

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0106.13
(445 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
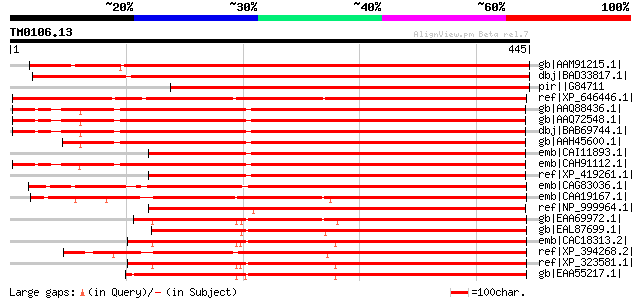
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM91215.1| unknown protein [Arabidopsis thaliana] gi|2019690... 667 0.0
dbj|BAD33817.1| putative tbc1 domain family protein [Oryza sativ... 622 e-177
pir||G84711 hypothetical protein At2g30710 [imported] - Arabidop... 566 e-160
ref|XP_646446.1| hypothetical protein DDB0190714 [Dictyostelium ... 360 5e-98
gb|AAQ88436.1| TBC domain-containing protein [Rattus norvegicus] 345 2e-93
gb|AAQ72548.1| TBC1 domain-containing protein [Homo sapiens] gi|... 344 4e-93
dbj|BAB69744.1| hypothetical protein [Macaca fascicularis] gi|25... 344 4e-93
gb|AAH45600.1| Unknown (protein for MGC:61359) [Mus musculus] gi... 343 5e-93
emb|CAI11893.1| novel protein [Danio rerio] gi|55962494|emb|CAI1... 343 8e-93
emb|CAH91112.1| hypothetical protein [Pongo pygmaeus] 341 2e-92
ref|XP_419261.1| PREDICTED: similar to hypothetical protein [Gal... 340 5e-92
emb|CAG83036.1| unnamed protein product [Yarrowia lipolytica CLI... 339 9e-92
emb|CAA19167.1| SPBC530.01 [Schizosaccharomyces pombe] gi|191121... 339 1e-91
ref|NP_999964.1| hypothetical protein LOC407720 [Danio rerio] gi... 338 2e-91
gb|EAA69972.1| hypothetical protein FG10274.1 [Gibberella zeae P... 338 3e-91
gb|EAL87699.1| GTPase activating protein (Gyp1), putative [Asper... 337 4e-91
emb|CAC18313.2| related to GTPase activating protein [Neurospora... 335 1e-90
ref|XP_394268.2| PREDICTED: similar to ENSANGP00000011734 [Apis ... 335 1e-90
ref|XP_323581.1| related to GTPase activating protein [MIPS] [Ne... 335 1e-90
gb|EAA55217.1| hypothetical protein MG06874.4 [Magnaporthe grise... 333 5e-90
>gb|AAM91215.1| unknown protein [Arabidopsis thaliana] gi|20196901|gb|AAC02742.2|
expressed protein [Arabidopsis thaliana]
gi|13877621|gb|AAK43888.1| Unknown protein [Arabidopsis
thaliana] gi|18402435|ref|NP_565706.1| RabGAP/TBC
domain-containing protein [Arabidopsis thaliana]
Length = 440
Score = 667 bits (1721), Expect = 0.0
Identities = 326/431 (75%), Positives = 374/431 (86%), Gaps = 6/431 (1%)
Query: 18 DSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPTYTRSNSYNDAGTSDHASE 77
DSRF+QTL++VQG LKGRS+PGK+LL++R DP SPTY RS S NDAG ++
Sbjct: 13 DSRFNQTLKNVQGFLKGRSIPGKVLLTRRSDPPPYP--ISPTYQRSLSENDAGRNELFES 70
Query: 78 TVEVEVHSSSGIPGEN---KLKISTSHVENPSEDVRKSSMGARATDSARVMKFTKVLSGT 134
VEVE H+SS KL+ S S E ++V+ +G R++DSARVMKF KVLS T
Sbjct: 71 PVEVEDHNSSKKHDNTYAGKLR-SNSSAERSVKEVQNLKIGVRSSDSARVMKFNKVLSET 129
Query: 135 VVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIP 194
VIL+KLRELAW+GVP YMRP VWRLLLGYAPPNSDRRE VLRRKRLEYL+ V Q+YD+P
Sbjct: 130 TVILEKLRELAWNGVPHYMRPDVWRLLLGYAPPNSDRREAVLRRKRLEYLESVGQFYDLP 189
Query: 195 DTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGIND 254
D+ERS+DEINMLRQIAVDCPRTVPDVSFFQQ+QVQKSLERILY WAIRHPASGYVQGIND
Sbjct: 190 DSERSDDEINMLRQIAVDCPRTVPDVSFFQQEQVQKSLERILYTWAIRHPASGYVQGIND 249
Query: 255 LVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGI 314
LVTPF V+FLSEYL+G +D+WSM DLS++++S+VEADCYWCL+KLLDGMQDHYTFAQPGI
Sbjct: 250 LVTPFLVIFLSEYLDGGVDSWSMDDLSAEKVSDVEADCYWCLTKLLDGMQDHYTFAQPGI 309
Query: 315 QRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAE 374
QRLVFKLKELVRRID+PVS HME GLEFLQFAFRW+NCLLIREIPFN++ RLWDTYLAE
Sbjct: 310 QRLVFKLKELVRRIDEPVSRHMEEHGLEFLQFAFRWYNCLLIREIPFNLINRLWDTYLAE 369
Query: 375 GDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHS 434
GDALPDFLVYI+ASFLLTWSD+L+KLDFQ++VMFLQHLPT +W+ QELEMVLSRA+MWHS
Sbjct: 370 GDALPDFLVYIYASFLLTWSDELKKLDFQEMVMFLQHLPTHNWSDQELEMVLSRAYMWHS 429
Query: 435 MFNNSPNHLAT 445
MFNNSPNHLA+
Sbjct: 430 MFNNSPNHLAS 440
>dbj|BAD33817.1| putative tbc1 domain family protein [Oryza sativa (japonica
cultivar-group)] gi|50725365|dbj|BAD34437.1| putative
tbc1 domain family protein [Oryza sativa (japonica
cultivar-group)]
Length = 444
Score = 622 bits (1603), Expect = e-177
Identities = 300/426 (70%), Positives = 345/426 (80%), Gaps = 3/426 (0%)
Query: 20 RFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPTYTRSNSYNDAGTSDHASETV 79
RF+QTLR+VQG+LKGRS PGK+LL++R +P+ S + Y S
Sbjct: 22 RFNQTLRNVQGMLKGRSFPGKVLLTRRSEPLSPPEYSPRYENDRDEYEQNEGSQEGKGQA 81
Query: 80 EVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARATDSARVMKFTKVLSGTVVILD 139
S N S++ N D + GARATDSAR+ KFT LS VILD
Sbjct: 82 SGNTADSMSAKKSNPPSTSST---NSLPDAQGLVSGARATDSARIAKFTNELSRPAVILD 138
Query: 140 KLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTERS 199
KLREL+WSGVP YMRP +WRLLLGYAPPN+DRREGVL RKRLEY++CVSQYYDIPDTERS
Sbjct: 139 KLRELSWSGVPPYMRPNIWRLLLGYAPPNADRREGVLTRKRLEYVECVSQYYDIPDTERS 198
Query: 200 EDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPF 259
++EINMLRQIAVDCPRTVPDV+FFQ Q+QKSLERILY WAIRHPASGYVQGINDL+TPF
Sbjct: 199 DEEINMLRQIAVDCPRTVPDVTFFQHPQIQKSLERILYTWAIRHPASGYVQGINDLLTPF 258
Query: 260 FVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVF 319
VVFLSE+LEG++D WSM LS ++SN+EADCYWCLSK LDGMQDHYTFAQPGIQRLVF
Sbjct: 259 LVVFLSEHLEGNMDTWSMEKLSPQDVSNIEADCYWCLSKFLDGMQDHYTFAQPGIQRLVF 318
Query: 320 KLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALP 379
+LKELV RID+PVS HME QGL+FLQFAFRWFNCL+IREIPF++VTRLWDTYLAEGD LP
Sbjct: 319 RLKELVHRIDEPVSKHMEEQGLDFLQFAFRWFNCLMIREIPFHLVTRLWDTYLAEGDYLP 378
Query: 380 DFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMFNNS 439
DFLVYI ASFLLTWSDKL+KLDFQ++VMFLQHLPT++W H ELEMVLSRA+MWH+MF +S
Sbjct: 379 DFLVYISASFLLTWSDKLKKLDFQEMVMFLQHLPTRNWAHHELEMVLSRAYMWHTMFKSS 438
Query: 440 PNHLAT 445
P+HLA+
Sbjct: 439 PSHLAS 444
>pir||G84711 hypothetical protein At2g30710 [imported] - Arabidopsis thaliana
Length = 343
Score = 567 bits (1460), Expect = e-160
Identities = 261/307 (85%), Positives = 292/307 (95%)
Query: 139 DKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTER 198
+KLRELAW+GVP YMRP VWRLLLGYAPPNSDRRE VLRRKRLEYL+ V Q+YD+PD+ER
Sbjct: 37 EKLRELAWNGVPHYMRPDVWRLLLGYAPPNSDRREAVLRRKRLEYLESVGQFYDLPDSER 96
Query: 199 SEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTP 258
S+DEINMLRQIAVDCPRTVPDVSFFQQ+QVQKSLERILY WAIRHPASGYVQGINDLVTP
Sbjct: 97 SDDEINMLRQIAVDCPRTVPDVSFFQQEQVQKSLERILYTWAIRHPASGYVQGINDLVTP 156
Query: 259 FFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLV 318
F V+FLSEYL+G +D+WSM DLS++++S+VEADCYWCL+KLLDGMQDHYTFAQPGIQRLV
Sbjct: 157 FLVIFLSEYLDGGVDSWSMDDLSAEKVSDVEADCYWCLTKLLDGMQDHYTFAQPGIQRLV 216
Query: 319 FKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDAL 378
FKLKELVRRID+PVS HME GLEFLQFAFRW+NCLLIREIPFN++ RLWDTYLAEGDAL
Sbjct: 217 FKLKELVRRIDEPVSRHMEEHGLEFLQFAFRWYNCLLIREIPFNLINRLWDTYLAEGDAL 276
Query: 379 PDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMFNN 438
PDFLVYI+ASFLLTWSD+L+KLDFQ++VMFLQHLPT +W+ QELEMVLSRA+MWHSMFNN
Sbjct: 277 PDFLVYIYASFLLTWSDELKKLDFQEMVMFLQHLPTHNWSDQELEMVLSRAYMWHSMFNN 336
Query: 439 SPNHLAT 445
SPNHLA+
Sbjct: 337 SPNHLAS 343
>ref|XP_646446.1| hypothetical protein DDB0190714 [Dictyostelium discoideum]
gi|60474404|gb|EAL72341.1| hypothetical protein
DDB0190714 [Dictyostelium discoideum]
Length = 544
Score = 360 bits (924), Expect = 5e-98
Identities = 188/442 (42%), Positives = 279/442 (62%), Gaps = 7/442 (1%)
Query: 3 NNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPTYTR 62
NN+ +NNNN N +++ + T + + S L + + S+ S T
Sbjct: 95 NNNNNNNNNNNNNNNNNQTTTTTTTTSKMTTSSSASNLNTLENNLSHLSLSSSSINTSVI 154
Query: 63 SNSYNDAGTSDHASETVEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARATDSA 122
+S + S S ++ ++SS+G G + S+S + + +++++ + +S
Sbjct: 155 GHSNSSTHLSASNSNSLSSSLNSSNG--GIDTSSSSSSLQQQQQQQQQQNNI--QNINSN 210
Query: 123 RVMKFTKVLS-GTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRL 181
R+ KF K+L+ G V ++ L+ L W G+PD RP W++LLGY P N +RR+ L RKR
Sbjct: 211 RIKKFEKLLNTGIHVDMESLKTLGWRGIPDRYRPMSWKILLGYLPSNCERRDEHLERKRK 270
Query: 182 EYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAI 241
EY D + QYY D +R E + L+QI +D PRT P V FFQQ +Q LERILY W I
Sbjct: 271 EYRDGLPQYYT-SDEKRGESDRRTLKQIQMDVPRTNPGVPFFQQPLIQDILERILYLWGI 329
Query: 242 RHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLD 301
RHP++GYVQGINDL TPF VFLSEY+E + N + + S ++ VEAD YWCL+KLLD
Sbjct: 330 RHPSTGYVQGINDLATPFIWVFLSEYVE-DVANCQVDQIDSTILAMVEADSYWCLTKLLD 388
Query: 302 GMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPF 361
G+QDHYTFAQPGIQR++ LK L+ +I++ + H+ +Q +F+ FAFRW NCLL+REIPF
Sbjct: 389 GIQDHYTFAQPGIQRMLASLKGLLEKINNSLCAHLADQDAQFITFAFRWMNCLLMREIPF 448
Query: 362 NMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQE 421
+V R+WDTYL+E + F VY+ A+FL+ WSD+L++ DF D+++FLQ PTQ+W ++
Sbjct: 449 PLVIRMWDTYLSEKEGFSVFHVYVCAAFLVLWSDELKQRDFPDIMIFLQKPPTQNWEERD 508
Query: 422 LEMVLSRAFMWHSMFNNSPNHL 443
+E + S AF + S++ + +HL
Sbjct: 509 IESLFSTAFYYRSLYEEAQSHL 530
>gb|AAQ88436.1| TBC domain-containing protein [Rattus norvegicus]
Length = 505
Score = 345 bits (884), Expect = 2e-93
Identities = 189/445 (42%), Positives = 277/445 (61%), Gaps = 21/445 (4%)
Query: 3 NNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPT--- 59
+++E++ ++P++ +S+ + L + +L+ S RV P + + +S
Sbjct: 74 DDEEEDLSSPSLQTLNSKVA--LATAAQVLENHS-------KLRVKPERSQSTTSDVPAN 124
Query: 60 YTRSNSYNDAGTSDHASET-VEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARA 118
Y S +DA S ++S+T + +H +P L+ V S+ +
Sbjct: 125 YKVIKSSSDAQLSRNSSDTCLRNTLHKQQSLP----LRPIIPLVARISDQNASGAPPMTV 180
Query: 119 TDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRR 178
+ R+ KF ++LS LD+LR+ +W GVP +RP WRLL GY P N++RR+ L+R
Sbjct: 181 REKTRLEKFRQLLSSHNTDLDELRKWSWPGVPREVRPVTWRLLSGYLPANTERRKLTLQR 240
Query: 179 KRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYA 238
KR EY + QYYD + E +D RQI +D PRT P + FQQ VQ+ ERIL+
Sbjct: 241 KREEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFI 297
Query: 239 WAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSK 298
WAIRHPASGYVQGINDLVTPFFVVFLSEY+E ++N+ +++LS D + ++EAD +WC+SK
Sbjct: 298 WAIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSK 357
Query: 299 LLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIRE 358
LLDG+QD+YTFAQPGIQ+ V L+ELV RID+ V +H +E+LQFAFRW N LL+RE
Sbjct: 358 LLDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHSHFRRYEVEYLQFAFRWMNNLLMRE 417
Query: 359 IPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFLQHLPTQDW 417
+P RLWDTY +E + F +Y+ A+FL+ W + L + DFQ L+M LQ+LPT W
Sbjct: 418 LPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLIKWRKEILDEEDFQGLLMLLQNLPTIHW 477
Query: 418 THQELEMVLSRAFMWHSMFNNSPNH 442
++E+ ++L+ A+ MF ++PNH
Sbjct: 478 GNEEIGLLLAEAYRLKYMFADAPNH 502
>gb|AAQ72548.1| TBC1 domain-containing protein [Homo sapiens]
gi|55664885|emb|CAH71194.1| TBC1 domain family, member
22B [Homo sapiens] gi|56205912|emb|CAI20318.1| TBC1
domain family, member 22B [Homo sapiens]
gi|56203822|emb|CAI22620.1| TBC1 domain family, member
22B [Homo sapiens] gi|47117913|sp|Q9NU19|TB22B_HUMAN
TBC1 domain family member 22B
gi|40068063|ref|NP_060242.2| TBC1 domain family, member
22B [Homo sapiens]
Length = 505
Score = 344 bits (882), Expect = 4e-93
Identities = 189/445 (42%), Positives = 275/445 (61%), Gaps = 21/445 (4%)
Query: 3 NNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPT--- 59
+++E++ ++P+ +S+ + L + +L+ S RV P + + +S
Sbjct: 74 DDEEEDFSSPSFQTLNSKVA--LATAAQVLENHS-------KLRVKPERSQSTTSDVPAN 124
Query: 60 YTRSNSYNDAGTSDHASET-VEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARA 118
Y S +DA S ++S+T + +H +P L+ V S+ +
Sbjct: 125 YKVIKSSSDAQLSRNSSDTCLRNPLHKQQSLP----LRPIIPLVARISDQNASGAPPMTV 180
Query: 119 TDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRR 178
+ R+ KF ++LS LD+LR+ +W GVP +RP WRLL GY P N++RR+ L+R
Sbjct: 181 REKTRLEKFRQLLSSQNTDLDELRKCSWPGVPREVRPITWRLLSGYLPANTERRKLTLQR 240
Query: 179 KRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYA 238
KR EY + QYYD + E +D RQI +D PRT P + FQQ VQ+ ERIL+
Sbjct: 241 KREEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFI 297
Query: 239 WAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSK 298
WAIRHPASGYVQGINDLVTPFFVVFLSEY+E ++N+ +++LS D + ++EAD +WC+SK
Sbjct: 298 WAIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSK 357
Query: 299 LLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIRE 358
LLDG+QD+YTFAQPGIQ+ V L+ELV RID+ V H +E+LQFAFRW N LL+RE
Sbjct: 358 LLDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMRE 417
Query: 359 IPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFLQHLPTQDW 417
+P RLWDTY +E + F +Y+ A+FL+ W + L + DFQ L+M LQ+LPT W
Sbjct: 418 LPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLIKWRKEILDEEDFQGLLMLLQNLPTIHW 477
Query: 418 THQELEMVLSRAFMWHSMFNNSPNH 442
++E+ ++L+ A+ MF ++PNH
Sbjct: 478 GNEEIGLLLAEAYRLKYMFADAPNH 502
>dbj|BAB69744.1| hypothetical protein [Macaca fascicularis]
gi|25008321|sp|Q95LL3|TB22B_MACFA TBC1 domain family
member 22B
Length = 505
Score = 344 bits (882), Expect = 4e-93
Identities = 189/445 (42%), Positives = 275/445 (61%), Gaps = 21/445 (4%)
Query: 3 NNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPT--- 59
+++E++ ++P+ +S+ + L + +L+ S RV P + + +S
Sbjct: 74 DDEEEDFSSPSFQTLNSKVA--LATAAQVLENHS-------KLRVKPERSQSTTSDVPAN 124
Query: 60 YTRSNSYNDAGTSDHASET-VEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARA 118
Y S +DA S ++S+T + +H +P L+ V S+ +
Sbjct: 125 YKVIKSSSDAQLSRNSSDTCLRNPLHKQQSLP----LRPIIPLVARISDQNASGAPPMTV 180
Query: 119 TDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRR 178
+ R+ KF ++LS LD+LR+ +W GVP +RP WRLL GY P N++RR+ L+R
Sbjct: 181 REKTRLEKFRQLLSSHNTDLDELRKCSWPGVPREVRPVTWRLLSGYLPANTERRKLTLQR 240
Query: 179 KRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYA 238
KR EY + QYYD + E +D RQI +D PRT P + FQQ VQ+ ERIL+
Sbjct: 241 KREEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFI 297
Query: 239 WAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSK 298
WAIRHPASGYVQGINDLVTPFFVVFLSEY+E ++N+ +++LS D + ++EAD +WC+SK
Sbjct: 298 WAIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSK 357
Query: 299 LLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIRE 358
LLDG+QD+YTFAQPGIQ+ V L+ELV RID+ V H +E+LQFAFRW N LL+RE
Sbjct: 358 LLDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMRE 417
Query: 359 IPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFLQHLPTQDW 417
+P RLWDTY +E + F +Y+ A+FL+ W + L + DFQ L+M LQ+LPT W
Sbjct: 418 LPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLIKWRKEILDEEDFQGLLMLLQNLPTIHW 477
Query: 418 THQELEMVLSRAFMWHSMFNNSPNH 442
++E+ ++L+ A+ MF ++PNH
Sbjct: 478 GNEEIGLLLAEAYRLKYMFADAPNH 502
>gb|AAH45600.1| Unknown (protein for MGC:61359) [Mus musculus]
gi|38348532|ref|NP_941049.1| TBC1 domain family, member
22B [Mus musculus]
Length = 505
Score = 343 bits (881), Expect = 5e-93
Identities = 183/402 (45%), Positives = 256/402 (63%), Gaps = 12/402 (2%)
Query: 46 RVDPIDNSNLSSPT---YTRSNSYNDAGTSDHASET-VEVEVHSSSGIPGENKLKISTSH 101
RV P + + +S Y S +DA S ++S+T + +H +P L+
Sbjct: 108 RVKPERSQSTTSDVPANYKVIKSSSDAQLSRNSSDTCLRNPLHKQQSLP----LRPIIPL 163
Query: 102 VENPSEDVRKSSMGARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLL 161
V S+ + + R+ KF ++LS LD+LR+ +W GVP +RP WRLL
Sbjct: 164 VARISDQNASGAPPMTVREKTRLEKFRQLLSSHNTDLDELRKWSWPGVPREVRPVTWRLL 223
Query: 162 LGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVS 221
GY P N++RR+ L+RKR EY + QYYD + E +D RQI +D PRT P +
Sbjct: 224 SGYLPANTERRKLTLQRKREEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIP 280
Query: 222 FFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLS 281
FQQ VQ+ ERIL+ WAIRHPASGYVQGINDLVTPFFVVFLSEY+E ++N+ +++LS
Sbjct: 281 LFQQPLVQEIFERILFIWAIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLS 340
Query: 282 SDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGL 341
D + ++EAD +WC+SKLLDG+QD+YTFAQPGIQ+ V L+ELV RID+ V +H +
Sbjct: 341 QDMLRSIEADSFWCMSKLLDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHSHFRRYEV 400
Query: 342 EFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKL 400
E+LQFAFRW N LL+RE+P RLWDTY +E + F +Y+ A+FL+ W + L +
Sbjct: 401 EYLQFAFRWMNNLLMRELPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLIKWRKEILDEE 460
Query: 401 DFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMFNNSPNH 442
DFQ L+M LQ+LPT W ++E+ ++L+ A+ MF ++PNH
Sbjct: 461 DFQGLLMLLQNLPTIHWGNEEIGLLLAEAYRLKYMFADAPNH 502
>emb|CAI11893.1| novel protein [Danio rerio] gi|55962494|emb|CAI11596.1| novel
protein [Danio rerio]
Length = 364
Score = 343 bits (879), Expect = 8e-93
Identities = 165/324 (50%), Positives = 230/324 (70%), Gaps = 4/324 (1%)
Query: 120 DSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRK 179
+++R+ KF +VL+G L++LR+L+WSG+P +RP W+LL GY P N++RRE L+RK
Sbjct: 41 EASRLDKFRQVLAGPNTDLEELRKLSWSGIPRQVRPITWKLLSGYLPANAERRESTLQRK 100
Query: 180 RLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAW 239
R EY + QYYD + E +D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 101 RQEYFGFIEQYYDSRNDEHHQDTY---RQIHIDIPRTNPLIPLFQQASVQEIFERILFIW 157
Query: 240 AIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKL 299
AIRHPASGYVQGINDLVTPFFVV++ EY+E ++N+++S L + + N+EAD +WC+SKL
Sbjct: 158 AIRHPASGYVQGINDLVTPFFVVYVFEYIEEEVENFNVSSLQEEVLRNIEADSFWCMSKL 217
Query: 300 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREI 359
LDG+QD+YTFAQPGIQR V L+ELV RID+ V M+ +E+LQFAFRW N LL+RE+
Sbjct: 218 LDGIQDNYTFAQPGIQRKVKALEELVSRIDETVHRQMQLYEVEYLQFAFRWMNNLLMREL 277
Query: 360 PFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFLQHLPTQDWT 418
P RLWDTY AE + F +Y+ A+FL+ W + L++ DFQ L++ LQ+LPT W
Sbjct: 278 PLRCTIRLWDTYQAEPEGFSRFHLYVCAAFLVRWRKEILEEKDFQGLMILLQNLPTMHWG 337
Query: 419 HQELEMVLSRAFMWHSMFNNSPNH 442
++E+ ++L+ A+ F ++PNH
Sbjct: 338 NEEVSVLLAEAYRLKFAFADAPNH 361
>emb|CAH91112.1| hypothetical protein [Pongo pygmaeus]
Length = 505
Score = 341 bits (875), Expect = 2e-92
Identities = 188/445 (42%), Positives = 275/445 (61%), Gaps = 21/445 (4%)
Query: 3 NNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSP---T 59
+++E++ ++P+ +S+ + L + +L+ S RV P + + +S +
Sbjct: 74 DDEEEDFSSPSFQTLNSKVA--LATAAQVLENHS-------KLRVKPERSQSTTSDVPAS 124
Query: 60 YTRSNSYNDAGTSDHASET-VEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARA 118
Y S +DA S ++S+T + +H +P L+ V S+ +
Sbjct: 125 YKVIKSSSDAQLSRNSSDTCLRNPLHKQQSLP----LRPIIPLVARISDQNASGAPPMTV 180
Query: 119 TDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRR 178
+ R+ KF ++LS LD+LR+ +W GVP +RP WRLL GY P N++RR+ L+R
Sbjct: 181 REKTRLEKFRQLLSSHNTDLDELRKYSWPGVPREVRPITWRLLSGYLPANTERRKLTLQR 240
Query: 179 KRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYA 238
KR EY + QYYD + E +D RQI +D PRT P + FQQ VQ+ ERIL+
Sbjct: 241 KREEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFI 297
Query: 239 WAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSK 298
WAIRHPASGYVQGINDLVTPFFVVFLSEY+E ++N+ +++LS D + ++ AD +WC+SK
Sbjct: 298 WAIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIGADSFWCMSK 357
Query: 299 LLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIRE 358
LLDG+QD+YTFAQPGIQ+ V L+ELV RID+ V H +E+LQFAFRW N LL+RE
Sbjct: 358 LLDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMRE 417
Query: 359 IPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFLQHLPTQDW 417
+P RLWDTY +E + F +Y+ A+FL+ W + L + DFQ L+M LQ+LPT W
Sbjct: 418 LPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLIKWRKEILDEEDFQGLLMLLQNLPTIHW 477
Query: 418 THQELEMVLSRAFMWHSMFNNSPNH 442
++E+ ++L+ A+ MF ++PNH
Sbjct: 478 GNEEIGLLLAEAYRLKYMFADAPNH 502
>ref|XP_419261.1| PREDICTED: similar to hypothetical protein [Gallus gallus]
Length = 571
Score = 340 bits (872), Expect = 5e-92
Identities = 167/324 (51%), Positives = 225/324 (68%), Gaps = 4/324 (1%)
Query: 120 DSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRK 179
+ R+ KF ++LS LD+LR+ +W GVP +RP WRLL GY P N +RR+ L+RK
Sbjct: 248 EKTRLEKFRQLLSSHNTDLDELRKCSWPGVPREVRPVTWRLLSGYLPANMERRKLTLQRK 307
Query: 180 RLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAW 239
R EY + QYYD + E +D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 308 REEYFGFIQQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIW 364
Query: 240 AIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKL 299
AIRHPASGYVQGINDLVTPFFVVFLSEY+E ++N+ +++LS D + ++EAD +WC+SKL
Sbjct: 365 AIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDVLRSIEADSFWCMSKL 424
Query: 300 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREI 359
LDG+QD+YTFAQPGIQ+ V L+ELV RID+ V H +E+LQFAFRW N LL+RE+
Sbjct: 425 LDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRKYEVEYLQFAFRWMNNLLMREL 484
Query: 360 PFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKLDFQDLVMFLQHLPTQDWT 418
P RLWDTY +E + F +Y+ A+FL+ W + L + DFQ L+M LQ+LPT W
Sbjct: 485 PLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLIKWRKEILDEEDFQGLLMLLQNLPTIHWG 544
Query: 419 HQELEMVLSRAFMWHSMFNNSPNH 442
++E+ ++L+ A+ MF ++PNH
Sbjct: 545 NEEIGLLLAEAYRLKYMFADAPNH 568
>emb|CAG83036.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50546633|ref|XP_500786.1| hypothetical protein
[Yarrowia lipolytica]
Length = 494
Score = 339 bits (870), Expect = 9e-92
Identities = 182/428 (42%), Positives = 264/428 (61%), Gaps = 23/428 (5%)
Query: 17 FDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPTYTRSNSYNDAGTSDHAS 76
+D+ LR+ Q + + + K+ S+R D I S S Y + +
Sbjct: 88 WDANVDNGLRNTQAV---KCVENKVEGSERPDLIKRS--SKDVYPSIRQISASNIKSQVF 142
Query: 77 ETVEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARATDSARVMKFTKVLSGTVV 136
+ +S + + GE+ L ++++ + KS+ TDS ++ KF VL+ V
Sbjct: 143 GILHDPSNSLNAVRGESDLSVNST--------LSKSA----PTDSIKLGKFKTVLAAPNV 190
Query: 137 ILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDT 196
+ +L++LAWSG+P +RP W+LLLGY P NSDRR L RKR EY D V +
Sbjct: 191 DIGELKKLAWSGIPLELRPLSWQLLLGYLPTNSDRRVDTLARKRQEYKDGVEHVFHKVAL 250
Query: 197 ERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLV 256
+++ M QI +D PRT P + + Q+SLERILY WA+RHPASGYVQGINDLV
Sbjct: 251 DQA-----MWHQIEIDVPRTNPHLKLYGFPATQRSLERILYLWAVRHPASGYVQGINDLV 305
Query: 257 TPFFVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQR 316
TPFF FLS Y++ +++ + L + + VEAD +WCLSKLL+G+QD+Y AQPGIQR
Sbjct: 306 TPFFQTFLSAYIDEDVESCDPAQLPREVMDVVEADSFWCLSKLLEGIQDNYVHAQPGIQR 365
Query: 317 LVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEG- 375
V L++L RID ++ H+E++ +EF+QF+FRW NCLL+RE+ R+WDTY+AEG
Sbjct: 366 QVAGLRDLTSRIDAKLAKHLESEQVEFMQFSFRWMNCLLMRELSVKNTIRMWDTYMAEGP 425
Query: 376 DALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSM 435
+ +F VY+ A+FL+ WS KL ++FQD+++FLQ LPT+DW E+E++LS AFMW S+
Sbjct: 426 NGFSEFHVYVCATFLVRWSAKLIHMEFQDIMIFLQSLPTKDWGEGEIELLLSEAFMWQSL 485
Query: 436 FNNSPNHL 443
F N+ HL
Sbjct: 486 FKNASAHL 493
>emb|CAA19167.1| SPBC530.01 [Schizosaccharomyces pombe] gi|19112106|ref|NP_595314.1|
GTPase activating protein [Schizosaccharomyces pombe
972h-] gi|7490611|pir||T40517 GTPase activating protein
- fission yeast (Schizosaccharomyces pombe)
Length = 514
Score = 339 bits (869), Expect = 1e-91
Identities = 189/437 (43%), Positives = 270/437 (61%), Gaps = 26/437 (5%)
Query: 19 SRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNL----SSPTYTRSNSYNDAGTSDH 74
+R S +L +V G K S LSQ D SN+ S P RS + N + S
Sbjct: 90 ARSSLSLEAVVG--KKVSAKFAKYLSQTDDDWGVSNIKASKSLPRMARSTTPNSSSRSLF 147
Query: 75 ASETVEV-----EVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARATDSARVMKFTK 129
V+ ++HSS P KL + VE + S R+ KF++
Sbjct: 148 PQNGVDTTTSRQKLHSSGRFPLPAKLAHRSVEVEVAESNALVS----------RIKKFSR 197
Query: 130 VLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRLEYLDCVSQ 189
+L +V L+ LR LAW+G+P RP VW+ LLGY P N+ RRE L+RKR EY
Sbjct: 198 ILDAPIVDLNALRTLAWNGIPSEHRPIVWKYLLGYLPCNASRREVTLKRKRDEYNAAKDS 257
Query: 190 YYDIPDTERSEDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYV 249
++ +TE + + RQI +D PRT P + +Q Q+ LERILY WA RHPASGYV
Sbjct: 258 CFNT-NTEPPPLDQTIWRQIVLDVPRTNPSILLYQNPLTQRMLERILYVWASRHPASGYV 316
Query: 250 QGINDLVTPFFVVFLSEYLEGSID--NWSMSDLSSDEISNVEADCYWCLSKLLDGMQDHY 307
QGI+DLVTPF VFLSEY+ G D + ++ L +++EAD YWCLSKLLDG+QD+Y
Sbjct: 317 QGISDLVTPFIQVFLSEYI-GDKDPMTYDIALLDETNRNDIEADAYWCLSKLLDGIQDNY 375
Query: 308 TFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRL 367
AQPGI+R V L+EL RID+P+ H++ +G++FLQF+FRW NCLL+RE+ + + R+
Sbjct: 376 IHAQPGIRRQVNNLRELTLRIDEPLVKHLQMEGVDFLQFSFRWMNCLLMRELSISNIIRM 435
Query: 368 WDTYLAEG-DALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVL 426
WDTY+AEG +F +Y+ A+FL+ WS +LQK++FQD+++FLQ +PT+DW+ +++E++L
Sbjct: 436 WDTYMAEGVQGFSEFHLYVCAAFLVKWSSELQKMEFQDILIFLQSIPTKDWSTKDIEILL 495
Query: 427 SRAFMWHSMFNNSPNHL 443
S AF+W S+++ + HL
Sbjct: 496 SEAFLWKSLYSGAGAHL 512
>ref|NP_999964.1| hypothetical protein LOC407720 [Danio rerio]
gi|46403241|gb|AAS92640.1| C22orf4-like protein [Danio
rerio]
Length = 567
Score = 338 bits (867), Expect = 2e-91
Identities = 164/342 (47%), Positives = 231/342 (66%), Gaps = 19/342 (5%)
Query: 120 DSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRK 179
+++R+ KF +VL+G L++LR+L+WSG+P +RP W+LL GY P N++RRE L+RK
Sbjct: 223 EASRLDKFRQVLAGPNTDLEELRKLSWSGIPRQVRPITWKLLSGYLPANAERRESTLQRK 282
Query: 180 RLEYLDCVSQYYDIPDTERSEDEINMLR------------------QIAVDCPRTVPDVS 221
R EY + QYYD + E +D + +I +D PRT P +
Sbjct: 283 RQEYFGFIEQYYDSRNDEHHQDTYRQIHIDIPRMSPESLVLQPKVTEIHIDIPRTNPLIP 342
Query: 222 FFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGSIDNWSMSDLS 281
FQQ VQ+ ERIL+ WAIRHPASGYVQGINDLVTPFFVV++ EY+E ++N+++S L
Sbjct: 343 LFQQASVQEIFERILFIWAIRHPASGYVQGINDLVTPFFVVYVFEYIEEEVENFNVSSLQ 402
Query: 282 SDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGL 341
+ + N+EAD +WC+SKLLDG+QD+YTFAQPGIQR V L+ELV RID+ V HM+ +
Sbjct: 403 EEVLRNIEADSFWCMSKLLDGIQDNYTFAQPGIQRKVKALEELVSRIDETVHRHMQLYEV 462
Query: 342 EFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW-SDKLQKL 400
E+LQFAFRW N LL+RE+P RLWDTY AE + F +Y+ A+FL+ W + L++
Sbjct: 463 EYLQFAFRWMNNLLMRELPLRCTIRLWDTYQAEPEGFSHFHLYVCAAFLVRWRKEILEEK 522
Query: 401 DFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMFNNSPNH 442
DFQ L++ LQ+LPT W ++E+ ++L+ A+ F ++PNH
Sbjct: 523 DFQGLMILLQNLPTMHWGNEEVSVLLAEAYRLKFAFADAPNH 564
>gb|EAA69972.1| hypothetical protein FG10274.1 [Gibberella zeae PH-1]
gi|46137517|ref|XP_390450.1| hypothetical protein
FG10274.1 [Gibberella zeae PH-1]
Length = 564
Score = 338 bits (866), Expect = 3e-91
Identities = 175/358 (48%), Positives = 233/358 (64%), Gaps = 23/358 (6%)
Query: 107 EDVRKSSMGARATDS-----ARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLL 161
+++ K++ + TD+ R+ KF K+L + + L LR+LAWSGVP +R W+LL
Sbjct: 195 KEILKANATPKETDAINSRITRINKFKKLLQASTISLPDLRQLAWSGVPQEVRAITWQLL 254
Query: 162 LGYAPPNSDRREGVLRRKRLEYLDCVSQYYDI---PDTE----------RSEDEINMLRQ 208
L Y P NS+RR L RKR EYLD V Q ++ P T R DE + Q
Sbjct: 255 LSYLPANSERRVATLERKRKEYLDGVRQAFERGGGPTTTSANTASTGRTRGLDEA-IWHQ 313
Query: 209 IAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYL 268
I++D PRT P + + + Q+SLERILY WA+RHPASGYVQGINDLVTPF+ VFL Y+
Sbjct: 314 ISIDVPRTNPHIELYSYEATQRSLERILYLWAVRHPASGYVQGINDLVTPFWQVFLGIYI 373
Query: 269 EGSIDNWSMSD---LSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELV 325
G D S D L + VEAD +WCL+KLLDG+QDHY AQPGIQR V L++L
Sbjct: 374 -GDPDIESGMDPGQLPKSVLDAVEADSFWCLTKLLDGIQDHYIVAQPGIQRQVTALRDLT 432
Query: 326 RRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYI 385
RID +S H+E +G+EF+QF+FRW NCLL+RE R+WDTYLAE +F +Y+
Sbjct: 433 ARIDANLSKHLEQEGIEFIQFSFRWMNCLLMREFSVKNTIRMWDTYLAEEQGFSEFHLYV 492
Query: 386 FASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMFNNSPNHL 443
A+FL+ WSDKL +DFQ+++MFLQ LPT+ WT +++E++LS AF+W S+F S HL
Sbjct: 493 CAAFLVKWSDKLLDMDFQEIMMFLQSLPTKGWTEKDIELLLSEAFIWQSLFKGSAAHL 550
>gb|EAL87699.1| GTPase activating protein (Gyp1), putative [Aspergillus fumigatus
Af293]
Length = 454
Score = 337 bits (864), Expect = 4e-91
Identities = 166/337 (49%), Positives = 226/337 (66%), Gaps = 16/337 (4%)
Query: 122 ARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLLLGYAPPNSDRREGVLRRKRL 181
+R+ KF ++L + V L +LR LAWSG+P +R W++LLGY P NS+RR L RKR
Sbjct: 106 SRINKFKRLLQTSTVSLSELRNLAWSGIPAEVRAMTWQILLGYLPTNSERRVSTLERKRK 165
Query: 182 EYLDCVSQYYDIPDTE-------------RSEDEINMLRQIAVDCPRTVPDVSFFQQQQV 228
EYLD V Q ++ T R DE + QI++D PRT P + + +
Sbjct: 166 EYLDGVRQAFERSTTPSPGNPQASSTGRGRGLDEA-IWHQISIDVPRTSPHIKLYGYEAT 224
Query: 229 QKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLE--GSIDNWSMSDLSSDEIS 286
Q+SLERILY WAIRHPASGYVQGINDLVTPF+ VFL Y+ ++ L +
Sbjct: 225 QRSLERILYVWAIRHPASGYVQGINDLVTPFWQVFLGMYMTDLNVEEDMDPGQLPRSVLD 284
Query: 287 NVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQF 346
VEAD +WCL+KLLDG+QD+Y +AQPGI R V L++L RID ++ H+EN+G+EF+QF
Sbjct: 285 AVEADSFWCLTKLLDGIQDNYIYAQPGIHRQVRALRDLTMRIDSTLAKHLENEGVEFMQF 344
Query: 347 AFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLV 406
+FRW NCLL+RE+ R+WDTY+AE F VY+ A+FL+ WSD+L K+DFQ+++
Sbjct: 345 SFRWMNCLLMREMSVQNTIRMWDTYMAEEQGFSRFHVYVCAAFLVKWSDQLIKMDFQEIM 404
Query: 407 MFLQHLPTQDWTHQELEMVLSRAFMWHSMFNNSPNHL 443
MFLQ LPT+DWT +++E++LS AF+W S+F +S HL
Sbjct: 405 MFLQALPTRDWTEKDIELLLSEAFIWQSLFQDSRAHL 441
>emb|CAC18313.2| related to GTPase activating protein [Neurospora crassa]
Length = 602
Score = 335 bits (860), Expect = 1e-90
Identities = 171/358 (47%), Positives = 232/358 (64%), Gaps = 17/358 (4%)
Query: 102 VENPSEDVRKSSMGARATDS--ARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWR 159
+ +PS S A +S R+ KF K+L + + L+ LR LAWSGVP+ +R W+
Sbjct: 231 INHPSVPPNASQKEIDAVNSRITRINKFKKILQASTIPLNDLRALAWSGVPEEVRAMTWQ 290
Query: 160 LLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDI------PDT------ERSEDEINMLR 207
LLL Y P +S+RR L RKR EYLD V Q ++ P T R DE +
Sbjct: 291 LLLSYLPTSSERRVATLERKRKEYLDGVRQAFERAGGAPPPSTGKGGGGNRGLDEA-IWH 349
Query: 208 QIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEY 267
QI++D PRT P + + + Q+SLERILY WA+RHPASGYVQGINDLVTPF+ VFL Y
Sbjct: 350 QISIDVPRTNPHIELYGYEATQRSLERILYVWAVRHPASGYVQGINDLVTPFWQVFLGTY 409
Query: 268 LEGSIDNWSMS--DLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELV 325
+ M L + VEAD +WCL+KLLDG+QDHY AQPGIQR V L++L
Sbjct: 410 ITDPDIESGMDPGQLPRAVLDAVEADTFWCLTKLLDGIQDHYIVAQPGIQRQVAALRDLT 469
Query: 326 RRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYI 385
+RID ++ H+E + +EF+QF+FRW NCLL+REI R+WDTY+AE +F +Y+
Sbjct: 470 QRIDAGLAKHLEEENVEFIQFSFRWMNCLLMREISVKNTIRMWDTYMAEEQGFSEFHLYV 529
Query: 386 FASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMFNNSPNHL 443
A+FL+ WSDKL K+DFQ+++MFLQ LPT++WT +++E++LS A++W S+F S HL
Sbjct: 530 CAAFLVKWSDKLVKMDFQEIMMFLQSLPTREWTEKDIELLLSEAYIWQSLFKGSSAHL 587
>ref|XP_394268.2| PREDICTED: similar to ENSANGP00000011734 [Apis mellifera]
Length = 501
Score = 335 bits (860), Expect = 1e-90
Identities = 182/410 (44%), Positives = 256/410 (62%), Gaps = 31/410 (7%)
Query: 47 VDPIDNSNLSSPTYTRSNSYNDAGTSDHASETVEVEVHSSS--------GIPGENKLKIS 98
V P+ N++ T+++ N+ S H+SE +++ +S G P + I+
Sbjct: 104 VQPLHLRNVNLSTHSQMNN------SQHSSEDTDIKYGASPQHKPTQFPGRPQPLRQTIA 157
Query: 99 TSHVENPSEDVRKSSMGARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVW 158
+ PS++ S ++ KF +L +V+ LD+LR+L+WSG+P +R W
Sbjct: 158 PTKFFIPSKEQDGES---------KIDKFQSLLEASVLNLDELRQLSWSGIPARLRSVTW 208
Query: 159 RLLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDIPDTERSEDEINMLRQIAVDCPRTVP 218
RLL Y P N +RR+ VL RKRL+Y + V QYYD TER E + RQI +D PR P
Sbjct: 209 RLLSEYLPANLERRQHVLERKRLDYWNLVKQYYD---TERDEGFQDTYRQIHIDIPRMSP 265
Query: 219 DVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEYLEGS----IDN 274
+S FQQ VQ ERILY WAIRHPASGYVQG+NDLVTPFF+VFL E + S ++N
Sbjct: 266 LISLFQQTTVQLIFERILYIWAIRHPASGYVQGMNDLVTPFFLVFLQEAVPVSAWQDLEN 325
Query: 275 WSMSDLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDDPVST 334
+ ++ L ++ +EAD +WCLSK LDG+QD+Y FAQ GIQ V +LKEL++RID P+
Sbjct: 326 YDVASLQKEQRDIIEADSFWCLSKFLDGIQDNYIFAQLGIQHKVNQLKELIQRIDAPLHQ 385
Query: 335 HMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTW- 393
H+ G+++LQF+FRW N LL REIP + RLWDTYLAE D F +Y+ A+FLL W
Sbjct: 386 HLHQHGVDYLQFSFRWMNNLLTREIPLHCTIRLWDTYLAESDRFASFQLYVCAAFLLRWR 445
Query: 394 SDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMFNNSPNHL 443
L + DFQ L++ LQ+LPTQ+WT E+ ++++ A+ F ++PNHL
Sbjct: 446 RHLLLQPDFQGLMLMLQNLPTQNWTDSEIGILVAEAYKLKFTFADAPNHL 495
>ref|XP_323581.1| related to GTPase activating protein [MIPS] [Neurospora crassa]
gi|28922774|gb|EAA31996.1| related to GTPase activating
protein [MIPS] [Neurospora crassa]
Length = 600
Score = 335 bits (860), Expect = 1e-90
Identities = 171/358 (47%), Positives = 232/358 (64%), Gaps = 17/358 (4%)
Query: 102 VENPSEDVRKSSMGARATDS--ARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWR 159
+ +PS S A +S R+ KF K+L + + L+ LR LAWSGVP+ +R W+
Sbjct: 229 INHPSVPPNASQKEIDAVNSRITRINKFKKILQASTIPLNDLRALAWSGVPEEVRAMTWQ 288
Query: 160 LLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDI------PDT------ERSEDEINMLR 207
LLL Y P +S+RR L RKR EYLD V Q ++ P T R DE +
Sbjct: 289 LLLSYLPTSSERRVATLERKRKEYLDGVRQAFERAGGAPPPSTGKGGGGNRGLDEA-IWH 347
Query: 208 QIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVVFLSEY 267
QI++D PRT P + + + Q+SLERILY WA+RHPASGYVQGINDLVTPF+ VFL Y
Sbjct: 348 QISIDVPRTNPHIELYGYEATQRSLERILYVWAVRHPASGYVQGINDLVTPFWQVFLGTY 407
Query: 268 LEGSIDNWSMS--DLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELV 325
+ M L + VEAD +WCL+KLLDG+QDHY AQPGIQR V L++L
Sbjct: 408 ITDPDIESGMDPGQLPRAVLDAVEADTFWCLTKLLDGIQDHYIVAQPGIQRQVAALRDLT 467
Query: 326 RRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYI 385
+RID ++ H+E + +EF+QF+FRW NCLL+REI R+WDTY+AE +F +Y+
Sbjct: 468 QRIDAGLAKHLEEENVEFIQFSFRWMNCLLMREISVKNTIRMWDTYMAEEQGFSEFHLYV 527
Query: 386 FASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMFNNSPNHL 443
A+FL+ WSDKL K+DFQ+++MFLQ LPT++WT +++E++LS A++W S+F S HL
Sbjct: 528 CAAFLVKWSDKLVKMDFQEIMMFLQSLPTREWTEKDIELLLSEAYIWQSLFKGSSAHL 585
>gb|EAA55217.1| hypothetical protein MG06874.4 [Magnaporthe grisea 70-15]
gi|39977979|ref|XP_370377.1| hypothetical protein
MG06874.4 [Magnaporthe grisea 70-15]
Length = 679
Score = 333 bits (855), Expect = 5e-90
Identities = 170/363 (46%), Positives = 231/363 (62%), Gaps = 21/363 (5%)
Query: 100 SHVENPSEDVRKSSMGARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWR 159
SH P D + A + R+ KF K+L + + L +LR LAWSGVP+ +R W+
Sbjct: 223 SHPSTP-HDATPKEIDAANSRITRINKFKKILQSSSIPLTELRALAWSGVPEEVRAMTWQ 281
Query: 160 LLLGYAPPNSDRREGVLRRKRLEYLDCVSQYYDI-----PDTERSE------------DE 202
LLL Y P +S+RR VL RKR EYLD V Q ++ P S DE
Sbjct: 282 LLLSYLPTSSERRVAVLERKRKEYLDGVRQAFESGSGVKPSNSSSNAASTPGRGGRGLDE 341
Query: 203 INMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVV 262
+ QI++D PRT P + + + Q+SLERILY WA+RHPASGYVQGINDLVTPF+ V
Sbjct: 342 A-IWHQISIDVPRTNPHIELYGYEATQRSLERILYVWAVRHPASGYVQGINDLVTPFWQV 400
Query: 263 FLSEYLEGSIDNWSMS--DLSSDEISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFK 320
FL Y+ M L + VEAD +WCL+KLLDG+QDHY AQPGIQR V
Sbjct: 401 FLGTYITDPDIERGMDPGQLPRAVLDAVEADTFWCLTKLLDGIQDHYIVAQPGIQRQVSA 460
Query: 321 LKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPD 380
L++L RID+ ++ H+E + +EF+QF+FRW NCLL+REI R+WDTY+AE + +
Sbjct: 461 LRDLTARIDEQLAKHLERENVEFIQFSFRWMNCLLMREISVKNTIRMWDTYMAEENGFSE 520
Query: 381 FLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAFMWHSMFNNSP 440
F +Y+ A+FL+ WS KL K+DFQ+++MFLQ LPT++WT +++E++LS AF+W S++ S
Sbjct: 521 FHLYVCAAFLVKWSAKLTKMDFQEIMMFLQSLPTKEWTEKDIELLLSEAFIWQSLYKGSS 580
Query: 441 NHL 443
HL
Sbjct: 581 AHL 583
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 778,581,292
Number of Sequences: 2540612
Number of extensions: 32395864
Number of successful extensions: 183539
Number of sequences better than 10.0: 923
Number of HSP's better than 10.0 without gapping: 531
Number of HSP's successfully gapped in prelim test: 393
Number of HSP's that attempted gapping in prelim test: 180355
Number of HSP's gapped (non-prelim): 2098
length of query: 445
length of database: 863,360,394
effective HSP length: 131
effective length of query: 314
effective length of database: 530,540,222
effective search space: 166589629708
effective search space used: 166589629708
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0106.13


