits (1726), Expect = 0.0
Identities = 373/828 (45%), Positives = 514/828 (62%), Gaps = 18/828 (2%)
Query: 88 VHKFGGTCMGSSQRIKNVGDIVLEDDSERKLVVVSAMSKVTDMMYELINKAQSRDESYVS 147
VHKFGGT + + R ++V ++L D ++ VVSAM VTD + EL A +
Sbjct: 33 VHKFGGTSVADADRYRHVAQLLLARDETVQVTVVSAMKGVTDALIELAELAAKDRPEWRD 92
Query: 148 SLDAVLEKHSLTAHDLLDGDGLATFLSQLHHDISNLKAMLRAIYIAGHATESFTDFVVGH 207
+H A LL G+ + L +L +L A+ + G D V G
Sbjct: 93 RWHETRARHRGAAVALL-GEHSGPTVEWLDERFEHLSQILGALAVIGELPREVLDRVQGL 151
Query: 208 GELWSAQMLSLVIRKNGADCKWMDTRDVLIVNPTSSNQVDPDYLESEKRLETWFSLNPCK 267
GE++SAQ+L R G DC +D RDVL+VN VD D+ S ++L TW +P
Sbjct: 152 GEVYSAQLLGDHFRAIGEDCAVLDARDVLVVNRGELG-VDVDWEVSAQQLATWRQAHPQT 210
Query: 268 VIIATGFIASTPQKIPTTLKRDGSDFSAAIMGALFRARQVTIWTDVDGVYSADPRKVSEA 327
++ TGF+A TTL R+GSD+S AI ALF A ++ IWTDVDGV SADPR V EA
Sbjct: 211 RVVVTGFVARDRADRITTLGRNGSDYSGAIFAALFDADELHIWTDVDGVLSADPRVVPEA 270
Query: 328 VILKTLSYQEAWEMSYFGANVLHPRTIIPVMQYGIPILIRNIFNLSAPGTKICHPSVNDN 387
V L+TLSY EA E++YFGA V+HP+T+ P ++ G+PI+IRN F PGT+I S
Sbjct: 271 VQLETLSYDEACELAYFGAKVVHPQTMSPAIERGLPIIIRNTFQPEHPGTRITASSA--- 327
Query: 388 EDMMNVKNFVKGFATIDNLALINVEGTGMAGVPGTASAIFGAVKDVGANVIMISQASSEH 447
V+ +KG +LA++N+EGTG+ GVPGTA +F +++ +V+MISQ SSEH
Sbjct: 328 -----VRGPIKGLTLSPDLAVLNLEGTGLIGVPGTAERVFASLRTAQVSVVMISQGSSEH 382
Query: 448 SICFAVPEKEVKAVAEALQSRFRHALDNGRLSQVAIIPNCSILAAVGQKMASTPGVSATL 507
SIC V + E + AL F H L G++ +V + S+LAAVG MA PGV+A L
Sbjct: 383 SICCVVKQHESERARNALLQAFAHELTVGQVQRVQLTTGISVLAAVGDGMAGQPGVAARL 442
Query: 508 FNALAKANINVRAIAQGCSEYNITVVLKREDCVKALRAVHSRFYLSRTTIAMGIIGPGLI 567
F +L +A++N+ AIAQG SE NI+V + KALRA H+ F+LS T ++G+IGPG +
Sbjct: 443 FESLGRAHVNILAIAQGSSERNISVAIDAAHATKALRAAHAGFWLSPQTFSVGVIGPGNV 502
Query: 568 GSTLLDQLRDQASILKEEFNIDLRVMGILGSKSMLLSDWGIDLARWRELRDESGEVANLE 627
G+ LLDQLR L + N+DLR+ ++ MLL + G+ + WR+ + +LE
Sbjct: 503 GAALLDQLRVAQPQLLGKANLDLRLRAVVSRGRMLLDERGL-VGDWRDAFASAATPTDLE 561
Query: 628 KFVQHVHGNHFIPNTAIVDCTADSVIAGHYYDWLRKGIHVVTPNKKANSGPLDQYLKLRA 687
+F H+ H +P+T I+DC+ + +A Y WL GIHVVTPNK+A SGPL +Y +RA
Sbjct: 562 RFTTHLLSAH-LPHTVIIDCSGSAEVADRYAGWLAAGIHVVTPNKQAGSGPLARYEAIRA 620
Query: 688 LQRQSYTHYFYEATVGAGLPIVSTLRGLLETGDRILQIEGIFSGTLSYIFNNFKDGRAFS 747
S + YEATVGAGLP+++TLR L++TGD + IEGIFSGTL+++FN + F+
Sbjct: 621 AADASGARFRYEATVGAGLPVITTLRDLVDTGDTVTSIEGIFSGTLAWLFNKYDGSVPFA 680
Query: 748 EVVQEAKEAGYTEPDPRDDLSGTDVARKVIILARESGLKLELSNIPVESLVPEPLQACAS 807
E+V +A+ GYTEPDPRDDLSG DVARK++ILARE+G + L ++ VESLVP L+ +S
Sbjct: 681 ELVTQARSMGYTEPDPRDDLSGVDVARKLVILAREAGRAISLEDVQVESLVPAALRQ-SS 739
Query: 808 AQEFMQRLPEFDQMFAKKLEEAENAGEVLRYVGVV--DVTSKKGVVELRRYKKDHPFAQL 865
++FM RLPE D FA++L A G VLRYV + D G+VEL DH FA L
Sbjct: 740 VEDFMARLPEVDAAFAQRLANARVRGNVLRYVAQLPPDRAPSVGLVEL---PADHAFANL 796
Query: 866 SGSDNIIAFTTRRYKDQPLIVRGPGAGAQVTAGGIFSDILRLASYLGA 913
+DN++ FTTRRY PL+V+GPGAG +VTA G+F+D+LR+A+ GA
Sbjct: 797 RLTDNVVQFTTRRYCQNPLVVQGPGAGPEVTAAGVFADLLRVAAGEGA 844
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,501,555,623
Number of Sequences: 2540612
Number of extensions: 62800834
Number of successful extensions: 163575
Number of sequences better than 10.0: 898
Number of HSP's better than 10.0 without gapping: 764
Number of HSP's successfully gapped in prelim test: 134
Number of HSP's that attempted gapping in prelim test: 160182
Number of HSP's gapped (non-prelim): 1322
length of query: 915
length of database: 863,360,394
effective HSP length: 137
effective length of query: 778
effective length of database: 515,296,550
effective search space: 400900715900
effective search space used: 400900715900
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0108.1


