

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0107.8
(408 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
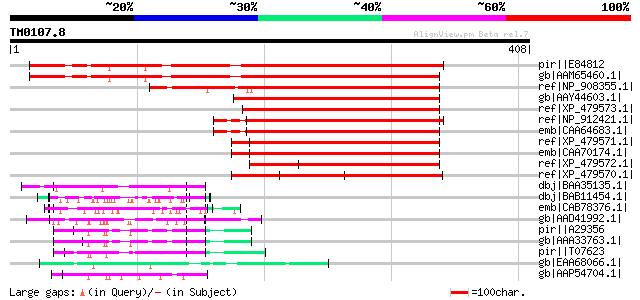
Score E
Sequences producing significant alignments: (bits) Value
pir||E84812 hypothetical protein At2g39050 [imported] - Arabidop... 304 3e-81
gb|AAM65460.1| unknown [Arabidopsis thaliana] gi|20197446|gb|AAC... 297 3e-79
ref|NP_908355.1| unknown protein [Oryza sativa (japonica cultiva... 234 4e-60
gb|AAY44603.1| stress responsive protein [Triticum aestivum] 226 8e-58
ref|XP_479573.1| r40g3 protein [Oryza sativa (japonica cultivar-... 226 1e-57
ref|NP_912421.1| Putative r40c1 protein - rice [Oryza sativa (ja... 223 6e-57
emb|CAA64683.1| osr40c1 [Oryza sativa] gi|7489571|pir||T03911 r4... 223 6e-57
ref|XP_479571.1| r40g2 protein [Oryza sativa (japonica cultivar-... 221 4e-56
emb|CAA70174.1| osr40g2 [Oryza sativa (indica cultivar-group)] g... 221 4e-56
ref|XP_479572.1| putative r40c2 protein [Oryza sativa (japonica ... 216 1e-54
ref|XP_479570.1| putative r40c1 protein [Oryza sativa (japonica ... 200 8e-50
dbj|BAA35135.1| Extensin [Adiantum capillus-veneris] 78 4e-13
dbj|BAB11454.1| unnamed protein product [Arabidopsis thaliana] 78 6e-13
emb|CAB78376.1| extensin-like protein [Arabidopsis thaliana] gi|... 75 3e-12
gb|AAD41992.1| hypothetical protein [Arabidopsis thaliana] gi|20... 75 5e-12
pir||A29356 hydroxyproline-rich glycoprotein (clone Hyp3.6) - ki... 71 5e-11
gb|AAA33763.1| hydroxyproline-rich glycoprotein 71 5e-11
pir||T07623 extensin homolog HRGP2 - soybean (fragment) gi|34745... 70 9e-11
gb|EAA68066.1| hypothetical protein FG10164.1 [Gibberella zeae P... 70 9e-11
gb|AAP54704.1| putative phospholipase [Oryza sativa (japonica cu... 70 2e-10
>pir||E84812 hypothetical protein At2g39050 [imported] - Arabidopsis thaliana
Length = 326
Score = 304 bits (779), Expect = 3e-81
Identities = 157/336 (46%), Positives = 209/336 (61%), Gaps = 30/336 (8%)
Query: 16 HTHHHNNRRDDDEEQHPNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPPF 75
H HH +++RDD E+ ++ PP +P P+ Q QP F +P+ P PQ P
Sbjct: 5 HQHHRHHQRDDGEDDRQSFGVPPPHVDAP----PQPHGLYQSQPHF--DPYAPTPQAPAP 58
Query: 76 YG-----EPHRPPPSQPETNVFHTSHDVTYGHVSH-----PPQSHHPDHSPNYGYPVPPA 125
Y EPH PPP + E + ++GHVSH P +S+ P+H GY
Sbjct: 59 YRSETQFEPHAPPPYRSEPYFETPAPPPSFGHVSHVGHQSPNESYPPEHHRYGGY----- 113
Query: 126 QQPPFSSYSAPPPPNAATVHHVSHEVHPPSNTNVLHVSHETNQFPNSVHHVGHQGPTALS 185
Q P +S ++ H H + P +++ ++ + N+ P+++ L+
Sbjct: 114 -QQPSNSLLESHGDHSGVTHVAHHSSNQPQSSSGVYHKPDENRLPDNL--------AGLA 164
Query: 186 GKPTYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLV 245
G+ T +V +KA P+Y LT+R G+VILAP +P++E Q WYKDEKYST+VKDA+ P F+LV
Sbjct: 165 GRATVKVYSKAEPNYNLTIRDGKVILAPADPSDEAQHWYKDEKYSTKVKDADGHPCFALV 224
Query: 246 NKATGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVRMVNNIHLNMDAY 305
NKATGEA+KHS+G +HPV LI Y PD LDES+LWTESKD GD YR++RMVNN LN+DAY
Sbjct: 225 NKATGEAMKHSVGATHPVHLIRYVPDKLDESVLWTESKDFGDGYRTIRMVNNTRLNVDAY 284
Query: 306 HGDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPYCKL 341
HGD SGGV DGTTIVLW WNKGDNQ WKI P+CKL
Sbjct: 285 HGDSKSGGVRDGTTIVLWDWNKGDNQLWKIFPFCKL 320
>gb|AAM65460.1| unknown [Arabidopsis thaliana] gi|20197446|gb|AAC79615.2| expressed
protein [Arabidopsis thaliana]
gi|20147403|gb|AAM10411.1| At2g39050/T7F6.22
[Arabidopsis thaliana] gi|15724197|gb|AAL06490.1|
At2g39050/T7F6.22 [Arabidopsis thaliana]
gi|18404934|ref|NP_565899.1| hydroxyproline-rich
glycoprotein family protein [Arabidopsis thaliana]
Length = 317
Score = 297 bits (761), Expect = 3e-79
Identities = 154/333 (46%), Positives = 206/333 (61%), Gaps = 30/333 (9%)
Query: 16 HTHHHNNRRDDDEEQHPNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPPF 75
H HH +++RDD E+ ++ PP +P P+ Q QP F +P+ P PQ P
Sbjct: 5 HQHHRHHQRDDGEDDRQSFGVPPPHVDAP----PQPHGLYQSQPHF--DPYAPTPQAPAP 58
Query: 76 YG-----EPHRPPPSQPETNVFHTSHDVTYGHVSH-----PPQSHHPDHSPNYGYPVPPA 125
Y EPH PPP + E + ++GHVSH P +S+ P+H GY
Sbjct: 59 YRSETQFEPHAPPPYRSEPYFETPAPPPSFGHVSHVGHQSPNESYPPEHHRYGGY----- 113
Query: 126 QQPPFSSYSAPPPPNAATVHHVSHEVHPPSNTNVLHVSHETNQFPNSVHHVGHQGPTALS 185
Q P +S ++ H H + P +++ ++ + N+ P+++ L+
Sbjct: 114 -QQPSNSLLESHGDHSGVTHVAHHSSNQPQSSSGVYHKPDENRLPDNL--------AGLA 164
Query: 186 GKPTYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLV 245
G+ T +V +KA P+Y LT+R G+VILAP +P++E Q WYKDEKYST+VKDA+ P F+LV
Sbjct: 165 GRATVKVYSKAEPNYNLTIRDGKVILAPADPSDEAQHWYKDEKYSTKVKDADGHPCFALV 224
Query: 246 NKATGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVRMVNNIHLNMDAY 305
NKATGEA+KHS+G +HPV LI Y PD LDES+LWTESKD GD YR++RMVNN LN+DAY
Sbjct: 225 NKATGEAMKHSVGATHPVHLIRYVPDKLDESVLWTESKDFGDGYRTIRMVNNTRLNVDAY 284
Query: 306 HGDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
HGD SGGV DGTTIVLW WNKGDNQ WKI P+
Sbjct: 285 HGDSKSGGVRDGTTIVLWDWNKGDNQLWKIFPF 317
>ref|NP_908355.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|10697200|dbj|BAB16331.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 268
Score = 234 bits (597), Expect = 4e-60
Identities = 125/250 (50%), Positives = 158/250 (63%), Gaps = 27/250 (10%)
Query: 111 HPDHSPNYGYPVPPAQQPPFSSYSAPPPPNAATVHHVSHE-VHPP------SNTNVLHVS 163
HP +P YG+ P PP APPP + H SH+ HPP NV+HVS
Sbjct: 24 HPAPAPAYGHDSAP---PPGPYGQAPPPADPYARHPPSHDYAHPPPAYGGGGYGNVVHVS 80
Query: 164 HETNQFPNSVHHVGHQGPTALS------------GKP--TYRVVTKANPD-YALTVRHGQ 208
HE + H G G +S G P T+R+ KA D Y+L VR G+
Sbjct: 81 HEVSDHQRPTPHYG--GSEYISPVQETRPYHGGGGAPPVTHRIYCKAGEDNYSLAVRDGK 138
Query: 209 VILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLVNKATGEAIKHSIGESHPVRLIPY 268
V L + ++ Q W KD KYSTRVKD E PA +LVNKATG+A+KHSIG+SHPVRL+ Y
Sbjct: 139 VCLVRSDRDDHTQHWVKDMKYSTRVKDEEGYPAMALVNKATGDALKHSIGQSHPVRLVRY 198
Query: 269 KPDYLDESILWTESKDLGDSYRSVRMVNNIHLNMDAYHGDKNSGGVHDGTTIVLWQWNKG 328
P+Y+DES+LWTES+D+G +R +RMVNNI+LN DA HGDK+ GGV DGTT+VLW+W +G
Sbjct: 199 NPEYMDESVLWTESRDVGSGFRCIRMVNNIYLNFDALHGDKDHGGVRDGTTLVLWEWCEG 258
Query: 329 DNQRWKIIPY 338
DNQRWKI+P+
Sbjct: 259 DNQRWKIVPW 268
>gb|AAY44603.1| stress responsive protein [Triticum aestivum]
Length = 195
Score = 226 bits (577), Expect = 8e-58
Identities = 98/162 (60%), Positives = 133/162 (81%)
Query: 177 GHQGPTALSGKPTYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDA 236
G+ G + +PT ++ +ANP+YA++VR+G+V+LAP NP +++Q W KD ++ST +KD
Sbjct: 33 GNCGGRPMPPQPTVKIYCRANPNYAMSVRNGKVVLAPANPKDDYQHWIKDMRWSTSIKDE 92
Query: 237 ERCPAFSLVNKATGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVRMVN 296
E PAF++VNKATG+AIKHS+G+SHPVRL+PY PDYLDES+LWTES+D+G+ +R VRMVN
Sbjct: 93 EGYPAFAMVNKATGQAIKHSLGQSHPVRLVPYNPDYLDESVLWTESRDVGNGFRCVRMVN 152
Query: 297 NIHLNMDAYHGDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
NI+LN DA +GDK GGV DGT +VLW+W +GDNQRWK+ PY
Sbjct: 153 NIYLNFDALNGDKYHGGVRDGTEVVLWKWCEGDNQRWKLQPY 194
>ref|XP_479573.1| r40g3 protein [Oryza sativa (japonica cultivar-group)]
gi|1658315|emb|CAA70175.1| osr40g3 [Oryza sativa (indica
cultivar-group)] gi|34394519|dbj|BAC83806.1| r40g3
protein [Oryza sativa (japonica cultivar-group)]
gi|7489573|pir||T03962 r40g3 protein - rice
Length = 204
Score = 226 bits (576), Expect = 1e-57
Identities = 99/155 (63%), Positives = 128/155 (81%)
Query: 184 LSGKPTYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFS 243
L +PT +V +ANP+YA+T R+G V+LAP NP +E+Q W KD ++ST +KD E PAF+
Sbjct: 49 LPPQPTVKVYCRANPNYAMTARNGAVVLAPANPKDEYQHWIKDMRWSTSIKDEEGYPAFA 108
Query: 244 LVNKATGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVRMVNNIHLNMD 303
LVNKATG+AIKHS+G+SHPVRL+PY P+ +DES+LWTES+D+G+ +R +RMVNNI+LN D
Sbjct: 109 LVNKATGQAIKHSLGQSHPVRLVPYNPEVMDESVLWTESRDVGNGFRCIRMVNNIYLNFD 168
Query: 304 AYHGDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
A+HGDK GGV DGT IVLW+W +GDNQRWKI PY
Sbjct: 169 AFHGDKYHGGVRDGTDIVLWKWCEGDNQRWKIQPY 203
>ref|NP_912421.1| Putative r40c1 protein - rice [Oryza sativa (japonica
cultivar-group)] gi|24899397|gb|AAN64997.1| Putative
r40c1 protein - rice [Oryza sativa (japonica
cultivar-group)]
Length = 373
Score = 223 bits (569), Expect = 6e-57
Identities = 102/178 (57%), Positives = 133/178 (74%), Gaps = 5/178 (2%)
Query: 161 HVSHETNQFPNSVHHVGHQGPTALSGKPTYRVVTKANPDYALTVRHGQVILAPFNPNNEH 220
H H +Q P HH G G + +PT+++ +A+ Y + VR G V+LAP NP +EH
Sbjct: 7 HGHHGQDQPPQ--HHGGGGGG---AHQPTFKIFCRADEGYCVAVREGNVVLAPTNPRDEH 61
Query: 221 QLWYKDEKYSTRVKDAERCPAFSLVNKATGEAIKHSIGESHPVRLIPYKPDYLDESILWT 280
Q WYKD ++S ++KD E PAF+LVNKATG AIKHS+G+ HPV+L P+ P+Y DES+LWT
Sbjct: 62 QHWYKDMRFSAKIKDEEGNPAFALVNKATGLAIKHSLGQGHPVKLAPFNPEYPDESVLWT 121
Query: 281 ESKDLGDSYRSVRMVNNIHLNMDAYHGDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
ES D+G S+R +RM+NNI LN DA+HGDK+ GGVHDGTTIVLW+W KGDNQ WKI+P+
Sbjct: 122 ESGDVGKSFRCIRMLNNIRLNFDAFHGDKDHGGVHDGTTIVLWEWAKGDNQCWKILPW 179
Score = 218 bits (555), Expect = 3e-55
Identities = 95/155 (61%), Positives = 122/155 (78%)
Query: 187 KPTYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLVN 246
+PT R+ KA+ +++TVR G V LAP NP +E+Q W KD ++S +KD E PAF+LVN
Sbjct: 197 EPTVRIFCKADEGFSVTVRGGSVCLAPTNPRDEYQHWIKDMRHSNSIKDEEGYPAFALVN 256
Query: 247 KATGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVRMVNNIHLNMDAYH 306
+ TGEAIKHS GE HPV+L+PY P Y DES+LWTES+D+G +R +RMVNNI+LN DA H
Sbjct: 257 RVTGEAIKHSQGEGHPVKLVPYNPGYQDESVLWTESRDVGHGFRCIRMVNNIYLNFDALH 316
Query: 307 GDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPYCKL 341
GDK+ GGV DGTT+ LW+W +GDNQRWKI+P+CKL
Sbjct: 317 GDKDHGGVRDGTTVALWKWCEGDNQRWKIVPWCKL 351
>emb|CAA64683.1| osr40c1 [Oryza sativa] gi|7489571|pir||T03911 r40c1 protein - rice
Length = 348
Score = 223 bits (569), Expect = 6e-57
Identities = 102/178 (57%), Positives = 133/178 (74%), Gaps = 5/178 (2%)
Query: 161 HVSHETNQFPNSVHHVGHQGPTALSGKPTYRVVTKANPDYALTVRHGQVILAPFNPNNEH 220
H H +Q P HH G G + +PT+++ +A+ Y + VR G V+LAP NP +EH
Sbjct: 7 HGHHGQDQPPQ--HHGGGGGG---AHQPTFKIFCRADEGYCVAVREGNVVLAPTNPRDEH 61
Query: 221 QLWYKDEKYSTRVKDAERCPAFSLVNKATGEAIKHSIGESHPVRLIPYKPDYLDESILWT 280
Q WYKD ++S ++KD E PAF+LVNKATG AIKHS+G+ HPV+L P+ P+Y DES+LWT
Sbjct: 62 QHWYKDMRFSAKIKDEEGNPAFALVNKATGLAIKHSLGQGHPVKLAPFNPEYPDESVLWT 121
Query: 281 ESKDLGDSYRSVRMVNNIHLNMDAYHGDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
ES D+G S+R +RM+NNI LN DA+HGDK+ GGVHDGTTIVLW+W KGDNQ WKI+P+
Sbjct: 122 ESGDVGKSFRCIRMLNNIRLNFDAFHGDKDHGGVHDGTTIVLWEWAKGDNQCWKILPW 179
Score = 211 bits (537), Expect = 3e-53
Identities = 92/152 (60%), Positives = 119/152 (77%)
Query: 187 KPTYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLVN 246
+PT R+ KA+ +++TVR G V LAP NP +E+Q W KD ++S +KD E PAF+LVN
Sbjct: 197 EPTVRIFCKADEGFSVTVRGGSVCLAPTNPRDEYQHWIKDMRHSNSIKDEEGYPAFALVN 256
Query: 247 KATGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVRMVNNIHLNMDAYH 306
+ TGEAIKHS GE HPV+L+PY P Y DES+LWTES+D+G +R +RMVNNI+LN DA H
Sbjct: 257 RVTGEAIKHSQGEGHPVKLVPYNPGYQDESVLWTESRDVGHGFRCIRMVNNIYLNFDALH 316
Query: 307 GDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
GDK+ GGV DGTT+ LW+W +GDNQRWKI+P+
Sbjct: 317 GDKDHGGVRDGTTVALWKWCEGDNQRWKIVPW 348
>ref|XP_479571.1| r40g2 protein [Oryza sativa (japonica cultivar-group)]
gi|34394517|dbj|BAC83804.1| r40g2 protein [Oryza sativa
(japonica cultivar-group)]
Length = 343
Score = 221 bits (562), Expect = 4e-56
Identities = 97/165 (58%), Positives = 128/165 (76%), Gaps = 1/165 (0%)
Query: 175 HVGHQGPTALSG-KPTYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRV 233
H +Q P A S +++ +AN +Y LTVR V+LAP NP +EHQ W+KD ++ST+V
Sbjct: 6 HHHNQAPAAPSDPNQIFKIFCRANENYCLTVRDSAVVLAPVNPKDEHQHWFKDMRFSTKV 65
Query: 234 KDAERCPAFSLVNKATGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVR 293
KD E PAF+LVNKATG A+KHS+G+SHPV+L+P+ P+Y D S+LWTESKD+G +R +R
Sbjct: 66 KDGEGMPAFALVNKATGLAVKHSLGQSHPVKLVPFNPEYEDASVLWTESKDVGKGFRCIR 125
Query: 294 MVNNIHLNMDAYHGDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
MVNN LN+DA+HGDK+ GGV DGTT+VLW+W KGDNQ WKI+P+
Sbjct: 126 MVNNTRLNLDAFHGDKDHGGVRDGTTVVLWEWCKGDNQSWKILPW 170
Score = 216 bits (549), Expect = 1e-54
Identities = 93/150 (62%), Positives = 120/150 (80%)
Query: 189 TYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLVNKA 248
T RV + A DY LTVR+G LAP NP +++Q W KD ++S +++D E PAF+LVNK
Sbjct: 194 TVRVFSAAGEDYCLTVRNGTACLAPKNPRDDYQHWIKDMRHSNKIRDEEGYPAFALVNKV 253
Query: 249 TGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVRMVNNIHLNMDAYHGD 308
TGEAIKHS G+ HPV+L+PY P+Y DES+LWTESKD+G +R +RMVNNI+LN DA+HGD
Sbjct: 254 TGEAIKHSTGQGHPVKLVPYNPEYQDESVLWTESKDVGKGFRCIRMVNNIYLNFDAFHGD 313
Query: 309 KNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
K+ GG+HDGT IVLW+W +GDNQRWKI+P+
Sbjct: 314 KDHGGIHDGTEIVLWKWCEGDNQRWKILPW 343
>emb|CAA70174.1| osr40g2 [Oryza sativa (indica cultivar-group)]
gi|7489572|pir||T03960 r40g2 protein - rice (fragment)
Length = 343
Score = 221 bits (562), Expect = 4e-56
Identities = 97/165 (58%), Positives = 128/165 (76%), Gaps = 1/165 (0%)
Query: 175 HVGHQGPTALSG-KPTYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRV 233
H +Q P A S +++ +AN +Y LTVR V+LAP NP +EHQ W+KD ++ST+V
Sbjct: 6 HHHNQAPAAPSDPNQIFKIFCRANENYCLTVRDSAVVLAPVNPKDEHQHWFKDMRFSTKV 65
Query: 234 KDAERCPAFSLVNKATGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVR 293
KD E PAF+LVNKATG A+KHS+G+SHPV+L+P+ P+Y D S+LWTESKD+G +R +R
Sbjct: 66 KDGEGMPAFALVNKATGLAVKHSLGQSHPVKLVPFNPEYEDASVLWTESKDVGKGFRCIR 125
Query: 294 MVNNIHLNMDAYHGDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
MVNN LN+DA+HGDK+ GGV DGTT+VLW+W KGDNQ WKI+P+
Sbjct: 126 MVNNTRLNLDAFHGDKDHGGVRDGTTVVLWEWCKGDNQSWKILPW 170
Score = 211 bits (538), Expect = 3e-53
Identities = 91/150 (60%), Positives = 118/150 (78%)
Query: 189 TYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLVNKA 248
T RV + A DY LTVR+G LAP NP +++Q W KD ++S +++D E PAF+LVNK
Sbjct: 194 TVRVFSPAGEDYCLTVRNGTACLAPKNPRDDYQYWIKDMRHSNKIRDEEGYPAFALVNKV 253
Query: 249 TGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVRMVNNIHLNMDAYHGD 308
TGEAIKHS G+ HPV+L+PY P+Y DES+LW ESK +G +R +RMVNNI+LN DA+HGD
Sbjct: 254 TGEAIKHSTGQGHPVKLVPYNPEYQDESVLWRESKHVGKGFRCIRMVNNIYLNFDAFHGD 313
Query: 309 KNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
K+ GG+HDGT IVLW+W +GDNQRWKI+P+
Sbjct: 314 KDHGGIHDGTEIVLWKWCEGDNQRWKILPW 343
>ref|XP_479572.1| putative r40c2 protein [Oryza sativa (japonica cultivar-group)]
gi|34394518|dbj|BAC83805.1| putative r40c2 protein
[Oryza sativa (japonica cultivar-group)]
Length = 285
Score = 216 bits (549), Expect = 1e-54
Identities = 93/150 (62%), Positives = 120/150 (80%)
Query: 189 TYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLVNKA 248
T RV + A DY LTVR+G LAP NP +++Q W KD ++S +++D E PAF+LVNK
Sbjct: 136 TVRVFSAAGEDYCLTVRNGTACLAPKNPRDDYQHWIKDMRHSNKIRDEEGYPAFALVNKV 195
Query: 249 TGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGDSYRSVRMVNNIHLNMDAYHGD 308
TGEAIKHS G+ HPV+L+PY P+Y DES+LWTESKD+G +R +RMVNNI+LN DA+HGD
Sbjct: 196 TGEAIKHSTGQGHPVKLVPYNPEYQDESVLWTESKDVGKGFRCIRMVNNIYLNFDAFHGD 255
Query: 309 KNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
K+ GG+HDGT IVLW+W +GDNQRWKI+P+
Sbjct: 256 KDHGGIHDGTEIVLWKWCEGDNQRWKILPW 285
Score = 174 bits (442), Expect = 3e-42
Identities = 73/111 (65%), Positives = 95/111 (84%)
Query: 228 KYSTRVKDAERCPAFSLVNKATGEAIKHSIGESHPVRLIPYKPDYLDESILWTESKDLGD 287
++ST+VKD E PAF+LVNKATG A+KHS+G+SHPV+L+P+ P+Y D S+LWTESKD+G
Sbjct: 2 RFSTKVKDGEGMPAFALVNKATGLAVKHSLGQSHPVKLVPFNPEYEDASVLWTESKDVGK 61
Query: 288 SYRSVRMVNNIHLNMDAYHGDKNSGGVHDGTTIVLWQWNKGDNQRWKIIPY 338
+R +RMVNN LN+DA+HGDK+ GGV DGTT+VLW+W KGDNQ WKI+P+
Sbjct: 62 GFRCIRMVNNTRLNLDAFHGDKDHGGVRDGTTVVLWEWCKGDNQSWKILPW 112
>ref|XP_479570.1| putative r40c1 protein [Oryza sativa (japonica cultivar-group)]
gi|34394516|dbj|BAC83803.1| putative r40c1 protein
[Oryza sativa (japonica cultivar-group)]
Length = 402
Score = 200 bits (508), Expect = 8e-50
Identities = 89/129 (68%), Positives = 107/129 (81%), Gaps = 1/129 (0%)
Query: 213 PFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLVNKATGEAIKHSIGESHPV-RLIPYKPD 271
P Q W KD ++ST +KD E PAF+LVNKATGEAIKHS+G+SHPV RL+PY P+
Sbjct: 274 PIPGMTSRQHWVKDMRHSTSIKDEEGYPAFALVNKATGEAIKHSLGQSHPVVRLVPYNPE 333
Query: 272 YLDESILWTESKDLGDSYRSVRMVNNIHLNMDAYHGDKNSGGVHDGTTIVLWQWNKGDNQ 331
YLDES+LWTESKD+G +R VRMVNNI+LN DA+HGDK+ GGVHDGTT+VLW+W KGDNQ
Sbjct: 334 YLDESVLWTESKDVGHGFRCVRMVNNIYLNFDAFHGDKDHGGVHDGTTVVLWEWCKGDNQ 393
Query: 332 RWKIIPYCK 340
RWKI+P+CK
Sbjct: 394 RWKILPWCK 402
Score = 100 bits (250), Expect = 6e-20
Identities = 49/90 (54%), Positives = 65/90 (71%), Gaps = 3/90 (3%)
Query: 175 HVGHQGP-TALSGKPTYRVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRV 233
H +Q P T +PT+++ +A+ Y LTVRH V+LAP NP +H WYKD ++STRV
Sbjct: 9 HGQYQPPATGPQHEPTFKIFCRADEGYCLTVRHDAVVLAPTNPREQH--WYKDMRHSTRV 66
Query: 234 KDAERCPAFSLVNKATGEAIKHSIGESHPV 263
KD E PAF+LVN+ATG A+KHS+G+SHPV
Sbjct: 67 KDEEGHPAFALVNRATGLAVKHSLGQSHPV 96
>dbj|BAA35135.1| Extensin [Adiantum capillus-veneris]
Length = 207
Score = 78.2 bits (191), Expect = 4e-13
Identities = 51/151 (33%), Positives = 68/151 (44%), Gaps = 16/151 (10%)
Query: 10 NNSHATHTHHHNNRRDDDEEQHPNYP---PPPGSTGSPYSSFNNPYPPPQQQPPF-YGEP 65
++SH H H+H++++ HP+YP PPP + PY + P P P PP+ Y P
Sbjct: 48 HDSHWRHPHYHHHKQ--RTPWHPHYPHKSPPPSPSSPPYKYKSPPPPSPSPPPPYIYKSP 105
Query: 66 HRPPPQQPPFYGEPHRPPPSQPETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPP- 124
P P PP Y PPPS S Y + S PP S P Y P PP
Sbjct: 106 PPPSPSPPPPYIYKSPPPPS--------PSPPPPYLYKSPPPPSPSPPPPYIYKSPPPPP 157
Query: 125 -AQQPPFSSYSAPPPPNAATVHHVSHEVHPP 154
PP YS+PPPP+ + ++ PP
Sbjct: 158 CTPSPPPYFYSSPPPPSPSPPPPYQYKSPPP 188
Score = 50.1 bits (118), Expect = 1e-04
Identities = 37/108 (34%), Positives = 42/108 (38%), Gaps = 25/108 (23%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQ---QPPFYGEPHRPPPSQPETNV 91
PPPP + P + +P PP PP Y PPP PP Y PPPS
Sbjct: 121 PPPPSPSPPPPYLYKSPPPPSPSPPPPYIYKSPPPPPCTPSPPPYFYSSPPPPS------ 174
Query: 92 FHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPP 139
S Y + S PP SH P P PP+ S PPPP
Sbjct: 175 --PSPPPPYQYKSPPPPSH----------PSP----PPYVYKSPPPPP 206
Score = 41.2 bits (95), Expect = 0.059
Identities = 39/146 (26%), Positives = 52/146 (34%), Gaps = 48/146 (32%)
Query: 13 HATHTHHHNNRRDDDEEQHPNYPPPPGSTGSPYSSFNNPYPP--PQQQPPFYGEPHRPPP 70
H + HHH D +HP+Y + P+ P P + PP P P
Sbjct: 41 HVPYHHHH-----DSHWRHPHYH---------HHKQRTPWHPHYPHKSPP-------PSP 79
Query: 71 QQPPF-YGEPHRPPPSQPETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPP 129
PP+ Y P P PS P ++ S PP S P PP PP
Sbjct: 80 SSPPYKYKSPPPPSPSPPPPYIYK----------SPPPPS-----------PSPP---PP 115
Query: 130 FSSYSAPPPPNAATVHHVSHEVHPPS 155
+ S PPP + ++ PPS
Sbjct: 116 YIYKSPPPPSPSPPPPYLYKSPPPPS 141
>dbj|BAB11454.1| unnamed protein product [Arabidopsis thaliana]
Length = 1289
Score = 77.8 bits (190), Expect = 6e-13
Identities = 51/125 (40%), Positives = 56/125 (44%), Gaps = 27/125 (21%)
Query: 31 HPNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPPFYGEPHRPPPSQPETN 90
H PPP S GSP PPP PP YG P PPP PP YG P PPP P
Sbjct: 930 HSVLSPPPPSYGSP--------PPPPPPPPSYGSP-PPPPPPPPSYGSPPPPPPPPP--- 977
Query: 91 VFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSA-PPPPNAATVHHVSH 149
G+ S PP P P+YG P PP PPFS S+ PPPP +H +
Sbjct: 978 ----------GYGSPPPP---PPPPPSYGSP-PPPPPPPFSHVSSIPPPPPPPPMHGGAP 1023
Query: 150 EVHPP 154
PP
Sbjct: 1024 PPPPP 1028
Score = 77.4 bits (189), Expect = 7e-13
Identities = 51/123 (41%), Positives = 57/123 (45%), Gaps = 27/123 (21%)
Query: 32 PNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPPFYGEPHRPPPSQPETNV 91
P PPPP P+S+ ++ PP PP YG P PPP PP YG P PPP P
Sbjct: 918 PPPPPPP-----PFSNAHSVLSPP---PPSYGSP-PPPPPPPPSYGSPPPPPPPPP---- 964
Query: 92 FHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPPNAATVHHVSHEV 151
+YG S PP P P YG P PP PP SY +PPPP HVS
Sbjct: 965 -------SYG--SPPPP---PPPPPGYGSPPPPPPPPP--SYGSPPPPPPPPFSHVSSIP 1010
Query: 152 HPP 154
PP
Sbjct: 1011 PPP 1013
Score = 76.3 bits (186), Expect = 2e-12
Identities = 48/123 (39%), Positives = 52/123 (42%), Gaps = 12/123 (9%)
Query: 32 PNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPPFYGEPHRPPPSQPETNV 91
P PPPP S GSP PPP PP YG P PPP PP YG P PPP P
Sbjct: 944 PPPPPPPPSYGSP--------PPPPPPPPSYGSPP-PPPPPPPGYGSPPPPPPPPPSYGS 994
Query: 92 FHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPPNAATVHHVSHEV 151
+ HVS P P P +G PP PP APPPP +H +
Sbjct: 995 PPPPPPPPFSHVSSIPPP--PPPPPMHGGAPPPPPPPPMHG-GAPPPPPPPPMHGGAPPP 1051
Query: 152 HPP 154
PP
Sbjct: 1052 PPP 1054
Score = 58.9 bits (141), Expect = 3e-07
Identities = 40/115 (34%), Positives = 45/115 (38%), Gaps = 25/115 (21%)
Query: 32 PNYPPPPGSTGSP-------YSSFNNPYPPPQQQPPFYGEPHRPPPQQPPFYGEPHRPPP 84
P PPPP S GSP +S ++ PPP P G P PPP PP +G PPP
Sbjct: 983 PPPPPPPPSYGSPPPPPPPPFSHVSSIPPPPPPPPMHGGAP--PPPPPPPMHGGAPPPPP 1040
Query: 85 SQPETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPP 139
P H PP P P +G PP P F PPPP
Sbjct: 1041 PPPM-------------HGGAPPP---PPPPPMHGGAPPPPPPPMFGGAQPPPPP 1079
Score = 52.4 bits (124), Expect = 3e-05
Identities = 51/165 (30%), Positives = 58/165 (34%), Gaps = 43/165 (26%)
Query: 32 PNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGE--------PHRPPPQQP-----PFYGE 78
P PPPP S+ P S P PPP PPF E P PPP P P G
Sbjct: 670 PPPPPPPFSSERPNSGTVLP-PPPPPPPPFSSERPNSGTVLPPPPPPPLPFSSERPNSGT 728
Query: 79 PHRPPPSQPETNVF----------HTSHDVTYGHVSHPP--------QSHHPDHSPNYGY 120
PPPS P +V+ TS T PP Q +
Sbjct: 729 VLPPPPSPPWKSVYASALAIPAICSTSQAPTSSPTPPPPPPAYYSVGQKSSDLQTSQLPS 788
Query: 121 PVPPAQQPPFSSY-----------SAPPPPNAATVHHVSHEVHPP 154
P PP PPF+S PPPP A+V S + PP
Sbjct: 789 PPPPPPPPPFASVRRNSETLLPPPPPPPPPPFASVRRNSETLLPP 833
Score = 52.4 bits (124), Expect = 3e-05
Identities = 43/158 (27%), Positives = 58/158 (36%), Gaps = 35/158 (22%)
Query: 32 PNYPPPP------GSTGSPYSSFNNPYPPPQQQPPFYGEPHR--------PPPQQPPFYG 77
P PPPP G S + P PPP PP + R PPP PP +
Sbjct: 762 PTPPPPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVRRNSETLLPPPPPPPPPPFA 821
Query: 78 EPHR--------PPPSQPETNVFHTS---HDVTYGHVSHPPQSHHPDHSP-------NYG 119
R PPP P +++ ++ H+ S PP P SP +Y
Sbjct: 822 SVRRNSETLLPPPPPPPPWKSLYASTFETHEACSTSSSPPPPPPPPPFSPLNTTKANDYI 881
Query: 120 YPVPPAQQPPFSSYSAPPPPNAATVHHVSHEVHPPSNT 157
P PP P++S + P +H +S PP T
Sbjct: 882 LPPPPL---PYTSIAPSPSVKILPLHGISSAPSPPVKT 916
Score = 51.2 bits (121), Expect = 6e-05
Identities = 37/118 (31%), Positives = 43/118 (36%), Gaps = 13/118 (11%)
Query: 32 PNYPPPPGSTGSPYSSFNNPY----PPPQQQPPFYGEPHRPPPQQPPFYGEPHRPPPSQP 87
P PPPP G+P P PPP PP +G PPP PP +G PPP
Sbjct: 1024 PPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHG--GAPPPPPPPMFGGAQPPPPPPM 1081
Query: 88 ETNVFHTSHDVTYGHVSHPPQ------SHHPDHSPNYGYPVPPAQQPPFSSYSAPPPP 139
G PP + P P +G PP PP + PPPP
Sbjct: 1082 RGGAPPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMHG-GAPPPPPPPMRGGAPPPPP 1138
Score = 50.8 bits (120), Expect = 7e-05
Identities = 49/175 (28%), Positives = 60/175 (34%), Gaps = 45/175 (25%)
Query: 23 RRDDDEEQHPNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYG------EPHR--------- 67
RR+ + P PPPP S + PPP PP+ E H
Sbjct: 802 RRNSETLLPPPPPPPPPPFASVRRNSETLLPPPPPPPPWKSLYASTFETHEACSTSSSPP 861
Query: 68 PPPQQPPF-------YGEPHRPPPSQPETNVFHT---------------SHDVTYGHVSH 105
PPP PPF + PPP P T++ + S V
Sbjct: 862 PPPPPPPFSPLNTTKANDYILPPPPLPYTSIAPSPSVKILPLHGISSAPSPPVKTAPPPP 921
Query: 106 PPQSHHPDHS------PNYGYPVPPAQQPPFSSYSAPPPPNAATVHHVSHEVHPP 154
PP HS P+YG P PP PP SY +PPPP + S PP
Sbjct: 922 PPPPFSNAHSVLSPPPPSYGSPPPPPPPPP--SYGSPPPPPPPPPSYGSPPPPPP 974
Score = 50.4 bits (119), Expect = 1e-04
Identities = 47/130 (36%), Positives = 51/130 (39%), Gaps = 28/130 (21%)
Query: 32 PNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHR-------PPPQQPPFYGE------ 78
P PPPP T S Y + P PPP PPF E PPP PPF E
Sbjct: 649 PPPPPPPLPTYSHYQTSQLP-PPPPPPPPFSSERPNSGTVLPPPPPPPPPFSSERPNSGT 707
Query: 79 --PHRPPPSQPETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVP-PA----QQPPFS 131
P PPP P +S G V PP S P Y + PA Q P S
Sbjct: 708 VLPPPPPPPLP-----FSSERPNSGTVLPPPPS--PPWKSVYASALAIPAICSTSQAPTS 760
Query: 132 SYSAPPPPNA 141
S + PPPP A
Sbjct: 761 SPTPPPPPPA 770
Score = 48.9 bits (115), Expect = 3e-04
Identities = 44/167 (26%), Positives = 58/167 (34%), Gaps = 40/167 (23%)
Query: 32 PNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHR----PPPQQPPFYG---------- 77
P PPPP S+ P S P PPP P P+ PPP PP+
Sbjct: 691 PPPPPPPFSSERPNSGTVLPPPPPPPLPFSSERPNSGTVLPPPPSPPWKSVYASALAIPA 750
Query: 78 ----------EPHRPPPSQPETNVFHTSHDVTYGHVSHPPQSHHPD-------HSPNYGY 120
P PPP +V S D+ + PP P +S
Sbjct: 751 ICSTSQAPTSSPTPPPPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVRRNSETLLP 810
Query: 121 PVPPAQQPPFSSYSA---------PPPPNAATVHHVSHEVHPPSNTN 158
P PP PPF+S PPPP +++ + E H +T+
Sbjct: 811 PPPPPPPPPFASVRRNSETLLPPPPPPPPWKSLYASTFETHEACSTS 857
Score = 44.3 bits (103), Expect = 0.007
Identities = 37/111 (33%), Positives = 41/111 (36%), Gaps = 16/111 (14%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQ---PPFYGEPHR---PPPQQPPFYGEPHRPPPSQPE 88
PPPP G+P P PPP + PP P R PPP PP +G PPP P
Sbjct: 1077 PPPPMRGGAP------PPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMHGGAP-PPPPPPM 1129
Query: 89 TNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPP 139
G P P G P PP +PP PPPP
Sbjct: 1130 RGGAPPPPPPPGGRGPGAPPPPPPPGGRAPGPPPPPGPRPP---GGGPPPP 1177
Score = 38.1 bits (87), Expect = 0.50
Identities = 24/64 (37%), Positives = 26/64 (40%), Gaps = 14/64 (21%)
Query: 32 PNYPPPPGSTGSPYSSFNNPYPPPQQQ--------PPFYGEPHRPPPQQPPFYGEPHRPP 83
P PPPP G+P P PPP + PP P PPP PP P PP
Sbjct: 1110 PPPPPPPMHGGAP------PPPPPPMRGGAPPPPPPPGGRGPGAPPPPPPPGGRAPGPPP 1163
Query: 84 PSQP 87
P P
Sbjct: 1164 PPGP 1167
>emb|CAB78376.1| extensin-like protein [Arabidopsis thaliana]
gi|4584539|emb|CAB40769.1| extensin-like protein
[Arabidopsis thaliana] gi|15235668|ref|NP_193070.1|
leucine-rich repeat family protein / extensin family
protein [Arabidopsis thaliana] gi|7484961|pir||T06291
extensin homolog T9E8.80 - Arabidopsis thaliana
Length = 760
Score = 75.5 bits (184), Expect = 3e-12
Identities = 44/106 (41%), Positives = 45/106 (41%), Gaps = 16/106 (15%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQP-PFYGEPHRPPPSQPETNVFH 93
PPPP YS P PPP PP Y P PPP P P Y P PPP P V+
Sbjct: 426 PPPPSPPPPVYS----PPPPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPPPPPPPPPVY- 480
Query: 94 TSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPP 139
S PP S P P Y P PP PP YS PPPP
Sbjct: 481 ----------SPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPP 516
Score = 69.7 bits (169), Expect = 2e-10
Identities = 47/132 (35%), Positives = 53/132 (39%), Gaps = 15/132 (11%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPPFYGEPHRPPPSQPETNVFHT 94
PPPP P S P PPP PP Y P PP PP P PPP P V+
Sbjct: 453 PPPPPPPPPPVYSPPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSP 512
Query: 95 SHDVTYGHVSHPPQSHHPDHSPNY--GYPVPPAQQPP---FSS-------YSAPPPPNAA 142
Y S PP P +P Y P PP PP FS YS+PPPP+++
Sbjct: 513 PPPPVY---SSPPPPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSSPPPPHSS 569
Query: 143 TVHHVSHEVHPP 154
H H P
Sbjct: 570 PPPHSPPPPHSP 581
Score = 67.8 bits (164), Expect = 6e-10
Identities = 48/136 (35%), Positives = 53/136 (38%), Gaps = 11/136 (8%)
Query: 32 PNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPPFYGEP-------HRPPP 84
P PPPP P S P PPP P + P PPP PP Y P PPP
Sbjct: 466 PPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSSPPPPP 525
Query: 85 SQPETNVFHTSHDVTYGHVSHPPQSHHPDHSP-NYGYPVPPAQQPPFSSYSAPPPPNAAT 143
S T V+ T H PPQ P P Y P PP PP S PPPP++
Sbjct: 526 SPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSSPPPPHSSPPPHS---PPPPHSPP 582
Query: 144 VHHVSHEVHPPSNTNV 159
+ PP T V
Sbjct: 583 PPIYPYLSPPPPPTPV 598
Score = 58.9 bits (141), Expect = 3e-07
Identities = 57/175 (32%), Positives = 68/175 (38%), Gaps = 39/175 (22%)
Query: 32 PNYPPPPGSTGSPYSSFNNPYPPPQQ-----QPPFYGEPH-----RPPPQQPPFYGEPHR 81
P+ PPPP S P + +P PPP P Y P PPP P P
Sbjct: 572 PHSPPPPHSPPPPIYPYLSPPPPPTPVSSPPPTPVYSPPPPPPCIEPPPPPPCIEYSPPP 631
Query: 82 PPP-----SQPETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPA---QQPPFSS- 132
PPP S P V+++S + S PP P +Y P PP PP S
Sbjct: 632 PPPVVHYSSPPPPPVYYSSPPPPPVYYSSPP----PPPPVHYSSPPPPEVHYHSPPPSPV 687
Query: 133 -YSAPPPPNAATVHHVSHEVHPPSNTNVLHVSHETNQFPNSVHH-----VGHQGP 181
YS+PPPP +A E PP V H + P VHH V HQ P
Sbjct: 688 HYSSPPPPPSAPC-----EESPPPAPVVHH-----SPPPPMVHHSPPPPVIHQSP 732
Score = 58.2 bits (139), Expect = 5e-07
Identities = 45/127 (35%), Positives = 52/127 (40%), Gaps = 24/127 (18%)
Query: 31 HPNYPPPP----GSTGSPYSSFNNPYPPPQQQPPFYGEPHRP------PPQQPPFYGEPH 80
H + PPPP S P +++P PPP P Y P P PP P Y P
Sbjct: 637 HYSSPPPPPVYYSSPPPPPVYYSSPPPPP---PVHYSSPPPPEVHYHSPPPSPVHYSSPP 693
Query: 81 RPP-----PSQPETNVFHTSHDVTYGHVSHPPQSHH---PDHSPNYGYPVPPAQQPPFSS 132
PP S P V H S H S PP H P SP Y P+PP S
Sbjct: 694 PPPSAPCEESPPPAPVVHHSPPPPMVHHSPPPPVIHQSPPPPSPEYEGPLPPVIG---VS 750
Query: 133 YSAPPPP 139
Y++PPPP
Sbjct: 751 YASPPPP 757
Score = 54.3 bits (129), Expect = 7e-06
Identities = 49/142 (34%), Positives = 59/142 (41%), Gaps = 26/142 (18%)
Query: 28 EEQHPNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPPFYGEP-----HRP 82
E + + PPPP S+ P+S PPP PP P+ PP P P + P
Sbjct: 556 EPYYYSSPPPPHSSPPPHS------PPPPHSPPPPIYPYLSPPPPPTPVSSPPPTPVYSP 609
Query: 83 PPSQPETNVFHTSHDVTYGHVSHPPQSHH--PDHSPNYGYPVPPAQQPPFSSYSAPPPPN 140
PP P + Y PP H+ P P Y Y PP PP YS+PPPP
Sbjct: 610 PPPPPCIEPPPPPPCIEYSPPPPPPVVHYSSPPPPPVY-YSSPP---PPPVYYSSPPPP- 664
Query: 141 AATVHHVS---HEVH----PPS 155
VH+ S EVH PPS
Sbjct: 665 -PPVHYSSPPPPEVHYHSPPPS 685
Score = 54.3 bits (129), Expect = 7e-06
Identities = 39/108 (36%), Positives = 42/108 (38%), Gaps = 20/108 (18%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQ---PPFYGEPHRPPPSQPETNV 91
PP P SP PPP PP PH PPP PP Y PPP P ++
Sbjct: 554 PPEPYYYSSP--------PPPHSSPP----PHSPPPPHSPPPPIYPYLSPPPPPTPVSSP 601
Query: 92 FHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPP 139
T Y PP P P Y PP PP YS+PPPP
Sbjct: 602 PPTP---VYSPPPPPPCIEPPPPPPCIEYSPPP--PPPVVHYSSPPPP 644
>gb|AAD41992.1| hypothetical protein [Arabidopsis thaliana]
gi|20197656|gb|AAM15181.1| hypothetical protein
[Arabidopsis thaliana] gi|25407889|pir||C84672
hypothetical protein At2g27380 [imported] - Arabidopsis
thaliana gi|15225875|ref|NP_180307.1| proline-rich
family protein [Arabidopsis thaliana]
Length = 761
Score = 74.7 bits (182), Expect = 5e-12
Identities = 55/137 (40%), Positives = 64/137 (46%), Gaps = 20/137 (14%)
Query: 14 ATHTHHHNNRRDDDEEQHPN--YPPPPGSTGSPYSSFNNP-YPPPQQQPPFYGEPHRPPP 70
AT + N D HP Y PP T P ++ P YPPP Q+PP Y P PPP
Sbjct: 27 ATVKQNFNKYETDSGHAHPPPIYGAPPSYTTPPPPIYSPPIYPPPIQKPPTYSPPIYPPP 86
Query: 71 -QQP--PFYGEPHRPPPSQ-PETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PP 124
Q+P P Y P PPP Q P T TY +PP P +P Y P+ PP
Sbjct: 87 IQKPPTPTYSPPIYPPPIQKPPT--------PTYSPPIYPPPIQKPP-TPTYSPPIYPPP 137
Query: 125 AQQPPFSSYSAP--PPP 139
Q+PP SYS P PPP
Sbjct: 138 IQKPPTPSYSPPVKPPP 154
Score = 70.9 bits (172), Expect = 7e-11
Identities = 62/189 (32%), Positives = 78/189 (40%), Gaps = 32/189 (16%)
Query: 32 PNYPPP--PGSTGSPYSSFNNPY-PPPQQQPPF--YGEPHRPPP-QQP--PFYGEPHRPP 83
P Y PP P P +++ P PPP Q+PP Y P +PPP Q+P P Y P +PP
Sbjct: 464 PTYSPPIKPPPVKPPTPTYSPPVQPPPVQKPPTPTYSPPVKPPPIQKPPTPTYSPPIKPP 523
Query: 84 PSQPETNVF---------HTSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSS 132
P +P T + H TY PP H P +P Y P+ PP +PP +
Sbjct: 524 PVKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPIHKPP-TPTYSPPIKPPPVHKPPTPT 582
Query: 133 YSAP--PPPNAATVHHVSHEVH-PPSNTNVLHVSHETNQFPNSVHHVGHQGPTALSGKPT 189
YS P PPP VH + PP +H P H+ PT PT
Sbjct: 583 YSPPIKPPP----VHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPPT-----PT 633
Query: 190 YRVVTKANP 198
Y K P
Sbjct: 634 YSPPIKPPP 642
Score = 65.9 bits (159), Expect = 2e-09
Identities = 46/120 (38%), Positives = 56/120 (46%), Gaps = 20/120 (16%)
Query: 32 PNYPPP---PGSTGSPYSSFNNPY-PPPQQQPPF--YGEPHRPPPQQP--PFYGEPHRPP 83
P Y PP P P +++ P PPP Q+PP Y P +PPP +P P Y P +PP
Sbjct: 296 PTYSPPVKSPPVQKPPTPTYSPPIKPPPVQKPPTPTYSPPIKPPPVKPPTPIYSPPVKPP 355
Query: 84 PSQPETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSSYSAP--PPP 139
P H Y PP H P +P Y PV PP Q+PP +YS P PPP
Sbjct: 356 P-------VHKPPTPIYSPPVKPPPVHKPP-TPIYSPPVKPPPIQKPPTPTYSPPIKPPP 407
Score = 63.5 bits (153), Expect = 1e-08
Identities = 50/134 (37%), Positives = 63/134 (46%), Gaps = 22/134 (16%)
Query: 32 PNYPPPPGSTGSPYSSFNNP-YPPPQQQPPF--YGEPHRPPP-QQPPF--YGEPHRPPPS 85
P YPPP +P +++ P YPPP Q+PP Y P PPP Q+PP Y P +PPP
Sbjct: 98 PIYPPPIQKPPTP--TYSPPIYPPPIQKPPTPTYSPPIYPPPIQKPPTPSYSPPVKPPPV 155
Query: 86 Q-PETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPV-PPAQQPP---FSSYSAPPPPN 140
Q P T TY PP H P +P Y P+ PP +PP +S PPP +
Sbjct: 156 QMPPTP--------TYSPPIKPPPVHKPP-TPTYSPPIKPPVHKPPTPIYSPPIKPPPVH 206
Query: 141 AATVHHVSHEVHPP 154
S + PP
Sbjct: 207 KPPTPIYSPPIKPP 220
Score = 62.8 bits (151), Expect = 2e-08
Identities = 49/137 (35%), Positives = 58/137 (41%), Gaps = 23/137 (16%)
Query: 32 PNYPPP--PGSTGSPYSSFNNP--YPPPQQQPP--FYGEPHRPPPQQ---PPFYGEPHRP 82
P Y PP P P + +P PPP +PP Y P +PPP Q P Y P +P
Sbjct: 228 PTYSPPVKPPPVHKPPTPIYSPPIKPPPVHKPPTPIYSPPVKPPPVQTPPTPIYSPPVKP 287
Query: 83 PPSQPETNVFHTSHDVTYG-HVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSSYSAP--P 137
PP H TY V PP P +P Y P+ PP Q+PP +YS P P
Sbjct: 288 PP-------VHKPPTPTYSPPVKSPPVQKPP--TPTYSPPIKPPPVQKPPTPTYSPPIKP 338
Query: 138 PPNAATVHHVSHEVHPP 154
PP S V PP
Sbjct: 339 PPVKPPTPIYSPPVKPP 355
Score = 61.2 bits (147), Expect = 5e-08
Identities = 42/120 (35%), Positives = 51/120 (42%), Gaps = 20/120 (16%)
Query: 32 PNYPPP---PGSTGSPYSSFNNPYPPPQQQPP--FYGEPHRPPP---QQPPFYGEPHRPP 83
P Y PP P P +++ P PP +PP Y P +PPP P Y P +PP
Sbjct: 161 PTYSPPIKPPPVHKPPTPTYSPPIKPPVHKPPTPIYSPPIKPPPVHKPPTPIYSPPIKPP 220
Query: 84 PSQPETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSSYSAP--PPP 139
P H TY PP H P +P Y P+ PP +PP YS P PPP
Sbjct: 221 P-------VHKPPTPTYSPPVKPPPVHKPP-TPIYSPPIKPPPVHKPPTPIYSPPVKPPP 272
Score = 60.5 bits (145), Expect = 9e-08
Identities = 59/189 (31%), Positives = 73/189 (38%), Gaps = 32/189 (16%)
Query: 32 PNYPPP---PGSTGSPYSSFNNPYPPPQQQPPF--YGEPHRPPPQQ---PPFYGEPHRPP 83
P Y PP P P +++ P PP +PP Y P +PPP Q P Y P +PP
Sbjct: 447 PIYSPPVKPPPVHKPPTPTYSPPIKPPPVKPPTPTYSPPVQPPPVQKPPTPTYSPPVKPP 506
Query: 84 PSQ-PETNVFH--------TSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSS 132
P Q P T + TY PP H P +P Y P+ PP +PP +
Sbjct: 507 PIQKPPTPTYSPPIKPPPVKPPTPTYSPPIKPPPVHKPP-TPTYSPPIKPPPIHKPPTPT 565
Query: 133 YSAP--PPPNAATVHHVSHEVH-PPSNTNVLHVSHETNQFPNSVHHVGHQGPTALSGKPT 189
YS P PPP VH + PP +H P H+ PT PT
Sbjct: 566 YSPPIKPPP----VHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPPT-----PT 616
Query: 190 YRVVTKANP 198
Y K P
Sbjct: 617 YSPPIKPPP 625
Score = 59.3 bits (142), Expect = 2e-07
Identities = 47/122 (38%), Positives = 55/122 (44%), Gaps = 23/122 (18%)
Query: 32 PNYPPP---PGSTGSPYSSFNNPY-PPPQQQPPF--YGEPHRPPPQQPP---FYGEPHRP 82
P Y PP P P +++ P PPP +PP Y P +PPP Q P Y P +P
Sbjct: 615 PTYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPVQKPPTPTYSPPVKP 674
Query: 83 PPSQ-PETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSSYSAP--P 137
PP Q P T TY PP P +P Y PV PP Q PP +YS P P
Sbjct: 675 PPVQLPPTP--------TYSPPVKPPPVQVPP-TPTYSPPVKPPPVQVPPTPTYSPPIKP 725
Query: 138 PP 139
PP
Sbjct: 726 PP 727
Score = 58.9 bits (141), Expect = 3e-07
Identities = 41/115 (35%), Positives = 48/115 (41%), Gaps = 16/115 (13%)
Query: 32 PNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPP---QQPPFYGEPHRPPPSQPE 88
P PPP +P S PP + P Y P +PPP P Y P +PPP
Sbjct: 402 PIKPPPLQKPPTPTYSPPIKLPPVKPPTPIYSPPVKPPPVHKPPTPIYSPPVKPPP---- 457
Query: 89 TNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSSYSAP--PPP 139
H TY PP P +P Y PV PP Q+PP +YS P PPP
Sbjct: 458 ---VHKPPTPTYSPPIKPPPVKPP--TPTYSPPVQPPPVQKPPTPTYSPPVKPPP 507
Score = 58.5 bits (140), Expect = 4e-07
Identities = 45/136 (33%), Positives = 58/136 (42%), Gaps = 22/136 (16%)
Query: 32 PNYPPP---PGSTGSPYSSFNNPYPPPQQQPP--FYGEPHRPPP---QQPPFYGEPHRPP 83
P Y PP P P +++ P PP +PP Y P +PPP P Y P +PP
Sbjct: 313 PTYSPPIKPPPVQKPPTPTYSPPIKPPPVKPPTPIYSPPVKPPPVHKPPTPIYSPPVKPP 372
Query: 84 P-SQPETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSSYSAP--PP 138
P +P T ++ V PP P +P Y P+ PP Q+PP +YS P P
Sbjct: 373 PVHKPPTPIYSPP-------VKPPPIQKPP--TPTYSPPIKPPPLQKPPTPTYSPPIKLP 423
Query: 139 PNAATVHHVSHEVHPP 154
P S V PP
Sbjct: 424 PVKPPTPIYSPPVKPP 439
Score = 58.2 bits (139), Expect = 5e-07
Identities = 46/134 (34%), Positives = 58/134 (42%), Gaps = 21/134 (15%)
Query: 32 PNYPPP---PGSTGSPYSSFNNPY-PPPQQQPPF--YGEPHRPPPQQ---PPFYGEPHRP 82
P Y PP P P +++ P PPP Q+PP Y P +PPP Q P Y P +P
Sbjct: 632 PTYSPPIKPPPVHKPPTPTYSPPIKPPPVQKPPTPTYSPPVKPPPVQLPPTPTYSPPVKP 691
Query: 83 PPSQ-PETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSSYSAPPPP 139
PP Q P T TY PP P +P Y P+ PP Q PP + +PP
Sbjct: 692 PPVQVPPTP--------TYSPPVKPPPVQVPP-TPTYSPPIKPPPVQVPPTPTTPSPPQG 742
Query: 140 NAATVHHVSHEVHP 153
T ++ HP
Sbjct: 743 GYGTPPPYAYLSHP 756
Score = 57.4 bits (137), Expect = 8e-07
Identities = 46/122 (37%), Positives = 54/122 (43%), Gaps = 23/122 (18%)
Query: 32 PNYPPP---PGSTGSPYSSFNNPY-PPPQQQPPF--YGEPHRPPP---QQPPFYGEPHRP 82
P Y PP P P +++ P PPP +PP Y P +PPP P Y P +P
Sbjct: 598 PTYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKPPPVHKPPTPTYSPPIKP 657
Query: 83 PPSQ-PETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPV--PPAQQPPFSSYSAP--P 137
PP Q P T TY PP P +P Y PV PP Q PP +YS P P
Sbjct: 658 PPVQKPPT--------PTYSPPVKPPPVQLPP-TPTYSPPVKPPPVQVPPTPTYSPPVKP 708
Query: 138 PP 139
PP
Sbjct: 709 PP 710
>pir||A29356 hydroxyproline-rich glycoprotein (clone Hyp3.6) - kidney bean
(fragment)
Length = 163
Score = 71.2 bits (173), Expect = 5e-11
Identities = 44/132 (33%), Positives = 59/132 (44%), Gaps = 13/132 (9%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPP--FYGEPHRPPPQQPP--FYGEPHRPPPSQPETN 90
PPPP + P + +P PP PP +Y P P P PP +Y P P PS P
Sbjct: 19 PPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPY 78
Query: 91 VFH-------TSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFS-SYSAPPPPNAA 142
+H T H Y + S PP + +P +Y P PP+ PP Y +PPPP+ A
Sbjct: 79 YYHSPPPPSPTPHP-PYYYKSPPPPTSYPPPPYHYVSPPPPSPSPPPPYYYKSPPPPSPA 137
Query: 143 TVHHVSHEVHPP 154
++ PP
Sbjct: 138 PAPKYIYKSPPP 149
Score = 63.9 bits (154), Expect = 8e-09
Identities = 41/111 (36%), Positives = 50/111 (44%), Gaps = 17/111 (15%)
Query: 35 PPPPGSTGSPYSSFNNPYPP-PQQQPPFYGEPHRPPPQQP----PFYGEPHRPPPSQPET 89
PPPP + P + +P PP P PP+Y H PPP P P+Y + PP S P
Sbjct: 51 PPPPSPSPPPPYYYKSPPPPSPSPPPPYYY--HSPPPPSPTPHPPYYYKSPPPPTSYPPP 108
Query: 90 NVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQP-PFSSYSAPPPP 139
Y +VS PP S P Y P PP+ P P Y +PPPP
Sbjct: 109 ---------PYHYVSPPPPSPSPPPPYYYKSPPPPSPAPAPKYIYKSPPPP 150
Score = 60.5 bits (145), Expect = 9e-08
Identities = 43/143 (30%), Positives = 55/143 (38%), Gaps = 23/143 (16%)
Query: 51 PYPPPQQQPPFYGEPHRPPPQQPP---FYGEPHRPPPSQPETNVFHTSHDVTYGHVSHPP 107
P P P +PP+Y + PP PP +Y P P PS P Y + S PP
Sbjct: 4 PPPSPVVKPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPP----------YYYKSPPP 53
Query: 108 QSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPPNAATVHHVSHEVHPPSNTNVLHVSHETN 167
S P Y P PP+ PP Y PPP + T H + PP T+
Sbjct: 54 PSPSPPPPYYYKSPPPPSPSPPPPYYYHSPPPPSPTPHPPYYYKSPPPPTS--------- 104
Query: 168 QFPNSVHHVGHQGPTALSGKPTY 190
+P +H P + S P Y
Sbjct: 105 -YPPPPYHYVSPPPPSPSPPPPY 126
Score = 58.5 bits (140), Expect = 4e-07
Identities = 40/107 (37%), Positives = 48/107 (44%), Gaps = 16/107 (14%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPP--FYGEPHRPPPSQPETNVF 92
PPP S PY + P P P PP+Y + PP PP ++ PPP P
Sbjct: 68 PPPSPSPPPPYYYHSPPPPSPTPHPPYYYKSPPPPTSYPPPPYHYV--SPPPPSP----- 120
Query: 93 HTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPP 139
S Y + S PP S P +P Y Y PP PP YS+PPPP
Sbjct: 121 --SPPPPYYYKSPPPPS--PAPAPKYIYKSPP---PPAYIYSSPPPP 160
>gb|AAA33763.1| hydroxyproline-rich glycoprotein
Length = 154
Score = 71.2 bits (173), Expect = 5e-11
Identities = 44/132 (33%), Positives = 59/132 (44%), Gaps = 13/132 (9%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPP--FYGEPHRPPPQQPP--FYGEPHRPPPSQPETN 90
PPPP + P + +P PP PP +Y P P P PP +Y P P PS P
Sbjct: 10 PPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPPY 69
Query: 91 VFH-------TSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFS-SYSAPPPPNAA 142
+H T H Y + S PP + +P +Y P PP+ PP Y +PPPP+ A
Sbjct: 70 YYHSPPPPSPTPHP-PYYYKSPPPPTSYPPPPYHYVSPPPPSPSPPPPYYYKSPPPPSPA 128
Query: 143 TVHHVSHEVHPP 154
++ PP
Sbjct: 129 PAPKYIYKSPPP 140
Score = 63.9 bits (154), Expect = 8e-09
Identities = 41/111 (36%), Positives = 50/111 (44%), Gaps = 17/111 (15%)
Query: 35 PPPPGSTGSPYSSFNNPYPP-PQQQPPFYGEPHRPPPQQP----PFYGEPHRPPPSQPET 89
PPPP + P + +P PP P PP+Y H PPP P P+Y + PP S P
Sbjct: 42 PPPPSPSPPPPYYYKSPPPPSPSPPPPYYY--HSPPPPSPTPHPPYYYKSPPPPTSYPPP 99
Query: 90 NVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQP-PFSSYSAPPPP 139
Y +VS PP S P Y P PP+ P P Y +PPPP
Sbjct: 100 ---------PYHYVSPPPPSPSPPPPYYYKSPPPPSPAPAPKYIYKSPPPP 141
Score = 58.5 bits (140), Expect = 4e-07
Identities = 40/107 (37%), Positives = 48/107 (44%), Gaps = 16/107 (14%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPPFYGEPHRPPPQQPP--FYGEPHRPPPSQPETNVF 92
PPP S PY + P P P PP+Y + PP PP ++ PPP P
Sbjct: 59 PPPSPSPPPPYYYHSPPPPSPTPHPPYYYKSPPPPTSYPPPPYHYV--SPPPPSP----- 111
Query: 93 HTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPP 139
S Y + S PP S P +P Y Y PP PP YS+PPPP
Sbjct: 112 --SPPPPYYYKSPPPPS--PAPAPKYIYKSPP---PPAYIYSSPPPP 151
Score = 55.5 bits (132), Expect = 3e-06
Identities = 40/136 (29%), Positives = 52/136 (37%), Gaps = 23/136 (16%)
Query: 58 QPPFYGEPHRPPPQQPP---FYGEPHRPPPSQPETNVFHTSHDVTYGHVSHPPQSHHPDH 114
+PP+Y + PP PP +Y P P PS P Y + S PP S P
Sbjct: 2 KPPYYYKSPPPPSPSPPPPYYYKSPPPPSPSPPPP----------YYYKSPPPPSPSPPP 51
Query: 115 SPNYGYPVPPAQQPPFSSYSAPPPPNAATVHHVSHEVHPPSNTNVLHVSHETNQFPNSVH 174
Y P PP+ PP Y PPP + T H + PP T+ +P +
Sbjct: 52 PYYYKSPPPPSPSPPPPYYYHSPPPPSPTPHPPYYYKSPPPPTS----------YPPPPY 101
Query: 175 HVGHQGPTALSGKPTY 190
H P + S P Y
Sbjct: 102 HYVSPPPPSPSPPPPY 117
>pir||T07623 extensin homolog HRGP2 - soybean (fragment)
gi|347455|gb|AAA33971.1| hydroxyproline-rich
glycoprotein
Length = 169
Score = 70.5 bits (171), Expect = 9e-11
Identities = 44/124 (35%), Positives = 59/124 (47%), Gaps = 13/124 (10%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPP---FYGEPHRPPPQQPPFYGEPHRPPPSQPETNV 91
PPPP + P +++P PPP PP +Y P P P PP Y PPPS
Sbjct: 41 PPPPSPSPPPPYYYHSP-PPPSPSPPSPYYYKSPPPPSPSPPPPYYYKSPPPPSP----- 94
Query: 92 FHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFS-SYSAPPPPNAATVHHVSHE 150
TSH Y + S PP + +P +Y P PP+ PP Y++PPPP+ A ++
Sbjct: 95 --TSHP-PYYYKSPPPPTSYPPPPYHYVSPPPPSPSPPPPYHYTSPPPPSPAPAPKYIYK 151
Query: 151 VHPP 154
PP
Sbjct: 152 SPPP 155
Score = 67.8 bits (164), Expect = 6e-10
Identities = 47/161 (29%), Positives = 60/161 (37%), Gaps = 23/161 (14%)
Query: 44 PYSSFNNPYPPPQQQPPFYGEPHRPPPQQPP---FYGEPHRPPPSQPETNVFHTSHDVTY 100
PY + P P P PP+Y + PP PP +Y P P PS P +H
Sbjct: 3 PYYYHSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYHSPPPPSPSPPPPYYYH------- 55
Query: 101 GHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPPNAATVHHVSHEVHPPSNTNVL 160
S PP S P Y P PP+ PP Y PPP + T H + PP T+
Sbjct: 56 ---SPPPPSPSPPSPYYYKSPPPPSPSPPPPYYYKSPPPPSPTSHPPYYYKSPPPPTS-- 110
Query: 161 HVSHETNQFPNSVHHVGHQGPTALSGKPTYRVVTKANPDYA 201
+P +H P + S P Y + P A
Sbjct: 111 --------YPPPPYHYVSPPPPSPSPPPPYHYTSPPPPSPA 143
Score = 65.9 bits (159), Expect = 2e-09
Identities = 40/108 (37%), Positives = 48/108 (44%), Gaps = 12/108 (11%)
Query: 35 PPPPGSTGSPYSSFNNPYPPPQQQPPFY--GEPHRPPPQQPPFYGEPHRPPPSQPETNVF 92
PPP S SPY + P P P PP+Y P P PP+Y + PP S P
Sbjct: 58 PPPSPSPPSPYYYKSPPPPSPSPPPPYYYKSPPPPSPTSHPPYYYKSPPPPTSYPPP--- 114
Query: 93 HTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQP-PFSSYSAPPPP 139
Y +VS PP S P +Y P PP+ P P Y +PPPP
Sbjct: 115 ------PYHYVSPPPPSPSPPPPYHYTSPPPPSPAPAPKYIYKSPPPP 156
Score = 61.2 bits (147), Expect = 5e-08
Identities = 39/107 (36%), Positives = 49/107 (45%), Gaps = 15/107 (14%)
Query: 35 PPPPGSTGSPYSSFNNPYPP-PQQQPPFYGEPHRPPPQQPPF-YGEPHRPPPSQPETNVF 92
PPPP + P + +P PP P PP+Y + PP PP Y PPPS +
Sbjct: 73 PPPPSPSPPPPYYYKSPPPPSPTSHPPYYYKSPPPPTSYPPPPYHYVSPPPPSPSPPPPY 132
Query: 93 HTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPPPP 139
H + S PP S P +P Y Y PP PP Y++PPPP
Sbjct: 133 H--------YTSPPPPS--PAPAPKYIYKSPP---PPVYIYASPPPP 166
>gb|EAA68066.1| hypothetical protein FG10164.1 [Gibberella zeae PH-1]
gi|46137297|ref|XP_390340.1| hypothetical protein
FG10164.1 [Gibberella zeae PH-1]
Length = 477
Score = 70.5 bits (171), Expect = 9e-11
Identities = 64/240 (26%), Positives = 85/240 (34%), Gaps = 34/240 (14%)
Query: 24 RDDDEEQHPNYPPPPGSTGSPYSSFNNPYPPPQQQPPFYGE----PHRPPPQQPPFYGEP 79
+ ++QH Y PPP S + + F P PP Q PP G+ P P PQ PP G+
Sbjct: 20 QQQQQQQHRQYAPPPTSPPANQTQFYPP-PPQPQSPPQAGQAMNYPPPPQPQSPPQTGQA 78
Query: 80 HRPPPSQPETNVFHTSHDVTYGHVSHPPQS---------HHPDHSPNYGYPVPPAQQPPF 130
PP T+ H T H PPQ +H S YP PP P +
Sbjct: 79 VSYPPPPQHTSSPQVPHQPTPVHYPAPPQQQATPSPQIRNHSHSSSQGNYPPPPMSAPAY 138
Query: 131 SSYSAPPPPNAATVHHVSHEVHPPSNTNVLHVSHETNQFPNSVHHVGHQGPTALSGKPTY 190
+ P A HPP T H + N F P A PT
Sbjct: 139 QQHFQFNQPQA---------THPPQQTQ-YHAPGQQNNFA--------MRPAATPPAPTP 180
Query: 191 RVVTKANPDYALTVRHGQVILAPFNPNNEHQLWYKDEKYSTRVKDAERCPAFSLVNKATG 250
V T T +H + AP +P+ H Y EK + ++ P + +G
Sbjct: 181 PVSTPPVATPQSTQQHFEPPPAPVSPH--HNDSYPPEKQGLEQEAPQQQPGTARAMTVSG 238
Score = 41.2 bits (95), Expect = 0.059
Identities = 54/247 (21%), Positives = 83/247 (32%), Gaps = 63/247 (25%)
Query: 4 PFNQGRNNSHATHTHHHNNRRDDDEEQHPNYPPPPGSTGS--PYSSFNNP---YPPPQQQ 58
P Q RN+SH++ NYPPPP S + + FN P +PP Q Q
Sbjct: 112 PSPQIRNHSHSS--------------SQGNYPPPPMSAPAYQQHFQFNQPQATHPPQQTQ 157
Query: 59 PPFYGEPHRPPPQQPPFYGEPHR--PPPSQPETNVFHTSHDVTYGHVSHPPQSHHPDHSP 116
+ P QQ F P P P+ P + + T H PP P H+
Sbjct: 158 -------YHAPGQQNNFAMRPAATPPAPTPPVSTPPVATPQSTQQHFEPPPAPVSPHHND 210
Query: 117 NYGYPVPPAQQPPFSSYSAPPPPNAATVHHVSHEVHPPSNTNVLHVSHETNQFPNSVHHV 176
+Y P ++ + P A VS + S + F + +
Sbjct: 211 SY-----PPEKQGLEQEAPQQQPGTARAMTVSGAPQAGNFVGAEATSDDIGTFNGGSYRI 265
Query: 177 GHQGPTA-----------LSGKP-------------------TYRVVTKANPDYALTVRH 206
H+ + +SGKP +++T A + +
Sbjct: 266 SHRDTNSILTIQLAMGCPMSGKPGAMIAMSPTVTLKGQIKFSVKKMITGAELSTSTYIGP 325
Query: 207 GQVILAP 213
G+V+LAP
Sbjct: 326 GEVLLAP 332
>gb|AAP54704.1| putative phospholipase [Oryza sativa (japonica cultivar-group)]
gi|37536230|ref|NP_922417.1| putative phospholipase
[Oryza sativa (japonica cultivar-group)]
gi|62733657|gb|AAX95771.1| putative phospholipase [Oryza
sativa (japonica cultivar-group)]
gi|27311294|gb|AAO00720.1| putative phospholipase [Oryza
sativa (japonica cultivar-group)]
Length = 1046
Score = 69.7 bits (169), Expect = 2e-10
Identities = 55/171 (32%), Positives = 69/171 (40%), Gaps = 49/171 (28%)
Query: 34 YP-PPPGSTGSPYSSFNNPYPPPQQQPP-----------FYGEPHRPPPQ-QPPFYG--- 77
YP PPP PY + YPPPQQQPP F+G P PPPQ QP Y
Sbjct: 11 YPYPPPQQYPYPYGQYPYQYPPPQQQPPPPSAYLSPSRSFHGYPSAPPPQPQPQPYAHHS 70
Query: 78 ---EPHRPPPSQ----------PETNVFHTSHDVTYGHVSHPPQSHHPDHSPN---YGYP 121
+P+ PPP P V+ H + S+P + P SP+ + +P
Sbjct: 71 APLQPYPPPPQHHAYPPPQPHPPSPYVYDPYHAPAAAYPSYPSPNPSPSISPSSSFHHHP 130
Query: 122 VPPAQQPPFSSYSA----------------PPPPNAATVHHVSH-EVHPPS 155
PP+ P SY + PPPP+ A V S V PPS
Sbjct: 131 EPPSPSPSAPSYPSIADGLANMHVSDRHDYPPPPSPAAVPAASSPSVLPPS 181
Score = 62.8 bits (151), Expect = 2e-08
Identities = 47/135 (34%), Positives = 58/135 (42%), Gaps = 24/135 (17%)
Query: 42 GSPYSSFNNPYPPPQQQPPFYGE-PHR-PPPQQPP------------FYGEPHRPPPS-Q 86
G + PYPPPQQ P YG+ P++ PPPQQ P F+G P PPP Q
Sbjct: 3 GGNHGGGGYPYPPPQQYPYPYGQYPYQYPPPQQQPPPPSAYLSPSRSFHGYPSAPPPQPQ 62
Query: 87 PETNVFHTSHDVTY-----GHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAP-PPPN 140
P+ H++ Y H PPQ H P Y P A P SY +P P P+
Sbjct: 63 PQPYAHHSAPLQPYPPPPQHHAYPPPQPHPPSPYVYDPYHAPAAAYP---SYPSPNPSPS 119
Query: 141 AATVHHVSHEVHPPS 155
+ H PPS
Sbjct: 120 ISPSSSFHHHPEPPS 134
Score = 36.6 bits (83), Expect = 1.4
Identities = 33/121 (27%), Positives = 40/121 (32%), Gaps = 33/121 (27%)
Query: 30 QHPNYPPPPGSTGSPY------------SSFNNPYPPPQQQPPFYGEPHRPPPQQPPFYG 77
QH YPPP SPY S+ +P P P P H PP
Sbjct: 81 QHHAYPPPQPHPPSPYVYDPYHAPAAAYPSYPSPNPSPSISPSSSFHHHPEPPS------ 134
Query: 78 EPHRPPPSQPETNVFHTSHDVTYGHVSHPPQSHHPDHSPNYGYPVPPAQQPPFSSYSAPP 137
P PS P +Y ++ + H +Y P PA P SS S P
Sbjct: 135 ----PSPSAP-----------SYPSIADGLANMHVSDRHDYPPPPSPAAVPAASSPSVLP 179
Query: 138 P 138
P
Sbjct: 180 P 180
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.137 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 942,153,720
Number of Sequences: 2540612
Number of extensions: 52046639
Number of successful extensions: 455584
Number of sequences better than 10.0: 8989
Number of HSP's better than 10.0 without gapping: 1305
Number of HSP's successfully gapped in prelim test: 8269
Number of HSP's that attempted gapping in prelim test: 301976
Number of HSP's gapped (non-prelim): 52284
length of query: 408
length of database: 863,360,394
effective HSP length: 130
effective length of query: 278
effective length of database: 533,080,834
effective search space: 148196471852
effective search space used: 148196471852
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0107.8


