

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
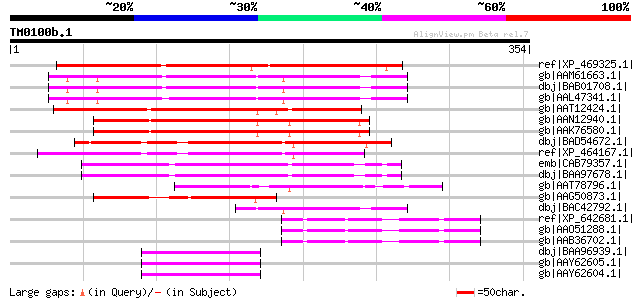
Query= TM0100b.1
(354 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_469325.1| putative zinc finger protein [Oryza sativa] gi|... 199 1e-49
gb|AAM61663.1| flowering protein CONSTANS, putative [Arabidopsis... 195 2e-48
dbj|BAB01708.1| unnamed protein product [Arabidopsis thaliana] g... 195 2e-48
gb|AAL47341.1| unknown protein [Arabidopsis thaliana] gi|1459605... 194 3e-48
gb|AAT12424.1| male-specific transcription factor M88B7.2 [March... 194 3e-48
gb|AAN12940.1| putative flowering protein CONSTANS [Arabidopsis ... 191 3e-47
gb|AAK76580.1| putative flowering protein CONSTANS [Arabidopsis ... 189 8e-47
dbj|BAD54672.1| putative zinc-finger protein [Oryza sativa (japo... 189 8e-47
ref|XP_464167.1| putative GATA zinc finger protein [Oryza sativa... 187 4e-46
emb|CAB79357.1| putative protein [Arabidopsis thaliana] gi|50517... 160 5e-38
dbj|BAA97678.1| ZIM [Arabidopsis thaliana] gi|11358938|pir||JC73... 160 5e-38
gb|AAT78796.1| putative zinc finger protein [Oryza sativa (japon... 148 3e-34
gb|AAG50873.1| hypothetical protein [Arabidopsis thaliana] gi|12... 114 4e-24
dbj|BAC42792.1| unknown protein [Arabidopsis thaliana] 98 4e-19
ref|XP_642681.1| putative GATA-binding transcription factor [Dic... 64 9e-09
gb|AAO51288.1| similar to Dictyostelium discoideum (Slime mold).... 64 9e-09
gb|AAB36702.1| putative transcription factor [Dictyostelium disc... 64 9e-09
dbj|BAA96939.1| unnamed protein product [Arabidopsis thaliana] g... 61 4e-08
gb|AAY62605.1| pseudo response regulator 3 [Arabidopsis thaliana] 61 4e-08
gb|AAY62604.1| pseudo response regulator 3 [Arabidopsis thaliana] 61 4e-08
>ref|XP_469325.1| putative zinc finger protein [Oryza sativa]
gi|13174240|gb|AAK14414.1| putative zinc finger protein
[Oryza sativa]
Length = 319
Score = 199 bits (505), Expect = 1e-49
Identities = 122/249 (48%), Positives = 157/249 (62%), Gaps = 17/249 (6%)
Query: 33 DEGQHAPATVAASASNASAPHPRA-GELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIP 91
DE + PA A+ SA P + +LT+ F+GEVYVF +VTPEKVQAVLLLLG+ ++P
Sbjct: 54 DEEELPPAEDPAAPEPVSALLPGSPNQLTLLFQGEVYVFESVTPEKVQAVLLLLGRSEMP 113
Query: 92 NSTPNSGLLQQKNFPQIKGINDPSQSSNL-SRRFASLVRFREKRKERCFEKKIRYTCRKE 150
N L Q+ + +G +D Q +++ ++R ASL+RFREKRKER F+KKIRY RKE
Sbjct: 114 PGLANMVLPNQR---ENRGYDDLLQRTDIPAKRVASLIRFREKRKERNFDKKIRYAVRKE 170
Query: 151 VAQRMHRKNGQFA---SLKEDYKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMR 207
VA RM R+ GQFA +++ + SP L +S G+ S E +CQ+CGTSEK TPAMR
Sbjct: 171 VALRMQRRKGQFAGRANMEGESLSPGCEL-ASQGSGQDFLSRESKCQNCGTSEKMTPAMR 229
Query: 208 RGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTSADMKPS--------TTEP 259
RGPAGPR+LCNACGLMWANKGTLR+ KA ++A + PS P
Sbjct: 230 RGPAGPRTLCNACGLMWANKGTLRNCPKAKVESSVVATEQSNAAVSPSGIDNKELVVPNP 289
Query: 260 ENSCADQAE 268
EN A E
Sbjct: 290 ENITASHGE 298
>gb|AAM61663.1| flowering protein CONSTANS, putative [Arabidopsis thaliana]
Length = 297
Score = 195 bits (496), Expect = 2e-48
Identities = 123/259 (47%), Positives = 151/259 (57%), Gaps = 24/259 (9%)
Query: 27 HFANGADEGQHA--PATVAASASNASAPHPRAGE----LTISFEGEVYVFPAVTPEKVQA 80
H G DEG P+ SA N R E LT+SF+G+VYVF V+PEKVQA
Sbjct: 42 HADGGMDEGVETDIPSHPGNSADNRGEVVDRGIENGDQLTLSFQGQVYVFDRVSPEKVQA 101
Query: 81 VLLLLGQKDIPNSTPNS-GLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCF 139
VLLLLG +++P++ P + G Q N ++ G++ Q ++ +R ASL+RFREKRK R F
Sbjct: 102 VLLLLGGREVPHTLPTTLGSPHQNN--RVLGLSGTPQRLSVPQRLASLLRFREKRKGRNF 159
Query: 140 EKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPC-------PESIERR 192
+K IRYT RKEVA RM RK GQF S K +G+ S G+ + E
Sbjct: 160 DKTIRYTVRKEVALRMQRKKGQFTSAKSS-NDDSGSTGSDWGSNQSWAVEGTETQKPEVL 218
Query: 193 CQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTSADM 252
C+HCGTSEKSTP MRRGP GPR+LCNACGLMWANKGTLRDLSK T +
Sbjct: 219 CRHCGTSEKSTPMMRRGPDGPRTLCNACGLMWANKGTLRDLSKVPPP-------QTPQHL 271
Query: 253 KPSTTEPENSCADQAEEGT 271
+ E N ADQ E T
Sbjct: 272 SVNKNEDANLEADQMMEVT 290
>dbj|BAB01708.1| unnamed protein product [Arabidopsis thaliana]
gi|18402914|ref|NP_566676.1| zinc finger (GATA type)
family protein [Arabidopsis thaliana]
gi|38603658|dbj|BAD02930.1| GATA-type zinc finger
protein [Arabidopsis thaliana]
Length = 297
Score = 195 bits (495), Expect = 2e-48
Identities = 123/259 (47%), Positives = 151/259 (57%), Gaps = 24/259 (9%)
Query: 27 HFANGADEGQHA--PATVAASASNASAPHPRAGE----LTISFEGEVYVFPAVTPEKVQA 80
H G DEG P+ SA N R E LT+SF+G+VYVF V+PEKVQA
Sbjct: 42 HADGGMDEGVETDIPSHPGNSADNRGEVVDRGIENGDQLTLSFQGQVYVFDRVSPEKVQA 101
Query: 81 VLLLLGQKDIPNSTPNS-GLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCF 139
VLLLLG +++P++ P + G Q N ++ G++ Q ++ +R ASL+RFREKRK R F
Sbjct: 102 VLLLLGGREVPHTLPTTLGSPHQNN--RVLGLSGTPQRLSVPQRLASLLRFREKRKGRNF 159
Query: 140 EKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPC-------PESIERR 192
+K IRYT RKEVA RM RK GQF S K +G+ S G+ + E
Sbjct: 160 DKTIRYTVRKEVALRMQRKKGQFTSAKSS-NDDSGSTGSDWGSNQSWAVEGTETQKPEVL 218
Query: 193 CQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTSADM 252
C+HCGTSEKSTP MRRGP GPR+LCNACGLMWANKGTLRDLSK T +
Sbjct: 219 CRHCGTSEKSTPMMRRGPDGPRTLCNACGLMWANKGTLRDLSKVPPP-------QTPQHL 271
Query: 253 KPSTTEPENSCADQAEEGT 271
+ E N ADQ E T
Sbjct: 272 SLNKNEDANLEADQMMEVT 290
>gb|AAL47341.1| unknown protein [Arabidopsis thaliana] gi|14596059|gb|AAK68757.1|
Unknown protein [Arabidopsis thaliana]
gi|30686115|ref|NP_850618.1| zinc finger (GATA type)
family protein [Arabidopsis thaliana]
Length = 295
Score = 194 bits (493), Expect = 3e-48
Identities = 123/259 (47%), Positives = 150/259 (57%), Gaps = 26/259 (10%)
Query: 27 HFANGADEGQHA--PATVAASASNASAPHPRAGE----LTISFEGEVYVFPAVTPEKVQA 80
H G DEG P+ SA N R E LT+SF+G+VYVF V+PEKVQA
Sbjct: 42 HADGGMDEGVETDIPSHPGNSADNRGEVVDRGIENGDQLTLSFQGQVYVFDRVSPEKVQA 101
Query: 81 VLLLLGQKDIPNSTPNS-GLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCF 139
VLLLLG +++P++ P + G Q N +G++ Q ++ +R ASL+RFREKRK R F
Sbjct: 102 VLLLLGGREVPHTLPTTLGSPHQNN----RGLSGTPQRLSVPQRLASLLRFREKRKGRNF 157
Query: 140 EKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPC-------PESIERR 192
+K IRYT RKEVA RM RK GQF S K +G+ S G+ + E
Sbjct: 158 DKTIRYTVRKEVALRMQRKKGQFTSAKSS-NDDSGSTGSDWGSNQSWAVEGTETQKPEVL 216
Query: 193 CQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTSADM 252
C+HCGTSEKSTP MRRGP GPR+LCNACGLMWANKGTLRDLSK T +
Sbjct: 217 CRHCGTSEKSTPMMRRGPDGPRTLCNACGLMWANKGTLRDLSKVPPP-------QTPQHL 269
Query: 253 KPSTTEPENSCADQAEEGT 271
+ E N ADQ E T
Sbjct: 270 SLNKNEDANLEADQMMEVT 288
>gb|AAT12424.1| male-specific transcription factor M88B7.2 [Marchantia polymorpha]
Length = 393
Score = 194 bits (493), Expect = 3e-48
Identities = 112/220 (50%), Positives = 138/220 (61%), Gaps = 14/220 (6%)
Query: 31 GADEGQ-HAPATVAASASNASAPHPRAG-ELTISFEGEVYVFPAVTPEKVQAVLLLLGQK 88
G DE + H+ A N A + +LT+S++GEVYVF V PEKVQAVLLLLG +
Sbjct: 108 GLDEAEMHSDGAHPGDAPNQLAVRNQGTTQLTLSYQGEVYVFDTVPPEKVQAVLLLLGGR 167
Query: 89 DIPNSTPNSGLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCR 148
+IP SG+ + KG+++ N+ +R ASL RFREKRKERC++KKIRYT R
Sbjct: 168 EIPPGM--SGVNVSGHHHTNKGVSELPARMNMPQRLASLTRFREKRKERCYDKKIRYTVR 225
Query: 149 KEVAQRMHRKNGQFASLKE--DYKSPAGNLDSSN------GTPPCPESIERRCQHCGTSE 200
KEVAQRM RK GQFAS + + P + D S GT + + C HCG E
Sbjct: 226 KEVAQRMQRKKGQFASSRTLGEEGGPVSSWDGSQIPGQQVGTGVGQQEVT--CVHCGIGE 283
Query: 201 KSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTP 240
+STP MRRGP+GPR+LCNACGLMWANKG LRDLSK P
Sbjct: 284 RSTPMMRRGPSGPRTLCNACGLMWANKGVLRDLSKNPPLP 323
>gb|AAN12940.1| putative flowering protein CONSTANS [Arabidopsis thaliana]
gi|42571823|ref|NP_974002.1| zinc finger (GATA type)
family protein [Arabidopsis thaliana]
gi|18403600|ref|NP_564593.1| zinc finger (GATA type)
family protein [Arabidopsis thaliana]
gi|38603660|dbj|BAD02931.1| GATA-type zinc finger
protein [Arabidopsis thaliana]
Length = 302
Score = 191 bits (485), Expect = 3e-47
Identities = 108/199 (54%), Positives = 130/199 (65%), Gaps = 12/199 (6%)
Query: 58 ELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPNSGLLQQKNFPQIKGINDPSQS 117
+LT+SF+G+VYVF +V PEKVQAVLLLLG +++P + P GL ++ + Q
Sbjct: 83 QLTLSFQGQVYVFDSVLPEKVQAVLLLLGGRELPQAAP-PGLGSPHQNNRVSSLPGTPQR 141
Query: 118 SNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKE--DYKSPAGN 175
++ +R ASLVRFREKRK R F+KKIRYT RKEVA RM R GQF S K D + AG+
Sbjct: 142 FSIPQRLASLVRFREKRKGRNFDKKIRYTVRKEVALRMQRNKGQFTSAKSNNDEAASAGS 201
Query: 176 LDSSNGTPPCPESI----ERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLR 231
SN T S E C+HCG EKSTP MRRGPAGPR+LCNACGLMWANKG R
Sbjct: 202 SWGSNQTWAIESSEAQHQEISCRHCGIGEKSTPMMRRGPAGPRTLCNACGLMWANKGAFR 261
Query: 232 DLSKAA-----KTPYEQNE 245
DLSKA+ P +NE
Sbjct: 262 DLSKASPQTAQNLPLNKNE 280
>gb|AAK76580.1| putative flowering protein CONSTANS [Arabidopsis thaliana]
Length = 302
Score = 189 bits (481), Expect = 8e-47
Identities = 108/199 (54%), Positives = 129/199 (64%), Gaps = 12/199 (6%)
Query: 58 ELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPNSGLLQQKNFPQIKGINDPSQS 117
+LT+SF+G+VYVF +V PEKVQAVLLLLG +++P + P GL ++ + Q
Sbjct: 83 QLTLSFQGQVYVFDSVLPEKVQAVLLLLGGRELPQAAP-PGLGSPHQNNRVSSLPGTPQR 141
Query: 118 SNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKE--DYKSPAGN 175
++ +R ASLVRFREKRK R F KKIRYT RKEVA RM R GQF S K D + AG+
Sbjct: 142 FSIPQRLASLVRFREKRKGRNFGKKIRYTVRKEVALRMQRNKGQFTSAKSNNDEAASAGS 201
Query: 176 LDSSNGTPPCPESI----ERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLR 231
SN T S E C+HCG EKSTP MRRGPAGPR+LCNACGLMWANKG R
Sbjct: 202 SWGSNQTWAIESSEAQHQEISCRHCGIGEKSTPMMRRGPAGPRTLCNACGLMWANKGAFR 261
Query: 232 DLSKAA-----KTPYEQNE 245
DLSKA+ P +NE
Sbjct: 262 DLSKASPQTAQNLPLNKNE 280
>dbj|BAD54672.1| putative zinc-finger protein [Oryza sativa (japonica
cultivar-group)] gi|53791868|dbj|BAD53990.1| putative
zinc-finger protein [Oryza sativa (japonica
cultivar-group)]
Length = 340
Score = 189 bits (481), Expect = 8e-47
Identities = 113/224 (50%), Positives = 139/224 (61%), Gaps = 20/224 (8%)
Query: 45 SASNASAPHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPNSGLLQQKN 104
+AS A P + +LT+SF+GEVYVF +V+P+KVQAVLLLLG +++ P G +
Sbjct: 115 AASGAVQPMA-SNQLTLSFQGEVYVFDSVSPDKVQAVLLLLGGREL---NPGLGSGASSS 170
Query: 105 FPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFAS 164
P K +N P R ASL+RFREKRKER F+KKIRY+ RKEVA RM R GQF S
Sbjct: 171 APYSKRLNFP-------HRVASLMRFREKRKERNFDKKIRYSVRKEVALRMQRNRGQFTS 223
Query: 165 LKEDYKSPAGNLDSSNGTPPCPESIERR------CQHCGTSEKSTPAMRRGPAGPRSLCN 218
K L +S+G+P S+E R C HCG + K+TP MRRGP GPR+LCN
Sbjct: 224 SKPKGDEATSELTASDGSPNWG-SVEGRPPSAAECHHCGINAKATPMMRRGPDGPRTLCN 282
Query: 219 ACGLMWANKGTLRDLSKAAKTPYE--QNERDTSADMKPSTTEPE 260
ACGLMWANKG LRDLSKA TP + + D + TTE E
Sbjct: 283 ACGLMWANKGMLRDLSKAPPTPIQVVASVNDGNGSAAAPTTEQE 326
>ref|XP_464167.1| putative GATA zinc finger protein [Oryza sativa (japonica
cultivar-group)] gi|45736034|dbj|BAD13061.1| putative
GATA zinc finger protein [Oryza sativa (japonica
cultivar-group)]
Length = 328
Score = 187 bits (475), Expect = 4e-46
Identities = 107/229 (46%), Positives = 134/229 (57%), Gaps = 17/229 (7%)
Query: 20 PMQVDGVHFANGADEGQHAPATVAASASNASAPHPRAGELTISFEGEVYVFPAVTPEKVQ 79
PM D A + VA + PH + LT+SF+GEVYVF +V+ E+VQ
Sbjct: 68 PMDADASAAAVAGMDPHGEMVPVAGGEAGGGYPHVASNTLTLSFQGEVYVFESVSAERVQ 127
Query: 80 AVLLLLGQKDIPNSTPNSGLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCF 139
AVLLLLG +++ P SG + + K +N P R ASL+RFREKRKER F
Sbjct: 128 AVLLLLGGREL---APGSGSVPSSSAAYSKKMNFP-------HRMASLMRFREKRKERNF 177
Query: 140 EKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPCPESIERR------C 193
+KKIRYT RKEVA RM R GQF S K + + SS G+P ++E R C
Sbjct: 178 DKKIRYTVRKEVALRMQRNRGQFTSSKSKAEEATSVITSSEGSPNWG-AVEGRPPSAAEC 236
Query: 194 QHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYE 242
HCG S STP MRRGP GPR+LCNACGLMWANKGT+R+++K P +
Sbjct: 237 HHCGISAASTPMMRRGPDGPRTLCNACGLMWANKGTMREVTKGPPVPLQ 285
>emb|CAB79357.1| putative protein [Arabidopsis thaliana] gi|5051789|emb|CAB45082.1|
putative protein [Arabidopsis thaliana]
gi|21554169|gb|AAM63248.1| ZIM [Arabidopsis thaliana]
gi|8918533|dbj|BAA97679.1| ZIM [Arabidopsis thaliana]
gi|15233844|ref|NP_194178.1| zinc finger (GATA type)
protein ZIM (ZIM) [Arabidopsis thaliana]
gi|30686540|ref|NP_849435.1| zinc finger (GATA type)
protein ZIM (ZIM) [Arabidopsis thaliana]
gi|7487328|pir||T09910 hypothetical protein T22A6.300 -
Arabidopsis thaliana
Length = 309
Score = 160 bits (405), Expect = 5e-38
Identities = 101/219 (46%), Positives = 122/219 (55%), Gaps = 16/219 (7%)
Query: 50 SAPHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLLG-QKDIPNSTPNSGLLQQKNFPQI 108
S P A +LTISF G+VYVF AV +KV AVL LLG ++ L QQ+N +
Sbjct: 75 SRPPEGANQLTISFRGQVYVFDAVGADKVDAVLSLLGGSTELAPGPQVMELAQQQNHMPV 134
Query: 109 KGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKED 168
+ +L +R SL RFR+KR RCFEKK+RY R+EVA RM R GQF S K
Sbjct: 135 V---EYQSRCSLPQRAQSLDRFRKKRNARCFEKKVRYGVRQEVALRMARNKGQFTSSKMT 191
Query: 169 YKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKG 228
+ D + I C HCG S K TP MRRGP+GPR+LCNACGL WAN+G
Sbjct: 192 DGAYNSGTDQDSAQDDAHPEIS--CTHCGISSKCTPMMRRGPSGPRTLCNACGLFWANRG 249
Query: 229 TLRDLSKAAKTPYEQNERDTSADMKPSTTEPENSCADQA 267
TLRDLSK + E + A MKP + S AD A
Sbjct: 250 TLRDLSK-------KTEENQLALMKP---DDGGSVADAA 278
>dbj|BAA97678.1| ZIM [Arabidopsis thaliana] gi|11358938|pir||JC7336 zinc-finger
protein - Arabidopsis thaliana
Length = 309
Score = 160 bits (405), Expect = 5e-38
Identities = 101/219 (46%), Positives = 122/219 (55%), Gaps = 16/219 (7%)
Query: 50 SAPHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLLG-QKDIPNSTPNSGLLQQKNFPQI 108
S P A +LTISF G+VYVF AV +KV AVL LLG ++ L QQ+N +
Sbjct: 75 SRPPEGANQLTISFRGQVYVFDAVGADKVDAVLSLLGGSTELAPGPQVMELAQQQNHMPV 134
Query: 109 KGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKED 168
+ +L +R SL RFR+KR RCFEKK+RY R+EVA RM R GQF S K
Sbjct: 135 V---EYQSRCSLPQRAQSLDRFRKKRNARCFEKKVRYGVRQEVALRMARNKGQFTSSKMT 191
Query: 169 YKSPAGNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKG 228
+ D + I C HCG S K TP MRRGP+GPR+LCNACGL WAN+G
Sbjct: 192 DGAYNSGTDQDSAQDDAHPEIS--CTHCGISSKCTPMMRRGPSGPRTLCNACGLFWANRG 249
Query: 229 TLRDLSKAAKTPYEQNERDTSADMKPSTTEPENSCADQA 267
TLRDLSK + E + A MKP + S AD A
Sbjct: 250 TLRDLSK-------KTEENQLALMKP---DDGGSVADAA 278
>gb|AAT78796.1| putative zinc finger protein [Oryza sativa (japonica
cultivar-group)]
Length = 201
Score = 148 bits (373), Expect = 3e-34
Identities = 88/191 (46%), Positives = 113/191 (59%), Gaps = 24/191 (12%)
Query: 113 DPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSP 172
D ++ +RR ASL+RFREKRKERCF+KKIRY+ RKEVAQ+M R+ GQFA + D+
Sbjct: 15 DEKSTTVAARRVASLMRFREKRKERCFDKKIRYSVRKEVAQKMKRRKGQFAG-RADFG-- 71
Query: 173 AGNLDSSNGTPPCPESI--------ERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMW 224
D S + PC + E CQ+CG S + TPAMRRGPAGPRSLCNACGLMW
Sbjct: 72 ----DGSCSSAPCGSTANGEDDHIRETHCQNCGISSRLTPAMRRGPAGPRSLCNACGLMW 127
Query: 225 ANKGTLRDLSKAAKTPYEQNERDTSADMKPSTTEPENSCADQAEEGTPEENKPVPMDSRE 284
ANKGTLR A K Q+ D S T + ++S A+ E + D+
Sbjct: 128 ANKGTLRSPLNAPKMTV-QHPADLS-----KTGDTDDSKANLCAE---HNQTTMKTDTEM 178
Query: 285 TPEKKNEQDIL 295
PE++ + D+L
Sbjct: 179 VPEQEQKADVL 189
>gb|AAG50873.1| hypothetical protein [Arabidopsis thaliana]
gi|12325361|gb|AAG52620.1| flowering protein CONSTANS,
putative; 7571-5495 [Arabidopsis thaliana]
gi|25405485|pir||F96554 hypothetical protein F19C24.17
[imported] - Arabidopsis thaliana
Length = 299
Score = 114 bits (285), Expect = 4e-24
Identities = 67/127 (52%), Positives = 83/127 (64%), Gaps = 16/127 (12%)
Query: 58 ELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPNSGLLQQKNFPQIKGINDPSQS 117
+LT+SF+G+VYVF +V PEKVQAVLLLLG +++P + P G+ P Q+
Sbjct: 83 QLTLSFQGQVYVFDSVLPEKVQAVLLLLGGRELPQAAP-------------PGLGSPHQN 129
Query: 118 SNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLK--EDYKSPAGN 175
+ +SR ASLVRFREKRK R F+KKIRYT RKEVA RM R GQF S K D + AG+
Sbjct: 130 NRVSR-LASLVRFREKRKGRNFDKKIRYTVRKEVALRMQRNKGQFTSAKSNNDEAASAGS 188
Query: 176 LDSSNGT 182
SN T
Sbjct: 189 SWGSNQT 195
>dbj|BAC42792.1| unknown protein [Arabidopsis thaliana]
Length = 123
Score = 97.8 bits (242), Expect = 4e-19
Identities = 59/124 (47%), Positives = 67/124 (53%), Gaps = 15/124 (12%)
Query: 155 MHRKNGQFASLKEDYKSPAGNLDSSNGTPPC-------PESIERRCQHCGTSEKSTPAMR 207
M RK GQF S K +G+ S G+ + E C+HCGTSEKSTP MR
Sbjct: 1 MQRKKGQFTSAKSS-NDDSGSTGSDWGSNQSWAVEGTETQKPEVLCRHCGTSEKSTPMMR 59
Query: 208 RGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTSADMKPSTTEPENSCADQA 267
RGP GPR+LCNACGLMWANKGTLRDLSK T + + E N ADQ
Sbjct: 60 RGPDGPRTLCNACGLMWANKGTLRDLSKVPPP-------QTPQHLSLNKNEDANLEADQM 112
Query: 268 EEGT 271
E T
Sbjct: 113 MEVT 116
>ref|XP_642681.1| putative GATA-binding transcription factor [Dictyostelium
discoideum] gi|60470787|gb|EAL68759.1| putative
GATA-binding transcription factor [Dictyostelium
discoideum]
Length = 872
Score = 63.5 bits (153), Expect = 9e-09
Identities = 40/136 (29%), Positives = 67/136 (48%), Gaps = 17/136 (12%)
Query: 186 PESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNE 245
P S R C+ CG+S+ TP RRGP+G SLCNACG+ W KG + K ++ + +
Sbjct: 287 PMSAPRSCEFCGSSQ--TPTWRRGPSGKGSLCNACGIKWRLKGK-DGIFKPSQKQQNRQK 343
Query: 246 RDTSADMKPSTTEPENSCADQAEEGTPEENKPVPMDSRETPEKKNEQDILGTAQTVTDNL 305
SA +P + ++ P++ +P + P+++N+Q Q +
Sbjct: 344 PIMSAQKQP-----------KQQQNQPQQQQPQQPQQPQQPQQQNQQQ---NQQQSIQSQ 389
Query: 306 SIQLENHALSLDEQDI 321
Q +N+ L L++Q I
Sbjct: 390 PQQRQNNHLILNQQTI 405
>gb|AAO51288.1| similar to Dictyostelium discoideum (Slime mold). Putative
transcription factor
Length = 844
Score = 63.5 bits (153), Expect = 9e-09
Identities = 40/136 (29%), Positives = 67/136 (48%), Gaps = 17/136 (12%)
Query: 186 PESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNE 245
P S R C+ CG+S+ TP RRGP+G SLCNACG+ W KG + K ++ + +
Sbjct: 259 PMSAPRSCEFCGSSQ--TPTWRRGPSGKGSLCNACGIKWRLKGK-DGIFKPSQKQQNRQK 315
Query: 246 RDTSADMKPSTTEPENSCADQAEEGTPEENKPVPMDSRETPEKKNEQDILGTAQTVTDNL 305
SA +P + ++ P++ +P + P+++N+Q Q +
Sbjct: 316 PIMSAQKQP-----------KQQQNQPQQQQPQQPQQPQQPQQQNQQQ---NQQQSIQSQ 361
Query: 306 SIQLENHALSLDEQDI 321
Q +N+ L L++Q I
Sbjct: 362 PQQRQNNHLILNQQTI 377
>gb|AAB36702.1| putative transcription factor [Dictyostelium discoideum]
Length = 872
Score = 63.5 bits (153), Expect = 9e-09
Identities = 40/136 (29%), Positives = 67/136 (48%), Gaps = 17/136 (12%)
Query: 186 PESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNE 245
P S R C+ CG+S+ TP RRGP+G SLCNACG+ W KG + K ++ + +
Sbjct: 287 PMSAPRSCEFCGSSQ--TPTWRRGPSGKGSLCNACGIKWRLKGK-DGIFKPSQKQQNRQK 343
Query: 246 RDTSADMKPSTTEPENSCADQAEEGTPEENKPVPMDSRETPEKKNEQDILGTAQTVTDNL 305
SA +P + ++ P++ +P + P+++N+Q Q +
Sbjct: 344 PIMSAQKQP-----------KQQQNQPQQQQPQQPQQPQQPQQQNQQQ---NQQQSIQSQ 389
Query: 306 SIQLENHALSLDEQDI 321
Q +N+ L L++Q I
Sbjct: 390 PQQRQNNHLILNQQTI 405
>dbj|BAA96939.1| unnamed protein product [Arabidopsis thaliana]
gi|18424319|ref|NP_568919.1| pseudo-response regulator 3
(APRR3) [Arabidopsis thaliana]
gi|52783244|sp|Q9LVG4|APRR3_ARATH Two-component response
regulator-like APRR3 (Pseudo-response regulator 3)
gi|10281008|dbj|BAB13744.1| pseudo-response regulator 3
[Arabidopsis thaliana]
Length = 495
Score = 61.2 bits (147), Expect = 4e-08
Identities = 32/81 (39%), Positives = 49/81 (59%)
Query: 91 PNSTPNSGLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKE 150
P+ +P + LL + S S ++R A+L++FR KRKERCFEKK+RY RK+
Sbjct: 410 PHDSPIAKLLGSSSSSDNPLKQQSSGSDRWAQREAALMKFRLKRKERCFEKKVRYHSRKK 469
Query: 151 VAQRMHRKNGQFASLKEDYKS 171
+A++ GQF ++D+KS
Sbjct: 470 LAEQRPHVKGQFIRKRDDHKS 490
>gb|AAY62605.1| pseudo response regulator 3 [Arabidopsis thaliana]
Length = 495
Score = 61.2 bits (147), Expect = 4e-08
Identities = 32/81 (39%), Positives = 49/81 (59%)
Query: 91 PNSTPNSGLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKE 150
P+ +P + LL + S S ++R A+L++FR KRKERCFEKK+RY RK+
Sbjct: 410 PHDSPIAKLLGSSSSSDNPLKQQSSGSDRWAQREAALMKFRLKRKERCFEKKVRYHSRKK 469
Query: 151 VAQRMHRKNGQFASLKEDYKS 171
+A++ GQF ++D+KS
Sbjct: 470 LAEQRPHVKGQFIRKRDDHKS 490
>gb|AAY62604.1| pseudo response regulator 3 [Arabidopsis thaliana]
Length = 495
Score = 61.2 bits (147), Expect = 4e-08
Identities = 32/81 (39%), Positives = 49/81 (59%)
Query: 91 PNSTPNSGLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKE 150
P+ +P + LL + S S ++R A+L++FR KRKERCFEKK+RY RK+
Sbjct: 410 PHDSPIAKLLGSSSSSDNPLKQQSSGSDRWAQREAALMKFRLKRKERCFEKKVRYHSRKK 469
Query: 151 VAQRMHRKNGQFASLKEDYKS 171
+A++ GQF ++D+KS
Sbjct: 470 LAEQRPHVKGQFIRKRDDHKS 490
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.311 0.128 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 633,411,262
Number of Sequences: 2540612
Number of extensions: 28376101
Number of successful extensions: 81128
Number of sequences better than 10.0: 856
Number of HSP's better than 10.0 without gapping: 362
Number of HSP's successfully gapped in prelim test: 495
Number of HSP's that attempted gapping in prelim test: 80334
Number of HSP's gapped (non-prelim): 1252
length of query: 354
length of database: 863,360,394
effective HSP length: 129
effective length of query: 225
effective length of database: 535,621,446
effective search space: 120514825350
effective search space used: 120514825350
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0100b.1


