

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0103.12
(1541 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
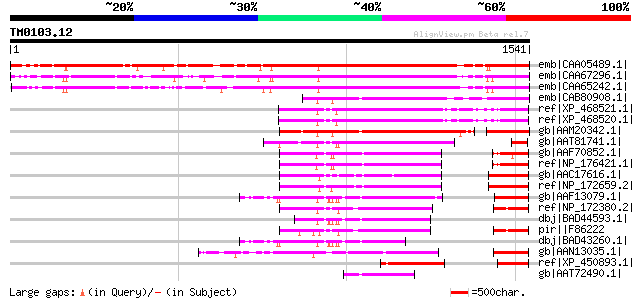
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA05489.1| ENBP1 [Medicago truncatula] gi|11358945|pir||T43... 1452 0.0
emb|CAA67296.1| chloroplast DNA-binding protein PD3 [Pisum sativ... 1399 0.0
emb|CAA65242.1| ENBP1 [Vicia sativa] gi|7488872|pir||T10955 earl... 1373 0.0
emb|CAB80908.1| putative protein (fragment) [Arabidopsis thalian... 602 e-170
ref|XP_468521.1| putative DNA-binding protein PD3, chloroplast [... 534 e-150
ref|XP_468520.1| putative DNA-binding protein PD3, chloroplast [... 533 e-149
gb|AAM20342.1| unknown protein [Arabidopsis thaliana] gi|1817633... 530 e-148
gb|AAT81741.1| jmjC domain containing protein [Oryza sativa (jap... 430 e-118
gb|AAF70852.1| F24O1.3 [Arabidopsis thaliana] gi|7486103|pir||T0... 418 e-115
ref|NP_176421.1| transcription factor jumonji (jmjC) domain-cont... 418 e-115
gb|AAC17616.1| Contains similarity to box helicases gb|U29097 fr... 410 e-112
ref|NP_172659.2| transcription factor jumonji (jmjC) domain-cont... 408 e-112
gb|AAF13079.1| hypothetical protein [Arabidopsis thaliana] gi|15... 347 2e-93
ref|NP_172380.2| transcription factor jumonji (jmjC) domain-cont... 297 2e-78
dbj|BAD44593.1| hypothetical protein [Arabidopsis thaliana] gi|5... 285 9e-75
pir||F86222 hypothetical protein [imported] - Arabidopsis thalia... 275 7e-72
dbj|BAD43260.1| hypothetical protein [Arabidopsis thaliana] 229 6e-58
gb|AAN13035.1| unknown protein [Arabidopsis thaliana] 223 5e-56
ref|XP_450893.1| DNA-binding protein PD3, chloroplast-like [Oryz... 196 7e-48
gb|AAT72490.1| AT1G62310 [Arabidopsis lyrata subsp. petraea] 192 8e-47
>emb|CAA05489.1| ENBP1 [Medicago truncatula] gi|11358945|pir||T43213 ENBP1 protein -
barrel medic
Length = 1701
Score = 1452 bits (3760), Expect = 0.0
Identities = 871/1723 (50%), Positives = 1048/1723 (60%), Gaps = 268/1723 (15%)
Query: 1 MDETGE-ECRRCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRS 59
MDETG+ E RRC R GWRC E+AL GK+ CERH Y +K S ++
Sbjct: 1 MDETGDDESRRCSRNGSGGWRCKEQALPGKTHCERHHEY-------YKSRNSSSFVEKNG 53
Query: 60 GRRKPVDNSENGVV----DDGCKELFG-DPNGTPTVVDEFTGLCGVSEGDAGVNLNLGCE 114
G R NGVV D+G K LFG D +G VV+ F G+ G E + GVN +G E
Sbjct: 54 GIR-------NGVVVDDHDNGGKGLFGGDDHG---VVEGFGGVFGDVEVNGGVNAGIGRE 103
Query: 115 SLNLQDKGEEGQQVHSGGFGEGCGRMGQVLGDYGVEYA----EDRNAVAGLG-------A 163
NL +G+ GQQ G F + G +GQ LGD GVE+ EDRN GLG
Sbjct: 104 RFNLWQQGD-GQQ--GGRFEQASGNLGQFLGD-GVEFVGGFVEDRNRAVGLGQQWGGVGV 159
Query: 164 FRN------VGNEDHGCVAGRNVCVNDRLGLPSEGIESLIGEEPGFG-----SFQALLCK 212
F N VG +DHG VC ND G S+GI LIGE FG SFQALL +
Sbjct: 160 FGNGGGVSGVGKDDHGNGVD-GVCGNDSPGFGSDGINGLIGEGGCFGNLYDRSFQALLSQ 218
Query: 213 DRGCAEDVIFIGDVTGFEGLSGENTHGFRDEVGGFVENPCFEGEND--SNKEGPGSNYKM 270
R C EDV G T F+GL GE+ + FR VG + FEGE + S P S+ KM
Sbjct: 219 GRVCDEDVNLTGGGTSFQGLGGESAYDFRG-VGNLSQCGKFEGEKNVGSILTVPESSNKM 277
Query: 271 SALGFEEEIGLLLSRGGTTNEEARCEALRPLSKRGRPKGSKNENDNKQLSTALDGQSVGG 330
A G EE + +LLS G + NEEAR EAL+PL+KRGRPKGSKN+ K++ +G++V G
Sbjct: 278 GAFGVEEGMEMLLSGGVSINEEARGEALKPLAKRGRPKGSKNKIKKKEVDLVTNGETVCG 337
Query: 331 DDNAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAEKSGRP------------ 378
N GT +V L E +V SG+ D+ E D+G+ AR +K G+P
Sbjct: 338 SANVGT----TVEILETEKSVFSGKADQ---EGVDMGDIARTKKRGQPEDWKRGRHIILA 390
Query: 379 ---------------------KVSKNKIRRVEHVGNVV--AVKIVGPKKHGRPKSSKCRK 415
K S N+ + VE V + V A +I PKK GR ++SKC K
Sbjct: 391 VGYEIDGVGEITGPMERGRKSKGSVNEEKNVEEVSSEVAGAGEIARPKKLGRMEASKCGK 450
Query: 416 KNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCNN---------EGSGAGEIVRPKK 466
+ ++E ++ GEI KK GRPKGS + ++ NN E +GAGEI RPKK
Sbjct: 451 EIVVEVSNDVGGEIVRRKKRGRPKGSKCGKEIVLEVNNEVVGAEVIIEVAGAGEIARPKK 510
Query: 467 RGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSAD 526
GR GSK ++EVS A +I PKK GRPKGSK ++ +V+V+ EVAG+
Sbjct: 511 LGRLEGSKCGKEIVVEVSNDVAG----EIVRPKKRGRPKGSKCGKEIVVEVNHEVAGAG- 565
Query: 527 CEIAGPKKCGRPKGSMKKRKSLVCASILEGAGGITREGLENKML--SNLCQEHIEYTQPV 584
EIA KK GRPKG +++ ++ +G ++ LE++ L L Q+ ++ +P
Sbjct: 566 -EIARSKKRGRPKGYKCQKEIVIKRGRPKGTKN-KKKILEDQELHVQTLVQDEVQNVKPK 623
Query: 585 VRGGRPKGSRNKKIKLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIG-----KPIG 639
+ GRPKGS+NKK +A +D NK + +E+K G G K I
Sbjct: 624 L--GRPKGSKNKKKNIAGED-------GNK--------LHKEKKRRGWPKGFCLKPKEIA 666
Query: 640 LDNDKATLASDRDQET---PNQTLAQDEVQNDKSSVKPKRGRPKGSKNKMKSIANKARNK 696
D+ R + + P +T Q + + + +RGRPKG+ K K I + K
Sbjct: 667 ARLDEKIERRGRPKGSGMKPKETAVQLDAKIE------RRGRPKGAGKKPKEIVVRLDTK 720
Query: 697 FGKVRNMRGRPKGSLRKKNETAYCLDSQNERNS---LDGRTSTEAAYRN----------- 742
+ RGRPKGS +K+ E A L Q E +DG ST +++
Sbjct: 721 IER----RGRPKGSGKKQKEVASQLALQIESQKSTRVDGALSTIVPHKHIQEESISPLKD 776
Query: 743 ------------------------------DVDLHRGHCSQEELLRMLSVEHKNIQGVGV 772
D+H+ CS E LR L +HKN Q V V
Sbjct: 777 PVNKEEKSDFVLECSKDSGIEKITKGLMSKSGDVHK-RCS--ERLRTLLTDHKNSQDVEV 833
Query: 773 EET---------IDYGLRSSGLMGDTERKKETRILRCHQCWRNSWSGVVICAKCKRKQYC 823
EET ID+ L SS LMG+ E KKE R LRCHQCW+ S +G+V+C KCKRK+YC
Sbjct: 834 EETFCENEVEEAIDHELESSDLMGEPETKKEPRNLRCHQCWKKSRTGIVVCTKCKRKKYC 893
Query: 824 YECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKDISVMTGSGEADTGVILQKLLYLLN 883
YECI KWY KTREEIE ACPFCL CNCRLCLKK IS M G+GEAD V LQKL YLL
Sbjct: 894 YECIAKWYQDKTREEIETACPFCLDYCNCRLCLKKTISTMNGNGEADADVKLQKLFYLLK 953
Query: 884 KTLPLLQHIQREQISEMEVEASMHGSPLMEED------------IQFDNCNTSIVNFHRS 931
KTLPLLQHIQREQ SE+EVEAS+HGS ++EE + DNCNTSIVNFHRS
Sbjct: 954 KTLPLLQHIQREQKSELEVEASIHGSLMVEEKDILQAAVDDDDRVYCDNCNTSIVNFHRS 1013
Query: 932 CPNPNCRYDLCLTCCMELRNGLHYEDIPAS-GNEETIDEPPITSAWRAEINGRIPCPPKA 990
C NP CRYDLCLTCC ELRNG+H +DIPAS GNEE ++ PP T AWRAE NG IPCPPKA
Sbjct: 1014 CVNPYCRYDLCLTCCTELRNGVHSKDIPASGGNEEMVNTPPETIAWRAETNGSIPCPPKA 1073
Query: 991 RGGCGTSILSLRRLFEANWVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSV 1050
RGGCGT+ LSLRRLF+ANW+ KL R+AEELTI+Y PP VDL + C +C F D A NS
Sbjct: 1074 RGGCGTATLSLRRLFKANWIEKLTRDAEELTIKYQPPIVDLSLECSECRSFEEDAAHNSA 1133
Query: 1051 RKAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMW 1110
RKAASRET HDN LYCPDA+++GDTE++HFQRHWIRGEPVIVRNV++K SGLSW PMVMW
Sbjct: 1134 RKAASRETGHDNLLYCPDAIEIGDTEFDHFQRHWIRGEPVIVRNVYKKGSGLSWDPMVMW 1193
Query: 1111 RAFRGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPP 1170
RAFR A ILK+E TFKAIDCLDWCEVQ+N FQFFKGYL GRRYRNGWPEMLKLKDWPP
Sbjct: 1194 RAFRLAKNILKDEADTFKAIDCLDWCEVQVNAFQFFKGYLTGRRYRNGWPEMLKLKDWPP 1253
Query: 1171 SNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPAVLKPDLGPKTYIAYGSLEEL 1230
+N FE+C PRHGAEF AMLPFSDYTHPK GILNLATKLP VLKPDLGPKTYIAYG+LEEL
Sbjct: 1254 TNFFEDCLPRHGAEFTAMLPFSDYTHPKSGILNLATKLPTVLKPDLGPKTYIAYGALEEL 1313
Query: 1231 SRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDLYGRINKTVV 1290
SRGDSVTKLHCDISDAVNILTHT +VK P WQ KIIKKL+KKYE EDMR+LYG +K
Sbjct: 1314 SRGDSVTKLHCDISDAVNILTHTADVKTPAWQSKIIKKLKKKYEVEDMRELYGLDSKAAG 1373
Query: 1291 SHRSKHKKCRTGISMDPKIPENDDTMGRNSNLRGSQSNEEIVVNELSTRSSSLGESRSDS 1350
S K KK R G+++D KI E +D GR+S L SQ E+ + D
Sbjct: 1374 SRGRKRKKRRVGVTVDLKISEKEDINGRDSTLLESQEKED----------------KLDR 1417
Query: 1351 AACVQGFSESSESKSVLNAGEQAILNMYKRFVKFDLNNHDSGYLFPGKDCEWMHYDVNNG 1410
ACVQ FSES++SK LN Q +++ RF +FDLN+ DS +L P DCE M YD N
Sbjct: 1418 EACVQEFSESTKSKLDLNVSNQEVIDS-PRFQQFDLNSLDSNFLVPRNDCESMLYD--NV 1474
Query: 1411 KQWCSSP-------------VMPC-------------------SKIQIDKTVPVKNDISS 1438
+Q CS P PC S I+ DK V+N++ S
Sbjct: 1475 EQRCSRPRDGSCKGNTSVIDNQPCGGTKETTFVNGLDSSDISSSDIETDKIESVENEMPS 1534
Query: 1439 NNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQI 1498
NN ND H+ETQ+GSAVWDIFRRQDVPKLTEYL KHH+EFRH +LPV+ V HPIHDQ
Sbjct: 1535 NNLCGNDVHLETQYGSAVWDIFRRQDVPKLTEYLNKHHREFRHITSLPVNFVIHPIHDQH 1594
Query: 1499 LYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRNR 1541
YLNEKHK+QLK EYG+EPWTFEQHLGEAVFIPAGCPHQVRNR
Sbjct: 1595 FYLNEKHKKQLKLEYGVEPWTFEQHLGEAVFIPAGCPHQVRNR 1637
>emb|CAA67296.1| chloroplast DNA-binding protein PD3 [Pisum sativum]
gi|7488797|pir||T06461 DNA-binding protein PD3,
chloroplast - garden pea
Length = 1629
Score = 1399 bits (3620), Expect = 0.0
Identities = 865/1717 (50%), Positives = 1017/1717 (58%), Gaps = 328/1717 (19%)
Query: 1 MDETG-EECRRCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRS 59
MD G EE RRC SG CE ++K R KE SG
Sbjct: 1 MDVGGDEEFRRCNSNG-----------SGNPQCEP-----ERKKNR-KESTSG------- 36
Query: 60 GRRKPVDNSENGVVDD-GCKELFGDPNGTPTVVDEFTGLCGVSEGDAGVNLNLGCESLNL 118
+RK +NS+NGVVDD G K LF D +G+ E + GV+L +G + NL
Sbjct: 37 RKRKAEENSQNGVVDDDGGKGLF----------DGGSGIFAEGEVNGGVDLGIGSGNFNL 86
Query: 119 QDKGEEGQQVHSGGFGEGCGRMGQVLGDYGVEY----AEDRNAV------AGLGAFRN-- 166
+G EGQQ+ FGEG G +G+ LGD GV++ EDRN V + +G F N
Sbjct: 87 WQQGGEGQQLV---FGEGSGNLGKFLGD-GVDFLGGFVEDRNGVGLGQPWSSVGVFGNAA 142
Query: 167 ----VGNEDHG-CVAGRNVCVNDRLGLPSEGIESLIGEEP-GFG-----SFQALLCKDRG 215
V EDHG CV G VC ND LG S+GIE LIGEE GFG SFQALLC+ +
Sbjct: 143 GVSGVAKEDHGKCVDG--VCGNDSLGFHSQGIEGLIGEEEAGFGNLYDRSFQALLCQGKV 200
Query: 216 CAEDVIFIGDVTGFEGLSGENTHGFRDEVGGFVEN-----PCFEGEN--DSNKEGPGSNY 268
C EDV IG TGF+GL GE+ + FR EVG V N F GE + E P S+
Sbjct: 201 CDEDVNLIGGGTGFQGLVGESAYDFRGEVGEGVGNLNESGGKFGGEKIVGNVLEAPNSSN 260
Query: 269 KMSALGFEEEIGLLLSRG-GTTNEEARCEALRPLSKR-GRPKGSKNENDNKQLSTALDGQ 326
K+ A+G EE I LL S G T NEEAR E L+PL++R GRPKGSKN+ K +S AL+G+
Sbjct: 261 KVKAIGVEEGIELLPSGGVSTPNEEARGEVLKPLTRRGGRPKGSKNK--KKGVSLALEGK 318
Query: 327 SVGGDDNAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAEKSGRPKVSKNKIR 386
+ G DNAG IGM +V L E +V SG+ D E D+GE AR + +++
Sbjct: 319 ADCGSDNAGAIGMGTVEVLENEKSVFSGKYD---GEGVDMGEIARTREC-------SQLE 368
Query: 387 RVEHVGNVVAVKIVGPKKHGRPKSSKCRKKNIMEAGDEAAG--EIGGDKKLG-RPKGSLN 443
+++ G +V AG E AG EI KL KG +N
Sbjct: 369 DLKYTGEIV-----------------------FAAGYEVAGVSEISRPCKLAPESKGLVN 405
Query: 444 KLKNTVDCNNEGSGAGEIVRPKKRGRPRGSKNKVNNIMEVSKKAA--------------- 488
K KN + + A EI RPKKRGR +GS NK+ SK+ A
Sbjct: 406 KEKNVEEVSTSSEVAVEIARPKKRGRRKGSTNKMVTYSACSKEGADDIETQGSEEKMLIV 465
Query: 489 ---SGGDCKIAGPKKCGRPKGSKNKQKNIVQVSQEVAGSADCEIAGPKKCGRPKGSMKKR 545
G D + K G ++NK ++I E G K P+ S R
Sbjct: 466 PYQKGADGFASPTNKLGPASSTRNKLRSI-------------EFEGYK---NPEMSSNVR 509
Query: 546 KSLVCASILEGAGGITREGLENKMLSNLCQ----------EHIE--YTQPVVRGGRPKGS 593
LE G T GLE L+ LC+ +HIE T P+++ GRPKGS
Sbjct: 510 --------LEDDG--TPSGLEITTLAPLCEMEKGMPFESAKHIENLTTPPIIKRGRPKGS 559
Query: 594 RNKKIKLAFQDMVDEVRFANKESDKATCAVGEEQKDHGSDIGKPIGLDNDKATLASDRDQ 653
+NKK +LA Q+ + H DI K IG++N +AT S DQ
Sbjct: 560 KNKKKELADQEHI----------------------GHEGDIIKLIGVENYEATAVSVGDQ 597
Query: 654 ETPNQTLAQDEVQNDKSSVKPKRGRPKGSKNKMKSIANKARNKFGKVRNMRGRPKGSLRK 713
E QTL QDEVQN VKPK GRPKGSKNK K+I +A NK + + RGRPKGS +K
Sbjct: 598 ELVVQTLGQDEVQN----VKPKIGRPKGSKNKKKNIDGEAENKLHE-KKKRGRPKGSGKK 652
Query: 714 KNETAYCLDSQNE---RNSLDGRTST----------------EAAYRNDVDLHRGHCSQ- 753
+ E A LD++ E + G ST + Y+ D CS+
Sbjct: 653 QKENASRLDAEIECENNTRVYGILSTTIQHKHIHEESVLLLEDQVYKKDDADFVPECSKE 712
Query: 754 ---EELLRMLSVEHKNIQ-----GVG-------VEETIDY-------------------- 778
E++ + L E N+ VG V+ET D+
Sbjct: 713 SGIEKIAKGLVSESDNLHKTQDVEVGDIFYEKEVKETRDHGLESDNVHKTQDVEMGDIFY 772
Query: 779 ----------GLRSSGLMGDTERKKETRILRCHQCWRNSWSGVVICAKCKRKQYCYECIT 828
GL SS +MGD E KKE R RCHQCW+ S +G+V+C+KCKRK+YCYECI
Sbjct: 773 EKEVKETRDHGLESSDMMGDCEAKKEPRNSRCHQCWKKSRTGLVVCSKCKRKKYCYECIA 832
Query: 829 KWYPGKTREEIEIACPFCLRNCNCRLCLKKDISVMTGSGEADTGVILQKLLYLLNKTLPL 888
KWY KTREEIE ACPFCL CNCR+CLKK IS M G+ EAD V L+KLLYLLNKTLPL
Sbjct: 833 KWYQDKTREEIETACPFCLDYCNCRMCLKKAISTMNGNDEADRDVKLRKLLYLLNKTLPL 892
Query: 889 LQHIQREQISEMEVEASMHGSPLMEED------------IQFDNCNTSIVNFHRSCPNPN 936
LQ IQREQ E+EVEASMHGS L+EE+ + DNCNTSIVNFHRSC NPN
Sbjct: 893 LQDIQREQRYELEVEASMHGSQLVEEEDIRKAEVDDDDRVYCDNCNTSIVNFHRSCSNPN 952
Query: 937 CRYDLCLTCCMELRNGLHYEDIPASGNEETIDEPPITSAWRAEINGRIPCPPKARGGCGT 996
C+YDLCLTCC ELR G+H +DIPASGNEE +D PP + WRAE NG IPCPPKARGGCG
Sbjct: 953 CQYDLCLTCCTELRIGVHCKDIPASGNEEMVDAPPESIPWRAETNGSIPCPPKARGGCGI 1012
Query: 997 SILSLRRLFEANWVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSVRKAASR 1056
+ LSLRRLFEANW++KL R EELT++Y PP DL +GC +C F D+AQNS RKAASR
Sbjct: 1013 ATLSLRRLFEANWIDKLTRGVEELTVKYQPPIADLSLGCSECRSFEEDVAQNSARKAASR 1072
Query: 1057 ETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAFRGA 1116
ET +DNFLYCPDAV++G+T ++HFQRHWIRGEPVIVRNV++KASGLSW PMVMWRAF GA
Sbjct: 1073 ETGYDNFLYCPDAVEIGETTFQHFQRHWIRGEPVIVRNVYKKASGLSWDPMVMWRAFMGA 1132
Query: 1117 NKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEE 1176
KILKE+ FKAIDCLDWCEV+IN FQFFKGYLEGRRYRNGWP MLKLKDWPPSN FEE
Sbjct: 1133 RKILKEDAVNFKAIDCLDWCEVEINAFQFFKGYLEGRRYRNGWPAMLKLKDWPPSNFFEE 1192
Query: 1177 CFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPAVLKPDLGPKTYIAYGSLEELSRGDSV 1236
C PRHGAEFIAMLPFSDYTHPK GILNLATKLPAVLKPDLGPKTYIAYG+ +ELSRGDSV
Sbjct: 1193 CLPRHGAEFIAMLPFSDYTHPKSGILNLATKLPAVLKPDLGPKTYIAYGTSDELSRGDSV 1252
Query: 1237 TKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDLYGRINKTVVSHRSKH 1296
TKLHCDISDAVNILTHT EVK P WQ +IIKKLQKKYE EDMR+LY V R +
Sbjct: 1253 TKLHCDISDAVNILTHTAEVKPPPWQSRIIKKLQKKYEVEDMRELYSHDKTAVGLPRKRG 1312
Query: 1297 KKCRTGISMDPKIPENDDTMGRNSNLRGSQSNEEIVVNELSTRSSSLGESRSDSAACVQG 1356
+K R G +DPKI E +DT GR+S L+GSQ EE L E S
Sbjct: 1313 RKRRVGFGVDPKISEKEDTNGRDSTLQGSQGKEE-----------KLDEQES-------- 1353
Query: 1357 FSESSESKSVLNAGEQAILNMYKRFVKFDLNNHDSGYLFPGKDCEWMHYDVNNGKQWCSS 1416
SE ++ K LNA EQ I N RF +FDLN+HDS +L PG DCE MHYD Q CSS
Sbjct: 1354 -SEPTKIKFDLNASEQEISNS-PRFQQFDLNSHDSSFLVPGNDCESMHYD---NVQQCSS 1408
Query: 1417 -------------PVMPCSKIQIDKTV-------------------PVKNDISSNNFFQN 1444
PCS + K V V+ D SNN Q+
Sbjct: 1409 QGDESYKGISSVIDDQPCSGTKETKIVNKLNSSDDFCQMVETNNIDSVEKDSLSNNSCQD 1468
Query: 1445 DDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQILYLNEK 1504
D H+ TQ GSAVWDIFRR DVPKLTEYLKKHH+EFRH NLPV+SV HPIHDQILYLNEK
Sbjct: 1469 DVHLGTQNGSAVWDIFRRHDVPKLTEYLKKHHREFRHIINLPVNSVIHPIHDQILYLNEK 1528
Query: 1505 HKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRNR 1541
HK+QLK EYG+EPWTFEQHLGEAVFIPAGCPHQVRNR
Sbjct: 1529 HKKQLKLEYGVEPWTFEQHLGEAVFIPAGCPHQVRNR 1565
>emb|CAA65242.1| ENBP1 [Vicia sativa] gi|7488872|pir||T10955 early nodulin binding
protein 1 - spring vetch
Length = 1641
Score = 1373 bits (3553), Expect = 0.0
Identities = 828/1670 (49%), Positives = 998/1670 (59%), Gaps = 234/1670 (14%)
Query: 6 EECRRCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRSGRRKPV 65
EE RRC R G RC + AL G CE+H L ++ + KE +SG ++R +
Sbjct: 8 EEFRRCNRNGS-GRRCKDGALPGNLQCEKHQLCAVERKKNRKETSSG---RKRKAEESLL 63
Query: 66 DNSENGVVDDGCKELFGDPNGTPTVVDEFTGLCGVSEG---DAGVNLNLGCESLNLQDKG 122
D DDG LF G G G G + GVN+ +G ES NL +G
Sbjct: 64 DE------DDGGNGLFAGGAG---------GAGGAGGGVFVEGGVNVGIGSESFNLWQEG 108
Query: 123 EEGQQVHSGGFGEGCGRMGQVLGDYGVEY----AEDRNAV------AGLGAFRN------ 166
E GQQV FGEG G +G+ LGD GVE+ EDRN V + +G F N
Sbjct: 109 E-GQQVEV--FGEGSGNLGKFLGD-GVEFLGGFVEDRNVVGLGQPWSSVGVFGNAGGVAG 164
Query: 167 VGNEDHGCVAGRNVCVNDRLGLPSEGIESLIGEEPG-FG-----SFQALLCKDRGCAEDV 220
VG EDHG VC ND L S+GIE LIGEE G FG SFQALLC+ + C EDV
Sbjct: 165 VGKEDHGSRVD-GVCGNDSLAFHSQGIEGLIGEEEGGFGNLYDRSFQALLCQGKVCDEDV 223
Query: 221 IFIGDVTGFEGLSGENTHGFRDEVGGFVENPC-----FEGENDSNK--EGPGSNYKMSAL 273
IG TGF+GL GE+ F + G V N F+GE + E P S+ KM A+
Sbjct: 224 NLIGGATGFQGLVGESAFNFSGQDGEGVGNLSQHDGKFDGEKNVGNILEAPSSSNKMKAI 283
Query: 274 GFEEEIGLLLSRGGTTNEEARCEALRPLSKRG-RPKGSKNENDNKQLSTALDGQSVGGDD 332
G EE + LL S G + NEEAR E L+PL+ RG RPKGSKN+ K +S LDG++V G D
Sbjct: 284 GVEEGVELLPSAGVSANEEARGEVLKPLTSRGGRPKGSKNKK-KKGVSLVLDGEAVCGSD 342
Query: 333 NAGTIGMSSVTDLGIEIAVLSGEKDKSSDEVADLGETARAEKSGRPKVSKNKIRRVEHVG 392
NAGTIGM +V L E +V G KS E D GE AR + P+ K+ G
Sbjct: 343 NAGTIGMGTVEVLENEKSVFCG---KSDGEGVDTGEIARTRECSPPENQKSTTEIFLAHG 399
Query: 393 NVVAVKIVGPKKHGRPKSSKCRKKNIMEAGDEAAGEIGGDKKLGRPKGSLNKLKNTVDCN 452
VA G + RP + ++KN+ E E A EI KK GRPKGS K V C+
Sbjct: 400 YEVA----GVGEISRPLN---KEKNVEEVSSEVAVEISRSKKRGRPKGSTKK---KVTCS 449
Query: 453 NEGSGAGEIVRPKKRGRPRGSKNKVNNIMEVSKKAASGGDCKIAGPKKCGRPKGSKNKQK 512
GAG+I +GS+ K+ I+ K A D + K G ++NK +
Sbjct: 450 ASRKGAGDIAT-------QGSEEKML-IVPYQKDA----DEFASLTSKLGPALSTRNKLR 497
Query: 513 NIVQVSQEVAGSADCEIAGPKKCGRPKGSMKKRKSLVCASILEGAGGITREGLENKMLSN 572
S E G ++ + G+ + A + E G+ E K + N
Sbjct: 498 -----STEFEGCKHPGMSSNVRLEENDGTASGLEITTLAPLREAEKGMPFESA--KHIEN 550
Query: 573 LCQEHIEYTQPVVRGGRPKGSRNKKIKLAFQDMV----DEVRFANKESDKATCAVGEEQ- 627
L T P+V+ GRPKGS+NKK LA Q+ + D ++ ES +A +VG+++
Sbjct: 551 LT------TPPIVKRGRPKGSKNKKKTLADQEHIGHGGDIIKLIGMESSEAAVSVGDQEL 604
Query: 628 ----------KDHGSDIGKPIGLDNDKATLASDRDQETPNQTLAQDEVQNDKSSVKPKRG 677
+ +G+P G N K + E +N KRG
Sbjct: 605 VVQPLVKVRFRMLNPKMGRPKGSKNKKKNV--------------DGEAENGLHKEGKKRG 650
Query: 678 RPKGSKNKMKSIANK--ARNKFGKVRNM-----RGRPKGSLRKKNETAYCLDSQNERNS- 729
RPKGS N K N+ A+ + N+ RGRPKGS E A LD++ ER
Sbjct: 651 RPKGSVNNPKETGNEKIAKGLVSESSNVHKIERRGRPKGSAPNPKENASRLDAEIEREKN 710
Query: 730 --LDGRTSTEAAYRN----DVDLHRGH------------CSQ----EELLRMLSVEHKNI 767
+ G ST +++ + L H CS+ E++ + L E N+
Sbjct: 711 THVYGILSTTMPHKHIHEESILLLEDHVNKKDDADFVLECSKESGIEKIAKGLVSESDNV 770
Query: 768 Q-----GVG-------VEETIDYGLRSSGLMGDTERKKETRILRCHQCWRNSWSGVVICA 815
VG V+ETID+ L S ++GD E KKE R RCHQCW+ S +G+V+C+
Sbjct: 771 HKTHDVEVGDIFYEKEVKETIDHRLEPSDMLGDCETKKEPRNSRCHQCWKKSRTGLVVCS 830
Query: 816 KCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKDISVMTGSGEADTGVIL 875
KCK+K+YCYEC+ KWY KTREEIE ACPFCL CNCR+CLKK IS M G+ EAD V L
Sbjct: 831 KCKKKKYCYECVAKWYQDKTREEIETACPFCLDYCNCRMCLKKAISSMNGNDEADRDVKL 890
Query: 876 QKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPLMEED------------IQFDNCNT 923
+KL YLL KTLPLLQ IQREQ E+EVEA+MHGS L+EE+ + DNCNT
Sbjct: 891 RKLFYLLKKTLPLLQDIQREQRYELEVEATMHGSQLVEEEDIRKAEVDDDDRVYCDNCNT 950
Query: 924 SIVNFHRSCPNPNCRYDLCLTCCMELRNGLHYEDIPASGNEETIDEPPITSAWRAEINGR 983
SIVNFHRSC NPNC YDLCLTCC ELR G+H +DIP SGNEE +D PP + AWRAE NG
Sbjct: 951 SIVNFHRSCSNPNCEYDLCLTCCTELRLGVHCKDIPTSGNEEMVDAPPESIAWRAETNGS 1010
Query: 984 IPCPPKARGGCGTSILSLRRLFEANWVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVV 1043
IPCPP+ARGGCGT+ILSLRRLFEANW++KL R EELT++Y PP +DL +GC +C F
Sbjct: 1011 IPCPPEARGGCGTAILSLRRLFEANWIDKLTRGVEELTVKYQPPIMDLALGCSECRSFEE 1070
Query: 1044 DLAQNSVRKAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLS 1103
D+AQNS RKAASRET +DNFLYCPDAV+ G+T +EHFQRHWIRGEPVIVRN ++KASGLS
Sbjct: 1071 DVAQNSARKAASRETGYDNFLYCPDAVETGETTFEHFQRHWIRGEPVIVRNAYKKASGLS 1130
Query: 1104 WHPMVMWRAFRGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEML 1163
W PMVMWRAF GA KILKE+ FKAIDCLDWCEV+IN FQFFKGYLEGRRYRNGWP ML
Sbjct: 1131 WDPMVMWRAFMGARKILKEDAVNFKAIDCLDWCEVEINAFQFFKGYLEGRRYRNGWPAML 1190
Query: 1164 KLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPAVLKPDLGPKTYIA 1223
KLKDWPPSN FEEC PRHGAEFIAMLPFSDYTHPK GILNLATKLPA KPDLGPKTYIA
Sbjct: 1191 KLKDWPPSNFFEECLPRHGAEFIAMLPFSDYTHPKSGILNLATKLPAASKPDLGPKTYIA 1250
Query: 1224 YGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDLYG 1283
YG+ +ELSRGDSVTKLHCDISDAVNILTHT EVK P WQ +II+KLQKKYE EDMR+LY
Sbjct: 1251 YGTSDELSRGDSVTKLHCDISDAVNILTHTAEVKPPPWQSRIIRKLQKKYEDEDMRELYS 1310
Query: 1284 RINKTVVSHRSKHKKCRTGISMDPKIPENDDTMGRNSNLRGSQSNEEIVVNELSTRSSSL 1343
+ K V R + +K R G S+DPK E +DT GR+S L+GSQ EE + ++ S
Sbjct: 1311 QDKKEVGLPRKRGRKRRVGFSVDPKTSEKEDTSGRDSTLQGSQGKEEKLDDQES------ 1364
Query: 1344 GESRSDSAACVQGFSESSESKSVLNAGEQAILNMYKRFVKFDLNNHDSGYLFPGKDCEWM 1403
SE ++ + LNA EQ I + RF +FDLN+HDS L PG DCE M
Sbjct: 1365 --------------SEPTKIEFDLNASEQEISDS-PRFQQFDLNSHDSSLLVPGNDCESM 1409
Query: 1404 HYDVNNGKQWCSS-------------PVMPCSKIQIDKTV-------------------P 1431
HYD N ++ CSS PCS + K V
Sbjct: 1410 HYD--NVQERCSSQGDGSYKGISSVIDDQPCSGTKETKNVNKLNSSDDNCSDIETNNIDS 1467
Query: 1432 VKNDISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVT 1491
V+ DI SN+ QND H+ TQ GSAVWDIFRR DVPKLTEYLKKHH+EFRH NLPV+SV
Sbjct: 1468 VEKDILSNSLCQNDVHLGTQNGSAVWDIFRRHDVPKLTEYLKKHHREFRHIVNLPVNSVI 1527
Query: 1492 HPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRNR 1541
HPIHDQILYLNEKHK+QLK EYG+EPWTFEQHLGEAVFIPAGCPHQVRNR
Sbjct: 1528 HPIHDQILYLNEKHKKQLKIEYGVEPWTFEQHLGEAVFIPAGCPHQVRNR 1577
>emb|CAB80908.1| putative protein (fragment) [Arabidopsis thaliana]
gi|5262153|emb|CAB45782.1| putative protein (fragment)
[Arabidopsis thaliana] gi|25372069|pir||B85013
hypothetical protein AT4g00990 [imported] - Arabidopsis
thaliana gi|7486356|pir||T10539 hypothetical protein
F3I3.10 - Arabidopsis thaliana (fragment)
Length = 730
Score = 602 bits (1551), Expect = e-170
Identities = 337/711 (47%), Positives = 428/711 (59%), Gaps = 86/711 (12%)
Query: 868 EADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPLME-----------EDI 916
E DT V L++L YLL K LP+L+ I EQ E+E+E+++ G P+ E E I
Sbjct: 4 EKDTDVKLKQLQYLLVKVLPVLKDIYTEQNRELEIESTIRGHPVTEANIKRCKLDPSERI 63
Query: 917 QFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHYED--------------IPASG 962
D C TSI NFHRSCPN NC D+CL+CC EL G H E IPA
Sbjct: 64 YCDLCRTSIANFHRSCPNKNCSVDICLSCCKELSEGFHQERDGKKNAEGKGYECRIPAGQ 123
Query: 963 NEETIDEPPIT-SAWRAEINGRIPCPPKARGGCGTSILSLRRLFEANWVNKLVRNAEELT 1021
+++ P+ S W+ + IPCPPK GGCGTS L LRRL++ +WV KL+ NAE+ T
Sbjct: 124 GKDSDAYVPLHFSTWKLNSDSSIPCPPKECGGCGTSTLELRRLWKRDWVEKLITNAEKCT 183
Query: 1022 IQYHPPSVDLLVGCLQCHRFVVDLAQNSVRK-AASRETNHDNFLYCPDAVDMGDTEYEHF 1080
+ + P VD++ C C +S+R+ AA R+ HDNFLY P+AVD+ + + HF
Sbjct: 184 LNFRPTDVDIVHECSSC-----STNSDSIRRQAAFRKNAHDNFLYSPNAVDLAEDDIAHF 238
Query: 1081 QRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAFRGAN---KILKEEPTTFKAIDCLDWCE 1137
Q HW++ EPVIVRNV EK SGLSW PMVMWRA R + K +EE T KA+DCLDWCE
Sbjct: 239 QFHWMKAEPVIVRNVLEKTSGLSWEPMVMWRACREMDPKRKGTEEETTKVKALDCLDWCE 298
Query: 1138 VQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHP 1197
V+IN+ QFF+GYLEGR ++NGWPEMLKLKDWPPS+ FE+ PRH AEFIA LPF DYT P
Sbjct: 299 VEINLHQFFEGYLEGRMHKNGWPEMLKLKDWPPSDLFEKRLPRHNAEFIAALPFFDYTDP 358
Query: 1198 KFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEV 1256
K GILNLAT+ P LKPDLGPKTYIAYG EEL+RGDSVTKLHCDISDAVN+LTHT +V
Sbjct: 359 KSGILNLATRFPEGSLKPDLGPKTYIAYGFHEELNRGDSVTKLHCDISDAVNVLTHTAKV 418
Query: 1257 KAPLWQPKIIKKLQKKY-EAEDMRDLYGRINKTVVSHRSKHKKCRTGISMDPKIPENDDT 1315
+ P + + IK QKKY EA + Y K +K K E D++
Sbjct: 419 EIPPVKYQNIKVHQKKYAEAMLQKQQYSGQVKEASELENKSMK------------EVDES 466
Query: 1316 MGRNSNLRGSQSNEEIVVNELSTRSSSLGESRSDSAACVQGFSESSESKSVLNAGEQAIL 1375
+L+ +NEE N S+R S GE+ K +++ G I
Sbjct: 467 ---KKDLKDKAANEEQSNN--SSRPSGSGEA----------------EKVIISKGIARIR 505
Query: 1376 NMYKRFV-KFDLNNHDSGYLFP----GKDCEWMHYDVNNGKQWCSSPVMPCSKIQIDKTV 1430
+ +V K L N ++G + P C+ +P P ++
Sbjct: 506 ELSHSYVYKHMLLNMENGLMMPTLLATPPCDTE-----------DNPTQPAVSTSVESIQ 554
Query: 1431 PVKNDISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSV 1490
K D ++ + G AVWDIFRR+DVPKL ++LK+H EFRH NN P++SV
Sbjct: 555 EQKLDAPKETDGNTNERSKAVHGGAVWDIFRREDVPKLIQFLKRHEHEFRHFNNEPLESV 614
Query: 1491 THPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRNR 1541
HPIHDQ ++L++ K+QLK+E+ IEPWTFEQHLGEAVFIPAGCPHQVRNR
Sbjct: 615 IHPIHDQTMFLSDSQKKQLKEEFDIEPWTFEQHLGEAVFIPAGCPHQVRNR 665
>ref|XP_468521.1| putative DNA-binding protein PD3, chloroplast [Oryza sativa (japonica
cultivar-group)] gi|48716466|dbj|BAD23073.1| putative
DNA-binding protein PD3, chloroplast [Oryza sativa
(japonica cultivar-group)] gi|48716323|dbj|BAD22935.1|
putative DNA-binding protein PD3, chloroplast [Oryza
sativa (japonica cultivar-group)]
Length = 868
Score = 534 bits (1376), Expect = e-150
Identities = 320/807 (39%), Positives = 417/807 (51%), Gaps = 123/807 (15%)
Query: 798 LRCHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLK 857
L CHQC RN VV C C K++C CI +WYP E CP+C +NCNC+ CL+
Sbjct: 38 LMCHQCQRNDKGRVVWCKTCNNKRFCVPCINQWYPDLPENEFAAKCPYCRKNCNCKACLR 97
Query: 858 -KDISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPL----- 911
+ + E ++ ++L P L +++EQ++E E+EA + G +
Sbjct: 98 MRGVEEQPPRKEISKENQIRYACHVLRLLRPWLIELRQEQMAEKELEAKIQGVSVDQIKV 157
Query: 912 ------MEEDIQFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHYEDIPASGNEE 965
++E + + C+TSIV+FHRSC + C YDLCLTCC ELR G +IP E
Sbjct: 158 EQAVCDLDERVYCNRCSTSIVDFHRSCKH--CFYDLCLTCCQELRKG----EIPGGEEVE 211
Query: 966 TID--------------------------------------------EPPITSAWRAEIN 981
+D + W+A N
Sbjct: 212 ILDPEERDKDYAFGKILSDGENQRDSLKCRSDTQNSESNKGMASDENQKKALLLWKANSN 271
Query: 982 GRIPCPPKARGGCGTSILSLRRLFEANWVNKLVRNAEEL----TIQYHPPSVDLLVGCLQ 1037
G IPCP K + C S L L+ LF + +L +E++ T L C
Sbjct: 272 GSIPCPRKEKEDCSFSSLDLKCLFPEKLLPELEDRSEKVFWSETFAKELGRTSELCPCFD 331
Query: 1038 CHRFVVDLAQNSVRKAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFE 1097
H + +R+AA+RE + DN+LYCP A D+ D + HFQ HW +GEPV+V + +
Sbjct: 332 -HSGKIRSDSKKLRQAANREDSSDNYLYCPVATDIQDADLLHFQMHWAKGEPVVVSDTLK 390
Query: 1098 KASGLSWHPMVMWRAFRGANK-ILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRY- 1155
SGLSW PMVMWRA R K ++E +A+DCLDWCEV+INI FF GY GR +
Sbjct: 391 LTSGLSWEPMVMWRAVRERTKGKAEDEQFAVRAVDCLDWCEVEINIHMFFMGYTRGRTHP 450
Query: 1156 RNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPA-VLKP 1214
R WPEMLKLKDWPPS+SF++ PRHGAEFI+ LPF +YT P++G LNLA KLP VLKP
Sbjct: 451 RTYWPEMLKLKDWPPSSSFDQRLPRHGAEFISALPFPEYTDPRYGPLNLAVKLPGGVLKP 510
Query: 1215 DLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYE 1274
DLGPKTYIAYG EEL RGDSVTKLHCD+SDAVNILTHT EV + IK QKK +
Sbjct: 511 DLGPKTYIAYGFSEELGRGDSVTKLHCDMSDAVNILTHTAEVPCETYDAVQIKNTQKKMK 570
Query: 1275 AEDMRDLYGRINKTVVSHRSKHKKCRTGISMDPKIPENDDTMGRNSNLRGSQSNEEIVVN 1334
+D ++YG I K C + + +G S+ N V+
Sbjct: 571 MQDDMEIYGMIES---GSELKPSAC--------PVELGNKAVGEAPKASCSKEN----VH 615
Query: 1335 ELSTRSSSLGESRS-DSAACVQGFSESSESKSVLNAGEQAILNMYKRFVKFDLNNHDSGY 1393
L +S+ L + S A E+ +SV+++ N + NN D
Sbjct: 616 TLKDKSNGLDINASPPDDAGGDARDEALSYESVVHSDVAQCPNH-----NHETNNSD--- 667
Query: 1394 LFPGKDCEWMHYDVNNGKQWCSSPVMPCSKIQIDKTVPVKNDISSNNFFQNDDHMETQFG 1453
D G Q C K P K + +H E+ G
Sbjct: 668 ------------DARIGAQRCQKKA---------KGRPPKTGSGVS------EHQES--G 698
Query: 1454 SAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQILYLNEKHKRQLKKEY 1513
A+WDIFRR+D KL ++L+KH EFRH + PV V HPIHDQ YL +HKR+LK+EY
Sbjct: 699 GALWDIFRREDSEKLQDFLRKHAPEFRHIHCNPVKQVIHPIHDQAFYLTAEHKRKLKEEY 758
Query: 1514 GIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
G+EPWTFEQ LGEAV IPAGCPHQVRN
Sbjct: 759 GVEPWTFEQKLGEAVLIPAGCPHQVRN 785
>ref|XP_468520.1| putative DNA-binding protein PD3, chloroplast [Oryza sativa (japonica
cultivar-group)] gi|48716465|dbj|BAD23072.1| putative
DNA-binding protein PD3, chloroplast [Oryza sativa
(japonica cultivar-group)] gi|48716322|dbj|BAD22934.1|
putative DNA-binding protein PD3, chloroplast [Oryza
sativa (japonica cultivar-group)]
Length = 995
Score = 533 bits (1373), Expect = e-149
Identities = 320/806 (39%), Positives = 415/806 (50%), Gaps = 122/806 (15%)
Query: 798 LRCHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLK 857
L CHQC RN VV C C K++C CI +WYP E CP+C +NCNC+ CL+
Sbjct: 166 LMCHQCQRNDKGRVVWCKTCNNKRFCVPCINQWYPDLPENEFAAKCPYCRKNCNCKACLR 225
Query: 858 KDISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPL------ 911
E ++ ++L P L +++EQ++E E+EA + G +
Sbjct: 226 MRGVEEPPRKEISKENQIRYACHVLRLLRPWLIELRQEQMAEKELEAKIQGVSVDQIKVE 285
Query: 912 -----MEEDIQFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHYEDIPASGNEET 966
++E + + C+TSIV+FHRSC + C YDLCLTCC ELR G +IP E
Sbjct: 286 QAVCDLDERVYCNRCSTSIVDFHRSCKH--CFYDLCLTCCQELRKG----EIPGGEEVEI 339
Query: 967 ID--------------------------------------------EPPITSAWRAEING 982
+D + W+A NG
Sbjct: 340 LDPEERDKDYAFGKILSDGENQRDSLKCRSDTQNSESNKGMASDENQKKALLLWKANSNG 399
Query: 983 RIPCPPKARGGCGTSILSLRRLFEANWVNKLVRNAEEL----TIQYHPPSVDLLVGCLQC 1038
IPCP K + C S L L+ LF + +L +E++ T L C
Sbjct: 400 SIPCPRKEKEDCSFSSLDLKCLFPEKLLPELEDRSEKVFWSETFAKELGRTSELCPCFD- 458
Query: 1039 HRFVVDLAQNSVRKAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEK 1098
H + +R+AA+RE + DN+LYCP A D+ D + HFQ HW +GEPV+V + +
Sbjct: 459 HSGKIRSDSKKLRQAANREDSSDNYLYCPVATDIQDADLLHFQMHWAKGEPVVVSDTLKL 518
Query: 1099 ASGLSWHPMVMWRAFRGANK-ILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRY-R 1156
SGLSW PMVMWRA R K ++E +A+DCLDWCEV+INI FF GY GR + R
Sbjct: 519 TSGLSWEPMVMWRAVRERTKGKAEDEQFAVRAVDCLDWCEVEINIHMFFMGYTRGRTHPR 578
Query: 1157 NGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPA-VLKPD 1215
WPEMLKLKDWPPS+SF++ PRHGAEFI+ LPF +YT P++G LNLA KLP VLKPD
Sbjct: 579 TYWPEMLKLKDWPPSSSFDQRLPRHGAEFISALPFPEYTDPRYGPLNLAVKLPGGVLKPD 638
Query: 1216 LGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEA 1275
LGPKTYIAYG EEL RGDSVTKLHCD+SDAVNILTHT EV + IK QKK +
Sbjct: 639 LGPKTYIAYGFSEELGRGDSVTKLHCDMSDAVNILTHTAEVPCETYDAVQIKNTQKKMKM 698
Query: 1276 EDMRDLYGRINKTVVSHRSKHKKCRTGISMDPKIPENDDTMGRNSNLRGSQSNEEIVVNE 1335
+D ++YG I K C + + +G S+ N V+
Sbjct: 699 QDDMEIYGMIES---GSELKPSAC--------PVELGNKAVGEAPKASCSKEN----VHT 743
Query: 1336 LSTRSSSLGESRS-DSAACVQGFSESSESKSVLNAGEQAILNMYKRFVKFDLNNHDSGYL 1394
L +S+ L + S A E+ +SV+++ N + NN D
Sbjct: 744 LKDKSNGLDINASPPDDAGGDARDEALSYESVVHSDVAQCPNH-----NHETNNSD---- 794
Query: 1395 FPGKDCEWMHYDVNNGKQWCSSPVMPCSKIQIDKTVPVKNDISSNNFFQNDDHMETQFGS 1454
D G Q C K P K + +H E+ G
Sbjct: 795 -----------DARIGAQRCQKKA---------KGRPPKTGSGVS------EHQES--GG 826
Query: 1455 AVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQILYLNEKHKRQLKKEYG 1514
A+WDIFRR+D KL ++L+KH EFRH + PV V HPIHDQ YL +HKR+LK+EYG
Sbjct: 827 ALWDIFRREDSEKLQDFLRKHAPEFRHIHCNPVKQVIHPIHDQAFYLTAEHKRKLKEEYG 886
Query: 1515 IEPWTFEQHLGEAVFIPAGCPHQVRN 1540
+EPWTFEQ LGEAV IPAGCPHQVRN
Sbjct: 887 VEPWTFEQKLGEAVLIPAGCPHQVRN 912
>gb|AAM20342.1| unknown protein [Arabidopsis thaliana] gi|18176335|gb|AAL60025.1|
unknown protein [Arabidopsis thaliana]
gi|42566216|ref|NP_192008.3| transcription factor jumonji
(jmjC) domain-containing protein [Arabidopsis thaliana]
Length = 840
Score = 530 bits (1365), Expect = e-148
Identities = 291/623 (46%), Positives = 382/623 (60%), Gaps = 52/623 (8%)
Query: 800 CHQCW-RNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKK 858
CH C S S ++ C+KC +K YC++CI + Y +T EE+ ACPFC+ C CR CL+
Sbjct: 80 CHHCKILTSESDLIFCSKCNKKCYCFDCIKRSYSERTHEEVRAACPFCMMTCICRACLRL 139
Query: 859 DISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPLME----- 913
+ + S E DT V L++L YLL K LP+L+ I EQ E+E+E+++ G P+ E
Sbjct: 140 PLVIKPPS-EKDTDVKLKQLQYLLVKVLPVLKDIYTEQNRELEIESTIRGHPVTEANIKR 198
Query: 914 ------EDIQFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHYED---------- 957
E I D C TSI NFHRSCPN NC D+CL+CC EL G H E
Sbjct: 199 CKLDPSERIYCDLCRTSIANFHRSCPNKNCSVDICLSCCKELSEGFHQERDGKKNAEGKG 258
Query: 958 ----IPASGNEETIDEPPIT-SAWRAEINGRIPCPPKARGGCGTSILSLRRLFEANWVNK 1012
IPA +++ P+ S W+ + IPCPPK GGCGTS L LRRL++ +WV K
Sbjct: 259 YECRIPAGQGKDSDAYVPLHFSTWKLNSDSSIPCPPKECGGCGTSTLELRRLWKRDWVEK 318
Query: 1013 LVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSVRK-AASRETNHDNFLYCPDAVD 1071
L+ NAE+ T+ + P VD++ C C +S+R+ AA R+ HDNFLY P+AVD
Sbjct: 319 LITNAEKCTLNFRPTDVDIVHECSSC-----STNSDSIRRQAAFRKNAHDNFLYSPNAVD 373
Query: 1072 MGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAFRGAN---KILKEEPTTFK 1128
+ + + HFQ HW++ EPVIVRNV EK SGLSW PMVMWRA R + K +EE T K
Sbjct: 374 LAEDDIAHFQFHWMKAEPVIVRNVLEKTSGLSWEPMVMWRACREMDPKRKGTEEETTKVK 433
Query: 1129 AIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAM 1188
A+DCLDWCEV+IN+ QFF+GYLEGR ++NGWPEMLKLKDWPPS+ FE+ PRH AEFIA
Sbjct: 434 ALDCLDWCEVEINLHQFFEGYLEGRMHKNGWPEMLKLKDWPPSDLFEKRLPRHNAEFIAA 493
Query: 1189 LPFSDYTHPKFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAV 1247
LPF DYT PK GILNLAT+ P LKPDLGPKTYIAYG EEL+RGDSVTKLHCDISDAV
Sbjct: 494 LPFFDYTDPKSGILNLATRFPEGSLKPDLGPKTYIAYGFHEELNRGDSVTKLHCDISDAV 553
Query: 1248 NILTHTEEVKAPLWQPKIIKKLQKKY-EAEDMRDLYGRINKTVVSHRSKHKKCRTGISMD 1306
N+LTHT +V+ P + + IK QKKY EA + Y K +K K D
Sbjct: 554 NVLTHTAKVEIPPVKYQNIKVHQKKYAEAMLQKQQYSGQVKEASELENKSMKEVDESKKD 613
Query: 1307 PK-IPENDDTMGRNSNLRGSQSNEEIVVNE--------LSTRSSSLGESRSDSAACVQGF 1357
K N++ +S GS E++++++ +ST S+ E + D+ G
Sbjct: 614 LKDKAANEEQSNNSSRPSGSGEAEKVIISKEDNPTQPAVSTSVESIQEQKLDAPKETDG- 672
Query: 1358 SESSESKSVLNAGEQAILNMYKR 1380
++E ++ G A+ ++++R
Sbjct: 673 -NTNERSKAVHGG--AVWDIFRR 692
Score = 155 bits (393), Expect = 8e-36
Identities = 69/126 (54%), Positives = 88/126 (69%)
Query: 1416 SPVMPCSKIQIDKTVPVKNDISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKH 1475
+P P ++ K D ++ + G AVWDIFRR+DVPKL ++LK+H
Sbjct: 646 NPTQPAVSTSVESIQEQKLDAPKETDGNTNERSKAVHGGAVWDIFRREDVPKLIQFLKRH 705
Query: 1476 HKEFRHANNLPVDSVTHPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCP 1535
EFRH NN P++SV HPIHDQ ++L++ K+QLK+E+ IEPWTFEQHLGEAVFIPAGCP
Sbjct: 706 EHEFRHFNNEPLESVIHPIHDQTMFLSDSQKKQLKEEFDIEPWTFEQHLGEAVFIPAGCP 765
Query: 1536 HQVRNR 1541
HQVRNR
Sbjct: 766 HQVRNR 771
>gb|AAT81741.1| jmjC domain containing protein [Oryza sativa (japonica
cultivar-group)]
Length = 1003
Score = 430 bits (1105), Expect = e-118
Identities = 246/640 (38%), Positives = 342/640 (53%), Gaps = 84/640 (13%)
Query: 754 EELLRMLSVEHKNIQGVGV--EETIDYGLRSSGLMGDTERKKETRILR------CHQCWR 805
+E + M + +G G EE + + M D KK ++L CHQC R
Sbjct: 196 DEAIMMKPSKESKKRGAGKKQEEEENNTISIEDEMCDANNKKGKKMLTGENALMCHQCQR 255
Query: 806 NSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKDISVMTG 865
N V+ C C K++C C+ +WYPG + + CP+C +NCNC+ CL+ M G
Sbjct: 256 NDKGRVIWCKSCNNKRFCEPCMKRWYPGLSEVDFAAKCPYCRKNCNCKACLR-----MIG 310
Query: 866 SGEADTGVILQK-----LLYLLNKTLPLLQHIQREQISEMEVEASMHGSPL--------- 911
+ I ++ +++ LP L+ +Q+EQ+ E E+E + G +
Sbjct: 311 VEKPPEKKISEENQRRYAFRIVDLLLPWLKELQQEQMKEKELEGRLQGVSMDEVKLEQAD 370
Query: 912 --MEEDIQFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHYEDIPASGNEETID- 968
M+E + D C TSIV+FHRSC C YDLCL CC ELR G +IP +++
Sbjct: 371 CDMDERVYCDRCKTSIVDFHRSCKA--CSYDLCLACCWELRKG----EIPGGEEAKSVQW 424
Query: 969 --------------------------EPPITSA----------------WRAEINGRIPC 986
E P T W+A +G IPC
Sbjct: 425 EERGQKYVFGNISKDEKKRVSSKRHMETPSTETCNDMAVAGDPNNPLLLWKANSDGSIPC 484
Query: 987 PPKARGGCGTSILSLRRLFEANWVNKLVRNAEE-LTIQYHPPSVDLLVGCLQCHRFVVDL 1045
PPK GGCG S L LR L +++L A + + + +++ C +
Sbjct: 485 PPKEIGGCGASSLVLRCLLPEIMLSELEHRANKVIKREAFDKAINETSDQCPCFYHTSKI 544
Query: 1046 AQNSVRKAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWH 1105
N+ R+AA+R+ + DN+LYCPDA ++ + + HFQ HW +GEPVIV + SGLSW
Sbjct: 545 RTNATREAANRKGSSDNYLYCPDANNIQEDDLSHFQMHWSKGEPVIVSDALRLTSGLSWE 604
Query: 1106 PMVMWRAFRG--ANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRY-RNGWPEM 1162
P+VMWRA R N +++E KA+DCLDW EV+INI FF GY+ GRR+ WPEM
Sbjct: 605 PLVMWRALREKKTNGDVEDEHFAVKAVDCLDWNEVEINIHMFFMGYMRGRRHPMTFWPEM 664
Query: 1163 LKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPA-VLKPDLGPKTY 1221
LKLKDWPPS+ F++ PRHGAEFI LPF +YT P++G LNLA +LPA VLKPDLGPKTY
Sbjct: 665 LKLKDWPPSSMFDQRLPRHGAEFITALPFPEYTDPRYGPLNLAVRLPAGVLKPDLGPKTY 724
Query: 1222 IAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDL 1281
IAYG EEL RGDSVTKLHCD+SDAVNIL HT EV Q I K++ K +D+ +L
Sbjct: 725 IAYGCYEELGRGDSVTKLHCDMSDAVNILMHTAEVSYDTEQLDKIAKIKMKMREQDLHEL 784
Query: 1282 YGRINKTVVSHRSKHKKCRTGISMDPKIPENDDTMGRNSN 1321
+G ++++ ++ + + +M+ K N T G + N
Sbjct: 785 FG-VSESGAKGKADDEASKISCNMENKHTSNQSTKGLDIN 823
Score = 87.8 bits (216), Expect = 3e-15
Identities = 36/48 (75%), Positives = 43/48 (89%)
Query: 1490 VTHPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQ 1537
V+HPIHDQ YL +HKR+LK+E+G+EPWTFEQ LG+AVFIPAGCPHQ
Sbjct: 845 VSHPIHDQTFYLTVEHKRKLKEEHGVEPWTFEQKLGDAVFIPAGCPHQ 892
>gb|AAF70852.1| F24O1.3 [Arabidopsis thaliana] gi|7486103|pir||T01440 hypothetical
protein F24O1.2 - Arabidopsis thaliana
Length = 906
Score = 418 bits (1075), Expect = e-115
Identities = 223/517 (43%), Positives = 294/517 (56%), Gaps = 45/517 (8%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKD 859
CHQC + ++IC++C++ +C +CI KWYP + +++ CP C +NCNC CL +
Sbjct: 209 CHQCLKGERITLLICSECEKTMFCLQCIRKWYPNLSEDDVVEKCPLCRQNCNCSKCLHLN 268
Query: 860 ISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGS---------- 909
+ T E L YL+ LP L + Q E+E EA++ G
Sbjct: 269 GLIETSKRELAKSERRHHLQYLITLMLPFLNKLSIFQKLEIEFEATVQGKLPSEVEITAA 328
Query: 910 -PLMEEDIQFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNG---------LHYEDI- 958
+E + D+C TSIV+ HRSCP C Y+LCL CC E+R G HY D
Sbjct: 329 ISYTDERVYCDHCATSIVDLHRSCPK--CSYELCLKCCQEIREGSLSERPEMKFHYVDRG 386
Query: 959 ------------PASGNEETIDEPPITSAWRAEINGRIPCPPKARGGCGTSILSLRRLFE 1006
S E + P + W NG I C P+ GGCG +L LRR+
Sbjct: 387 HRYMHGLDAAEPSLSSTFEDEEANPSDAKWSLGENGSITCAPEKLGGCGERMLELRRILP 446
Query: 1007 ANWVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSVRKAASRETNHDNFLYC 1066
W++ L AE Y+ ++ C +C +L RK+ASR T+ DN+L+C
Sbjct: 447 LTWMSDLEHKAETFLSSYNISP--RMLNC-RCSSLETELT----RKSASRTTSSDNYLFC 499
Query: 1067 PDAVD-MGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAF-RGANKILKEEP 1124
P+++ + + E HFQ HW +GEPVIVRN + GLSW PMVMWRA N E
Sbjct: 500 PESLGVLKEEELLHFQEHWAKGEPVIVRNALDNTPGLSWEPMVMWRALCENVNSTSSSEM 559
Query: 1125 TTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAE 1184
+ KAIDCL CEV+IN QFF+GY +GR Y N WPEMLKLKDWPPS+ FE+ PRH E
Sbjct: 560 SQVKAIDCLANCEVEINTRQFFEGYSKGRTYENFWPEMLKLKDWPPSDKFEDLLPRHCDE 619
Query: 1185 FIAMLPFSDYTHPKFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDI 1243
FI+ LPF +Y+ P+ GILN+ATKLP +KPDLGPKTYIAYG +EL RGDSVTKLHCD+
Sbjct: 620 FISALPFQEYSDPRTGILNIATKLPEGFIKPDLGPKTYIAYGIPDELGRGDSVTKLHCDM 679
Query: 1244 SDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRD 1280
SDAVNILTHT EV Q +K L++K++ ++ D
Sbjct: 680 SDAVNILTHTAEVTLSQEQISSVKALKQKHKLQNKVD 716
Score = 134 bits (338), Expect = 2e-29
Identities = 74/129 (57%), Positives = 82/129 (63%), Gaps = 31/129 (24%)
Query: 1435 DISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVT--- 1491
+ISSN ++ ET GSA+WDIFRR+DVPKL EYL+KH KEFRH PV VT
Sbjct: 738 EISSN------ENEET--GSALWDIFRREDVPKLEEYLRKHCKEFRHTYCSPVTKVTTPT 789
Query: 1492 --------------------HPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIP 1531
HPIHDQ YL +HKR+LK EYGIEPWTF Q LGEAVFIP
Sbjct: 790 CINFMTNLFPVLTVSSFQVYHPIHDQSCYLTLEHKRKLKAEYGIEPWTFVQKLGEAVFIP 849
Query: 1532 AGCPHQVRN 1540
AGCPHQVRN
Sbjct: 850 AGCPHQVRN 858
>ref|NP_176421.1| transcription factor jumonji (jmjC) domain-containing protein
[Arabidopsis thaliana]
Length = 883
Score = 418 bits (1075), Expect = e-115
Identities = 223/517 (43%), Positives = 294/517 (56%), Gaps = 45/517 (8%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKD 859
CHQC + ++IC++C++ +C +CI KWYP + +++ CP C +NCNC CL +
Sbjct: 209 CHQCLKGERITLLICSECEKTMFCLQCIRKWYPNLSEDDVVEKCPLCRQNCNCSKCLHLN 268
Query: 860 ISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGS---------- 909
+ T E L YL+ LP L + Q E+E EA++ G
Sbjct: 269 GLIETSKRELAKSERRHHLQYLITLMLPFLNKLSIFQKLEIEFEATVQGKLPSEVEITAA 328
Query: 910 -PLMEEDIQFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNG---------LHYEDI- 958
+E + D+C TSIV+ HRSCP C Y+LCL CC E+R G HY D
Sbjct: 329 ISYTDERVYCDHCATSIVDLHRSCPK--CSYELCLKCCQEIREGSLSERPEMKFHYVDRG 386
Query: 959 ------------PASGNEETIDEPPITSAWRAEINGRIPCPPKARGGCGTSILSLRRLFE 1006
S E + P + W NG I C P+ GGCG +L LRR+
Sbjct: 387 HRYMHGLDAAEPSLSSTFEDEEANPSDAKWSLGENGSITCAPEKLGGCGERMLELRRILP 446
Query: 1007 ANWVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSVRKAASRETNHDNFLYC 1066
W++ L AE Y+ ++ C +C +L RK+ASR T+ DN+L+C
Sbjct: 447 LTWMSDLEHKAETFLSSYNISP--RMLNC-RCSSLETELT----RKSASRTTSSDNYLFC 499
Query: 1067 PDAVD-MGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAF-RGANKILKEEP 1124
P+++ + + E HFQ HW +GEPVIVRN + GLSW PMVMWRA N E
Sbjct: 500 PESLGVLKEEELLHFQEHWAKGEPVIVRNALDNTPGLSWEPMVMWRALCENVNSTSSSEM 559
Query: 1125 TTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAE 1184
+ KAIDCL CEV+IN QFF+GY +GR Y N WPEMLKLKDWPPS+ FE+ PRH E
Sbjct: 560 SQVKAIDCLANCEVEINTRQFFEGYSKGRTYENFWPEMLKLKDWPPSDKFEDLLPRHCDE 619
Query: 1185 FIAMLPFSDYTHPKFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDI 1243
FI+ LPF +Y+ P+ GILN+ATKLP +KPDLGPKTYIAYG +EL RGDSVTKLHCD+
Sbjct: 620 FISALPFQEYSDPRTGILNIATKLPEGFIKPDLGPKTYIAYGIPDELGRGDSVTKLHCDM 679
Query: 1244 SDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRD 1280
SDAVNILTHT EV Q +K L++K++ ++ D
Sbjct: 680 SDAVNILTHTAEVTLSQEQISSVKALKQKHKLQNKVD 716
Score = 145 bits (365), Expect = 1e-32
Identities = 73/106 (68%), Positives = 81/106 (75%), Gaps = 8/106 (7%)
Query: 1435 DISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPI 1494
+ISSN ++ ET GSA+WDIFRR+DVPKL EYL+KH KEFRH PV V HPI
Sbjct: 738 EISSN------ENEET--GSALWDIFRREDVPKLEEYLRKHCKEFRHTYCSPVTKVYHPI 789
Query: 1495 HDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
HDQ YL +HKR+LK EYGIEPWTF Q LGEAVFIPAGCPHQVRN
Sbjct: 790 HDQSCYLTLEHKRKLKAEYGIEPWTFVQKLGEAVFIPAGCPHQVRN 835
>gb|AAC17616.1| Contains similarity to box helicases gb|U29097 from C. elegans and to
the ENBP1 gene product gb|X95995 from Vicia sativa.
[Arabidopsis thaliana] gi|25372073|pir||D86254
hypothetical protein [imported] - Arabidopsis thaliana
Length = 851
Score = 410 bits (1053), Expect = e-112
Identities = 220/496 (44%), Positives = 290/496 (58%), Gaps = 31/496 (6%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKD 859
CHQC + + IC C+ + YC+ CI KWYP + ++I CPFC CNC CL
Sbjct: 193 CHQCSKGERRYLFICTFCEVRLYCFPCIKKWYPHLSTDDILEKCPFCRGTCNCCTCLHSS 252
Query: 860 ISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPLMEEDIQ-- 917
+ T + D L +L+ LP L+ + + Q E+E EA + S + DI
Sbjct: 253 GLIETSKRKLDKYERFYHLRFLIVAMLPFLKKLCKAQDQEIETEAKVQDSMASQVDISES 312
Query: 918 ---------FDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHYEDIPASGNEETID 968
++C TSIV+ HRSCP C Y+LCL CC E+R E +S +E+
Sbjct: 313 LCSNEERVFCNHCATSIVDLHRSCPK--CSYELCLNCCQEIRG----EPSSSSVSEDETK 366
Query: 969 EPPITSAWRAEINGRIPCPPKARGGCGTSILSLRRLFEANWVNKLVRNAEELTIQYHPPS 1028
P I W A+ NG I C PK GGCG S+L L+R+ W++ L + AE Y S
Sbjct: 367 TPSIK--WNADENGSIRCAPKELGGCGDSVLELKRILPVTWMSDLEQKAETFLASY---S 421
Query: 1029 VDLLVGCLQCHRFVVDLAQNSVRKAASRETNHDNFLYCPDAVD-MGDTEYEHFQRHWIRG 1087
+ + +C + + RKAASR+ + DN+LY PD++D + E HFQ HW +G
Sbjct: 422 IKPPMSYCRCSSDMSSMK----RKAASRDGSSDNYLYSPDSLDVLKQEELLHFQEHWSKG 477
Query: 1088 EPVIVRNVFEKASGLSWHPMVMWRAF-RGANKILKEEPTTFKAIDCLDWCEVQINIFQFF 1146
EPVIVRN +GLSW PMVMWRA + + + KAIDCL CE IN FF
Sbjct: 478 EPVIVRNALNNTAGLSWEPMVMWRALCENVDSAISSNMSDVKAIDCLANCE--INTLCFF 535
Query: 1147 KGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLAT 1206
+GY +GR Y N WPEMLKLKDWPPS+ FE PRH EFI+ LPF +Y+ P+ GILN+AT
Sbjct: 536 EGYSKGRTYENFWPEMLKLKDWPPSDKFENLLPRHCDEFISALPFQEYSDPRSGILNIAT 595
Query: 1207 KLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKI 1265
KLP +LKPDLGPKTY+AYG+ +EL RGDSVTKLHCD+SDAVNIL HT EV Q
Sbjct: 596 KLPEGLLKPDLGPKTYVAYGTSDELGRGDSVTKLHCDMSDAVNILMHTAEVTLSEEQRSA 655
Query: 1266 IKKLQKKYEAEDMRDL 1281
I L++K++ ++ ++L
Sbjct: 656 IADLKQKHKQQNEKEL 671
Score = 128 bits (321), Expect = 2e-27
Identities = 64/122 (52%), Positives = 80/122 (65%), Gaps = 4/122 (3%)
Query: 1423 KIQIDKTVPVKNDISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHA 1482
K Q +K + +N + ++ + + A+WDIF+R+DVPKL EYL+KH EFRH
Sbjct: 664 KQQNEKELQEQNGLEEEEVVSDEIVVYDETSGALWDIFKREDVPKLEEYLRKHCIEFRHT 723
Query: 1483 NNLPVDSVTHPIHDQILYLNEKHKRQLKKEYG----IEPWTFEQHLGEAVFIPAGCPHQV 1538
V V HPIHDQ +L +HKR+LK E+G IEPWTF Q LGEAVFIPAGCPHQV
Sbjct: 724 YCSRVTKVYHPIHDQSYFLTVEHKRKLKAEFGMVTWIEPWTFVQKLGEAVFIPAGCPHQV 783
Query: 1539 RN 1540
RN
Sbjct: 784 RN 785
>ref|NP_172659.2| transcription factor jumonji (jmjC) domain-containing protein
[Arabidopsis thaliana]
Length = 880
Score = 408 bits (1049), Expect = e-112
Identities = 223/521 (42%), Positives = 294/521 (55%), Gaps = 48/521 (9%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKD 859
CHQC + + IC C+ + YC+ CI KWYP + ++I CPFC CNC CL
Sbjct: 193 CHQCSKGERRYLFICTFCEVRLYCFPCIKKWYPHLSTDDILEKCPFCRGTCNCCTCLHSS 252
Query: 860 ISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPLMEEDIQ-- 917
+ T + D L +L+ LP L+ + + Q E+E EA + S + DI
Sbjct: 253 GLIETSKRKLDKYERFYHLRFLIVAMLPFLKKLCKAQDQEIETEAKVQDSMASQVDISES 312
Query: 918 ---------FDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNG---------------- 952
++C TSIV+ HRSCP C Y+LCL CC E+R G
Sbjct: 313 LCSNEERVFCNHCATSIVDLHRSCPK--CSYELCLNCCQEIRGGWLSDRPECQLQFEYRG 370
Query: 953 ---LHYEDIPASGNEETIDEPPITSA-WRAEINGRIPCPPKARGGCGTSILSLRRLFEAN 1008
+H E S + + DE S W A+ NG I C PK GGCG S+L L+R+
Sbjct: 371 TRYIHGEAAEPSSSSVSEDETKTPSIKWNADENGSIRCAPKELGGCGDSVLELKRILPVT 430
Query: 1009 WVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSVRKAASRETNHDNFLYCPD 1068
W++ L + AE Y S+ + +C + + RKAASR+ + DN+LY PD
Sbjct: 431 WMSDLEQKAETFLASY---SIKPPMSYCRCSSDMSSMK----RKAASRDGSSDNYLYSPD 483
Query: 1069 AVD-MGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAF-RGANKILKEEPTT 1126
++D + E HFQ HW +GEPVIVRN +GLSW PMVMWRA + + +
Sbjct: 484 SLDVLKQEELLHFQEHWSKGEPVIVRNALNNTAGLSWEPMVMWRALCENVDSAISSNMSD 543
Query: 1127 FKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFI 1186
KAIDCL CEV+IN FF+GY +GR Y N WPEMLKLKDWPPS+ FE PRH EFI
Sbjct: 544 VKAIDCLANCEVKINTLCFFEGYSKGRTYENFWPEMLKLKDWPPSDKFENLLPRHCDEFI 603
Query: 1187 AMLPFSDYTHPKFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISD 1245
+ LPF +Y+ P+ GILN+ATKLP +LKPDLGPKTY+AYG+ +EL RGDSVTKLHCD+SD
Sbjct: 604 SALPFQEYSDPRSGILNIATKLPEGLLKPDLGPKTYVAYGTSDELGRGDSVTKLHCDMSD 663
Query: 1246 A-----VNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDL 1281
A VNIL HT EV Q I L++K++ ++ ++L
Sbjct: 664 AVSFRIVNILMHTAEVTLSEEQRSAIADLKQKHKQQNEKEL 704
Score = 134 bits (336), Expect = 3e-29
Identities = 64/118 (54%), Positives = 80/118 (67%)
Query: 1423 KIQIDKTVPVKNDISSNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHA 1482
K Q +K + +N + ++ + + A+WDIF+R+DVPKL EYL+KH EFRH
Sbjct: 697 KQQNEKELQEQNGLEEEEVVSDEIVVYDETSGALWDIFKREDVPKLEEYLRKHCIEFRHT 756
Query: 1483 NNLPVDSVTHPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
V V HPIHDQ +L +HKR+LK E+GIEPWTF Q LGEAVFIPAGCPHQVRN
Sbjct: 757 YCSRVTKVYHPIHDQSYFLTVEHKRKLKAEFGIEPWTFVQKLGEAVFIPAGCPHQVRN 814
>gb|AAF13079.1| hypothetical protein [Arabidopsis thaliana]
gi|15231487|ref|NP_187418.1| transcription factor jumonji
(jmjC) domain-containing protein [Arabidopsis thaliana]
Length = 1027
Score = 347 bits (890), Expect = 2e-93
Identities = 244/715 (34%), Positives = 336/715 (46%), Gaps = 149/715 (20%)
Query: 681 GSKNKMKSIANKARNKFGKVRNMRGRPKGSLRKKNETAYCLDSQNERNSL--DGRTSTEA 738
G K K K N+A K K K +KNE +D N L + R +T+
Sbjct: 46 GPKRKGKRGGNRAPKKTPK--------KDEEMQKNE----IDEANRVTGLVKEKRAATKI 93
Query: 739 AYRNDVDLHRGHCSQEELLRMLSVEHKNIQGVGVEETIDYGLRSSGLMGDTERKK----- 793
R D + G S K ++G+ + G R + G+ K
Sbjct: 94 LNRKDSIIEVGEASGSM--------PKEVKGIRI------GKRKGEIDGEIPTKPGKKPK 139
Query: 794 ---ETRIL------RCHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACP 844
+ RI+ CHQC ++ V C C K+YC+ C+ WYP +E++ C
Sbjct: 140 TTVDPRIIGYRPDNMCHQCQKSDRI-VERCQTCNSKRYCHPCLDTWYPLIAKEDVAKKCM 198
Query: 845 FCLRNCNCRLCLKKDISV--MTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEV 902
FC CNCR CL+ D + + + +Q ++L LP L+ I EQ++E EV
Sbjct: 199 FCSSTCNCRACLRLDTKLKGINSNLIVSEEEKVQASKFILQSLLPHLKGINDEQVAEKEV 258
Query: 903 EASMHGSPLME-----------EDIQFDNCNTSIVNFHRSCPNPNCRYDL----CLT--- 944
EA ++G E E + D C TSI + HR+C + C +D+ CL
Sbjct: 259 EAKIYGLKFEEVRPQDAKAFPDERLYCDICKTSIYDLHRNCKS--CSFDICLSCCLEIRN 316
Query: 945 ----CCME------LRNGLHYE--------DIPASGNEETI------------------- 967
C E + GL YE + PA+ ++ +
Sbjct: 317 GKALACKEDVSWNYINRGLEYEHGQEGKVIEKPANKLDDKLKDKLDGKPDDKPKGKPKGR 376
Query: 968 -------------------------DEPPIT-------SAWRAEINGRIPCPPKARGGCG 995
DE P+ S W+A G I C CG
Sbjct: 377 PKGKPDDKPKGKLKGKQDDKPDDKPDEKPVNTDHMKYPSLWKANEAGIITCC------CG 430
Query: 996 TSILSLRRLFEANWVNKLVRNAEELT----IQYHPPSVDLLVGCLQCHRFVVDLAQNSVR 1051
L L+RL W+++LV E+ + P +V C R + D+ ++
Sbjct: 431 AGELVLKRLLPDGWISELVNRVEKTAEAGELLNLPETVLERCPCSNSDRHI-DIDSCNLL 489
Query: 1052 KAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWR 1111
KAA RE + DN+LY P D+ + +HFQ HW++GEPVIVRNV E SGLSW PMVM R
Sbjct: 490 KAACREGSEDNYLYSPSVWDVQQDDLKHFQHHWVKGEPVIVRNVLEATSGLSWEPMVMHR 549
Query: 1112 AFRGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPS 1171
A R + + A+DCLD+CEV++N+ +FF GY +GR R GWP +LKLKDWPP+
Sbjct: 550 ACRQISHVQHGSLKDVVAVDCLDFCEVKVNLHEFFTGYTDGRYDRMGWPLVLKLKDWPPA 609
Query: 1172 NSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPA-VLKPDLGPKTYIAYGSLEEL 1230
F++ PRH EF+ LP YTHP G LNLA KLP LKPD+GPKTY+A G +EL
Sbjct: 610 KVFKDNLPRHAEEFLCSLPLKHYTHPVNGPLNLAVKLPQNCLKPDMGPKTYVASGFAQEL 669
Query: 1231 SRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDMRDLYGRI 1285
RGDSVTKLHCD+SDAVNILTH EV P QP I L+KK+ +D+++LY +
Sbjct: 670 GRGDSVTKLHCDMSDAVNILTHISEV--PNMQPG-IGNLKKKHAEQDLKELYSSV 721
Score = 136 bits (342), Expect = 6e-30
Identities = 63/101 (62%), Positives = 73/101 (71%), Gaps = 1/101 (0%)
Query: 1440 NFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQIL 1499
N Q ++ET G A+WDIFRR+D+PKL Y++KHHKEFRH PV V HPIHDQ
Sbjct: 733 NSRQQVQNVETDDG-ALWDIFRREDIPKLESYIEKHHKEFRHLYCCPVSQVVHPIHDQNF 791
Query: 1500 YLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
YL H +LK+EYGIEPWTF Q LG+AV IP GCPHQVRN
Sbjct: 792 YLTRYHIMKLKEEYGIEPWTFNQKLGDAVLIPVGCPHQVRN 832
>ref|NP_172380.2| transcription factor jumonji (jmjC) domain-containing protein
[Arabidopsis thaliana] gi|42571415|ref|NP_973798.1|
transcription factor jumonji (jmjC) domain-containing
protein [Arabidopsis thaliana]
Length = 930
Score = 297 bits (760), Expect = 2e-78
Identities = 163/477 (34%), Positives = 248/477 (51%), Gaps = 37/477 (7%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKD 859
CHQC R ++ C KC ++ +C+ C++ Y + EE+E CP C C+C+ CL+ D
Sbjct: 206 CHQCQRKDRERIISCLKCNQRAFCHNCLSARYSEISLEEVEKVCPACRGLCDCKSCLRSD 265
Query: 860 ISVMTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEVEASMHGSPL-------- 911
++ E LQ L LL+ LP+++ I EQ E+E+E + +
Sbjct: 266 NTIKVRIREIPVLDKLQYLYRLLSAVLPVIKQIHLEQCMEVELEKRLREVEIDLVRARLK 325
Query: 912 MEEDIQFDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHYEDIPASGNEETIDE-- 969
+E + + C +V+++R CPN C YDLCL CC +LR + SG + + +
Sbjct: 326 ADEQMCCNVCRIPVVDYYRHCPN--CSYDLCLRCCQDLREE---SSVTISGTNQNVQDRK 380
Query: 970 --PPIT-------SAWRAEINGRIPCPPKARGGCGTSILSLRRLFEANWVNKLVRNAEEL 1020
P + W A +G IPCPPK GGCG+ L+L R+F+ NWV KLV+NAEE+
Sbjct: 381 GAPKLKLNFSYKFPEWEANGDGSIPCPPKEYGGCGSHSLNLARIFKMNWVAKLVKNAEEI 440
Query: 1021 TIQYHPPSVDLLVGCLQCHRFVVDLAQNSVRKAASRETNHDNFLYCPDAVDMGDTEYEHF 1080
+ GC D+ + K A RE + DN++Y P + F
Sbjct: 441 -----------VSGCKLSDLLNPDMCDSRFCKFAEREESGDNYVYSPSLETIKTDGVAKF 489
Query: 1081 QRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAFRG-ANKILKEEPTTFKAIDCLDWCEVQ 1139
++ W G V V+ V + +S W P +WR +++ L+E KAI+CLD EV
Sbjct: 490 EQQWAEGRLVTVKMVLDDSSCSRWDPETIWRDIDELSDEKLREHDPFLKAINCLDGLEVD 549
Query: 1140 INIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKF 1199
+ + +F + Y +G+ G P + KLKDWP ++ EE EFI PF +Y HP+
Sbjct: 550 VRLGEFTRAYKDGKNQETGLPLLWKLKDWPSPSASEEFIFYQRPEFIRSFPFLEYIHPRL 609
Query: 1200 GILNLATKLPAV-LKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEE 1255
G+LN+A KLP L+ D GPK Y++ G+ +E+S GDS+T +H ++ D V +L HT E
Sbjct: 610 GLLNVAAKLPHYSLQNDSGPKIYVSCGTYQEISAGDSLTGIHYNMRDMVYLLVHTSE 666
Score = 119 bits (297), Expect = 1e-24
Identities = 56/110 (50%), Positives = 78/110 (70%), Gaps = 9/110 (8%)
Query: 1436 ISSNNFFQNDDHMETQF-----GSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSV 1490
++ N +N D+ME+ G A WD+FRRQDVPKL+ YL++ F+ +N+ D V
Sbjct: 726 VNPENLTENGDNMESSCTSSCAGGAQWDVFRRQDVPKLSGYLQR---TFQKPDNIQTDFV 782
Query: 1491 THPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
+ P+++ L+LNE HKRQL+ E+G+EPWTFEQH GEA+FIPAGCP Q+ N
Sbjct: 783 SRPLYEG-LFLNEHHKRQLRDEFGVEPWTFEQHRGEAIFIPAGCPFQITN 831
Score = 42.4 bits (98), Expect = 0.13
Identities = 22/66 (33%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query: 10 RCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRSGRRKPVDNSE 69
RC R WRCT +++ K+VCE+H++ +K+ A+ KRRS + D
Sbjct: 29 RCKRSDGKQWRCTAMSMADKTVCEKHYIQAKKRAANSAFRANQKKAKRRSSLGE-TDTYS 87
Query: 70 NGVVDD 75
G +DD
Sbjct: 88 EGKMDD 93
>dbj|BAD44593.1| hypothetical protein [Arabidopsis thaliana]
gi|51971845|dbj|BAD44587.1| hypothetical protein
[Arabidopsis thaliana]
Length = 535
Score = 285 bits (729), Expect = 9e-75
Identities = 182/498 (36%), Positives = 245/498 (48%), Gaps = 103/498 (20%)
Query: 845 FCLRNCNCRLCLKKDISV--MTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEV 902
FC CNCR CL+ D + + + +Q ++L LP L+ I EQ++E EV
Sbjct: 2 FCSSTCNCRACLRLDTKLKGINSNLIVSEEEKVQASKFILQSLLPHLKGINDEQVAEKEV 61
Query: 903 EASMHGSPLME-----------EDIQFDNCNTSIVNFHRSCPNPNCRYDL----CLT--- 944
EA ++G E E + D C TSI + HR+C + C +D+ CL
Sbjct: 62 EAKIYGLKFEEVRPQDAKAFPDERLYCDICKTSIYDLHRNCKS--CSFDICLSCCLEIRN 119
Query: 945 ----CCME------LRNGLHYE--------DIPASGNEETI------------------- 967
C E + GL YE + PA+ ++ +
Sbjct: 120 GKALACKEDVSWNYINRGLEYEHGQEGKVIEKPANKLDDKLKDKLDGKPDDKPKGKPKGR 179
Query: 968 -------------------------DEPPIT-------SAWRAEINGRIPCPPKARGGCG 995
DE P+ S W+A G I C CG
Sbjct: 180 PKGKPDDKPKGKLKGKQDDKPDDKPDEKPVNTDHMKYPSLWKANEAGIITCC------CG 233
Query: 996 TSILSLRRLFEANWVNKLVRNAEELT----IQYHPPSVDLLVGCLQCHRFVVDLAQNSVR 1051
L L+RL W+++LV E+ + P +V C R + D+ ++
Sbjct: 234 AGELVLKRLLPDGWISELVNRVEKTAEAGELLNLPETVLERCPCSNSDRHI-DIDSCNLL 292
Query: 1052 KAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWR 1111
KAA RE + DN+LY P D+ + +HFQ HW++GEPVIVRNV E SGLSW PMVM R
Sbjct: 293 KAACREGSEDNYLYSPSVWDVQQDDLKHFQHHWVKGEPVIVRNVLEATSGLSWEPMVMHR 352
Query: 1112 AFRGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPS 1171
A R + + A+DCLD+CEV++N+ +FF GY +GR R GWP +LKLKDWPP+
Sbjct: 353 ACRQISHVQHGSLKDVVAVDCLDFCEVKVNLHEFFTGYTDGRYDRMGWPLVLKLKDWPPA 412
Query: 1172 NSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPA-VLKPDLGPKTYIAYGSLEEL 1230
F++ PRH EF+ LP YTHP G LNLA KLP LKPD+GPKTY+A G +EL
Sbjct: 413 KVFKDNLPRHAEEFLCSLPLKHYTHPVNGPLNLAVKLPQNCLKPDMGPKTYVASGFAQEL 472
Query: 1231 SRGDSVTKLHCDISDAVN 1248
RGDSVTKLHCD+SDAV+
Sbjct: 473 GRGDSVTKLHCDMSDAVS 490
>pir||F86222 hypothetical protein [imported] - Arabidopsis thaliana
gi|2342679|gb|AAB70402.1| Similar to Vicia sativa ENBP1
(gb|X95995). [Arabidopsis thaliana]
Length = 950
Score = 275 bits (704), Expect = 7e-72
Identities = 165/503 (32%), Positives = 250/503 (48%), Gaps = 68/503 (13%)
Query: 800 CHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCNCRLCLKKD 859
CHQC R ++ C KC ++ +C+ C++ Y + EE+E CP C C+C+ CL+ D
Sbjct: 220 CHQCQRKDRERIISCLKCNQRAFCHNCLSARYSEISLEEVEKVCPACRGLCDCKSCLRSD 279
Query: 860 ----------ISVMTGSGEADTGV----ILQKLLYL---LNKTLPLLQHIQREQISEME- 901
I + T + + +L KL YL L+ LP+++ I EQ++ E
Sbjct: 280 NTIKVWSRVIIKIQTSATYFGVRIREIPVLDKLQYLYRLLSAVLPVIKQIHLEQLTIDEL 339
Query: 902 --------VEASMHGSPLMEEDI-------------QFDNCNTSIVNFHRSCPNPNCRYD 940
V S+ S +E D+ F+ C +V+++R CPN C YD
Sbjct: 340 YILVTFFAVNTSLFYSIEVEIDLVRARLKADEQMCWYFNVCRIPVVDYYRHCPN--CSYD 397
Query: 941 LCLTCCMELRNGLHYEDIPASGNEETIDE----PPIT-------SAWRAEINGRIPCPPK 989
LCL CC +LR + SG + + + P + W A +G IPCPPK
Sbjct: 398 LCLRCCQDLREE---SSVTISGTNQNVQDRKGAPKLKLNFSYKFPEWEANGDGSIPCPPK 454
Query: 990 ARGGCGTSILSLRRLFEANWVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNS 1049
GGCG+ L+L R+F+ NWV KLV+NAEE+ + GC D+ +
Sbjct: 455 EYGGCGSHSLNLARIFKMNWVAKLVKNAEEI-----------VSGCKLSDLLNPDMCDSR 503
Query: 1050 VRKAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVM 1109
K A RE + DN++Y P + F++ W G V V+ V + +S W P +
Sbjct: 504 FCKFAEREESGDNYVYSPSLETIKTDGVAKFEQQWAEGRLVTVKMVLDDSSCSRWDPETI 563
Query: 1110 WRAFRG-ANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDW 1168
WR +++ L+E KAI+CLD EV + + +F + Y +G+ G P + KLKDW
Sbjct: 564 WRDIDELSDEKLREHDPFLKAINCLDGLEVDVRLGEFTRAYKDGKNQETGLPLLWKLKDW 623
Query: 1169 PPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLPAV-LKPDLGPKTYIAYGSL 1227
P ++ EE EFI PF +Y HP+ G+LN+A KLP L+ D GPK Y++ G+
Sbjct: 624 PSPSASEEFIFYQRPEFIRSFPFLEYIHPRLGLLNVAAKLPHYSLQNDSGPKIYVSCGTY 683
Query: 1228 EELSRGDSVTKLHCDISDAVNIL 1250
+E+S GDS+T +H ++ D N L
Sbjct: 684 QEISAGDSLTGIHYNMRDMGNWL 706
Score = 119 bits (297), Expect = 1e-24
Identities = 56/110 (50%), Positives = 78/110 (70%), Gaps = 9/110 (8%)
Query: 1436 ISSNNFFQNDDHMETQF-----GSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSV 1490
++ N +N D+ME+ G A WD+FRRQDVPKL+ YL++ F+ +N+ D V
Sbjct: 783 VNPENLTENGDNMESSCTSSCAGGAQWDVFRRQDVPKLSGYLQR---TFQKPDNIQTDFV 839
Query: 1491 THPIHDQILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
+ P+++ L+LNE HKRQL+ E+G+EPWTFEQH GEA+FIPAGCP Q+ N
Sbjct: 840 SRPLYEG-LFLNEHHKRQLRDEFGVEPWTFEQHRGEAIFIPAGCPFQITN 888
Score = 42.4 bits (98), Expect = 0.13
Identities = 22/66 (33%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query: 10 RCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRSGRRKPVDNSE 69
RC R WRCT +++ K+VCE+H++ +K+ A+ KRRS + D
Sbjct: 43 RCKRSDGKQWRCTAMSMADKTVCEKHYIQAKKRAANSAFRANQKKAKRRSSLGE-TDTYS 101
Query: 70 NGVVDD 75
G +DD
Sbjct: 102 EGKMDD 107
>dbj|BAD43260.1| hypothetical protein [Arabidopsis thaliana]
Length = 628
Score = 229 bits (584), Expect = 6e-58
Identities = 180/604 (29%), Positives = 259/604 (42%), Gaps = 145/604 (24%)
Query: 681 GSKNKMKSIANKARNKFGKVRNMRGRPKGSLRKKNETAYCLDSQNERNSL--DGRTSTEA 738
G K K K N+A K K K +KNE +D N L + R +T+
Sbjct: 46 GPKRKGKRGGNRAPKKTPK--------KDEEMQKNE----IDEANRVTGLVKEKRAATKI 93
Query: 739 AYRNDVDLHRGHCSQEELLRMLSVEHKNIQGVGVEETIDYGLRSSGLMGDTERKK----- 793
R D + G S K ++G+ + G R + G+ K
Sbjct: 94 LNRKDSIIEVGEASGSM--------PKEVKGIRI------GKRKGEIDGEIPTKPGKKPK 139
Query: 794 ---ETRIL------RCHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACP 844
+ RI+ CHQC ++ V C C K+YC+ C+ WYP +E++ C
Sbjct: 140 TTVDPRIIGYRPDNMCHQCQKSDRI-VERCQTCNSKRYCHPCLDTWYPLIAKEDVAKKCM 198
Query: 845 FCLRNCNCRLCLKKDISV--MTGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEMEV 902
FC CNCR CL+ D + + + +Q ++L LP L+ I EQ++E EV
Sbjct: 199 FCSSTCNCRACLRLDTKLKGINSNLIVSEEEKVQASKFILQSLLPHLKGINDEQVAEKEV 258
Query: 903 EASMHGSPLME-----------EDIQFDNCNTSIVNFHRSCPNPNCRYDL----CLT--- 944
EA ++G E E + D C TSI + HR+C + C +D+ CL
Sbjct: 259 EAKIYGLKFEEVRPQDAKAFPDERLYCDICKTSIYDLHRNCKS--CSFDICLSCCLEIRN 316
Query: 945 ----CCME------LRNGLHYE--------DIPASGNEETI------------------- 967
C E + GL YE + PA+ ++ +
Sbjct: 317 GKALACKEDVSWNYINRGLEYEHGQEGKVIEKPANKLDDKLKDKLDGKPDDKPKGKPKGR 376
Query: 968 -------------------------DEPPIT-------SAWRAEINGRIPCPPKARGGCG 995
DE P+ S W+A G I C CG
Sbjct: 377 PKGKPDDKPKGKLKGKQDDKPDDKPDEKPVNTDHMKYPSLWKANEAGIITCC------CG 430
Query: 996 TSILSLRRLFEANWVNKLVRNAEELT----IQYHPPSVDLLVGCLQCHRFVVDLAQNSVR 1051
L L+RL W+++LV E+ + P +V C R + D+ ++
Sbjct: 431 AGELVLKRLLPDGWISELVNRVEKTAEAGELLNLPETVLERCPCSNSDRHI-DIDSCNLL 489
Query: 1052 KAASRETNHDNFLYCPDAVDMGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWR 1111
KAA RE + DN+LY P D+ + +HFQ HW++GEPVIVRNV E SGLSW PMVM R
Sbjct: 490 KAACREGSEDNYLYSPSVWDVQQDDLKHFQHHWVKGEPVIVRNVLEATSGLSWEPMVMHR 549
Query: 1112 AFRGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPS 1171
A R + + A+DCLD+CEV++N+ +FF GY +GR R GWP +LKLKDWPP+
Sbjct: 550 ACRQISHVQHGSLKDVVAVDCLDFCEVKVNLHEFFTGYTDGRYDRMGWPLVLKLKDWPPA 609
Query: 1172 NSFE 1175
F+
Sbjct: 610 KVFK 613
>gb|AAN13035.1| unknown protein [Arabidopsis thaliana]
Length = 927
Score = 223 bits (567), Expect = 5e-56
Identities = 200/741 (26%), Positives = 318/741 (41%), Gaps = 101/741 (13%)
Query: 562 REGLENKMLSNLCQEHIEYTQPVVRGGRPKGSRNKKIKLAFQDMVDEVRFANKESDKATC 621
R LE K +C+ H ++Q ++ + K + + K+ + + DEV + E +++
Sbjct: 25 RRALEGK---KMCESH--HSQQSLKRSKQKVAESSKLVRSRRGGGDEVASSEIEPNESRI 79
Query: 622 AVGEEQKDHGSDIGKPIGLDNDKATL--ASDRDQETPNQTLAQDEVQND------KSSVK 673
K +G K + A D+ L + ++Q D K V+
Sbjct: 80 R------------SKRLGKSKRKRVMGEAEAMDEAVKKMKLKRGDLQLDLIRMVLKREVE 127
Query: 674 PKRGRPKGSKNKMKSIANKARNKFGKVRNMRGRPKGSLRKKNETAYCLDSQNERNSLDGR 733
KR R S NK KS N ++F R P G + ++
Sbjct: 128 -KRKRLPNSNNKKKS--NGGFSEFVGEELTRVLPNGIM-----------------AISPP 167
Query: 734 TSTEAAYRNDVDLHRGHCSQEELLRMLS--VEHKNIQGVGVEETIDYGLRSSGLMGDTER 791
+ T + + D+ G EE + M+ KNI+ + + + ++ GD
Sbjct: 168 SPTTSNVSSPCDVKVG----EEPISMIKRRFRSKNIEPLPIGK-----MQVVPFKGDLVN 218
Query: 792 KKETRILRCHQCWRNSWSGVVICAKCKRKQYCYECITKWYPGKTREEIEIACPFCLRNCN 851
++ + +RCH C + ++ C C+R+ +C +CI K G ++EE+E CP C +C
Sbjct: 219 GRKEKKMRCHWCGTRGFGDLISCLSCEREFFCIDCIEKRNKG-SKEEVEKKCPVCRGSCR 277
Query: 852 CRLCLKKDISVM----TGSGEADTGVILQKLLYLLNKTLPLLQHIQREQISEME------ 901
C++C + V + S +D +L L Y + LP+L+ I E E+E
Sbjct: 278 CKVCSVTNSGVTECKDSQSVRSDIDRVLH-LHYAVCMLLPVLKEINAEHKVEVENDAEKK 336
Query: 902 ----VEASMHGSPLMEEDIQ--FDNCNTSIVNFHRSCPNPNCRYDLCLTCCMELRNGLHY 955
E +H S L +D Q + + ++V+ R C + L
Sbjct: 337 EGNPAEPQIHSSELTSDDRQPCSNGRDFAVVDLQRMCTRSSSVLRL-------------- 382
Query: 956 EDIPASGNEETIDEPPITSAWRAEINGRIPCPPKARGGCGTSILSLRRLFEANWVNKLVR 1015
+ ++E++ + I C K GC ++ LF +KL
Sbjct: 383 -NSDQDQSQESLSRKVGSVKCSNGIKSPKVCKRKEVKGCSNNLFL--SLFPLELTSKLEI 439
Query: 1016 NAEELTIQYHPPSV-DLLVGCLQCHRFVVDLAQNSVRKAASRETNHD---NFLYCPDAVD 1071
+AEE+ Y P + D GC C + + + +T D NFLY P +D
Sbjct: 440 SAEEVVSCYELPEILDKYSGCPFCIGMETQSSSSDSHLKEASKTREDGTGNFLYYPTVLD 499
Query: 1072 MGDTEYEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMWRAFRGANKILKEEPTTFKAID 1131
EHFQ HW +G PVIVR+V + S L+W P+ ++ + ++ T D
Sbjct: 500 FHQNNLEHFQTHWSKGHPVIVRSVIKSGSSLNWDPVALF-----CHYLMNRNNKTGNTTD 554
Query: 1132 CLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWPPSNSFEECFPRHGAEFIAMLPF 1191
C+DW EV+I + QFF G L G+ N E LKL+ W S+ F+E FP H AE + +LP
Sbjct: 555 CMDWFEVEIGVKQFFLGSLRGKAETNTCQERLKLEGWLSSSLFKEQFPNHYAEILNILPI 614
Query: 1192 SDYTHPKFGILNLATKLP-AVLKPDLGPKTYIAYGSLEELSRGDSVTKLHCDISDAVNIL 1250
S Y PK G+LN+A LP V PD GP I+Y S EE ++ DSV KL + D V+IL
Sbjct: 615 SHYMDPKRGLLNIAANLPDTVQPPDFGPCLNISYRSGEEYAQPDSVKKLGFETCDMVDIL 674
Query: 1251 THTEEVKAPLWQPKIIKKLQK 1271
+ E Q I+KL K
Sbjct: 675 LYVTETPVSTNQICRIRKLMK 695
Score = 99.8 bits (247), Expect = 7e-19
Identities = 45/102 (44%), Positives = 70/102 (68%), Gaps = 2/102 (1%)
Query: 1438 SNNFFQNDDHMETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQ 1497
S N+ ++ + +G A WD+F++QDV KL EY+K H E ++ V+HP+ +Q
Sbjct: 753 SCNYSCEEESLSNTYG-AQWDVFQKQDVSKLLEYIKNHSLELESMDSSK-KKVSHPLLEQ 810
Query: 1498 ILYLNEKHKRQLKKEYGIEPWTFEQHLGEAVFIPAGCPHQVR 1539
YL+E HK +LK+E+ +EPW+F+Q +GEAV +PAGCP+Q+R
Sbjct: 811 SYYLDEYHKARLKEEFDVEPWSFDQCVGEAVILPAGCPYQIR 852
Score = 42.0 bits (97), Expect = 0.16
Identities = 22/60 (36%), Positives = 26/60 (42%)
Query: 10 RCGRKAPPGWRCTERALSGKSVCERHFLYNQKKTERWKEGASGITPKRRSGRRKPVDNSE 69
RC R WRC RAL GK +CE H K + K S + R G V +SE
Sbjct: 12 RCNRSDGKQWRCKRRALEGKKMCESHHSQQSLKRSKQKVAESSKLVRSRRGGGDEVASSE 71
>ref|XP_450893.1| DNA-binding protein PD3, chloroplast-like [Oryza sativa (japonica
cultivar-group)] gi|49389234|dbj|BAD26544.1| DNA-binding
protein PD3, chloroplast-like [Oryza sativa (japonica
cultivar-group)]
Length = 379
Score = 196 bits (497), Expect = 7e-48
Identities = 100/193 (51%), Positives = 124/193 (63%), Gaps = 3/193 (1%)
Query: 1100 SGLSWHPMVMWRAFRGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGW 1159
S LSW P MW G E KAIDCL CEV+I FF GY EGR Y+N W
Sbjct: 2 SCLSWEPPDMWSKVHGTGT--SPEMKNVKAIDCLSCCEVEICTQDFFNGYYEGRMYQNLW 59
Query: 1160 PEMLKLKDWPPSNSFEECFPRHGAEFIAMLPFSDYTHPKFGILNLATKLP-AVLKPDLGP 1218
PEMLKLKDWP SN FEE P HG +++ LPF YT+ K G+LN++T LP +LK D+GP
Sbjct: 60 PEMLKLKDWPTSNHFEELLPSHGVKYMNSLPFQPYTNLKSGLLNVSTLLPDDILKLDMGP 119
Query: 1219 KTYIAYGSLEELSRGDSVTKLHCDISDAVNILTHTEEVKAPLWQPKIIKKLQKKYEAEDM 1278
K+YIAYG +EL RGDSVTKLHCD+SDAVN+L HT EV Q IK L++++ A++
Sbjct: 120 KSYIAYGYAQELGRGDSVTKLHCDLSDAVNVLMHTAEVDPSEEQIDAIKSLKRRHTAQNE 179
Query: 1279 RDLYGRINKTVVS 1291
++ G + S
Sbjct: 180 KECSGNADGNYTS 192
Score = 138 bits (347), Expect = 2e-30
Identities = 63/92 (68%), Positives = 72/92 (77%)
Query: 1449 ETQFGSAVWDIFRRQDVPKLTEYLKKHHKEFRHANNLPVDSVTHPIHDQILYLNEKHKRQ 1508
ET G A+WDIFRR+DVPKL YL KH KEFRH V V +P+HD+ YL E+HKR+
Sbjct: 209 ETNKGGALWDIFRREDVPKLKLYLDKHSKEFRHIYCSAVQKVCNPVHDETFYLTEEHKRK 268
Query: 1509 LKKEYGIEPWTFEQHLGEAVFIPAGCPHQVRN 1540
LK+E+GIEPWTF Q LGEAVFIPAGCPHQVRN
Sbjct: 269 LKEEHGIEPWTFVQKLGEAVFIPAGCPHQVRN 300
>gb|AAT72490.1| AT1G62310 [Arabidopsis lyrata subsp. petraea]
Length = 205
Score = 192 bits (488), Expect = 8e-47
Identities = 98/212 (46%), Positives = 125/212 (58%), Gaps = 9/212 (4%)
Query: 992 GGCGTSILSLRRLFEANWVNKLVRNAEELTIQYHPPSVDLLVGCLQCHRFVVDLAQNSVR 1051
GGCG +L L+R+ ++ L AE Y+ L C L R
Sbjct: 1 GGCGDCVLELKRILPLTLMSDLEHKAETFLSSYNISPRMLNCRCSS-------LETEMTR 53
Query: 1052 KAASRETNHDNFLYCPDAVDMGDTE-YEHFQRHWIRGEPVIVRNVFEKASGLSWHPMVMW 1110
KAASR + DN+L+CP+++ + E HFQ HW +GEPVIVRN + GLSW PMVMW
Sbjct: 54 KAASRTKSSDNYLFCPESLGVLKEEGLLHFQEHWAKGEPVIVRNTLDNTPGLSWEPMVMW 113
Query: 1111 RAF-RGANKILKEEPTTFKAIDCLDWCEVQINIFQFFKGYLEGRRYRNGWPEMLKLKDWP 1169
RA N + + KAIDCL CEV+IN FF+GY +GR Y N WPEMLKLKDWP
Sbjct: 114 RALCENVNSTASSQMSQVKAIDCLANCEVEINTRHFFEGYSKGRTYENFWPEMLKLKDWP 173
Query: 1170 PSNSFEECFPRHGAEFIAMLPFSDYTHPKFGI 1201
PS+ FE+ PRH EFI+ LPF +Y++P+ GI
Sbjct: 174 PSDKFEDLLPRHCDEFISALPFQEYSNPRTGI 205
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,826,178,032
Number of Sequences: 2540612
Number of extensions: 132777304
Number of successful extensions: 253223
Number of sequences better than 10.0: 443
Number of HSP's better than 10.0 without gapping: 99
Number of HSP's successfully gapped in prelim test: 370
Number of HSP's that attempted gapping in prelim test: 250729
Number of HSP's gapped (non-prelim): 1412
length of query: 1541
length of database: 863,360,394
effective HSP length: 141
effective length of query: 1400
effective length of database: 505,134,102
effective search space: 707187742800
effective search space used: 707187742800
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 82 (36.2 bits)
Lotus: description of TM0103.12


