

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0101.5
(967 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
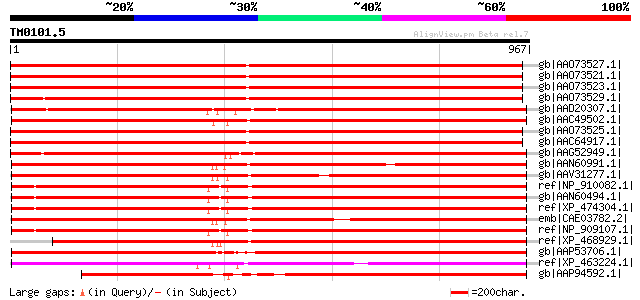
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO73527.1| gag-pol polyprotein [Glycine max] 885 0.0
gb|AAO73521.1| gag-pol polyprotein [Glycine max] 884 0.0
gb|AAO73523.1| gag-pol polyprotein [Glycine max] 879 0.0
gb|AAO73529.1| gag-pol polyprotein [Glycine max] 878 0.0
gb|AAD20307.1| copia-type pol polyprotein [Zea mays] 877 0.0
gb|AAC49502.1| Pol [Zea mays] gi|7489803|pir||T04112 pol protein... 872 0.0
gb|AAO73525.1| gag-pol polyprotein [Glycine max] 867 0.0
gb|AAC64917.1| gag-pol polyprotein [Glycine max] 866 0.0
gb|AAG52949.1| gag/pol polyprotein [Arabidopsis thaliana] 853 0.0
gb|AAN60991.1| Putative Zea mays retrotransposon Opie-2 [Oryza s... 844 0.0
gb|AAV31277.1| putative polyprotein [Oryza sativa (japonica cult... 842 0.0
ref|NP_910082.1| putative gag-pol polyprotein [Oryza sativa (jap... 839 0.0
gb|AAN60494.1| Putative Zea mays retrotransposon Opie-2 [Oryza s... 837 0.0
ref|XP_474304.1| OSJNBb0004A17.2 [Oryza sativa (japonica cultiva... 837 0.0
emb|CAE03782.2| OSJNBa0063G07.6 [Oryza sativa (japonica cultivar... 837 0.0
ref|NP_909107.1| unnamed protein product [Oryza sativa (japonica... 834 0.0
ref|XP_468929.1| putative polyprotein [Oryza sativa (japonica cu... 832 0.0
gb|AAP53706.1| hypothetical protein [Oryza sativa (japonica cult... 827 0.0
ref|XP_463224.1| putative integrase [Oryza sativa (japonica cult... 758 0.0
gb|AAP94592.1| retrotransposon Opie-2 [Zea mays] 728 0.0
>gb|AAO73527.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 885 bits (2287), Expect = 0.0
Identities = 445/956 (46%), Positives = 631/956 (65%), Gaps = 6/956 (0%)
Query: 1 PCIDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIR 60
P ++ VLLV GL NL+SISQL D+G+++ F + C ++ ++ S+ ++N Y
Sbjct: 612 PSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWT 671
Query: 61 LFELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEAC 120
E CL S +E + H+R GH +R + + VRG+P LK +C C
Sbjct: 672 PQETSYSST-CLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGEC 730
Query: 121 QKGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFI 180
Q GK K+S + +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV FI
Sbjct: 731 QIGKQVKMSHQKLRHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFI 790
Query: 181 SRKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTP 240
K E+ VF +++Q E C I R+RSDHG FEN +F S GI ++FS TP
Sbjct: 791 REKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITP 850
Query: 241 QQNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWK 300
QQNG+VERKNRTLQE +R ML ++ + WAEA+NT+CYI NR+++R T YE+WK
Sbjct: 851 QQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWK 910
Query: 301 KVKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEY 360
KP++ +FH FG CY L +++ K D KS + LGYS S+ +R++N+ +T+ E
Sbjct: 911 GRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMES 970
Query: 361 IHVRFDDKLDSDQSKLVEKFVDMSINVSDKGKAPEEAEPEEDSPEEVGPSDPQPQKKSRI 420
I+V DD + + + E + NV+D K+ E AE + + +E + P + +RI
Sbjct: 971 INVVVDDLSPARKKDVEEDVRTLGDNVADAAKSGENAENSDSATDESNINQPDKRSSTRI 1030
Query: 421 VASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEE 480
HPKELI+G+ + V TRS E ++S VS IEPK++ EAL D+ WI AM+E
Sbjct: 1031 QKMHPKELIIGDPNRGVTTRS----REVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQE 1086
Query: 481 ELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYT 540
EL QF +N+VW +V +P+G ++IGTKW+F+NK NE+G + RNKARLVAQGY Q EG+D+
Sbjct: 1087 ELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFD 1146
Query: 541 ETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDH 600
ETFAPVARLE+IRLL+ + L+QMDVKS FLNGY++EEVYV QP GF D +PDH
Sbjct: 1147 ETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDH 1206
Query: 601 VFNLKKSLYGLKQAPRAWYERL-RFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDII 659
V+ LKK+LYGLKQAPRAWYERL FL + + +G +D TLF K ++++I QIYVDDI+
Sbjct: 1207 VYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIV 1266
Query: 660 FGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNM 719
FG ++ + + F + MQ+EFEMS++GEL YFLG+QV Q ++ ++ QS+Y K ++KKF M
Sbjct: 1267 FGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGM 1326
Query: 720 TESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSD 779
+ +T L K++A V Q LYR MI S LYLTASRPDI ++ +CAR+Q++
Sbjct: 1327 ENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQAN 1386
Query: 780 PRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLG 839
P+ +HLT VKRIL+Y+ GT++ G++Y S L GYCDAD+AG +RKSTSG C +LG
Sbjct: 1387 PKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKSTSGGCFYLG 1446
Query: 840 SNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAI 899
+NL+SW SK+Q+ ++LSTAEAEYI+ + ++WMK L++Y + + + +YCDN +AI
Sbjct: 1447 NNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAI 1506
Query: 900 SLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFK 955
++SKNP+ HSR K+I++++H+IRD V + ADIFTK L ++F+
Sbjct: 1507 NISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQFE 1562
>gb|AAO73521.1| gag-pol polyprotein [Glycine max]
Length = 1574
Score = 884 bits (2283), Expect = 0.0
Identities = 445/956 (46%), Positives = 630/956 (65%), Gaps = 6/956 (0%)
Query: 1 PCIDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIR 60
P ++ VLLV GL NL+SISQL D+G+++ F + C ++ ++ S+ ++N Y
Sbjct: 610 PSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWT 669
Query: 61 LFELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEAC 120
E CL S +E + H+R GH +R + + VRG+P LK +C C
Sbjct: 670 PQETSYSST-CLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGEC 728
Query: 121 QKGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFI 180
Q GK K+S + +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWVKFI
Sbjct: 729 QIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVKFI 788
Query: 181 SRKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTP 240
K E+ VF +++Q E C I R+RSDHG FEN + S GI ++FS TP
Sbjct: 789 REKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRLTEFCTSEGITHEFSAAITP 848
Query: 241 QQNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWK 300
QQNG+VERKNRTLQE +R ML ++ + WAEA+NT+CYI NR+++R T YE+WK
Sbjct: 849 QQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWK 908
Query: 301 KVKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEY 360
KP++ +FH FG CY L +++ K D KS + LGYS S+ +R++N+ +T+ E
Sbjct: 909 GRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMES 968
Query: 361 IHVRFDDKLDSDQSKLVEKFVDMSINVSDKGKAPEEAEPEEDSPEEVGPSDPQPQKKSRI 420
I+V DD + + + E NV+D K+ E AE + + +E + P + +RI
Sbjct: 969 INVVVDDLSPARKKDVEEDVRTSGDNVADAAKSGENAENSDSATDESNINQPDKRSSTRI 1028
Query: 421 VASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEE 480
HPKELI+G+ + V TRS E ++S VS IEPK++ EAL D+ WI AM+E
Sbjct: 1029 QKMHPKELIIGDPNRGVTTRS----REVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQE 1084
Query: 481 ELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYT 540
EL QF +N+VW +V +P+G ++IGTKW+F+NK NE+G + RNKARLVAQGY Q EG+D+
Sbjct: 1085 ELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFD 1144
Query: 541 ETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDH 600
ETFAPVARLE+IRLL+ + L+QMDVKS FLNGY++EEVYV QP GF D +PDH
Sbjct: 1145 ETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDH 1204
Query: 601 VFNLKKSLYGLKQAPRAWYERL-RFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDII 659
V+ LKK+LYGLKQAPRAWYERL FL + + +G +D TLF K ++++I QIYVDDI+
Sbjct: 1205 VYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIV 1264
Query: 660 FGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNM 719
FG ++ + + F + MQ+EFEMS++GEL YFLG+QV Q ++ ++ QS+Y K ++KKF M
Sbjct: 1265 FGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGM 1324
Query: 720 TESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSD 779
+ +T L K++A V Q LYR MI S LYLTASRPDI ++ +CAR+Q++
Sbjct: 1325 ENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQAN 1384
Query: 780 PRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLG 839
P+ +HLT VKRIL+Y+ GT++ G++Y S L GYCDAD+AG +RKSTSG C +LG
Sbjct: 1385 PKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKSTSGGCFYLG 1444
Query: 840 SNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAI 899
+NL+SW SK+Q+ ++LSTAEAEYI+ + ++WMK L++Y + + + +YCDN +AI
Sbjct: 1445 NNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAI 1504
Query: 900 SLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFK 955
++SKNP+ HSR K+I++++H+IRD V + ADIFTK L ++F+
Sbjct: 1505 NISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQFE 1560
>gb|AAO73523.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 879 bits (2272), Expect = 0.0
Identities = 443/956 (46%), Positives = 628/956 (65%), Gaps = 6/956 (0%)
Query: 1 PCIDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIR 60
P ++ VLLV GL NL+SISQL D+G+++ F + C ++ ++ S+ ++N Y
Sbjct: 612 PSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWT 671
Query: 61 LFELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEAC 120
E CL S +E + H+R GH +R + + VRG+P LK +C C
Sbjct: 672 PQETSYSST-CLSSKEDEVRIWHQRFGHLHLRGMKKILDKSAVRGIPNLKIEEGRICGEC 730
Query: 121 QKGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFI 180
Q GK K+S + +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV FI
Sbjct: 731 QIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFI 790
Query: 181 SRKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTP 240
K + VF +++Q E C I R+RSDHG FEN +F S GI ++FS TP
Sbjct: 791 REKSGTFEVFKKLSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITP 850
Query: 241 QQNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWK 300
QQNG+VERKNRTLQE +R ML ++ + WAEA+NT+CYI NR+++R T YE+WK
Sbjct: 851 QQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWK 910
Query: 301 KVKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEY 360
KP++ +FH FG CY L +++ K D KS + LGYS S+ +R++N+ +T+ E
Sbjct: 911 GRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMES 970
Query: 361 IHVRFDDKLDSDQSKLVEKFVDMSINVSDKGKAPEEAEPEEDSPEEVGPSDPQPQKKSRI 420
I+V DD + + + E NV+D K+ E AE + + +E + P + +RI
Sbjct: 971 INVVVDDLSPARKKDVEEDVRTSGDNVADAAKSGENAENSDSATDESNINQPDKRSSTRI 1030
Query: 421 VASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEE 480
HPKELI+G+ + V TRS E ++S VS IEPK++ EAL D+ WI AM+E
Sbjct: 1031 QKMHPKELIIGDPNRGVTTRS----REVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQE 1086
Query: 481 ELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYT 540
EL QF +N+VW +V +P+G ++IGTKW+F+NK NE+G + RNKARLVAQGY Q EG+D+
Sbjct: 1087 ELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFD 1146
Query: 541 ETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDH 600
ETFAPVARLE+IRLL+ + L+QMDVKS FLNGY++EEVYV QP GF D +PDH
Sbjct: 1147 ETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDH 1206
Query: 601 VFNLKKSLYGLKQAPRAWYERL-RFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDII 659
V+ LKK+LYGLKQAPRAWYERL FL + + +G +D TLF K ++++I QIYVDDI+
Sbjct: 1207 VYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIV 1266
Query: 660 FGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNM 719
FG ++ + + F + MQ+EFEMS++GEL YFLG+QV Q ++ ++ QS+Y K ++KKF M
Sbjct: 1267 FGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGM 1326
Query: 720 TESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSD 779
+ +T L K++A V QK YR MI S LYLTASRPDI ++ +CAR+Q++
Sbjct: 1327 ENASHKRTPAPTHLKLSKDEAGTSVDQKPYRSMIGSLLYLTASRPDITYAVGVCARYQAN 1386
Query: 780 PRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLG 839
P+ +HL VKRIL+Y+ GT++ G++Y S L GYCDAD+AG +RKSTSG C +LG
Sbjct: 1387 PKISHLNQVKRILKYVNGTSDYGIMYCHCSSSMLVGYCDADWAGSADDRKSTSGGCFYLG 1446
Query: 840 SNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAI 899
+NL+SW SK+Q+ ++LSTAEAEYI+ + ++WMK L++Y + + + +YCDN +AI
Sbjct: 1447 NNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAI 1506
Query: 900 SLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFK 955
++SKNP+ HSR K+I++++H+IRD V + ADIFTK L ++F+
Sbjct: 1507 NISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQFE 1562
>gb|AAO73529.1| gag-pol polyprotein [Glycine max]
Length = 1577
Score = 878 bits (2268), Expect = 0.0
Identities = 443/956 (46%), Positives = 626/956 (65%), Gaps = 6/956 (0%)
Query: 1 PCIDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIR 60
P ++ VLLV GL NL+SISQL D+G+++ F + C ++ ++ S+ ++N Y
Sbjct: 613 PSLNKVLLVKGLTVNLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWT 672
Query: 61 LFELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEAC 120
E + CL S +E + H+R GH +R + + VRG+P LK +C C
Sbjct: 673 PQE-SSHSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGEC 731
Query: 121 QKGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFI 180
Q GK K+S + +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV FI
Sbjct: 732 QIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFI 791
Query: 181 SRKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTP 240
K ++ VF +++Q E C I R+RSDHG FEN KF S GI ++FS TP
Sbjct: 792 REKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITP 851
Query: 241 QQNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWK 300
QQNG+VERKNRTLQE +R ML ++ + WAEA+NT+CYI NR+++R T YE+WK
Sbjct: 852 QQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWK 911
Query: 301 KVKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEY 360
KP + +FH FG CY L +++ K D KS + LGYS S+ +R++N+ +T+ E
Sbjct: 912 GRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMES 971
Query: 361 IHVRFDDKLDSDQSKLVEKFVDMSINVSDKGKAPEEAEPEEDSPEEVGPSDPQPQKKSRI 420
I+V DD + + + E NV+D K+ E AE + + +E + P + RI
Sbjct: 972 INVVVDDLTPARKKDVEEDVRTSGDNVADTAKSAENAENSDSATDEPNINQPDKRPSIRI 1031
Query: 421 VASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEE 480
HPKELI+G+ + V TRS E ++S VS IEPK++ EAL D+ WI AM+E
Sbjct: 1032 QKMHPKELIIGDPNRGVTTRS----REIEIVSNSCFVSKIEPKNVKEALTDEFWINAMQE 1087
Query: 481 ELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYT 540
EL QF +N+VW +V +P+G ++IGTKW+F+NK NE+G + RNKARLVAQGY Q EG+D+
Sbjct: 1088 ELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFD 1147
Query: 541 ETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDH 600
ETFAPVARLE+IRLL+ + L+QMDVKS FLNGY++EE YV QP GF D +PDH
Sbjct: 1148 ETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDH 1207
Query: 601 VFNLKKSLYGLKQAPRAWYERL-RFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDII 659
V+ LKK+LYGLKQAPRAWYERL FL + + +G +D TLF K ++++I QIYVDDI+
Sbjct: 1208 VYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIV 1267
Query: 660 FGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNM 719
FG ++ + + F + MQ+EFEMS++GEL YFLG+QV Q ++ ++ QSKY K ++KKF M
Sbjct: 1268 FGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGM 1327
Query: 720 TESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSD 779
+ +T L K++A V Q LYR MI S LYLTASRPDI ++ +CAR+Q++
Sbjct: 1328 ENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQAN 1387
Query: 780 PRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLG 839
P+ +HL VKRIL+Y+ GT++ G++Y S L GYCDAD+AG +RKSTSG C +LG
Sbjct: 1388 PKISHLNQVKRILKYVNGTSDYGIMYCHCSGSMLVGYCDADWAGSADDRKSTSGGCFYLG 1447
Query: 840 SNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAI 899
+NL+SW SK+Q+ ++LSTAEAEYI+ + ++WMK L++Y + + + +YCDN +AI
Sbjct: 1448 NNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAI 1507
Query: 900 SLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFK 955
++SKNP+ HSR K+I++++H+IR+ V + ADIFTK L +F+
Sbjct: 1508 NISKNPVQHSRTKHIDIRHHYIRELVDDKVITLEHVDTEEQIADIFTKALDAKQFE 1563
>gb|AAD20307.1| copia-type pol polyprotein [Zea mays]
Length = 1063
Score = 877 bits (2266), Expect = 0.0
Identities = 477/1019 (46%), Positives = 622/1019 (60%), Gaps = 66/1019 (6%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
I NV LVD L++NLLS+SQL GY+ +F + D S+ F Y +
Sbjct: 45 ISNVFLVDSLDYNLLSVSQLCQMGYNCLFTDIGVTVFRRSDDSIAFKGVLEGQLYLVDFD 104
Query: 63 ELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQK 122
E CL++ W+ HRRL H M+ + L K + + GL + F D +C ACQ
Sbjct: 105 RAELDT--CLIAKTNMGWLWHRRLAHVGMKNLHKLLKGEHILGLTNVHFEKDRICSACQA 162
Query: 123 GKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFISR 182
GK KN+++T RPLELLH+DLFGP+ SIGG +Y +VIVDDYSR+TWV F+
Sbjct: 163 GKQVGTHHPHKNIMTTDRPLELLHMDLFGPIAYISIGGSKYCLVIVDDYSRFTWVFFLQE 222
Query: 183 KDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQQ 242
K ++ F+ + QNE RI ++RSD+G F+N + ES + GI ++FS P TPQQ
Sbjct: 223 KSQTQETLKGFLRRAQNEFGLRIKKIRSDNGTEFKNSQIESFLEEEGIKHEFSSPYTPQQ 282
Query: 243 NGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKKV 302
NGVVERKNRTL +M+RTML E FWAEAVNT+CY NR+ + IL KT YEL
Sbjct: 283 NGVVERKNRTLLDMARTMLDEYKTPDRFWAEAVNTACYAINRLYLHRILKKTSYELLTGK 342
Query: 303 KPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYIH 362
KPNISYF FG C+ L + R KF K+ + LLGY ++ +R++N +E
Sbjct: 343 KPNISYFRVFGSKCFILIKRGRKSKFAPKTVEGFLLGYDSNTRAYRVFNKSTGLVEVSCD 402
Query: 363 VRFD------------DKLDSDQSKLVEKFVDMSI------------------NVSDKGK 392
V FD D++ +Q+ + +MSI + S +
Sbjct: 403 VVFDETNGSQVEQVDLDEIGEEQAPCIA-LRNMSIGDVCPKESEEPPSTQDQPSSSMQAS 461
Query: 393 APEEAEPEEDSPEEVGPSDPQPQKKSR---------------------------IVASHP 425
P + E E + EE D PQ S I HP
Sbjct: 462 PPTQNEDEAQNDEEQNQEDEPPQDDSNDQGGDTNDQEKEDEEEPRPPHPRVHQAIQRDHP 521
Query: 426 KELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEEELNQF 485
+ ILG+ + V TRS E VS IEP ++EALQD DW++AM+EELN F
Sbjct: 522 VDTILGDIHKGVTTRSRVAHFCEHY----SFVSSIEPHRVEEALQDSDWVVAMQEELNNF 577
Query: 486 SKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYTETFAP 545
++N+VW++V +P +++GTKWVFRNK +E G V RNKARLVA+GY Q EG+D+ ET+AP
Sbjct: 578 TRNEVWHLVPRP-NQNVVGTKWVFRNKQDEHGVVTRNKARLVAKGYSQVEGLDFGETYAP 636
Query: 546 VARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDHVFNLK 605
VARLE+IR+L++++ H L+QMDVKS FLNG I EEVYV QPPGFED + P+HV+ L
Sbjct: 637 VARLESIRILLAYATYHGFKLYQMDVKSAFLNGPIKEEVYVEQPPGFEDSEYPNHVYRLS 696
Query: 606 KSLYGLKQAPRAWYERLR-FLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSAN 664
K+LYGLKQAPRAWYE LR FL+ N F GK D TLF KT ++D+ + QIYVDDIIFGS N
Sbjct: 697 KALYGLKQAPRAWYECLRDFLIANGFKVGKADPTLFTKTLENDLFVCQIYVDDIIFGSTN 756
Query: 665 SSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNMTESII 724
S C+EFS +M +FEMSMMGELKYFLG QV Q E T+I Q+KYT+++L KF M ++
Sbjct: 757 KSTCEEFSRIMTQKFEMSMMGELKYFLGFQVKQLQEGTFICQTKYTQDILTKFGMKDAKP 816
Query: 725 AKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSDPRETH 784
KT M L+ + V QK+YR MI S LYL ASRPDI+ S +CARFQSDP+E+H
Sbjct: 817 IKTPMGTNGHLDLDTGGKSVDQKVYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKESH 876
Query: 785 LTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVS 844
LT VKRILRYL T GL Y + S + L GY DAD+AG + RKSTSG CQFLG +LVS
Sbjct: 877 LTAVKRILRYLAYTPKFGLWYPRGSTFDLIGYSDADWAGCKINRKSTSGTCQFLGRSLVS 936
Query: 845 WASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAISLSKN 904
WASK+Q+++ALSTAEAEYI+ C +LWM+ L DY + +P+ CDN +AI ++ N
Sbjct: 937 WASKKQNSVALSTAEAEYIAAGHCCAQLLWMRQTLLDYGYKLTKVPLLCDNESAIKMADN 996
Query: 905 PILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
P+ HSR K+I ++YHF+RD+ QKG S + ADIFTKPL E F + LN+
Sbjct: 997 PVEHSRTKHIAIRYHFLRDHQQKGDIEISYINTKDQLADIFTKPLDEQSFTRLRHELNI 1055
>gb|AAC49502.1| Pol [Zea mays] gi|7489803|pir||T04112 pol protein homolog - maize
retrotransposon Opie-2
Length = 1068
Score = 872 bits (2254), Expect = 0.0
Identities = 469/1002 (46%), Positives = 616/1002 (60%), Gaps = 46/1002 (4%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
I NV LV+ L +NLLS+SQL + GY+ +F + DGS+ F Y +
Sbjct: 65 ISNVFLVESLGYNLLSVSQLCNMGYNCLFTNIDVSVFRRCDGSLAFKGVLDGKLYLVDFA 124
Query: 63 ELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQK 122
+ E CL++ W+ HRRL H M+ + L K + V GL ++F D C ACQ
Sbjct: 125 KEEAGLDACLIAKTSMGWLWHRRLAHVGMKNLHKLLKGEHVIGLTNVQFEKDRPCAACQA 184
Query: 123 GKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFISR 182
GK S KNV++TSRPLE+LH+DLFGPV SIGG +YG+VIVDD+SR+TWV F+
Sbjct: 185 GKQVGGSHHTKNVMTTSRPLEMLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQE 244
Query: 183 KDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQQ 242
K E+ F+ + QNE ++ ++RSD+G F+N + E + GI ++FS P TPQQ
Sbjct: 245 KSETQGTLKRFLRRAQNEFELKVKKIRSDNGSEFKNLQVEEFLEEEGIKHEFSAPYTPQQ 304
Query: 243 NGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKKV 302
NGVVERKNRTL +M+RTML E + FW EAVNT+C+ NR+ + IL T YEL
Sbjct: 305 NGVVERKNRTLIDMARTMLGEFKTPECFWTEAVNTACHAINRVYLHRILKNTSYELLTGN 364
Query: 303 KPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYIH 362
KPN+SYF FG CY L K R KF K+ + LLGY +K +R++N + +E
Sbjct: 365 KPNVSYFRVFGSKCYILVKKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSGD 424
Query: 363 VRFDDKLDSDQSKLV-------EKFVDMSINVSDKGKAPEEAEPEEDSP----------- 404
V FD+ S + ++V E +I G+ + + E + P
Sbjct: 425 VVFDETNGSPREQVVDCDDVDEEDIPTAAIRTMAIGEVRPQEQDEREQPSPSTMVHPPTQ 484
Query: 405 ----------------------EEVGPSDPQPQKKSRIVASHPKELILGNKDEPVRTRSA 442
EE P Q ++ I HP + ILG+ + V TRS
Sbjct: 485 DDEQVHQQEVCDQGGAQDDHVLEEEAQPAPPTQVRAMIQRDHPVDQILGDISKGVTTRSR 544
Query: 443 FRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHI 502
VS IEP ++EAL D DW+LAM+EELN F +N+VW +V +P+ ++
Sbjct: 545 L----VNFCEHNSFVSSIEPFRVEEALLDPDWVLAMQEELNNFKRNEVWTLVPRPK-QNV 599
Query: 503 IGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNH 562
+GTKWVFRNK +E+G V RNKARLVA+GY Q G+D+ ETFAPVARLE+IR+L++++ +H
Sbjct: 600 VGTKWVFRNKQDERGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLESIRILLAYAAHH 659
Query: 563 NIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERL 622
+ L+QMDVKS FLNG I EEVYV QPPGFEDE+ PDHV L K+LYGLKQAPRAWYE L
Sbjct: 660 SFRLYQMDVKSAFLNGPIKEEVYVEQPPGFEDERYPDHVCKLSKALYGLKQAPRAWYECL 719
Query: 623 R-FLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEM 681
R FL+ N F GK D TLF KT D+ + QIYVDDIIFGS N C+EFS +M +FEM
Sbjct: 720 RDFLIANAFKVGKADPTLFTKTCDGDLFVCQIYVDDIIFGSTNQKSCEEFSRVMTQKFEM 779
Query: 682 SMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDAS 741
SMMGEL YFLG QV Q + T+I Q+KYT++LLK+F M ++ AKT M +
Sbjct: 780 SMMGELNYFLGFQVKQLKDGTFISQTKYTQDLLKRFGMKDAKPAKTPMGTDGHTDLNKGG 839
Query: 742 GKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNL 801
V QK YR MI S LYL ASRPDI+ S +CARFQSDP+E HL VKRILRYL T
Sbjct: 840 KSVDQKAYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKECHLVAVKRILRYLVATPCF 899
Query: 802 GLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAE 861
GL Y K S + L GY D+DYAG + +RKSTSG CQFLG +LVSW SK+Q+++ALSTAEAE
Sbjct: 900 GLWYPKGSTFDLVGYSDSDYAGCKVDRKSTSGTCQFLGRSLVSWNSKKQTSVALSTAEAE 959
Query: 862 YISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFI 921
Y++ C +LWM+ L D+ S +P+ CDN +AI +++NP+ HSR K+I++++HF+
Sbjct: 960 YVAAGQCCAQLLWMRQTLRDFGYNLSKVPLLCDNESAIRMAENPVEHSRTKHIDIRHHFL 1019
Query: 922 RDYVQKGYFF*SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
RD+ QKG + ADIFTKPL E F + LN+
Sbjct: 1020 RDHQQKGDIEVFHVSTENQLADIFTKPLDEKTFCRLRSELNV 1061
>gb|AAO73525.1| gag-pol polyprotein [Glycine max]
Length = 1576
Score = 867 bits (2241), Expect = 0.0
Identities = 440/956 (46%), Positives = 621/956 (64%), Gaps = 6/956 (0%)
Query: 1 PCIDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIR 60
P ++ VLLV GL NL+SISQL D+G+++ F + C ++ ++ S+ ++N Y
Sbjct: 612 PSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWT 671
Query: 61 LFELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEAC 120
E CL S +E + H+R GH +R + + VRG+P LK +C C
Sbjct: 672 PQETSYSST-CLSSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGEC 730
Query: 121 QKGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFI 180
Q GK K+S + +TS LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV FI
Sbjct: 731 QIGKQVKMSHQKLQHQTTSMVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFI 790
Query: 181 SRKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTP 240
K ++ VF +++Q E C I R+RSDHG FEN KF S GI ++FS TP
Sbjct: 791 REKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITP 850
Query: 241 QQNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWK 300
QQNG+VERKNRTLQE +R ML ++ + WAEA+NT+CYI NR+++R T YE+WK
Sbjct: 851 QQNGIVERKNRTLQEATRVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWK 910
Query: 301 KVKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEY 360
KP + +FH FG CY L +++ K D KS + LGYS S+ +R++N+ +T+ E
Sbjct: 911 GRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMES 970
Query: 361 IHVRFDDKLDSDQSKLVEKFVDMSINVSDKGKAPEEAEPEEDSPEEVGPSDPQPQKKSRI 420
I+V DD + + + E NV+D K+ E AE + + +E + P RI
Sbjct: 971 INVVVDDLTPARKKDVEEDVRTSEDNVADTAKSAENAEKSDSTTDEPNINQPDKSPFIRI 1030
Query: 421 VASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEE 480
PKELI+G+ + V TRS E ++S VS IEPK++ EAL D+ WI AM+E
Sbjct: 1031 QKMQPKELIIGDPNRGVTTRS----REIEIVSNSCFVSKIEPKNVKEALTDEFWINAMQE 1086
Query: 481 ELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYT 540
EL QF +N+VW +V +P+G ++IGTKW+F+NK NE+G + RNKARLVAQGY Q EG+D+
Sbjct: 1087 ELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFD 1146
Query: 541 ETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDH 600
ETFAPVARLE+IRLL+ + L+QMDVKS FLNGY++EE YV QP GF D + DH
Sbjct: 1147 ETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHLDH 1206
Query: 601 VFNLKKSLYGLKQAPRAWYERL-RFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDII 659
V+ LKK+LYGLKQAPRAWYERL FL + + +G +D TLF K ++++I QIYVDDI+
Sbjct: 1207 VYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIV 1266
Query: 660 FGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNM 719
FG ++ + + F MQ+EFEMS++GEL YFLG+QV Q ++ ++ QSKY K ++KKF M
Sbjct: 1267 FGGMSNEMLRHFVPQMQSEFEMSLVGELHYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGM 1326
Query: 720 TESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSD 779
+ +T L K++A V Q LYR MI S LYLTASRPDI F+ +CAR+Q++
Sbjct: 1327 ENASHKRTPAPTHLKLSKDEAGTSVDQNLYRSMIGSLLYLTASRPDITFAVGVCARYQAN 1386
Query: 780 PRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLG 839
P+ +HL VKRIL+Y+ GT++ G++Y S+ L GYCDAD+AG +RK TSG C +LG
Sbjct: 1387 PKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSADDRKCTSGGCFYLG 1446
Query: 840 SNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAI 899
+NL+SW SK+Q+ ++LSTAEAEYI+ + ++WMK L++Y + + + +YCDN +AI
Sbjct: 1447 TNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAI 1506
Query: 900 SLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFK 955
++SKNP+ H+R K+I++++H+IRD V + ADIFTK L ++F+
Sbjct: 1507 NISKNPVQHNRTKHIDIRHHYIRDLVDDKIITLEHVDTEEQVADIFTKALDANQFE 1562
>gb|AAC64917.1| gag-pol polyprotein [Glycine max]
Length = 1550
Score = 866 bits (2237), Expect = 0.0
Identities = 439/956 (45%), Positives = 621/956 (64%), Gaps = 6/956 (0%)
Query: 1 PCIDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIR 60
P ++ VLLV GL NL+SISQL D+G+++ F + C ++ ++ S+ ++N Y
Sbjct: 586 PSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWT 645
Query: 61 LFELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEAC 120
E CL S +E + H+R GH +R + + VRG+P LK +C C
Sbjct: 646 PQETSYSST-CLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGEC 704
Query: 121 QKGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFI 180
Q GK K+S + +TSR LELLH+DL GP++ ES+G KRY V+VDD+SR+TWV FI
Sbjct: 705 QIGKQVKMSNQKLQHQTTSRVLELLHMDLMGPMQVESLGRKRYAYVVVDDFSRFTWVNFI 764
Query: 181 SRKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTP 240
K ++ VF +++Q E C I R+RSDHG FEN KF S GI ++FS TP
Sbjct: 765 REKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITP 824
Query: 241 QQNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWK 300
QQNG+VERKNRTLQE +R ML ++ + WAEA+NT+CYI NR+++R T YE+WK
Sbjct: 825 QQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWK 884
Query: 301 KVKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEY 360
KP + +FH G CY L +++ K D KS + LGYS S+ +R++N+ +T+ E
Sbjct: 885 GRKPTVKHFHICGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMES 944
Query: 361 IHVRFDDKLDSDQSKLVEKFVDMSINVSDKGKAPEEAEPEEDSPEEVGPSDPQPQKKSRI 420
I+V DD + + + E NV+D K+ E AE + + +E + P + RI
Sbjct: 945 INVVVDDLTPARKKDVEEDVRTSGDNVADTAKSAENAENSDSATDEPNINQPDKRPSIRI 1004
Query: 421 VASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEE 480
HPKELI+G+ + V TRS E ++S VS IEPK++ EAL D+ WI AM+E
Sbjct: 1005 QKMHPKELIIGDPNRGVTTRS----REIEIISNSCFVSKIEPKNVKEALTDEFWINAMQE 1060
Query: 481 ELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYT 540
EL QF +N+VW +V +P+G ++IGTKW+F+NK NE+G + RNKARLVAQGY Q EG+D+
Sbjct: 1061 ELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFD 1120
Query: 541 ETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDH 600
ETFAP ARLE+IRLL+ + L+QMDVKS FLNGY++EE YV QP GF D +PDH
Sbjct: 1121 ETFAPGARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDH 1180
Query: 601 VFNLKKSLYGLKQAPRAWYERL-RFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDII 659
V+ LKK+LYGLKQAPRAWYERL FL + + +G +D TLF K ++++I QIYVDDI+
Sbjct: 1181 VYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIV 1240
Query: 660 FGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNM 719
FG ++ + + F + MQ+EFEMS++GEL YFLG+QV Q ++ ++ QSKY K ++KKF M
Sbjct: 1241 FGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGM 1300
Query: 720 TESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSD 779
+ +T L K++A V Q LYR MI S LYLTASRPDI ++ CAR+Q++
Sbjct: 1301 ENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGGCARYQAN 1360
Query: 780 PRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLG 839
P+ +HL VKRIL+Y+ GT++ G++Y S+ L GYCDAD+AG +RKST G C +LG
Sbjct: 1361 PKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSVDDRKSTFGGCFYLG 1420
Query: 840 SNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAI 899
+N +SW SK+Q+ ++LSTAEAEYI+ + ++WMK L++Y + + + +YCDN +AI
Sbjct: 1421 TNFISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNLSAI 1480
Query: 900 SLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFK 955
++SKNP+ HSR K+I++++H+IRD V + ADIFTK L ++F+
Sbjct: 1481 NISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLEHVDTEEQIADIFTKALDANQFE 1536
>gb|AAG52949.1| gag/pol polyprotein [Arabidopsis thaliana]
Length = 1643
Score = 853 bits (2204), Expect = 0.0
Identities = 453/987 (45%), Positives = 636/987 (63%), Gaps = 34/987 (3%)
Query: 1 PCIDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIR 60
P + NV V+GL NL+S+SQL D+G + FN+ C A ++ + + L + NN Y
Sbjct: 664 PHLTNVYFVEGLTANLISVSQLCDEGLTVSFNKVKCWATNERNQNTLTGVRTGNNCYM-- 721
Query: 61 LFELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEAC 120
E K+ CL + E+ + H+RLGH + R +S L ++VRG+P LK +C AC
Sbjct: 722 ---WEEPKI-CLRAEKEDPVLWHQRLGHMNARSMSKLVNKEMVRGVPELKHIEKIVCGAC 777
Query: 121 QKGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFI 180
+GK +V K + T++ L+L+H+DL GP++TESI GKRY V+VDD+SR+ WV+FI
Sbjct: 778 NQGKQIRVQHKRVEGIQTTQVLDLIHMDLMGPMQTESIAGKRYVFVLVDDFSRYAWVRFI 837
Query: 181 SRKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTP 240
K E+ + F +Q++NE I ++RSD GG F N+ F S +S GI + +S PRTP
Sbjct: 838 REKSETANSFKILALQLKNEKKMGIKQIRSDRGGEFMNEAFNSFCESQGIFHQYSAPRTP 897
Query: 241 QQNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWK 300
Q NGVVERKNRTLQEM+R M+ + + FWAEA++T+CY+ NR+ VR +KTPYE+WK
Sbjct: 898 QSNGVVERKNRTLQEMARAMIHGNGVPEKFWAEAISTACYVINRVYVRLGSDKTPYEIWK 957
Query: 301 KVKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEY 360
KPN+SYF FGCVCY +N KD+L KFDS+S + LGY+ S +R++N IEE
Sbjct: 958 GKKPNLSYFRVFGCVCYIMNDKDQLGKFDSRSEEGFFLGYATNSLAYRVWNKQRGKIEES 1017
Query: 361 IHVRFDDKLDSDQSKLVEKFVDMSINVSDK-GKAPEEAEPE--------EDSPEEVGPS- 410
++V FDD + +V + ++S+ G+ + + + E+S EEV P+
Sbjct: 1018 MNVVFDDGSMPELQIIVRNRNEPQTSISNNHGEERNDNQFDNGDINKSGEESDEEVPPAQ 1077
Query: 411 ------------DPQPQKKSRIVASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVS 458
DP ++ +R V ++L G K + R ++F EE + S VS
Sbjct: 1078 VHRDHASKDIIGDPSGERVTRGVKQDYRQL-AGIKQKH-RVMASFACFEEIMFSC--FVS 1133
Query: 459 LIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGD 518
++EPK++ EAL+D WILAMEEEL +FS++ VW++V +P V++IGTKW+F+NK +E G+
Sbjct: 1134 IVEPKNVKEALEDHFWILAMEEELEEFSRHQVWDLVPRPPQVNVIGTKWIFKNKFDEVGN 1193
Query: 519 VVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNG 578
+ RNKARLVAQGY Q EG+D+ ETFAPVARLE IR L+ + LHQMDVK FLNG
Sbjct: 1194 ITRNKARLVAQGYTQVEGLDFDETFAPVARLECIRFLLGTACGMGFKLHQMDVKCAFLNG 1253
Query: 579 YISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERL-RFLLENEFVRGKVDT 637
I EEVYV QP GFE+ + P++V+ LKK+LYGLKQAPRAWYERL FL+ + RG VD
Sbjct: 1254 IIEEEVYVEQPKGFENLEFPEYVYKLKKALYGLKQAPRAWYERLTTFLIVQGYTRGSVDK 1313
Query: 638 TLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQ 697
TLF K I+I+QIYVDDI+FG + L K F + M EF MSM+GELKYFLG+Q++Q
Sbjct: 1314 TLFVKNDVHGIIIIQIYVDDIVFGGTSDKLVKTFVKTMTTEFRMSMVGELKYFLGLQINQ 1373
Query: 698 TPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFL 757
T E I QS Y + L+K+F M S A T M T L K++ KV +KLYRGMI S L
Sbjct: 1374 TDEGITISQSTYAQNLVKRFGMCSSKPAPTPMSTTTKLFKDEKGVKVDEKLYRGMIGSLL 1433
Query: 758 YLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYC 817
YLTA+RPD+ S LCAR+QS+P+ +HL VKRI++Y+ GT N GL Y + + L GYC
Sbjct: 1434 YLTATRPDLCLSVGLCARYQSNPKASHLLAVKRIIKYVSGTINYGLNYTRDTSLVLVGYC 1493
Query: 818 DADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKH 877
DAD+ G+ +R+ST+G FLGSNL+SW SK+Q+ ++LS+ ++EYI++ C T +LWM+
Sbjct: 1494 DADWGGNLDDRRSTTGGVFFLGSNLISWHSKKQNCVSLSSTQSEYIALGSCCTQLLWMRQ 1553
Query: 878 QLEDY-QILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLI 936
DY + + CDN +AI++SKNP+ HS K+I +++HF+R+ V++ +
Sbjct: 1554 MGLDYGMTFPDPLLVKCDNESAIAISKNPVQHSVTKHIAIRHHFVRELVEEKQITVEHVP 1613
Query: 937 LTINSADIFTKPLAEDRFKFILKNLNM 963
I DIFTKPL + F + K+L +
Sbjct: 1614 TEIQLVDIFTKPLDLNTFVNLQKSLGI 1640
>gb|AAN60991.1| Putative Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)] gi|34902324|ref|NP_912508.1| Putative
Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)]
Length = 2011
Score = 844 bits (2181), Expect = 0.0
Identities = 462/988 (46%), Positives = 609/988 (60%), Gaps = 45/988 (4%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
IDNV LV LN NLLS++Q+ D F + S +D S +F R N Y
Sbjct: 612 IDNVSLVKSLNFNLLSVAQICDLVLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYLGDFN 671
Query: 63 ELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQK 122
E CL++ W+ HRRL H M ++S L K DLV GL +KF D LC ACQ
Sbjct: 672 SSEANLKTCLVAKTSLGWLWHRRLAHVGMNQLSKLSKRDLVVGLKDVKFEKDKLCSACQA 731
Query: 123 GKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFISR 182
GK S K+++STSRPLELLH++ FGP +SIGG + +VIVDDYSR+TW+ F+
Sbjct: 732 GKQVACSHPTKSIMSTSRPLELLHMEFFGPTTYKSIGGNSHCLVIVDDYSRYTWMFFLHD 791
Query: 183 KDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQQ 242
K +F F + QNE C ++++RSD+G F+N E D GI ++ S +PQQ
Sbjct: 792 KSIVAELFKKFAKRGQNEFNCTLVKIRSDNGSKFKNTNIEDYCDDLGIKHELSATYSPQQ 851
Query: 243 NGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKKV 302
NGVVE KNRTL EM+RTML E ++ FWAEA+NT+C+ NR+ + +L KT YEL
Sbjct: 852 NGVVEMKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHRLLKKTSYELIVGR 911
Query: 303 KPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYIH 362
KPN++YF FGC CY RL KF+S+ + LLGY+ SK +R+YN + +EE
Sbjct: 912 KPNVAYFRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKAYRVYNKNKGIVEETAD 971
Query: 363 VRFDDKLDSDQSK----------LVEKFVDMS------INVSDKGKAPEEAEP------- 399
V+FD+ S + L+ +MS I V DK + EP
Sbjct: 972 VQFDETNGSQEGHENLDDVGDEGLMRAMKNMSIGDVKPIEVEDKPSTSTQDEPSTSATPS 1031
Query: 400 --EEDSPEEVGPSDPQPQK-KSRIVASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGL 456
+ + EE P P + + + HP + +LG+ + V+TRS ++
Sbjct: 1032 QAQVEVEEEKAQDLPMPPRIHTALSKDHPIDQVLGDISKGVQTRSRV----ASICEHYSF 1087
Query: 457 VSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEK 516
VS +EPK +DEAL D DWI AM +ELN F++N VW +V++ + ++IGTKWVFRNK +E
Sbjct: 1088 VSCLEPKHVDEALCDPDWINAMHKELNNFARNKVWTLVERLRDHNVIGTKWVFRNKQDEN 1147
Query: 517 GDVVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFL 576
G VVRNKAR VAQG+ Q EG+D+ ETFAPV RLEAI +L++F+ NI L QMDVKS FL
Sbjct: 1148 GLVVRNKARFVAQGFTQVEGLDFGETFAPVTRLEAICILLAFASCFNIKLFQMDVKSAFL 1207
Query: 577 NGYISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERLR-FLLENEFVRGKV 635
NG I+E V+V QPPGFED K P+HV+ L K+LYGLKQAPRAWYERLR FLL +F GKV
Sbjct: 1208 NGEIAELVFVEQPPGFEDPKYPNHVYKLSKALYGLKQAPRAWYERLRDFLLSKDFKIGKV 1267
Query: 636 DTTLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQV 695
DTTLF K DD + QIYVDDIIFG N CKEF +MM EFEMSM+GEL +F G+Q+
Sbjct: 1268 DTTLFTKIIGDDFFVCQIYVDDIIFGCTNEVFCKEFGDMMSREFEMSMIGELSFFHGLQI 1327
Query: 696 DQTPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICS 755
Q + T F + ++ KT M L+ ++ V KLYR MI S
Sbjct: 1328 KQLKDGT--------------FGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGS 1373
Query: 756 FLYLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSG 815
LYLTASRPDI+FS +CARFQ+ P+E HL VKRILRYLK ++ +GL Y K +++KL G
Sbjct: 1374 LLYLTASRPDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTIGLWYPKGAKFKLVG 1433
Query: 816 YCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWM 875
Y D+DYAG + +RKSTSG+CQ LG +LVSW+SK+Q+ +AL AEAEY+S C +LWM
Sbjct: 1434 YSDSDYAGCKVDRKSTSGSCQMLGRSLVSWSSKKQNFVALFIAEAEYVSAGSCCAQLLWM 1493
Query: 876 KHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSL 935
K L DY I + P+ C+N +AI ++ NP+ HSR K+I++++HF+RD+V K S +
Sbjct: 1494 KQILLDYGISFTKTPLLCENDSAIKIANNPVQHSRTKHIDIRHHFLRDHVAKCDIVISHI 1553
Query: 936 ILTINSADIFTKPLAEDRFKFILKNLNM 963
ADIFTKPL E RF + LN+
Sbjct: 1554 RTEDQLADIFTKPLDETRFCKLRNELNL 1581
>gb|AAV31277.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1577
Score = 842 bits (2175), Expect = 0.0
Identities = 457/988 (46%), Positives = 607/988 (61%), Gaps = 47/988 (4%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
IDNV LV LN NLLS++Q+ D F + S +D S +F R N Y +
Sbjct: 603 IDNVSLVKSLNFNLLSVAQICDLSLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYLVDFN 662
Query: 63 ELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQK 122
E CL++ W+ HRRL H M ++S K DLV GL +KF D LC ACQ
Sbjct: 663 SSEANLKTCLVAKTSLGWLWHRRLAHVGMNQLSKFSKRDLVMGLKDVKFEKDKLCSACQA 722
Query: 123 GKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFISR 182
GK S K+++STS+PLELLH+DLF P +SIGG + +VIVDDYSR+TWV F+
Sbjct: 723 GKQVACSHPTKSIMSTSKPLELLHMDLFDPTTYKSIGGNSHCLVIVDDYSRYTWVFFLHD 782
Query: 183 KDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQQ 242
K +F F + QNE +C ++++RS+ G F+N E D GI ++ +PQQ
Sbjct: 783 KSIVADLFKKFAKRAQNEFSCTLVKIRSNIGSEFKNTNIEDYCDDLGIKHELFATYSPQQ 842
Query: 243 NGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKKV 302
NGVVERKNRTL EM+RTML E ++ FWAEA+NT+C+ NR+ + +L KT YE+
Sbjct: 843 NGVVERKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHRVLKKTSYEVIVGR 902
Query: 303 KPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYIH 362
KPNI+YF FGC CY RL KF+S+ + LLGY+ +SK +R+YN + +EE
Sbjct: 903 KPNIAYFRVFGCKCYIHRKGVRLTKFESRCDEGFLLGYASKSKAYRVYNKNKGIVEETAD 962
Query: 363 VRFDDKLDSDQSK----------LVEKFVDMSIN------VSDKGKAPEEAEPEEDS--- 403
V+FD+ S + L+ +MSI V DK + EP +
Sbjct: 963 VQFDETNGSQEGHENLDDVGDEGLMRVMKNMSIGDVKPIEVEDKPSTSTQDEPSTSAMPS 1022
Query: 404 ------PEEVGPSDPQPQK-KSRIVASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGL 456
EE P P + + + HP + +LG+ + V+TRS ++
Sbjct: 1023 QAQVEVEEEKAQEPPMPPRIHTALSKDHPIDQVLGDISKGVQTRSRVT----SICEHYSF 1078
Query: 457 VSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEK 516
VS +E K +DEAL D DW+ AM EEL F++N VW +V++P+ ++IGTKWVFRNK +E
Sbjct: 1079 VSCLERKHVDEALCDPDWMNAMHEELKNFARNKVWTLVERPRDHNVIGTKWVFRNKQDEN 1138
Query: 517 GDVVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFL 576
G VVRNKARLVAQG+ Q EG+D+ ETFAPVARLEAI +L++F+ +I L QMDVKS FL
Sbjct: 1139 GLVVRNKARLVAQGFTQVEGLDFGETFAPVARLEAICILLAFASCFDIKLFQMDVKSAFL 1198
Query: 577 NGYISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERLR-FLLENEFVRGKV 635
N D K P+HV+ L K+LYGL+QAPRAWYERLR FLL +F GKV
Sbjct: 1199 N----------------DTKYPNHVYKLSKALYGLRQAPRAWYERLRDFLLSKDFKIGKV 1242
Query: 636 DTTLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQV 695
D TLF K DD + QIYVDDIIFGS N CKEF +MM EFEMSM+GEL +FLG+Q+
Sbjct: 1243 DITLFTKIIGDDFFVYQIYVDDIIFGSTNEVFCKEFGDMMSREFEMSMIGELSFFLGLQI 1302
Query: 696 DQTPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICS 755
Q T++ Q+KY K+LLK+F + ++ KT M L+ ++ V KLYR MI S
Sbjct: 1303 KQLKNGTFVSQTKYIKDLLKRFGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGS 1362
Query: 756 FLYLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSG 815
LYLT SRPDI+FS +CARFQ+ P+E HL VKRILRYLK ++ +GL Y K +++KL G
Sbjct: 1363 LLYLTVSRPDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTIGLWYPKGAKFKLVG 1422
Query: 816 YCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWM 875
Y D DYAG + +RKSTS +CQ LG +LVSW+SK+Q+++ALSTAE EY+S C +LWM
Sbjct: 1423 YSDPDYAGCKVDRKSTSSSCQMLGRSLVSWSSKKQNSVALSTAETEYVSAGSCCAQLLWM 1482
Query: 876 KHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSL 935
K L DY I + P+ CDN AI ++ NP+ HSR K+I++++HF+RD+V K S +
Sbjct: 1483 KQTLLDYGISFTKTPLLCDNDGAIKIANNPVQHSRTKHIDIRHHFLRDHVAKCDIVISHI 1542
Query: 936 ILTINSADIFTKPLAEDRFKFILKNLNM 963
ADIFTKPL E RF + LN+
Sbjct: 1543 RTEDQLADIFTKPLDETRFCKLRNELNI 1570
>ref|NP_910082.1| putative gag-pol polyprotein [Oryza sativa (japonica cultivar-group)]
gi|28269414|gb|AAO37957.1| putative gag-pol polyprotein
[Oryza sativa (japonica cultivar-group)]
Length = 1969
Score = 839 bits (2167), Expect = 0.0
Identities = 447/991 (45%), Positives = 613/991 (61%), Gaps = 39/991 (3%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
+ +V LV L +NLLS+SQL D+ ++ F + R + + V F+ R +
Sbjct: 807 LKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFD 865
Query: 63 ELETQKVKCLL-SVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQ 121
+CL+ S N + + HRRLGH +S + +DL+RGLP LK D +C C+
Sbjct: 866 SSAPGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVQKDLVCAPCR 925
Query: 122 KGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFIS 181
GK T S K +V T P +LLH+D GP + +S+GGK Y +V+VDD+SR++WV F+
Sbjct: 926 HGKMTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLE 985
Query: 182 RKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQ 241
K+E+ F + + E + +RSD+G F+N FES DS G+ + FS P PQ
Sbjct: 986 SKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQ 1045
Query: 242 QNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKK 301
QNGVVERKNRTL EM+RTML E + FW EA++ +C+I NR+ +R IL+KTPYEL
Sbjct: 1046 QNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFG 1105
Query: 302 VKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYI 361
+P +S+ FGC C+ L + + L KF+S+S + LGY+ S+ +R+Y I E
Sbjct: 1106 RRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETC 1164
Query: 362 HVRFDDKL---------------------DSDQSKLVEKFVDMSINVSDKGKAPEEAEPE 400
V FD+ D D + +D + V + G +P P
Sbjct: 1165 EVTFDEASPGARPEISGVPDESIFVDEDSDDDDDDSIPPPLDSTPPVQETG-SPSTTSPS 1223
Query: 401 EDSP-------EEVGPSDPQPQKKSRIVASHPKELILGNKDEPVRTRSAFRPSEETLLSL 453
D+P EE+ P I HP + ++G E V ++ L
Sbjct: 1224 GDAPTTSSSAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYE------LVN 1277
Query: 454 KGLVSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKL 513
V+ EPK++ AL D++W+ AM EEL F +N VW++V+ P G ++IGTKWVF+NKL
Sbjct: 1278 SAFVASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKL 1337
Query: 514 NEKGDVVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKS 573
E G +VRNKARLVAQG+ Q EG+D+ ETFAPVARLEAIR+L++F+ + L QMDVKS
Sbjct: 1338 GEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKS 1397
Query: 574 VFLNGYISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERLR-FLLENEFVR 632
FLNG I EEVYV QPPGFE+ K P+HVF L+K+LYGLKQAPRAWYERL+ FLL+N F
Sbjct: 1398 AFLNGVIEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGFEM 1457
Query: 633 GKVDTTLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLG 692
G VD TLF D L+VQIYVDDIIFG ++ +L +FS++M EFEMSMMGEL +FLG
Sbjct: 1458 GAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLG 1517
Query: 693 IQVDQTPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGM 752
+Q+ QT E ++HQ+KY+KELLKKF+M + T M T L ++ +V Q+ YR M
Sbjct: 1518 LQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSM 1577
Query: 753 ICSFLYLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYK 812
I S LYLTASRPDI FS LCARFQ+ PR +H VKRI RY+K T G+ Y +S
Sbjct: 1578 IGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRIFRYIKSTLEYGIWYSCSSALS 1637
Query: 813 LSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*M 872
+ + DAD+AG + +RKSTSG C FLG++LVSW+S++QS++A STAEAEY++ A + +
Sbjct: 1638 VRAFSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAASACSQV 1697
Query: 873 LWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF* 932
LWM L+DY + S +P+ CDNT+AI+++KNP+ HSR K+IE++YHF+RD V+KG
Sbjct: 1698 LWMISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEKGTIVL 1757
Query: 933 SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
+ ADIFTKPL RF+F+ L +
Sbjct: 1758 EFVESEKQLADIFTKPLDRSRFEFLRSELGV 1788
>gb|AAN60494.1| Putative Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)] gi|34902378|ref|NP_912535.1| Putative
Zea mays retrotransposon Opie-2 [Oryza sativa (japonica
cultivar-group)]
Length = 2145
Score = 837 bits (2163), Expect = 0.0
Identities = 446/991 (45%), Positives = 613/991 (61%), Gaps = 39/991 (3%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
+ +V LV L +NLLS+SQL D+ ++ F + R + + V F+ R +
Sbjct: 807 LKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFD 865
Query: 63 ELETQKVKCLL-SVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQ 121
+CL+ S N + + HRRLGH +S + +DL+RGLP LK D +C C+
Sbjct: 866 SSAPGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKDLVCAPCR 925
Query: 122 KGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFIS 181
GK T S K +V T P +LLH+D GP + +S+GGK Y +V+VDD+SR++WV F+
Sbjct: 926 HGKMTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLE 985
Query: 182 RKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQ 241
K+E+ F + + E + +RSD+G F+N FES DS G+ + FS P PQ
Sbjct: 986 SKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQ 1045
Query: 242 QNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKK 301
QNGVVERKNRTL EM+RTML E + FW EA++ +C+I NR+ +R IL+KTPYEL
Sbjct: 1046 QNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFG 1105
Query: 302 VKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYI 361
+P +S+ FGC C+ L + + L KF+S+S + LGY+ S+ +R+Y I E
Sbjct: 1106 RRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETC 1164
Query: 362 HVRFDDKL---------------------DSDQSKLVEKFVDMSINVSDKGKAPEEAEPE 400
V FD+ D D + +D + V + G +P P
Sbjct: 1165 EVTFDEASPGARPEISGVPDESIFVDEDSDDDDDDSIPPPLDSTPPVQETG-SPSTTSPS 1223
Query: 401 EDSP-------EEVGPSDPQPQKKSRIVASHPKELILGNKDEPVRTRSAFRPSEETLLSL 453
D+P EE+ P I HP + ++G E V ++ L
Sbjct: 1224 GDAPTTSSSAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYE------LVN 1277
Query: 454 KGLVSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKL 513
V+ EPK++ AL D++W+ AM EEL F +N VW++V+ P G ++IGTKWVF+NKL
Sbjct: 1278 SAFVASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKL 1337
Query: 514 NEKGDVVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKS 573
E G +VRNKARLVAQG+ Q EG+D+ ETFAPVARLEAIR+L++F+ + L QMDVKS
Sbjct: 1338 GEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKS 1397
Query: 574 VFLNGYISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERLR-FLLENEFVR 632
FLNG I EEVYV QPPGFE+ K P+HVF L+K+LYGLKQAPRAWYERL+ FLL+N F
Sbjct: 1398 AFLNGVIEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGFEM 1457
Query: 633 GKVDTTLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLG 692
G VD TLF D L+VQIYVDDIIFG ++ +L +FS++M EFEMSMMGEL +FLG
Sbjct: 1458 GAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLG 1517
Query: 693 IQVDQTPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGM 752
+Q+ QT E ++HQ+KY+KELLKKF+M + T M T L ++ +V Q+ YR M
Sbjct: 1518 LQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSM 1577
Query: 753 ICSFLYLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYK 812
I S LYLTASRPDI FS LCARFQ+ PR +H VKR+ RY+K T G+ Y +S
Sbjct: 1578 IGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRMFRYIKSTLEYGIWYSCSSALS 1637
Query: 813 LSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*M 872
+ + DAD+AG + +RKSTSG C FLG++LVSW+S++QS++A STAEAEY++ A + +
Sbjct: 1638 VRAFSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAASACSQV 1697
Query: 873 LWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF* 932
LWM L+DY + S +P+ CDNT+AI+++KNP+ HSR K+IE++YHF+RD V+KG
Sbjct: 1698 LWMISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEKGTIVL 1757
Query: 933 SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
+ ADIFTKPL RF+F+ L +
Sbjct: 1758 EFVESEKQLADIFTKPLDRSRFEFLRSELGV 1788
>ref|XP_474304.1| OSJNBb0004A17.2 [Oryza sativa (japonica cultivar-group)]
gi|32488723|emb|CAE03600.1| OSJNBb0004A17.2 [Oryza sativa
(japonica cultivar-group)]
Length = 1877
Score = 837 bits (2163), Expect = 0.0
Identities = 446/991 (45%), Positives = 613/991 (61%), Gaps = 39/991 (3%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
+ +V LV L +NLLS+SQL D+ ++ F + R + + V F+ R +
Sbjct: 889 LKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFD 947
Query: 63 ELETQKVKCLL-SVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQ 121
+CL+ S N + + HRRLGH +S + +DL+RGLP LK D +C C+
Sbjct: 948 SSAPGPSRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKDLVCAPCR 1007
Query: 122 KGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFIS 181
GK T S K +V T P +LLH+D GP + +S+GGK Y +V+VDD+SR++WV F+
Sbjct: 1008 HGKMTSSSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLE 1067
Query: 182 RKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQ 241
K+E+ F + + E + +RSD+G F+N FES DS G+ + FS P PQ
Sbjct: 1068 SKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQ 1127
Query: 242 QNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKK 301
QNGVVERKNRTL EM+RTML E + FW EA++ +C+I NR+ +R IL+KTPYEL
Sbjct: 1128 QNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFG 1187
Query: 302 VKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYI 361
+P +S+ FGC C+ L + + L KF+S+S + LGY+ S+ +R+Y I E
Sbjct: 1188 RRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETC 1246
Query: 362 HVRFDDKL---------------------DSDQSKLVEKFVDMSINVSDKGKAPEEAEPE 400
V FD+ D D + +D + V + G +P P
Sbjct: 1247 EVTFDEASPGARPEISGVPDESIFVDEDSDDDDDDSIPPPLDSTPPVQETG-SPSTTSPS 1305
Query: 401 EDSP-------EEVGPSDPQPQKKSRIVASHPKELILGNKDEPVRTRSAFRPSEETLLSL 453
D+P EE+ P I HP + ++G E V ++ L
Sbjct: 1306 GDAPTTSSSAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYE------LVN 1359
Query: 454 KGLVSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKL 513
V+ EPK++ AL D++W+ AM EEL F +N VW++V+ P G ++IGTKWVF+NKL
Sbjct: 1360 SAFVASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKL 1419
Query: 514 NEKGDVVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKS 573
E G +VRNKARLVAQG+ Q EG+D+ ETFAPVARLEAIR+L++F+ + L QMDVKS
Sbjct: 1420 GEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKS 1479
Query: 574 VFLNGYISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERLR-FLLENEFVR 632
FLNG I EEVYV QPPGFE+ K P+HVF L+K+LYGLKQAPRAWYERL+ FLL+N F
Sbjct: 1480 AFLNGVIEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGFEM 1539
Query: 633 GKVDTTLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLG 692
G VD TLF D L+VQIYVDDIIFG ++ +L +FS++M EFEMSMMGEL +FLG
Sbjct: 1540 GAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLG 1599
Query: 693 IQVDQTPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGM 752
+Q+ QT E ++HQ+KY+KELLKKF+M + T M T L ++ +V Q+ YR M
Sbjct: 1600 LQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSM 1659
Query: 753 ICSFLYLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYK 812
I S LYLTASRPDI FS LCARFQ+ PR +H VKR+ RY+K T G+ Y +S
Sbjct: 1660 IGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRMFRYIKSTLEYGIWYSCSSALS 1719
Query: 813 LSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*M 872
+ + DAD+AG + +RKSTSG C FLG++LVSW+S++QS++A STAEAEY++ A + +
Sbjct: 1720 VRAFSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAASACSQV 1779
Query: 873 LWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF* 932
LWM L+DY + S +P+ CDNT+AI+++KNP+ HSR K+IE++YHF+RD V+KG
Sbjct: 1780 LWMISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEKGTIVL 1839
Query: 933 SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
+ ADIFTKPL RF+F+ L +
Sbjct: 1840 EFVESEKQLADIFTKPLDRSRFEFLRSELGV 1870
>emb|CAE03782.2| OSJNBa0063G07.6 [Oryza sativa (japonica cultivar-group)]
gi|50923031|ref|XP_471876.1| OSJNBa0063G07.6 [Oryza
sativa (japonica cultivar-group)]
Length = 1539
Score = 837 bits (2161), Expect = 0.0
Identities = 455/987 (46%), Positives = 604/987 (61%), Gaps = 58/987 (5%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
IDNV V LN NLLS++Q+ D G F + S +D S +F R N Y +
Sbjct: 578 IDNVSFVKSLNFNLLSVAQICDLGLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYFVDFN 637
Query: 63 ELETQKVKCLLSVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQK 122
E CL++ W+ HRRL H M ++S L K DLV GL +KF D LC ACQ
Sbjct: 638 SSEANLKTCLVAKTSLGWLWHRRLAHVGMNQLSKLSKRDLVVGLKDVKFEKDKLCSACQA 697
Query: 123 GKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFISR 182
GK S K+++STSRPLELLH+DLFGP +SIGG + +VIVDDYSR+TWV F+
Sbjct: 698 GKQVACSHPTKSIMSTSRPLELLHMDLFGPTTYKSIGGNSHCLVIVDDYSRYTWVFFLHD 757
Query: 183 KDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQQ 242
K +F + QNE +C ++++RSD+G F+N E D GI ++ S +PQQ
Sbjct: 758 KSIVAELFKKIAKRAQNEFSCTLVKIRSDNGSEFKNTNIEDYCDDLGIKHELSATYSPQQ 817
Query: 243 NGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKKV 302
NGVVERKNRTL EM+RTML E ++ FWAEA+NT+C+ NR + +L T YEL
Sbjct: 818 NGVVERKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRFYLHRLLKNTSYELIVGR 877
Query: 303 KPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYIH 362
KPN++YF FGC CY RL KF+S+ + LLGY+ SK +R+YN + T+EE
Sbjct: 878 KPNVAYFRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKAYRVYNKNKGTVEETAD 937
Query: 363 VRFDDKLDSDQSK----------LVEKFVDMS------INVSDKGKAPEEAEPE------ 400
V+FD+ S + L+ +MS I V DK + EP
Sbjct: 938 VQFDETNGSQEGHENLDDVGDEGLIRAMKNMSFGDVKPIEVEDKPSTSTQDEPSTFATPS 997
Query: 401 --EDSPEEVGPSDP--QPQKKSRIVASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGL 456
+ EE DP P+ + + HP + +LG+ + V+T S ++
Sbjct: 998 QAQVEVEEEKAQDPPIPPRIHTTLSKDHPIDQVLGDISKGVQTLSRV----ASICEHYSF 1053
Query: 457 VSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEK 516
VS +EPK +DEAL D DW+ AM EELN F++N VW +V++P+ ++IGTKWVFRNK +E
Sbjct: 1054 VSCLEPKHVDEALCDPDWMNAMHEELNNFARNKVWTLVERPRDHNVIGTKWVFRNKQDEN 1113
Query: 517 GDVVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFL 576
G VVRNKARLVAQG+ Q EG+D+ ETFAPVARLEAI +L++F+ +I L QMDVKS FL
Sbjct: 1114 GLVVRNKARLVAQGFTQVEGLDFGETFAPVARLEAICILLAFASWFDIKLFQMDVKSAFL 1173
Query: 577 NGYISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERLRFLLENEFVRGKVD 636
NG I+E V+V QPPGFED K P+H F + GKVD
Sbjct: 1174 NGEIAELVFVEQPPGFEDPKYPNHDFKI----------------------------GKVD 1205
Query: 637 TTLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVD 696
TTLF K DD + QIYVDDIIFGS N CKEF +MM EFEMSM+GEL +FLG+Q+
Sbjct: 1206 TTLFTKIIGDDFFVCQIYVDDIIFGSTNEVFCKEFGDMMSREFEMSMIGELSFFLGLQIK 1265
Query: 697 QTPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSF 756
Q + T++ Q+KY K+LLK+F + ++ KT M L+ ++ V KLYR MI S
Sbjct: 1266 QLKDGTFVSQTKYIKDLLKRFGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGSL 1325
Query: 757 LYLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGY 816
LYLTASRPDI+FS +CA FQ+ P+E HL VKRILRYLK ++ +GL Y K +++KL GY
Sbjct: 1326 LYLTASRPDIMFSVCMCAWFQAAPKECHLVAVKRILRYLKYSSTIGLWYPKGAKFKLVGY 1385
Query: 817 CDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMK 876
D+DYAG + +R STSG+CQ LG +LVSW+SK+Q+++ALSTAEAEY+S C +LWMK
Sbjct: 1386 SDSDYAGCKVDRNSTSGSCQMLGRSLVSWSSKKQNSVALSTAEAEYVSAGSCCAQLLWMK 1445
Query: 877 HQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLI 936
L DY I + P+ CDN +AI ++ NP+ HSR K+I++++HF+RD+V K S +
Sbjct: 1446 QTLLDYGISFTKTPLLCDNDSAIKIANNPVQHSRTKHIDIRHHFLRDHVAKCDIVISHIR 1505
Query: 937 LTINSADIFTKPLAEDRFKFILKNLNM 963
ADIFTKPL E RF + LN+
Sbjct: 1506 TEDQLADIFTKPLDETRFCKLRNELNV 1532
>ref|NP_909107.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 1410
Score = 834 bits (2155), Expect = 0.0
Identities = 446/991 (45%), Positives = 612/991 (61%), Gaps = 39/991 (3%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
+ +V LV L +NLLS+SQL D+ ++ F + R + + V F+ R +
Sbjct: 422 LKDVALVSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFD 480
Query: 63 ELETQKVKCLL-SVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQ 121
+CL+ S N + + HRRLGH +S + +DL+RGLP LK D +C C+
Sbjct: 481 SSAPGPSRCLVASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKAPKDLVCAPCR 540
Query: 122 KGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFIS 181
GK T S K +V T P +LLH++ GP + +S+GGK Y +V+VDD+SR++WV F+
Sbjct: 541 HGKMTSSSHKPVTMVMTDGPGQLLHMNTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLE 600
Query: 182 RKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQ 241
K+E+ F + + E + +RSD+G F+N FES DS G+ + FS P PQ
Sbjct: 601 SKEETFGFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQ 660
Query: 242 QNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKK 301
QNGVVERKNRTL EM+RTML E + FW EA++ +C+I NR+ +R IL+KTPYEL
Sbjct: 661 QNGVVERKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFG 720
Query: 302 VKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYI 361
+P +S+ FGC C+ L + + L KF+S+S + LGY+ S+ +R+Y I E
Sbjct: 721 RRPKVSHLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETC 779
Query: 362 HVRFDDKL---------------------DSDQSKLVEKFVDMSINVSDKGKAPEEAEPE 400
V FD+ D D + +D + V + G +P P
Sbjct: 780 EVTFDEASPGARPEISGVLDESIFVDEDSDDDDDDSIPPPLDSTPPVQETG-SPSTTSPS 838
Query: 401 EDSP-------EEVGPSDPQPQKKSRIVASHPKELILGNKDEPVRTRSAFRPSEETLLSL 453
D+P EE+ P I HP + ++G E V ++ L
Sbjct: 839 GDAPTTSSSAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYD------LVN 892
Query: 454 KGLVSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKL 513
V+ EPK++ AL D++W+ AM EEL F +N VW++V+ P G ++IGTKWVF+NKL
Sbjct: 893 SAFVASFEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKL 952
Query: 514 NEKGDVVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKS 573
E G +VRNKARLVAQG+ Q EG+D+ ETFAPVARLEAIR+L++F+ + L QMDVKS
Sbjct: 953 GEDGSIVRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKS 1012
Query: 574 VFLNGYISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERLR-FLLENEFVR 632
FLNG I EEVYV QPPGFE+ K P+HVF L K+LYGLKQAPRAWYERL+ FLL+N F
Sbjct: 1013 AFLNGVIEEEVYVKQPPGFENPKFPNHVFKLDKALYGLKQAPRAWYERLKTFLLQNGFEM 1072
Query: 633 GKVDTTLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLG 692
G VD TLF D L+VQIYVDDIIFG ++ +L +FS++M EFEMSMMGEL +FLG
Sbjct: 1073 GAVDKTLFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLG 1132
Query: 693 IQVDQTPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGM 752
+Q+ QT E ++HQ+KY+KELLKKF+M + T M T L ++ +V Q+ YR M
Sbjct: 1133 LQIKQTKEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSM 1192
Query: 753 ICSFLYLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYK 812
I S LYLTASRPDI FS LCARFQ+ PR +H VKRI RY+K T G+ Y +S
Sbjct: 1193 IGSLLYLTASRPDIHFSVCLCARFQASPRTSHRQAVKRIFRYIKSTLEYGIWYSCSSALS 1252
Query: 813 LSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*M 872
+ + DAD+AG + +RKSTSG C FLG++LVSW+S++QS++A STAEAEY++ A + +
Sbjct: 1253 VRAFSDADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAASACSQV 1312
Query: 873 LWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF* 932
LWM L+DY + S +P+ CDNT+AI+++KNP+ HSR K+IE++YHF+RD V+KG
Sbjct: 1313 LWMISTLKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEKGTIVL 1372
Query: 933 SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
+ ADIFTKPL RF+F+ L +
Sbjct: 1373 EFVESEKQLADIFTKPLDRSRFEFLRSELGV 1403
>ref|XP_468929.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|28209450|gb|AAO37468.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1128
Score = 832 bits (2148), Expect = 0.0
Identities = 443/914 (48%), Positives = 591/914 (64%), Gaps = 35/914 (3%)
Query: 80 WV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQKGKFTKVSFKAKNVVSTS 139
W+ HRRL H M ++S L K DLV GL +KF D LC ACQ K S K+++STS
Sbjct: 213 WLWHRRLAHVGMNQLSKLSKRDLVVGLKDVKFEKDKLCSACQASKQVACSHPTKSIMSTS 272
Query: 140 RPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFISRKDESHSVFSTFIVQVQN 199
RPLELLH+DLFGP +SIGG + +VIVDDYS +TWV F+ K +F F + QN
Sbjct: 273 RPLELLHMDLFGPTTYKSIGGNSHCLVIVDDYSCYTWVFFLHDKCIVAELFKKFAKRAQN 332
Query: 200 ENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQQNGVVERKNRTLQEMSRT 259
E +C ++++RSD+G F+N E D I ++ S +PQQNGVVERKNRTL EM+RT
Sbjct: 333 EFSCTLVKIRSDNGSKFKNTNIEDYCDDLSIKHELSATYSPQQNGVVERKNRTLIEMART 392
Query: 260 MLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKKVKPNISYFHPFGCVCYAL 319
ML E ++ FWAEA+NT+C+ NR+ + +L KT YEL KPN++YF FGC CY
Sbjct: 393 MLDEYGVSDSFWAEAINTACHATNRLYLHRLLKKTSYELIVGRKPNVAYFRVFGCKCYIY 452
Query: 320 NTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYIHVRFDDKLDSDQSK---- 375
RL KF+S+ + LLGY+ SK +R+YN + +EE V+FD+ S +
Sbjct: 453 RKGVRLTKFESRCDEGFLLGYASNSKAYRVYNKNKGIVEETADVQFDETNGSQEGHENLD 512
Query: 376 ------LVEKFVDMSIN------VSDK-----------GKAPEEAEPEEDSPEEVGPSDP 412
L+ +MSI V DK +P +A+ E + + P P
Sbjct: 513 DVGDEGLMRAMKNMSIGDVKPIEVEDKPSTSTQDEPSTSASPSQAQVEVEKEKAQDPPMP 572
Query: 413 QPQKKSRIVASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDK 472
P+ + + HP + +LG+ + V+TRS ++ VS +EPK +DEAL D
Sbjct: 573 -PRIYTALSKDHPIDQVLGDISKGVQTRSPVA----SICEHYSFVSCLEPKHVDEALYDP 627
Query: 473 DWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYR 532
DW+ A+ EELN F++N VW +V++P+ ++IGTKWVFRNK +E VVRNKARLVAQG+
Sbjct: 628 DWMNAIHEELNNFARNKVWTLVERPRDHNVIGTKWVFRNKQDENRLVVRNKARLVAQGFT 687
Query: 533 QQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGF 592
Q E +D+ ETF PVARLEAIR+L++F+ +I L QMDVKS FLNG I+E V+V QPPGF
Sbjct: 688 QVEDLDFGETFGPVARLEAIRILLAFASCFDIKLFQMDVKSAFLNGEIAELVFVEQPPGF 747
Query: 593 EDEKNPDHVFNLKKSLYGLKQAPRAWYERLR-FLLENEFVRGKVDTTLFCKTYKDDILIV 651
+D K P+HV+ L K+LYGLKQAPRAWYERLR FLL +F GKVDTTLF K DD +
Sbjct: 748 DDPKYPNHVYKLSKALYGLKQAPRAWYERLRDFLLSKDFKIGKVDTTLFTKIIGDDFFVC 807
Query: 652 QIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTK 711
QIYVDDIIFGS N CKEF +MM EFEMSM+ EL +FLG+Q+ Q + T++ Q+KY K
Sbjct: 808 QIYVDDIIFGSTNEVFCKEFGDMMSREFEMSMIEELSFFLGLQIKQLKDGTFVSQTKYIK 867
Query: 712 ELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEH 771
+LLK+F + ++ KT M L+ ++ V KLYR MI S LYLTASRPDI+FS
Sbjct: 868 DLLKRFGLEDAKPIKTPMATNWHLDLDEGGKPVDLKLYRSMIGSLLYLTASRPDIMFSVC 927
Query: 772 LCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKST 831
+ ARFQ+ P+E HL VKRILRYLK ++ + L Y K +++KL GY D+DYAG + +RKST
Sbjct: 928 MYARFQAAPKECHLVAVKRILRYLKHSSTISLWYPKGAKFKLVGYSDSDYAGYKVDRKST 987
Query: 832 SGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQI--LESNI 889
SG+CQ LG +LVSW+SK+Q+++ALSTAEAEYIS C +LWMK L DY I E+
Sbjct: 988 SGSCQMLGRSLVSWSSKKQNSVALSTAEAEYISAGSCCAQLLWMKQILLDYGISFTETQT 1047
Query: 890 PIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPL 949
P+ C+N + I ++ NP+ H R K+I++++HF+ D+V K S + ADIFTKPL
Sbjct: 1048 PLLCNNDSTIKIANNPVQHFRTKHIDIRHHFLTDHVAKCDIVISHIRTEDQLADIFTKPL 1107
Query: 950 AEDRFKFILKNLNM 963
E RF + LN+
Sbjct: 1108 DETRFCKLRNELNV 1121
>gb|AAP53706.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|37534234|ref|NP_921419.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 1419
Score = 827 bits (2135), Expect = 0.0
Identities = 437/963 (45%), Positives = 607/963 (62%), Gaps = 34/963 (3%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
+ NV LV L +NLLS+SQ+ D+ +++ F + + SVL N R +K
Sbjct: 482 LKNVALVKDLKYNLLSVSQIVDENFEVHFKKTGSKVFDSCGDSVL-NISRYERVFKADFE 540
Query: 63 ELETQKVKCLLS-VNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQ 121
+ + CL++ +++ HRRLGH ++ L LDLVRGL LK D +C C+
Sbjct: 541 NSVSLVITCLVAKFDKDVMFWHRRLGHVGFDHLTRLSGLDLVRGLSKLKKDLDLICTPCR 600
Query: 122 KGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFIS 181
K S V T P +LLH+D GP + +S+GGK Y +VIVDD+SR++WV F++
Sbjct: 601 HAKMVSTSHAPIVSVMTDAPGQLLHMDTVGPARVQSVGGKWYVLVIVDDFSRYSWVFFMT 660
Query: 182 RKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQ 241
KDE+ F ++++ E + R+RSD+GG F+N FE + G+ ++FS PR PQ
Sbjct: 661 TKDEAFQHFRGLFLRLELEFPGSLKRIRSDNGGEFKNASFEQFCNERGLEHEFSSPRVPQ 720
Query: 242 QNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKK 301
QN VVERKNR L EM+RTML E + FWAEA+NT+CYI NR+ +R L KT YEL
Sbjct: 721 QNSVVERKNRVLVEMARTMLDEYKTTRKFWAEAINTACYISNRVFLRSKLGKTSYELRFG 780
Query: 302 VKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYI 361
+P +S+ FGC C+ L + + L KF+++S+ L LGY ++G+R+ I E
Sbjct: 781 HQPKVSHLRVFGCKCFVLKSGN-LDKFEARSTDGLFLGYPAHTRGYRVLILGTNKIVETC 839
Query: 362 HVRFDDKLDSDQSKLVEKFVDMSINVSDKGKAPEEAEPEEDSPEEVGPSDPQPQKKSRIV 421
V FD+ + + + + G+ E+ E + D +EVG + + + V
Sbjct: 840 EVSFDEASPGTRPDIAGTLSQVQ---GEDGRIFED-ESDYDDDDEVGSAGERTTRSK--V 893
Query: 422 ASHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEEE 481
+H V SAF V+ EPK + AL D+ WI AM EE
Sbjct: 894 TTHD-----------VCANSAF-------------VASFEPKDVSHALTDESWINAMHEE 929
Query: 482 LNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYTE 541
L F +N VW +V+ P G ++IGTKWVF+NK NE G +VRNKARLVAQG+ Q EG+D+ E
Sbjct: 930 LENFERNKVWTLVEPPSGHNVIGTKWVFKNKQNEDGLIVRNKARLVAQGFTQVEGLDFDE 989
Query: 542 TFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDHV 601
TFAPVAR+EAIRLL++F+ + L+QMDVKS FLNG+I EEVYV QPPGFE+ P+HV
Sbjct: 990 TFAPVARIEAIRLLLAFAASKGFKLYQMDVKSAFLNGFIQEEVYVKQPPGFENPDFPNHV 1049
Query: 602 FNLKKSLYGLKQAPRAWYERLR-FLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIF 660
F L K+LYGLKQAPRAWY+RL+ FLL F GKVD TLF + D+ L VQIYVDDIIF
Sbjct: 1050 FKLSKALYGLKQAPRAWYDRLKNFLLAKGFTMGKVDKTLFVLKHGDNQLFVQIYVDDIIF 1109
Query: 661 GSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNMT 720
G + ++ +F+E M+ EFEMSMMGEL YFLG+Q+ QTP+ T++HQ+KYTK+LL++F M
Sbjct: 1110 GCSTHAVVVDFAETMRREFEMSMMGELSYFLGLQIKQTPQGTFVHQTKYTKDLLRRFKME 1169
Query: 721 ESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSDP 780
T + T +L+ ++ V QK YR MI S LYLTASRPDI F+ LCARFQ+ P
Sbjct: 1170 NCKPISTPIGSTAVLDPDEDGEAVDQKEYRSMIGSLLYLTASRPDIQFAVCLCARFQASP 1229
Query: 781 RETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGS 840
R +H VKRI+RYL T G+ Y +S LSGY DAD+ G R +RKSTSG C FLG+
Sbjct: 1230 RASHRQAVKRIMRYLNHTLEFGIWYSTSSSICLSGYSDADFGGCRIDRKSTSGTCHFLGT 1289
Query: 841 NLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAIS 900
+L++W+S++QS++A STAE+EY++ A C + +LW+ L+DY I +P++CDNT+AI+
Sbjct: 1290 SLIAWSSRKQSSVAQSTAESEYVAAASCCSQILWLLSTLKDYGITFEKVPLFCDNTSAIN 1349
Query: 901 LSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFKFILKN 960
++KNP+ HSR K+I++++HF+RD+V+KG L + ADIFTKPL +RF F+
Sbjct: 1350 IAKNPVQHSRTKHIDIRFHFLRDHVEKGDVELQFLDTKLQIADIFTKPLDSNRFAFLRGE 1409
Query: 961 LNM 963
L +
Sbjct: 1410 LGV 1412
>ref|XP_463224.1| putative integrase [Oryza sativa (japonica cultivar-group)]
gi|40737034|gb|AAR89047.1| putative integrase [Oryza
sativa (japonica cultivar-group)]
Length = 1507
Score = 758 bits (1958), Expect = 0.0
Identities = 420/986 (42%), Positives = 593/986 (59%), Gaps = 55/986 (5%)
Query: 3 IDNVLLVDGLNHNLLSISQLADKGYDIIFNQKSCRAVSQIDGSVLFNSKRRNNTYKIRLF 62
+ NV LV+ LN+NLLS+ Q+ + +++ F + + SVL N R +K
Sbjct: 545 LKNVALVEDLNYNLLSVLQIVYENFEVHFKKTRSKVFDSCGDSVL-NISRYGRVFKADFE 603
Query: 63 ELETQKVKCLLS-VNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQ 121
+ + CL++ +++ HRRLGH ++ L LDLVRGLP LK D C C
Sbjct: 604 NPVSPVITCLVAKFDKDVMFWHRRLGHVGFDHLTRLSGLDLVRGLPKLKKDLDLDCAPCH 663
Query: 122 KGKFTKVSFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFIS 181
K S V T P +LLH+D GP + +S+GGK Y +VI+DD+SR++WV F++
Sbjct: 664 HAKMVASSHAPIVSVMTDAPRQLLHMDTVGPARVQSVGGKWYVLVIIDDFSRYSWVFFMA 723
Query: 182 RKDESHSVFSTFIVQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQ 241
KDE+ F ++++ E + R+RSD+G ++N FE + G+ ++FS PR PQ
Sbjct: 724 TKDEAFQHFRGLFLRLELEFPGSLKRIRSDNGSEYKNASFEQFCNERGLEHEFSSPRVPQ 783
Query: 242 QNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKK 301
QNGVVERKN L EM RTML E + FWAEA+NT+ YI NR+ +R L K+ YEL
Sbjct: 784 QNGVVERKNHVLVEMVRTMLDEYKTPRKFWAEAINTAYYISNRVFLRSKLGKSSYELRFG 843
Query: 302 VKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFR----IYNTDAKTI 357
+P +S+ FGC C+ L + + L KF+++S+ L LGY ++G+R + D +
Sbjct: 844 HQPKVSHLRVFGCKCFVLKSGN-LDKFEARSTDGLFLGYPAHTRGYRTLSQVQGEDGRIF 902
Query: 358 EEYIHVRFDDKLDS--------DQSKLVEKFVDMSINVSDKGKAPEEAEPE--EDSPEEV 407
E+ DD++ S Q+ SD+ E + D P E+
Sbjct: 903 EDESDDNDDDEVGSAGQTGRQAGQTAGTPPVRPAHEERSDRPGLSAEGSVDAVRDGPLEI 962
Query: 408 GPS---DPQPQKKSRIVAS------HPKELILGNKDEPVRTRSAFRPSEETLLSLKGLVS 458
S D + S +VA HP E I+GN E TRS + + + + V+
Sbjct: 963 TTSTSTDTEHGSTSEVVAPLHIQQRHPPEQIIGNIGERT-TRS--KVTTHDVCANYAFVA 1019
Query: 459 LIEPKSIDEALQDKDWILAMEEELNQFSKNDVWNIVKKPQGVHIIGTKWVFRNKLNEKGD 518
EPK + AL D+ WI AM EEL F +N VW +V+ P G +IIGTKWVF+NK NE
Sbjct: 1020 SFEPKDVSHALTDESWINAMHEELENFERNKVWTLVEPPSGHNIIGTKWVFKNKQNEDDL 1079
Query: 519 VVRNKARLVAQGYRQQEGIDYTETFAPVARLEAIRLLISFSVNHNIILHQMDVKSVFLNG 578
+VRNKARLVAQG+ Q EG+D+ ETFAPVAR+EAIRL ++F+ + L+QMDVKS FLNG
Sbjct: 1080 IVRNKARLVAQGFTQVEGLDFDETFAPVARIEAIRLFLAFASSKGFKLYQMDVKSAFLNG 1139
Query: 579 YISEEVYVHQPPGFEDEKNPDHVFNLKKSLYGLKQAPRAWYERLR-FLLENEFVRGKVDT 637
+I EEVYV QPPGFE+ P++VF L K+LYGLKQAPRAWY+RL+ FLL F GKVD
Sbjct: 1140 FIQEEVYVKQPPGFENPDFPNYVFKLSKALYGLKQAPRAWYDRLKNFLLAKGFTMGKVDK 1199
Query: 638 TLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQ 697
TLF L +F+E M+ EFEMSMMGEL YFLG+Q+ Q
Sbjct: 1200 TLFV-------------------------LKHDFAETMRREFEMSMMGELSYFLGLQIKQ 1234
Query: 698 TPEATYIHQSKYTKELLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFL 757
TP+ T++HQ+KYTK+LL++F M T + T +L+ ++ V QK YR MI S L
Sbjct: 1235 TPQGTFVHQTKYTKDLLRRFKMENCKPISTPIDSTAVLDPDEDGEAVDQKEYRSMIGSLL 1294
Query: 758 YLTASRPDILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTSEYKLSGYC 817
YLTASRP+I F+ LCARFQ+ PR +H VKRI+RYL T G+ Y +S LSGY
Sbjct: 1295 YLTASRPEIQFAVCLCARFQASPRASHRQAVKRIMRYLNHTLEFGIWYSTSSSLCLSGYS 1354
Query: 818 DADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTIALSTAEAEYISIAICNT*MLWMKH 877
DA++ G R +RKSTSG C FLG++L++W+S++QS++A STAE+EY++ A C + +LW+
Sbjct: 1355 DANFGGCRIDRKSTSGTCHFLGTSLIAWSSRKQSSVAQSTAESEYVAAASCCSQILWLLS 1414
Query: 878 QLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLIL 937
L++Y + +P++CDNT+AI+++KN + HSR K++++++HF+RD+V+KG L
Sbjct: 1415 TLKNYGLTFEKVPLFCDNTSAINIAKNLVQHSRTKHVDIRFHFLRDHVEKGDVELQFLDT 1474
Query: 938 TINSADIFTKPLAEDRFKFILKNLNM 963
+ ADIFTKPL + F F+ L +
Sbjct: 1475 KLQIADIFTKPLDSNCFTFLCGELGI 1500
>gb|AAP94592.1| retrotransposon Opie-2 [Zea mays]
Length = 1512
Score = 728 bits (1878), Expect = 0.0
Identities = 393/836 (47%), Positives = 521/836 (62%), Gaps = 68/836 (8%)
Query: 135 VVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFISRKDESHSVFSTFI 194
+V PLELLH+DLFGPV SIGG +YG+VIVDD+SR+TWV F+ K E+ F+
Sbjct: 731 IVFLQIPLELLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQDKSETQGTLKRFL 790
Query: 195 VQVQNENACRIMRVRSDHGGVFENDKFESLFDSYGIAYDFSCPRTPQQNGVVERKNRTLQ 254
+ QNE ++ ++RSD+G F+N + E + GI ++FS P TPQQNGVVERKNRTL
Sbjct: 791 RRAQNEFELKVKKIRSDNGSEFKNLQVEEFLEEEGIKHEFSAPYTPQQNGVVERKNRTLI 850
Query: 255 EMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKKVKPNISYFHPFGC 314
+M+RTML E + FW+EAVNT+C+ NR+ + +L KT YEL KPN+SYF FG
Sbjct: 851 DMARTMLGEFKTPECFWSEAVNTACHAINRVYLHRLLKKTSYELLTGNKPNVSYFRVFGS 910
Query: 315 VCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTIEEYIHVRFDDKLDSDQS 374
CY L K R KF K+ + LLGY +K +R++N + +E V FD+ S +
Sbjct: 911 KCYILVKKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSSDVVFDETNGSPRE 970
Query: 375 KLVEKFVDMSINVSDKGKAPEEAEPEEDSPEE------VGPSDPQPQKKSRIVASHPKEL 428
++V+ + EED P +G PQ Q +
Sbjct: 971 QVVDC----------------DDVDEEDVPTAAIRTMAIGEVRPQEQDE----------- 1003
Query: 429 ILGNKDEPVRTRSAFRPSEETLLSLKGLVSLIEPKSIDEALQDKDWILAMEEELNQFSKN 488
+D+P + + P+++ +P ++EAL D DW+LAM+EELN F +N
Sbjct: 1004 ----RDQPSSSTTVHPPTQDDE----------QPFRVEEALLDLDWVLAMQEELNNFKRN 1049
Query: 489 DVWNIVKKPQGVHIIGTKWVFRNKLNEKGDVVRNKARLVAQGYRQQEGIDYTETFAPVAR 548
+VW+ L+E G V RNKARLVA+GY Q G+D+ ETFAPVAR
Sbjct: 1050 EVWS--------------------LDEHGVVTRNKARLVAKGYAQVAGLDFEETFAPVAR 1089
Query: 549 LEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDHVFNLKKSL 608
LE+IR+L++++ +H+ L+QMDVKS FLNG I EEVYV QPPGFEDE+ PDHV L K+L
Sbjct: 1090 LESIRILLAYAAHHSFRLYQMDVKSAFLNGPIKEEVYVEQPPGFEDERYPDHVCKLAKAL 1149
Query: 609 YGLKQAPRAWYERLR-FLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANSSL 667
YGLKQAPRAWYE LR FL+ N F GK D TLF KT D+ + QI+VDDIIFGS N
Sbjct: 1150 YGLKQAPRAWYECLRDFLIANAFKVGKADPTLFTKTCNGDLFVCQIFVDDIIFGSTNQKS 1209
Query: 668 CKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKELLKKFNMTESIIAKT 727
C+EFS +M +FEMSMMG+L YFLG QV Q + T+I Q KYT++LLK+F M ++ AKT
Sbjct: 1210 CEEFSRVMTQKFEMSMMGKLNYFLGFQVKQLKDGTFISQMKYTQDLLKRFGMKDAKPAKT 1269
Query: 728 LMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRPDILFSEHLCARFQSDPRETHLTV 787
M + V QK YR MI S LYL ASRPDI+ S +CARFQS+P+E HL
Sbjct: 1270 PMGTDGHTDLNKGGKSVDQKAYRSMIGSLLYLCASRPDIMLSVCMCARFQSEPKECHLVA 1329
Query: 788 VKRILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWAS 847
VKRILRYL T GL Y K S + L GY D+D AG + +RKSTSG CQFLG +LVSW S
Sbjct: 1330 VKRILRYLVATPCFGLWYPKGSTFDLVGYSDSDNAGCKVDRKSTSGTCQFLGRSLVSWNS 1389
Query: 848 KRQSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPIL 907
K+Q+++ALSTAEAEY++ C +LWM+ L D+ S +P+ CDN +AI +++NP+
Sbjct: 1390 KKQTSVALSTAEAEYVAAGQCCAQLLWMRQTLRDFGYNLSKVPLLCDNESAIRMAENPVE 1449
Query: 908 HSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
H+R K+I++++HF+RD+ QKG + ADIFTKPL E F + LN+
Sbjct: 1450 HNRTKHIDIRHHFLRDHQQKGDIEVFHVSTENQLADIFTKPLDEKTFCRLRSELNV 1505
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,609,863,126
Number of Sequences: 2540612
Number of extensions: 68040955
Number of successful extensions: 176362
Number of sequences better than 10.0: 3227
Number of HSP's better than 10.0 without gapping: 1903
Number of HSP's successfully gapped in prelim test: 1325
Number of HSP's that attempted gapping in prelim test: 168759
Number of HSP's gapped (non-prelim): 4777
length of query: 967
length of database: 863,360,394
effective HSP length: 138
effective length of query: 829
effective length of database: 512,755,938
effective search space: 425074672602
effective search space used: 425074672602
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0101.5


