

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100a.5
(201 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
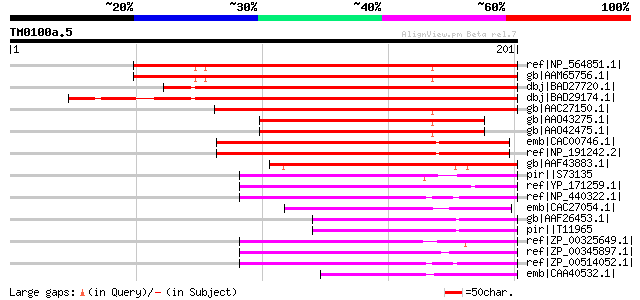
ref|NP_564851.1| expressed protein [Arabidopsis thaliana] 208 8e-53
gb|AAM65756.1| antigen receptor, putative [Arabidopsis thaliana] 207 1e-52
dbj|BAD27720.1| antigen receptor-like [Oryza sativa (japonica cu... 202 6e-51
dbj|BAD29174.1| antigen receptor-like protein [Oryza sativa (jap... 191 8e-48
gb|AAC27150.1| T8F5.20 [Arabidopsis thaliana] gi|7430888|pir||T0... 186 3e-46
gb|AAO43275.1| putative antigen receptor [Arabidopsis thaliana] ... 145 5e-34
gb|AAO42475.1| putative antigen receptor [Arabidopsis lyrata] 145 5e-34
emb|CAC00746.1| putative protein [Arabidopsis thaliana] gi|45752... 105 6e-22
ref|NP_191242.2| expressed protein [Arabidopsis thaliana] 105 6e-22
gb|AAF43883.1| hypothetical chloroplast RF20 [Mesostigma viride]... 96 6e-19
pir||S73135 probable allophycocyanin linker protein - red alga (... 72 7e-12
ref|YP_171259.1| hypothetical protein YCF20 [Synechococcus elong... 69 8e-11
ref|NP_440322.1| hypothetical protein sll1509 [Synechocystis sp.... 67 2e-10
emb|CAC27054.1| hypothetical protein [Guillardia theta] gi|13812... 60 3e-08
gb|AAF26453.1| unknown [Cyanidium caldarium] gi|11465590|ref|NP_... 59 6e-08
pir||T11965 hypothetical protein ORF96 - red alga (Cyanidium cal... 59 6e-08
ref|ZP_00325649.1| hypothetical protein Tery02004394 [Trichodesm... 56 5e-07
ref|ZP_00345897.1| hypothetical protein Npun02000315 [Nostoc pun... 56 5e-07
ref|ZP_00514052.1| Protein of unknown function DUF565 [Crocospha... 54 2e-06
emb|CAA40532.1| apcB L9.5 [Cyanidium caldarium] gi|256563|gb|AAB... 53 6e-06
>ref|NP_564851.1| expressed protein [Arabidopsis thaliana]
Length = 197
Score = 208 bits (529), Expect = 8e-53
Identities = 107/155 (69%), Positives = 128/155 (82%), Gaps = 3/155 (1%)
Query: 50 FPQSGFLGRKAVWKIAFALNTGG-VPGN-GEQQSLNDTSSSFGGTRLGRILSAGGRQLLD 107
+P F R+ KIAFAL+TG +PG+ GE Q +N + G TRLGRI AGG+QLL
Sbjct: 43 YPMKSFQIRRPNRKIAFALDTGSSIPGDSGEGQEMNGDRTGLGSTRLGRIAIAGGKQLLG 102
Query: 108 KLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLLYRKP- 166
K+NSARKN PMKIFLLLLGFYTANALATILGQTGDWDVLVAG+VVAAIEGIGML+Y+KP
Sbjct: 103 KINSARKNFPMKIFLLLLGFYTANALATILGQTGDWDVLVAGIVVAAIEGIGMLMYKKPS 162
Query: 167 PTVRTGRLQSFLLMLNYWKAGICLGLFVDAFKLGS 201
++ +G+LQSF++ +N+WKAG+CLGLFVDAFKLGS
Sbjct: 163 SSMFSGKLQSFVVFMNFWKAGVCLGLFVDAFKLGS 197
>gb|AAM65756.1| antigen receptor, putative [Arabidopsis thaliana]
Length = 197
Score = 207 bits (528), Expect = 1e-52
Identities = 107/155 (69%), Positives = 128/155 (82%), Gaps = 3/155 (1%)
Query: 50 FPQSGFLGRKAVWKIAFALNTGG-VPGN-GEQQSLNDTSSSFGGTRLGRILSAGGRQLLD 107
+P F R+ KIAFAL+TG +PG+ GE Q +N + G TRLGRI AGG+QLL
Sbjct: 43 YPMKSFQIRRPNRKIAFALDTGSSIPGDSGEGQEMNGDRTGLGSTRLGRIAIAGGKQLLA 102
Query: 108 KLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLLYRKP- 166
K+NSARKN PMKIFLLLLGFYTANALATILGQTGDWDVLVAG+VVAAIEGIGML+Y+KP
Sbjct: 103 KINSARKNFPMKIFLLLLGFYTANALATILGQTGDWDVLVAGIVVAAIEGIGMLMYKKPS 162
Query: 167 PTVRTGRLQSFLLMLNYWKAGICLGLFVDAFKLGS 201
++ +G+LQSF++ +N+WKAG+CLGLFVDAFKLGS
Sbjct: 163 SSMFSGKLQSFVVFMNFWKAGVCLGLFVDAFKLGS 197
>dbj|BAD27720.1| antigen receptor-like [Oryza sativa (japonica cultivar-group)]
Length = 217
Score = 202 bits (513), Expect = 6e-51
Identities = 100/140 (71%), Positives = 111/140 (78%), Gaps = 1/140 (0%)
Query: 62 WKIAFALNTGGVPGNGEQQSLNDTSSSFGGTRLGRILSAGGRQLLDKLNSARKNVPMKIF 121
WK FAL TGG P N + Q D G TRLGR++ A GR+LL+KLNSAR N P KIF
Sbjct: 79 WKPVFALETGG-PSNADSQDFEDDGGFLGRTRLGRLIQAAGRELLEKLNSARSNSPTKIF 137
Query: 122 LLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLLYRKPPTVRTGRLQSFLLML 181
L+L GFYTANALATILGQTGDWDVLVAGVVVAAIEGIGML+YRKP + GR QS + M+
Sbjct: 138 LVLFGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLMYRKPMSRPPGRFQSLIAMV 197
Query: 182 NYWKAGICLGLFVDAFKLGS 201
NYWKAG+CLGLFVDAFKLGS
Sbjct: 198 NYWKAGVCLGLFVDAFKLGS 217
>dbj|BAD29174.1| antigen receptor-like protein [Oryza sativa (japonica
cultivar-group)] gi|50251186|dbj|BAD29674.1| antigen
receptor-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 196
Score = 191 bits (486), Expect = 8e-48
Identities = 100/178 (56%), Positives = 118/178 (66%), Gaps = 10/178 (5%)
Query: 24 ASRVHGCCCEPILSGLTSNAGGKSSHFPQSGFLGRKAVWKIAFALNTGGVPGNGEQQSLN 83
A+ +H C P G T A G S ++ WK FAL TGG P N + Q
Sbjct: 29 AALLHPTCLSP--PGTTGRALGLSCQM-------KRTRWKPVFALETGG-PSNADNQDFE 78
Query: 84 DTSSSFGGTRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGDW 143
D G TRLGR++ A R+LL+KLNSAR P KIFL+LLGFYTANALAT+LGQTGDW
Sbjct: 79 DDGGFLGRTRLGRLIQAAARELLEKLNSARNKSPTKIFLVLLGFYTANALATVLGQTGDW 138
Query: 144 DVLVAGVVVAAIEGIGMLLYRKPPTVRTGRLQSFLLMLNYWKAGICLGLFVDAFKLGS 201
DV VA +VVA IEGIGML+YRKP + GR S + M+NYWKAG+CLG FVDAFK+GS
Sbjct: 139 DVFVAAIVVATIEGIGMLMYRKPASRPPGRFWSMITMVNYWKAGVCLGFFVDAFKVGS 196
>gb|AAC27150.1| T8F5.20 [Arabidopsis thaliana] gi|7430888|pir||T02365 hypothetical
protein T8F5.20 - Arabidopsis thaliana
gi|6136502|sp|O80813|Y20L_ARATH Ycf20-like protein
Length = 121
Score = 186 bits (473), Expect = 3e-46
Identities = 91/121 (75%), Positives = 107/121 (88%), Gaps = 1/121 (0%)
Query: 82 LNDTSSSFGGTRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTG 141
+N + G TRLGRI AGG+QLL K+NSARKN PMKIFLLLLGFYTANALATILGQTG
Sbjct: 1 MNGDRTGLGSTRLGRIAIAGGKQLLGKINSARKNFPMKIFLLLLGFYTANALATILGQTG 60
Query: 142 DWDVLVAGVVVAAIEGIGMLLYRKP-PTVRTGRLQSFLLMLNYWKAGICLGLFVDAFKLG 200
DWDVLVAG+VVAAIEGIGML+Y+KP ++ +G+LQSF++ +N+WKAG+CLGLFVDAFKLG
Sbjct: 61 DWDVLVAGIVVAAIEGIGMLMYKKPSSSMFSGKLQSFVVFMNFWKAGVCLGLFVDAFKLG 120
Query: 201 S 201
S
Sbjct: 121 S 121
>gb|AAO43275.1| putative antigen receptor [Arabidopsis thaliana]
gi|28436414|gb|AAO43274.1| putative antigen receptor
[Arabidopsis thaliana] gi|28436412|gb|AAO43273.1|
putative antigen receptor [Arabidopsis thaliana]
gi|28436410|gb|AAO43272.1| putative antigen receptor
[Arabidopsis thaliana] gi|28436408|gb|AAO43271.1|
putative antigen receptor [Arabidopsis thaliana]
gi|28436406|gb|AAO43270.1| putative antigen receptor
[Arabidopsis thaliana] gi|28436404|gb|AAO43269.1|
putative antigen receptor [Arabidopsis thaliana]
gi|28436402|gb|AAO43268.1| putative antigen receptor
[Arabidopsis thaliana] gi|28436400|gb|AAO43267.1|
putative antigen receptor [Arabidopsis thaliana]
Length = 92
Score = 145 bits (367), Expect = 5e-34
Identities = 70/90 (77%), Positives = 84/90 (92%), Gaps = 1/90 (1%)
Query: 100 AGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIG 159
AGG+QLL K+NSARKN PMKIFLLLLGFYTANALATILGQTGDWDVLVAG+VVAAIEGIG
Sbjct: 3 AGGKQLLGKINSARKNFPMKIFLLLLGFYTANALATILGQTGDWDVLVAGIVVAAIEGIG 62
Query: 160 MLLYRKP-PTVRTGRLQSFLLMLNYWKAGI 188
ML+Y+KP ++ +G+LQSF++ +N+WKAG+
Sbjct: 63 MLMYKKPSSSMFSGKLQSFVVFMNFWKAGV 92
>gb|AAO42475.1| putative antigen receptor [Arabidopsis lyrata]
Length = 92
Score = 145 bits (367), Expect = 5e-34
Identities = 70/90 (77%), Positives = 84/90 (92%), Gaps = 1/90 (1%)
Query: 100 AGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIG 159
AGG+QLL K+NSARKN PMKIFLLLLGFYTANALATILGQTGDWDVLVAG+VVAAIEGIG
Sbjct: 3 AGGKQLLGKINSARKNFPMKIFLLLLGFYTANALATILGQTGDWDVLVAGIVVAAIEGIG 62
Query: 160 MLLYRKP-PTVRTGRLQSFLLMLNYWKAGI 188
ML+Y+KP ++ +G+LQSF++ +N+WKAG+
Sbjct: 63 MLMYKKPSSSMFSGKLQSFVVFMNFWKAGV 92
>emb|CAC00746.1| putative protein [Arabidopsis thaliana] gi|45752730|gb|AAS76263.1|
At3g56830 [Arabidopsis thaliana] gi|11358429|pir||T51271
hypothetical protein T8M16_160 - Arabidopsis thaliana
Length = 188
Score = 105 bits (263), Expect = 6e-22
Identities = 51/116 (43%), Positives = 78/116 (66%), Gaps = 1/116 (0%)
Query: 83 NDTSSSFGGTRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGD 142
+ +++S G RL + + +LL K+ +K++P K+F LL+GFY+A A +T +GQTGD
Sbjct: 73 SSSNNSSRGLRLIKAIQVLRSKLLVKIQEIKKDLPKKLFFLLVGFYSATAFSTFIGQTGD 132
Query: 143 WDVLVAGVVVAAIEGIGMLLYRKPPTVRTGRLQSFLLMLNYWKAGICLGLFVDAFK 198
WDVL AG+ V +E IG L+YR + +++S + M NYWK G+ LGLF+D+FK
Sbjct: 133 WDVLSAGLAVLVVECIGALMYRASIPL-INKMRSTITMFNYWKTGLALGLFLDSFK 187
>ref|NP_191242.2| expressed protein [Arabidopsis thaliana]
Length = 230
Score = 105 bits (263), Expect = 6e-22
Identities = 51/116 (43%), Positives = 78/116 (66%), Gaps = 1/116 (0%)
Query: 83 NDTSSSFGGTRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGD 142
+ +++S G RL + + +LL K+ +K++P K+F LL+GFY+A A +T +GQTGD
Sbjct: 73 SSSNNSSRGLRLIKAIQVLRSKLLVKIQEIKKDLPKKLFFLLVGFYSATAFSTFIGQTGD 132
Query: 143 WDVLVAGVVVAAIEGIGMLLYRKPPTVRTGRLQSFLLMLNYWKAGICLGLFVDAFK 198
WDVL AG+ V +E IG L+YR + +++S + M NYWK G+ LGLF+D+FK
Sbjct: 133 WDVLSAGLAVLVVECIGALMYRASIPL-INKMRSTITMFNYWKTGLALGLFLDSFK 187
>gb|AAF43883.1| hypothetical chloroplast RF20 [Mesostigma viride]
gi|11466439|ref|NP_038445.1| hypothetical chloroplast
RF20 [Mesostigma viride]
gi|12230812|sp|Q9MUL5|YC20_MESVI Hypothetical 13.0 kDa
protein ycf20 (RF20)
Length = 116
Score = 95.9 bits (237), Expect = 6e-19
Identities = 56/102 (54%), Positives = 69/102 (66%), Gaps = 4/102 (3%)
Query: 104 QLLD-KLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLL 162
QLLD K+ + N P+KI LLLLGF+ A LAT+ GQTGDWDVLVAG++VA IE +G +
Sbjct: 15 QLLDSKIQNYSINFPIKILLLLLGFFIATVLATVFGQTGDWDVLVAGILVAMIEILGNKM 74
Query: 163 YRKPPTVRTGRLQ-SFLLM--LNYWKAGICLGLFVDAFKLGS 201
Y K + SFL + +NY K G+ GLFVDAFKLGS
Sbjct: 75 YSKKYISKKQVFDISFLSLIGINYIKIGLIFGLFVDAFKLGS 116
>pir||S73135 probable allophycocyanin linker protein - red alga (Porphyra
purpurea) chloroplast gi|1276680|gb|AAC08100.1|
hypothetical chloroplast ORF 20. [Porphyra purpurea]
gi|11465680|ref|NP_053824.1| ORF20 [Porphyra purpurea]
gi|1723324|sp|P51214|YC20_PORPU HYPOTHETICAL 11.9 KD
PROTEIN YCF20 (ORF108)
Length = 108
Score = 72.4 bits (176), Expect = 7e-12
Identities = 45/112 (40%), Positives = 64/112 (56%), Gaps = 9/112 (8%)
Query: 92 TRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVV 151
T+L S + L +L + + + LLLGF+ + L+TI GQTGDW ++ A ++
Sbjct: 4 TKLSIFFSYFVQNLSSRLYYSLNELTAGLISLLLGFFISTGLSTIPGQTGDWGIIAASLI 63
Query: 152 VAAIEGIGMLLY--RKPPTVRTGRLQSFLLMLNYWKAGICLGLFVDAFKLGS 201
VAAIE + ++Y +K VR + +LN K GI GLFVDAFKLGS
Sbjct: 64 VAAIELVSKIVYSNKKKYGVR-------INLLNNLKIGITYGLFVDAFKLGS 108
>ref|YP_171259.1| hypothetical protein YCF20 [Synechococcus elongatus PCC 6301]
gi|56685517|dbj|BAD78739.1| hypothetical protein YCF20
[Synechococcus elongatus PCC 6301]
gi|46129623|ref|ZP_00164134.2| hypothetical protein
Selo03000279 [Synechococcus elongatus PCC 7942]
Length = 112
Score = 68.9 bits (167), Expect = 8e-11
Identities = 38/110 (34%), Positives = 58/110 (52%), Gaps = 1/110 (0%)
Query: 92 TRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVV 151
TRL +++S G ++L + + ++I LL GF+ N L+ + GQ+G WD A V+
Sbjct: 4 TRLNQLVSEAGGRVLQWFQQPWRRLSLQIIFLLFGFWLTNTLSLVAGQSGVWDPSFAAVI 63
Query: 152 VAAIEGIGMLLYRKPPTVRTGRLQSFLLMLNYWKAGICLGLFVDAFKLGS 201
++E L YR P + R FLL+ N K G GL ++ KLGS
Sbjct: 64 TISVETCSWLYYRGPGAGKPDRSLPFLLLQNL-KLGALYGLALETLKLGS 112
>ref|NP_440322.1| hypothetical protein sll1509 [Synechocystis sp. PCC 6803]
gi|1652077|dbj|BAA17002.1| ycf20 [Synechocystis sp. PCC
6803] gi|6136503|sp|P72983|YC20L_SYNY3 Ycf20-like
protein
Length = 109
Score = 67.4 bits (163), Expect = 2e-10
Identities = 38/110 (34%), Positives = 59/110 (53%), Gaps = 4/110 (3%)
Query: 92 TRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVV 151
TRL I+ G+QL + + + + + L GF+ A+AT GQ WDV+ A +
Sbjct: 4 TRLNTIVEVRGQQLSQFFRNPWRRISLSLLSFLFGFFVGTAVATTAGQNSQWDVVCAAFI 63
Query: 152 VAAIEGIGMLLYRKPPTVRTGRLQSFLLMLNYWKAGICLGLFVDAFKLGS 201
+ E + YR+ V+ G LQ+ +LN +K G+ LF++AFKLGS
Sbjct: 64 LLFCELVNRWFYRR--GVKMGDLQA--EVLNIFKMGVSYSLFLEAFKLGS 109
>emb|CAC27054.1| hypothetical protein [Guillardia theta]
gi|13812367|ref|NP_113485.1| hypothetical protein
[Guillardia theta]
Length = 125
Score = 60.5 bits (145), Expect = 3e-08
Identities = 32/90 (35%), Positives = 49/90 (53%), Gaps = 6/90 (6%)
Query: 110 NSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLLYRKPPTV 169
N+ ++++ F LL GF++A + TI+G DWD L A V++ IE + Y+
Sbjct: 40 NNLKRDIYYGSFFLLFGFFSATSAITIIGSVADWDPLAAAVMLCWIEVFNKIFYKNISK- 98
Query: 170 RTGRLQSFLLMLNYWKAGICLGLFVDAFKL 199
+ +N +K GI LG+FVDAFKL
Sbjct: 99 -----KKIFNKINIFKIGINLGIFVDAFKL 123
>gb|AAF26453.1| unknown [Cyanidium caldarium] gi|11465590|ref|NP_045069.1|
hypothetical protein CycaCp052 [Cyanidium caldarium]
gi|14548001|sp|Q9MVP1|YC20_CYACA Hypothetical 11.5 kDa
protein ycf20
Length = 101
Score = 59.3 bits (142), Expect = 6e-08
Identities = 30/81 (37%), Positives = 47/81 (57%), Gaps = 1/81 (1%)
Query: 121 FLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLLYRKPPTVRTGRLQSFLLM 180
F+ L GF+ ++A +TIL QT +W +L A ++++ +E L ++ R F
Sbjct: 22 FIFLFGFFLSSATSTILIQTNEWSILTAAILISIVELFNYLKHKFQFNDRKSGYNCF-FF 80
Query: 181 LNYWKAGICLGLFVDAFKLGS 201
+N K G+ GLF+DAFKLGS
Sbjct: 81 INLAKLGLLYGLFIDAFKLGS 101
>pir||T11965 hypothetical protein ORF96 - red alga (Cyanidium caldarium)
chloroplast
Length = 96
Score = 59.3 bits (142), Expect = 6e-08
Identities = 30/81 (37%), Positives = 47/81 (57%), Gaps = 1/81 (1%)
Query: 121 FLLLLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLLYRKPPTVRTGRLQSFLLM 180
F+ L GF+ ++A +TIL QT +W +L A ++++ +E L ++ R F
Sbjct: 17 FIFLFGFFLSSATSTILIQTNEWSILTAAILISIVELFNYLKHKFQFNDRKSGYNCF-FF 75
Query: 181 LNYWKAGICLGLFVDAFKLGS 201
+N K G+ GLF+DAFKLGS
Sbjct: 76 INLAKLGLLYGLFIDAFKLGS 96
>ref|ZP_00325649.1| hypothetical protein Tery02004394 [Trichodesmium erythraeum IMS101]
Length = 110
Score = 56.2 bits (134), Expect = 5e-07
Identities = 38/112 (33%), Positives = 60/112 (52%), Gaps = 7/112 (6%)
Query: 92 TRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVV 151
TRL R+ + ++ L + + + + + LL G + A+A++TI GQ G DVL
Sbjct: 4 TRLNRLFNVILKRFEQWLVNPWRRISILLISLLFGNFAASAVSTIAGQEGYLDVLYTLAC 63
Query: 152 VAAIEGIGMLLYRKPPTVRTGRLQSFLL--MLNYWKAGICLGLFVDAFKLGS 201
+ E + L+Y R G++ FL+ +LN K G GLF++AFKLGS
Sbjct: 64 LFITEALNWLVYG-----RRGKVARFLIIDILNSLKIGFIYGLFLEAFKLGS 110
>ref|ZP_00345897.1| hypothetical protein Npun02000315 [Nostoc punctiforme PCC 73102]
Length = 110
Score = 56.2 bits (134), Expect = 5e-07
Identities = 36/110 (32%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 92 TRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVV 151
TRL +L A +L + + + + I L GF+ NA++T GQ + D++VAG +
Sbjct: 4 TRLNNLLDAIATRLGQWFLNPWRRLSLLIISFLFGFFLGNAVSTTAGQRAELDIVVAGFL 63
Query: 152 VAAIEGIGMLLYRKPPTVRTGRLQSFLLMLNYWKAGICLGLFVDAFKLGS 201
V E + Y + R F+ LN K G LF++AFKLGS
Sbjct: 64 VVLTEVTSRIFYSQSFFTRR---SLFVDSLNLLKVGFTYSLFLEAFKLGS 110
>ref|ZP_00514052.1| Protein of unknown function DUF565 [Crocosphaera watsonii WH 8501]
gi|67858016|gb|EAM53255.1| Protein of unknown function
DUF565 [Crocosphaera watsonii WH 8501]
Length = 110
Score = 54.3 bits (129), Expect = 2e-06
Identities = 32/110 (29%), Positives = 58/110 (52%), Gaps = 3/110 (2%)
Query: 92 TRLGRILSAGGRQLLDKLNSARKNVPMKIFLLLLGFYTANALATILGQTGDWDVLVAGVV 151
TRL ++ + + ++ + + + + +LLGF+ A++ GQ WD+ V +
Sbjct: 4 TRLNTLVEVTQTKFNETFSNPWRRISLSLISVLLGFFVGQAVSITAGQQTYWDITVGIFL 63
Query: 152 VAAIEGIGMLLYRKPPTVRTGRLQSFLLMLNYWKAGICLGLFVDAFKLGS 201
+ EGI + Y + + + GR S L +LN +K G+ L+V+A KLGS
Sbjct: 64 LIFTEGISRITYSR--SKKEGRSLS-LDILNLFKIGVTYSLYVEALKLGS 110
>emb|CAA40532.1| apcB L9.5 [Cyanidium caldarium] gi|256563|gb|AAB50632.1| putative
9.5 kda allophycocyanin linker protein [Cyanidium
caldarium] gi|282698|pir||S25307 probable
allophycocyanin linker protein - red alga (Cyanidium
caldarium) chloroplast gi|1351745|sp|P48409|YC20_GALSU
Hypothetical 9.5 kDa protein ycf20
Length = 83
Score = 52.8 bits (125), Expect = 6e-06
Identities = 28/78 (35%), Positives = 44/78 (55%), Gaps = 3/78 (3%)
Query: 124 LLGFYTANALATILGQTGDWDVLVAGVVVAAIEGIGMLLYRKPPTVRTGRLQSFLLMLNY 183
LLG +++N L TI QTGDW + + ++A E I + Y + ++ + N
Sbjct: 9 LLGIFSSNLLCTIYTQTGDWSLYLTSCIIALYEIISYISYNQ---FIKKTHKNIINCFNG 65
Query: 184 WKAGICLGLFVDAFKLGS 201
+K G+ GL++DAFKLGS
Sbjct: 66 FKIGLIYGLYLDAFKLGS 83
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 347,678,427
Number of Sequences: 2540612
Number of extensions: 14265078
Number of successful extensions: 31615
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 31554
Number of HSP's gapped (non-prelim): 56
length of query: 201
length of database: 863,360,394
effective HSP length: 122
effective length of query: 79
effective length of database: 553,405,730
effective search space: 43719052670
effective search space used: 43719052670
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0100a.5


