

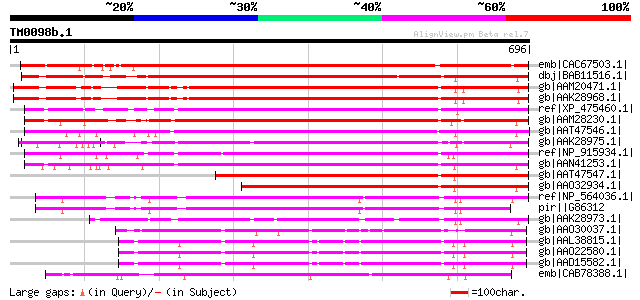
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098b.1
(696 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC67503.1| SET-domain-containing protein [Nicotiana tabacum] 784 0.0
dbj|BAB11516.1| SET-domain protein-like [Arabidopsis thaliana] g... 615 e-174
gb|AAM20471.1| unknown protein [Arabidopsis thaliana] gi|1841026... 613 e-174
gb|AAK28968.1| SUVH3 [Arabidopsis thaliana] 613 e-174
ref|XP_475460.1| 'unknown protein, conatins SET domain' [Oryza s... 586 e-166
gb|AAM28230.1| SET domain protein 105 [Zea mays] 575 e-162
gb|AAT47546.1| SET domain protein [Triticum aestivum] 556 e-157
gb|AAK28975.1| SET1 [Oryza sativa] 554 e-156
ref|NP_915934.1| similar to SET1 [Oryza sativa (japonica cultiva... 552 e-155
gb|AAN41253.1| SET domain protein 113 [Zea mays] 536 e-151
gb|AAT47547.1| SET domain protein [Triticum aestivum] 447 e-124
gb|AAO32934.1| SET domain protein SDG111 [Zea mays] 424 e-117
ref|NP_564036.1| SET domain-containing protein (SUVH7) [Arabidop... 416 e-114
pir||G86312 hypothetical protein F2H15.1 - Arabidopsis thaliana ... 397 e-109
gb|AAK28973.1| SUVH8 [Arabidopsis thaliana] gi|30580523|sp|Q9C5P... 377 e-103
gb|AAO30037.1| putative SET-domain protein [Arabidopsis thaliana... 372 e-101
gb|AAL38815.1| putative mammalian MHC III region protein G9a [Ar... 359 2e-97
gb|AAO22580.1| putative mammalian MHC III region protein G9a [Ar... 357 7e-97
gb|AAD15582.1| similar to mammalian MHC III region protein G9a [... 357 7e-97
emb|CAB78388.1| putative protein [Arabidopsis thaliana] gi|46783... 351 4e-95
>emb|CAC67503.1| SET-domain-containing protein [Nicotiana tabacum]
Length = 704
Score = 784 bits (2025), Expect = 0.0
Identities = 403/706 (57%), Positives = 505/706 (71%), Gaps = 41/706 (5%)
Query: 15 SFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGVAPFYPFFVS 74
S DK+RVL+VKPLR L PVFPSP+ SS +TPQ +PFVCV P+GPFP GVAPFYPF
Sbjct: 14 SIDKTRVLDVKPLRCLAPVFPSPNGMSSVSTPQP-SPFVCVPPTGPFPPGVAPFYPFVAP 72
Query: 75 PESQRLSEQNAQTPTAQR--------AAPISAAVPINSFRTPTGATNGDVGSSRRK---- 122
+S R E + QTP+ A PIS VP+NSFRTPT A NG+ G SRR
Sbjct: 73 NDSGRPGESSQQTPSGVPNQGGPFGFAQPISP-VPLNSFRTPTTA-NGNSGRSRRAVDDD 130
Query: 123 --SRGQLPEDDNF-----VDLSEVDGEGGTGDGKRRKPQKRIREKRCSS----DVDPDAV 171
S Q ++D F V ++ V+ + GTG KR +P+K R ++ +VD + +
Sbjct: 131 DYSNSQ-DQNDQFASGFSVHVNNVE-DSGTGK-KRGRPKKPRRAQQAEGLTPVEVDVEPL 187
Query: 172 ANEILKTINPGVFEILNQPDGSRDAVAYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLK 231
++L + + + + DG ++ L+++++ RR++ QIDE G SG +RPDLK
Sbjct: 188 LTQLLTSFKLVDLDQVKKADGDKELAGRVLLVFDLFRRRMTQIDESRDGPGSG-RRPDLK 246
Query: 232 AGTLMNTKGIRANSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEE 291
A ++ TKG+R N KRIG PG+EVGDIFFFR ELCLVGLHAP+MAGIDY+ K++ +E
Sbjct: 247 ASNMLMTKGVRTNQTKRIGNAPGIEVGDIFFFRMELCLVGLHAPTMAGIDYMSVKLTMDE 306
Query: 292 EPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTSREKG-ASDQKLERGNLALERSLHRGNDV 350
EPLAVSIVSSGGY+D+ DGDVLIY+GQGG R+ G DQKLERGNLALE+S+HR N+V
Sbjct: 307 EPLAVSIVSSGGYDDDGGDGDVLIYTGQGGVQRKDGQVFDQKLERGNLALEKSVHRANEV 366
Query: 351 RVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSI 410
RVIRG++D A+PTGK+Y+YDGLYKIQ SW EK K G NVFKYKL+R+PGQP+A+ +WKSI
Sbjct: 367 RVIRGVKDVAYPTGKIYIYDGLYKIQESWAEKNKVGCNVFKYKLLRVPGQPEAFKVWKSI 426
Query: 411 LQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESS 470
QW D ASRVGVILPDLTSGAE PVCLVNDVD+EKGPAYFTY P+LK S
Sbjct: 427 QQWKDGVASRVGVILPDLTSGAESQPVCLVNDVDDEKGPAYFTYIPSLKYSKPFVMPRPS 486
Query: 471 EGCTCNGGCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQ 530
C C GGCQPG C C Q NGG+LPYS+ G+L K++++ECG +C CPP+CRNR+SQ
Sbjct: 487 PSCHCVGGCQPGDSNCACIQSNGGFLPYSSLGVLLSYKTLIHECGSACSCPPNCRNRMSQ 546
Query: 531 GGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDST 590
GG K RLEVF+TK +GWGLRSWDPIR G FICEYAGEVID ++D+YIFD+T
Sbjct: 547 GGPKARLEVFKTKNRGWGLRSWDPIRGGGFICEYAGEVID------AGNYSDDNYIFDAT 600
Query: 591 RIYQQLEVFSS-DVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADL 649
RIY LE + E+ K+P PL I+A+N GN++RFMNHSC+PNV W+ VVR++ NEA
Sbjct: 601 RIYAPLEAERDYNDESRKVPFPLVISAKNGGNISRFMNHSCSPNVYWQLVVRQSNNEATY 660
Query: 650 HVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKCLCGSVKCRGYF 695
H+AF+AIRHIPPM ELT+DYG+ K ++KKCLCGS+ CRGYF
Sbjct: 661 HIAFFAIRHIPPMQELTFDYGMD---KADHRRKKCLCGSLNCRGYF 703
>dbj|BAB11516.1| SET-domain protein-like [Arabidopsis thaliana]
gi|30680715|ref|NP_850767.1| SET domain-containing
protein (SUVH1) [Arabidopsis thaliana]
gi|15238375|ref|NP_196113.1| SET domain-containing
protein (SUVH1) [Arabidopsis thaliana]
gi|13517743|gb|AAK28966.1| SUVH1 [Arabidopsis thaliana]
gi|30580528|sp|Q9FF80|SUV1_ARATH Histone-lysine
N-methyltransferase, H3 lysine-9 specific 1 (Histone
H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor
of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1)
Length = 670
Score = 615 bits (1587), Expect = e-174
Identities = 325/696 (46%), Positives = 434/696 (61%), Gaps = 53/696 (7%)
Query: 17 DKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGVAPFYPFFVSPE 76
DK+RVL++KPLRTL PVFPS + PFVC P GPFP G + FYPF S
Sbjct: 10 DKTRVLDIKPLRTLRPVFPSGNQ---------APPFVCAPPFGPFPPGFSSFYPFSSSQA 60
Query: 77 SQRLSEQNAQTPTAQRAAPISAAVPINSFRTPTGATNGDVGSSRRKSRGQLPEDDNFVDL 136
+Q + N Q P + P+ + P A+ + + R R D+
Sbjct: 61 NQHTPDLNQAQYPPQHQQPQNPP-PVYQQQPPQHASEPSLVTPLRSFRSP--------DV 111
Query: 137 SEVDGEGGTGDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDA 196
S + E KRR P+KR ++ G+ + ++ +G+R+
Sbjct: 112 SNGNAELEGSTVKRRIPKKR-------------PISRPENMNFESGI-NVADRENGNREL 157
Query: 197 VAYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVE 256
V LM ++ +RR+ Q+++ + KRPDLK+G+ +G+R N++KR G+VPGVE
Sbjct: 158 VLSVLMRFDALRRRFAQLEDAKEAVSGIIKRPDLKSGSTCMGRGVRTNTKKRPGIVPGVE 217
Query: 257 VGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIY 316
+GD+FFFRFE+CLVGLH+PSMAGIDYL K EEEP+A SIVSSG Y+++ + DVLIY
Sbjct: 218 IGDVFFFRFEMCLVGLHSPSMAGIDYLVVKGETEEEPIATSIVSSGYYDNDEGNPDVLIY 277
Query: 317 SGQGGTS-REKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKI 375
+GQGG + ++K +SDQKLERGNLALE+SL R + VRVIRG+++ +H K+Y+YDGLY+I
Sbjct: 278 TGQGGNADKDKQSSDQKLERGNLALEKSLRRDSAVRVIRGLKEASH-NAKIYIYDGLYEI 336
Query: 376 QNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKL 435
+ SWVEK KSG N FKYKLVR PGQP A+ W +I +W SR G+ILPD+TSG E +
Sbjct: 337 KESWVEKGKSGHNTFKYKLVRAPGQPPAFASWTAIQKWKTGVPSRQGLILPDMTSGVESI 396
Query: 436 PVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNGGY 495
PV LVN+VD + GPAYFTYS T+K ++ S GC C C+PG+ C C +KNGG
Sbjct: 397 PVSLVNEVDTDNGPAYFTYSTTVKYSESFKLMQPSFGCDCANLCKPGNLDCHCIRKNGGD 456
Query: 496 LPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPI 555
PY+ G+L K ++YEC PSC C +C+N+V+Q G+K+RLEVF+T +GWGLRSWD I
Sbjct: 457 FPYTGNGILVSRKPMIYECSPSCPC-STCKNKVTQMGVKVRLEVFKTANRGWGLRSWDAI 515
Query: 556 RAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL-----------EVFSSDVE 604
RAG+FIC Y GE D ++V++ DDY FD+T +Y + E
Sbjct: 516 RAGSFICIYVGEAKDKSKVQQTMA--NDDYTFDTTNVYNPFKWNYEPGLADEDACEEMSE 573
Query: 605 APKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMME 664
+IP PL I+A+N GNVARFMNHSC+PNV W+PV EN ++ +HVAF+AI HIPPM E
Sbjct: 574 ESEIPLPLIISAKNVGNVARFMNHSCSPNVFWQPVSYENNSQLFVHVAFFAISHIPPMTE 633
Query: 665 LTYDYGIVLPLKVGQ-----KKKKCLCGSVKCRGYF 695
LTYDYG+ P K+KC CGS CRG F
Sbjct: 634 LTYDYGVSRPSGTQNGNPLYGKRKCFCGSAYCRGSF 669
>gb|AAM20471.1| unknown protein [Arabidopsis thaliana] gi|18410265|ref|NP_565056.1|
SET domain-containing protein (SUVH3) [Arabidopsis
thaliana] gi|5903099|gb|AAD55657.1| Unknown protein
[Arabidopsis thaliana] gi|25083988|gb|AAN72148.1|
unknown protein [Arabidopsis thaliana]
gi|30580525|sp|Q9C5P4|SUVH3_ARATH Histone-lysine
N-methyltransferase, H3 lysine-9 specific 3 (Histone
H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Suppressor
of variegation 3-9 homolog 3) (Su(var)3-9 homolog 3)
gi|25406251|pir||F96756 hypothetical protein F3N23.30
[imported] - Arabidopsis thaliana
Length = 669
Score = 613 bits (1582), Expect = e-174
Identities = 337/709 (47%), Positives = 437/709 (61%), Gaps = 65/709 (9%)
Query: 6 GQDPAPAAGSFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGV 65
G + P +DKS VL++KPLR+L PVFP+ + G PFV P GP S
Sbjct: 6 GFNTVPNPNHYDKSIVLDIKPLRSLKPVFPNGNQ---------GPPFVGCPPFGPSSSEY 56
Query: 66 APFYPFFVSPESQRLSEQNAQTPTAQRAAPISAAVP-INSFRTPTGATNGDVGSSRRKSR 124
+ F+PF + + N T PI + VP + S+RTPT TNG SS K
Sbjct: 57 SSFFPFGAQQPTHDTPDLNQTQNT-----PIPSFVPPLRSYRTPT-KTNGPSSSSGTKR- 109
Query: 125 GQLPEDDNFVDLSEVDGEGGTGDGKRRKPQKRIREKRCSSDVDPDA-VANEILKTINPGV 183
G G K K+ +K +++ + D V + + G+
Sbjct: 110 -------------------GVGRPKGTTSVKKKEKKTVANEPNLDVQVVKKFSSDFDSGI 150
Query: 184 FEILNQPDGSRDAVAYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKGIRA 243
+ DG+ V+ LM ++ +RR+L Q+ E K + S A AGTLM+ G+R
Sbjct: 151 -SAAEREDGNAYLVSSVLMRFDAVRRRLSQV-EFTKSATSKA------AGTLMSN-GVRT 201
Query: 244 NSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGG 303
N +KR+G VPG+EVGDIFF R E+CLVGLH +MAGIDY+ +K +EE LA SIVSSG
Sbjct: 202 NMKKRVGTVPGIEVGDIFFSRIEMCLVGLHMQTMAGIDYIISKAGSDEESLATSIVSSGR 261
Query: 304 YEDNVEDGDVLIYSGQGGTS-REKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHP 362
YE +D + LIYSGQGG + + + ASDQKLERGNLALE SL +GN VRV+RG D A
Sbjct: 262 YEGEAQDPESLIYSGQGGNADKNRQASDQKLERGNLALENSLRKGNGVRVVRGEEDAASK 321
Query: 363 TGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVG 422
TGK+Y+YDGLY I SWVEK KSG N FKYKLVR PGQP A+ WKS+ +W + +R G
Sbjct: 322 TGKIYIYDGLYSISESWVEKGKSGCNTFKYKLVRQPGQPPAFGFWKSVQKWKEGLTTRPG 381
Query: 423 VILPDLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPG 482
+ILPDLTSGAE PV LVNDVD +KGPAYFTY+ +LK + GC+C+G C PG
Sbjct: 382 LILPDLTSGAESKPVSLVNDVDEDKGPAYFTYTSSLKYSETFKLTQPVIGCSCSGSCSPG 441
Query: 483 SHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRT 542
+H C C +KN G LPY +L + V+YECGP+C C SC+NRV Q GLK RLEVF+T
Sbjct: 442 NHNCSCIRKNDGDLPYLNGVILVSRRPVIYECGPTCPCHASCKNRVIQTGLKSRLEVFKT 501
Query: 543 KGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL------ 596
+ +GWGLRSWD +RAG+FICEYAGEV DN + + ED Y+FD++R++
Sbjct: 502 RNRGWGLRSWDSLRAGSFICEYAGEVKDNGNLR--GNQEEDAYVFDTSRVFNSFKWNYEP 559
Query: 597 EVFSSD--VEAPK---IPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHV 651
E+ D E P+ +PSPL I+A+ GNVARFMNHSC+PNV W+PV+RE E+ +H+
Sbjct: 560 ELVDEDPSTEVPEEFNLPSPLLISAKKFGNVARFMNHSCSPNVFWQPVIREGNGESVIHI 619
Query: 652 AFYAIRHIPPMMELTYDYGIVLPLKVGQK-----KKKCLCGSVKCRGYF 695
AF+A+RHIPPM ELTYDYGI + + ++ CLCGS +CRG F
Sbjct: 620 AFFAMRHIPPMAELTYDYGISPTSEARDESLLHGQRTCLCGSEQCRGSF 668
>gb|AAK28968.1| SUVH3 [Arabidopsis thaliana]
Length = 669
Score = 613 bits (1580), Expect = e-174
Identities = 337/709 (47%), Positives = 436/709 (60%), Gaps = 65/709 (9%)
Query: 6 GQDPAPAAGSFDKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGV 65
G + P +DKS VL++KPLR+L PVFP+ + G PFV P GP S
Sbjct: 6 GFNTVPNPNHYDKSIVLDIKPLRSLKPVFPNGNQ---------GPPFVGCPPFGPSSSEY 56
Query: 66 APFYPFFVSPESQRLSEQNAQTPTAQRAAPISAAVP-INSFRTPTGATNGDVGSSRRKSR 124
+ F+PF + + N T PI + VP + S+RTPT TNG SS K
Sbjct: 57 SSFFPFGAQQPTHDTPDLNQTQNT-----PIPSFVPPLRSYRTPT-KTNGPSSSSGTKR- 109
Query: 125 GQLPEDDNFVDLSEVDGEGGTGDGKRRKPQKRIREKRCSSDVDPDA-VANEILKTINPGV 183
G G K K+ +K +++ + D V + + G+
Sbjct: 110 -------------------GVGRPKGTTSVKKKEKKTVANEPNLDVQVVKKFSSDFDSGI 150
Query: 184 FEILNQPDGSRDAVAYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKGIRA 243
+ DG+ V+ LM ++ +RR+L Q+ E K + S A AGTLM+ G+R
Sbjct: 151 -SAAEREDGNAYLVSSVLMRFDAVRRRLSQV-EFTKSATSKA------AGTLMSN-GVRT 201
Query: 244 NSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGG 303
N +KR+G VPG+EVGDIFF R E+CLVGLH +MAGIDY+ +K +EE LA SIVSSG
Sbjct: 202 NMKKRVGTVPGIEVGDIFFSRIEMCLVGLHMQTMAGIDYIISKAGSDEESLATSIVSSGR 261
Query: 304 YEDNVEDGDVLIYSGQGGTS-REKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHP 362
YE +D + LIYSGQGG + + + ASDQKLERGNLALE SL +GN VRV+RG D A
Sbjct: 262 YEGEAQDPESLIYSGQGGNADKNRQASDQKLERGNLALENSLRKGNGVRVVRGEEDAASK 321
Query: 363 TGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVG 422
TGK+Y+YDGLY I SWVEK KSG N FKYKLVR PGQP A+ WKS+ +W + +R G
Sbjct: 322 TGKIYIYDGLYSISESWVEKGKSGCNTFKYKLVRQPGQPPAFGFWKSVQKWKEGLTTRPG 381
Query: 423 VILPDLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPG 482
+ILPDLTSGAE PV LVNDVD +KGPAYFTY+ LK + GC+C+G C PG
Sbjct: 382 LILPDLTSGAESKPVSLVNDVDEDKGPAYFTYTSPLKYSETFKLTQPVIGCSCSGSCSPG 441
Query: 483 SHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRT 542
+H C C +KN G LPY +L + V+YECGP+C C SC+NRV Q GLK RLEVF+T
Sbjct: 442 NHNCSCIRKNDGDLPYLNGVILVSRRPVIYECGPTCPCHASCKNRVIQTGLKSRLEVFKT 501
Query: 543 KGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL------ 596
+ +GWGLRSWD +RAG+FICEYAGEV DN + + ED Y+FD++R++
Sbjct: 502 RNRGWGLRSWDSLRAGSFICEYAGEVKDNGNLR--GNQEEDAYVFDTSRVFNSFKWNYEP 559
Query: 597 EVFSSD--VEAPK---IPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHV 651
E+ D E P+ +PSPL I+A+ GNVARFMNHSC+PNV W+PV+RE E+ +H+
Sbjct: 560 ELVDEDPSTEVPEEFNLPSPLLISAKKFGNVARFMNHSCSPNVFWQPVIREGNGESVIHI 619
Query: 652 AFYAIRHIPPMMELTYDYGIVLPLKVGQK-----KKKCLCGSVKCRGYF 695
AF+A+RHIPPM ELTYDYGI + + ++ CLCGS +CRG F
Sbjct: 620 AFFAMRHIPPMAELTYDYGISPTSEARDESLLHGQRTCLCGSEQCRGSF 668
>ref|XP_475460.1| 'unknown protein, conatins SET domain' [Oryza sativa (japonica
cultivar-group)] gi|50080305|gb|AAT69639.1| 'unknown
protein, conatins SET domain' [Oryza sativa (japonica
cultivar-group)]
Length = 672
Score = 586 bits (1511), Expect = e-166
Identities = 317/691 (45%), Positives = 420/691 (59%), Gaps = 49/691 (7%)
Query: 21 VLNVKPLRTLVPVFPSPS--NPSSSATPQGGAPFVCVSPSGPFPSGVAP-FYPFFVSPES 77
+++ KP+R+L P+FP+P N + S+TP P VCV+P G FP G P F S +
Sbjct: 15 LVDAKPIRSLAPMFPAPLGINVNQSSTP----PLVCVTPVGQFPVGFGSGILPTFGSTTA 70
Query: 78 QRLSEQNAQTPTAQRAAPISAAVPINSFRTPTGATNGDVGSSRRKSRGQLPEDDNFVDLS 137
+ + I A PI++++T G + D G P +
Sbjct: 71 FTTTANGVSYTSYTNNGAIDAT-PISAYKTRPGIVSLD---------GDEPYSGSASGRK 120
Query: 138 EVDGEGGTGDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAV 197
G DG KR + V + VA + L + P + R+ V
Sbjct: 121 SKRSSGSAADGSNGVKFKRPKP------VYKNFVAGKELAFLPPSSSD-------PREVV 167
Query: 198 AYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEV 257
M +E +RR+ Q+DE + S KR DLKAG +M IRAN KR+G+VPGVE+
Sbjct: 168 EAVHMTFEALRRRHLQLDEIQETS----KRADLKAGAIMMASNIRANVGKRVGLVPGVEI 223
Query: 258 GDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYS 317
GDIF+FR ELC++GLHAPSM GIDY+ K +E+ +A+ IV++GGYE+ +D D L+YS
Sbjct: 224 GDIFYFRMELCIIGLHAPSMGGIDYMSAKFGSDEDSVAICIVAAGGYENVDDDTDTLVYS 283
Query: 318 GQGGTSRE-KGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQ 376
G GG SR + DQKLERGNLALERSLHR N++RV+RG RD TGK+Y+YDGLYKIQ
Sbjct: 284 GSGGNSRNSEERHDQKLERGNLALERSLHRKNEIRVVRGFRDPFCLTGKIYIYDGLYKIQ 343
Query: 377 NSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKLP 436
SW E+ KSG N FKYKL+R PGQP +WK W D ASR VILPDL+S AE LP
Sbjct: 344 ESWKERTKSGINCFKYKLLREPGQPDGAALWKMTQGWIDNPASRGRVILPDLSSAAEALP 403
Query: 437 VCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNGGYL 496
VCLVN+VD+EKGP +FTY+ +K L L+ ++ +GC C C PG C C Q NGG L
Sbjct: 404 VCLVNEVDHEKGPGHFTYASQVKYLRPLSSMKPLQGCGCQSVCLPGDPNCACGQHNGGDL 463
Query: 497 PYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIR 556
PYS++GLLA K ++YECG +CHC +CRNRV+Q G++ EVFRT +GWGLR WDPIR
Sbjct: 464 PYSSSGLLACRKPIIYECGDACHCTTNCRNRVTQKGVRFHFEVFRTANRGWGLRCWDPIR 523
Query: 557 AGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQLEVF----------SSDVEAP 606
AG FICEY GEVID +V ++EDDYIF + ++ F S+ V A
Sbjct: 524 AGAFICEYTGEVIDELKVN--LDDSEDDYIFQTVCPGEKTLKFNFGPELIGEESTYVSAD 581
Query: 607 KI-PSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMEL 665
+ P P+ I+A+ GNV+RFMNHSC+PNV W+PV ++ +++ H+ F+A++HIPPM EL
Sbjct: 582 EFEPLPIKISAKKMGNVSRFMNHSCSPNVFWQPVQHDHGDDSHPHIMFFALKHIPPMTEL 641
Query: 666 TYDYGIVLPLKVGQKK-KKCLCGSVKCRGYF 695
T+DYG+ G ++ K C CGS CRG F
Sbjct: 642 TFDYGVAGSESSGSRRTKNCFCGSSNCRGVF 672
>gb|AAM28230.1| SET domain protein 105 [Zea mays]
Length = 678
Score = 575 bits (1482), Expect = e-162
Identities = 318/705 (45%), Positives = 428/705 (60%), Gaps = 71/705 (10%)
Query: 21 VLNVKPLRTLVPVFPSPS--NPSSSATPQGGAPFVCVSPSGPFPSGVA----PFYPFFV- 73
+L+ PLR+L P+FP+P N + S+TP P VCV+P G FP G P + F
Sbjct: 15 LLDANPLRSLAPMFPAPMGVNVNQSSTP----PLVCVTPVGQFPIGFGAGNLPAFGSFTT 70
Query: 74 -SPESQRLSEQNAQTPTAQRAAPISA-----AVPINSFRTPTGATNGDVGSSRRKSRGQL 127
S + +S A A PISA ++ ++ + N + S R+ RG+
Sbjct: 71 FSATTNGVSYTGTSGNGAIDATPISAYKTRSSISLDDDDDEPYSGNQGLASERKARRGRP 130
Query: 128 PEDDNFVDLSEVDGEGGTGDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEIL 187
P +G G KR KP + + VA + L VF L
Sbjct: 131 PGSGR---------DGSNGKLKRPKPTYK------------NFVAGKEL------VF--L 161
Query: 188 NQPDGSRDAVAYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKGIRANSRK 247
+ R+ V M +E +RR+ Q+DE + ++R DLKAG +M IRANS K
Sbjct: 162 SSTSDPREFVESVHMTFEALRRRHLQMDE----TQDASRRADLKAGAIMMASNIRANSGK 217
Query: 248 RIGVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDN 307
R+G VPGVE+GDIF+FR ELC++GLHAPSM GIDY+ TK ++E+ +A+ IVS+GGYE+
Sbjct: 218 RVGTVPGVEIGDIFYFRMELCVIGLHAPSMGGIDYMTTKFGKDEDSVAICIVSAGGYENE 277
Query: 308 VEDGDVLIYSGQGGTSRE-KGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKV 366
+D DVL+YSGQGG +R + DQKLERGNLALERSLHR N++RV+RG +D TGK+
Sbjct: 278 DDDTDVLVYSGQGGNNRNTEERHDQKLERGNLALERSLHRKNEIRVVRGFKDPFCLTGKI 337
Query: 367 YVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILP 426
Y+YDGLYKI SW E+ K G N FKYKL+R PGQ +WK +W D A+R V+L
Sbjct: 338 YIYDGLYKIHESWKERTKYGVNCFKYKLLREPGQRDGAALWKMTQRWIDNPATRGRVLLA 397
Query: 427 DLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKC 486
DL+S AE LPVCLVN+VD+EKGP +FTY+ +K L L+ ++ +GC C C PG C
Sbjct: 398 DLSSKAEILPVCLVNEVDHEKGPVHFTYTNQVKYLRPLSSMKKLQGCGCQSVCLPGDASC 457
Query: 487 GCTQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKG 546
C Q NGG LP+S++GLL+ K +VYECG SC+C +CRNRV+Q G +L EVFRT +G
Sbjct: 458 ACGQHNGGDLPFSSSGLLSCRKPIVYECGESCNCSTNCRNRVTQKGSRLHFEVFRTTNRG 517
Query: 547 WGLRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDST-------------RIY 593
WGLR W+PIRAG+FICEYAGEVID + ++EDDYIF + +
Sbjct: 518 WGLRCWEPIRAGSFICEYAGEVIDELKFN--LNDSEDDYIFQTVCPGEKTLKWNYGPELI 575
Query: 594 QQLEVFSSDVEAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAF 653
++ + S E P P+ I+A+N GNV+RFMNHSC+PNV W+PV ++ ++ H+ F
Sbjct: 576 GEVSTYVSPDEFE--PLPVKISAKNMGNVSRFMNHSCSPNVFWQPVQYDHGDDGHPHIMF 633
Query: 654 YAIRHIPPMMELTYDYGIVLPLKVG---QKKKKCLCGSVKCRGYF 695
+A++HIPPM ELTYDYG+ G ++ K C+CGS CRG F
Sbjct: 634 FALKHIPPMTELTYDYGVAGAESSGSGSRRTKNCVCGSQNCRGLF 678
>gb|AAT47546.1| SET domain protein [Triticum aestivum]
Length = 745
Score = 556 bits (1433), Expect = e-157
Identities = 313/736 (42%), Positives = 428/736 (57%), Gaps = 65/736 (8%)
Query: 21 VLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGV-APFYPFFVS----- 74
VL+++PLRTL P+FP+P ++ P P + V+ +G FP GV AP +P F S
Sbjct: 15 VLDIQPLRTLAPMFPAPLGVNTFNVPNTTPPLIFVTSAGQFPGGVGAPGHPAFRSFAAAF 74
Query: 75 ---------PESQRLSEQNAQTPTAQR--------AAPISAAVPINSFRTPT-GATNGD- 115
P S P + R A + N +T GATN
Sbjct: 75 GAQDAYGGQPASSGGQHDGGGHPASSRRQDGGGGQATTFADKYAANGGQTAANGATNLGA 134
Query: 116 ----------VGSSRRKSRGQLPEDDNFVDLSEVDGEGGTGDGKRRKPQKRIREKRCSSD 165
+ + R +P DD+ D G + G++ K + + S
Sbjct: 135 SADSPIDVIPISAYRPTPPPVIPLDDDDDDNEHFTGNQTSASGRKIKRPSHLSGYKMSGG 194
Query: 166 V---DPDAVANEILKTINPGVF---EILNQPDGS---RDAVAYTLMIYEVMRRKLGQIDE 216
+ D + V + KT + E + P S R+AV LM +E +RR+ Q+D
Sbjct: 195 LGSDDNNGVKTKRNKTSHRKAGADNEFTSVPLSSSNPREAVEEVLMNFEALRRRHLQLD- 253
Query: 217 KAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPS 276
+ KRPDLK G +M +RAN RKRIGVVPGVE+GDIF+FR ELC++GLHAP+
Sbjct: 254 ---AAQESTKRPDLKIGAVMMANNLRANIRKRIGVVPGVEIGDIFYFRMELCIIGLHAPT 310
Query: 277 MAGIDYLG-TKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTSRE-KGASDQKLE 334
MAGIDY+ T ++++ +AV IV++G YE+ + D L+YSG GG+S+ + DQKLE
Sbjct: 311 MAGIDYMTHTFGDKDDDSVAVCIVAAGVYENEDDATDTLVYSGSGGSSKNNEEMHDQKLE 370
Query: 335 RGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKL 394
RGNLAL+ SL R N +RV+RG +D GKVY+YDGLYKI SW E+ K+G N FKYKL
Sbjct: 371 RGNLALQMSLSRKNVIRVVRGFKDPGSLGGKVYMYDGLYKIHESWKERTKTGINCFKYKL 430
Query: 395 VRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDNEKGPAYFTY 454
+R PGQP+ IWK +W + A+R V+ PDL+SG E LPVCLVNDVD+EKGP FTY
Sbjct: 431 LREPGQPEGMSIWKMSRKWVENPATRGRVLHPDLSSGTENLPVCLVNDVDSEKGPGLFTY 490
Query: 455 SPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYEC 514
+K L+ ++ +GC+C C P C C + NGG LPYS+ GLL K+ +YEC
Sbjct: 491 ITQVKYPKPLSSMKPLQGCSCLNACLPSDTDCDCAEFNGGNLPYSSTGLLVCRKNRLYEC 550
Query: 515 GPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARV 574
G SC C +CRNRV+Q G+++ E+FRT +GWGLRSWDPIRAG+FICEY GEVID ++
Sbjct: 551 GESCQCSVNCRNRVTQKGIRVHFEIFRTGNRGWGLRSWDPIRAGSFICEYVGEVIDESK- 609
Query: 575 EELSGENEDDYIFDSTRIYQQL-----------EVFSSDVEAPKIPSPLYITARNEGNVA 623
L GE+EDDY+F + R ++ E +++ P P+ I+A+ GN++
Sbjct: 610 RNLDGEDEDDYLFQTVRPGEKTLKWDYVPELMGEQITNNSADTFEPLPIKISAKKMGNIS 669
Query: 624 RFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVG---QK 680
RFMNHSC+PN W+PV ++ ++ H+ F+A++HIPPM ELTYDYG + G
Sbjct: 670 RFMNHSCSPNAFWQPVQFDHGDDGHPHIMFFALKHIPPMTELTYDYGEIGADSGGIGSPG 729
Query: 681 KKKCLCGSVKCRGYFC 696
K+CLCGS CRGYFC
Sbjct: 730 AKRCLCGSSNCRGYFC 745
>gb|AAK28975.1| SET1 [Oryza sativa]
Length = 812
Score = 554 bits (1428), Expect = e-156
Identities = 320/734 (43%), Positives = 432/734 (58%), Gaps = 89/734 (12%)
Query: 13 AGSFDKSRVLNVKPLRTLVPVFPS----PSNPSSSATPQGGAPFVCVSPSGPFPSGV--- 65
AG+ + ++ +P R PS PS P + T P SG G
Sbjct: 116 AGTVESAKRKRGRPKRVQDSSVPSAHLVPSAPGGNITAVQTPPSATTDESGKKKRGRPKR 175
Query: 66 APFYPFFVSPESQRLSEQNAQTP--------TAQRAAPI-------SAAVPINS------ 104
P +P + ++ QTP T +R P ++A PI S
Sbjct: 176 VQDVPVLSTPSAPQVDSTVFQTPASAVNESVTRKRGRPRRVQDGADTSAPPIQSKYNEPV 235
Query: 105 FRTPTGATNGDVGSSRRKSRGQLPEDDNFVDLSEV-----DGEGGTGDGKRRKPQKRIRE 159
+TP+ T + G +R ++P D + LS D G GKR +P+K
Sbjct: 236 LQTPSAVTLPEDGKRKRGRPKRVP-DGALIPLSHSGVSIDDDSGEIITGKRGRPRK---- 290
Query: 160 KRCSSDVDPDAVANEILKTIN-PGVFEILNQPDGSRDAVAYTLMIYEVMRRKLGQIDEKA 218
+D + +N P +F D R++V LM+++ +RR+L Q+DE
Sbjct: 291 ------ID--------VNLLNLPSLFS-----DDPRESVDNVLMMFDALRRRLMQLDEVK 331
Query: 219 KGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMA 278
+G+ ++ +LKAG++M + +RAN KRIG VPGVEVGD+F+FR E+CLVGL++ SM+
Sbjct: 332 QGAK---QQHNLKAGSIMMSAELRANKNKRIGEVPGVEVGDMFYFRIEMCLVGLNSQSMS 388
Query: 279 GIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTSREKGASDQKLERGNL 338
GIDY+ K EE+P+A+SIVS+G YE+ +D DVL+Y+GQG + G DQKLERGNL
Sbjct: 389 GIDYMSAKFGNEEDPVAISIVSAGVYENTEDDPDVLVYTGQGMS----GKDDQKLERGNL 444
Query: 339 ALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLP 398
ALERSLHRGN +RV+R +RD PTGK+Y+YDGLYKI+ +WVEK K+GFNVFK+KL+R P
Sbjct: 445 ALERSLHRGNQIRVVRSVRDLTCPTGKIYIYDGLYKIREAWVEKGKTGFNVFKHKLLREP 504
Query: 399 GQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDNEKGPAYFTYSPTL 458
GQP +WK +W + +SR VIL D++ GAE PVCLVN+VD+EKGP++F Y+ L
Sbjct: 505 GQPDGIAVWKKTEKWRENPSSRDHVILRDISYGAESKPVCLVNEVDDEKGPSHFNYTTKL 564
Query: 459 KNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYECGPSC 518
N L+ + +GC C C PG + C CT +N G LPYSA+G+L ++YEC SC
Sbjct: 565 NYRNSLSSMRKMQGCNCASVCLPGDNNCSCTHRNAGDLPYSASGILVSRMPMLYECNDSC 624
Query: 519 HCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELS 578
C +CRNRV Q G ++ EVF+T +GWGLRSWDPIRAGTFICEYAGEVID +
Sbjct: 625 TCSHNCRNRVVQKGSQIHFEVFKTGDRGWGLRSWDPIRAGTFICEYAGEVIDRNSI---- 680
Query: 579 GENEDDYIFDSTRIYQQLE-----------VFSSDVEAPKIPSPLYITARNEGNVARFMN 627
EDDYIF+ T Q L S E PK P+ I+A+ GN+ARFMN
Sbjct: 681 -IGEDDYIFE-TPSEQNLRWNYAPELLGEPSLSDSSETPK-QLPIIISAKRTGNIARFMN 737
Query: 628 HSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYG-----IVLPLKVG-QKK 681
HSC+PNV W+PV+ ++ +E H+AF+AI+HIPPM ELTYDYG + L + G +K
Sbjct: 738 HSCSPNVFWQPVLYDHGDEGYPHIAFFAIKHIPPMTELTYDYGQSQGNVQLGINSGCRKS 797
Query: 682 KKCLCGSVKCRGYF 695
K CLC S KCRG F
Sbjct: 798 KNCLCWSRKCRGSF 811
Score = 53.5 bits (127), Expect = 2e-05
Identities = 36/118 (30%), Positives = 55/118 (46%), Gaps = 17/118 (14%)
Query: 17 DKSRVLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGV-----APFYPF 71
D + V++ KPLRTL P+FP+ + + + VC++P GP+ G A P
Sbjct: 13 DNAAVVDAKPLRTLTPMFPAALGLHTFTAKENSSSIVCITPFGPYAGGTEQAMPASIPPM 72
Query: 72 FVSPESQRLSEQNAQTPTAQRAAPISAAVPINSFRTPTG-------ATNGDVGSSRRK 122
F SP + ++ N + P A ++ A P N TG A G V S++RK
Sbjct: 73 FASPAAP--ADPNQRQP---YAVHLNGAAPANGTANNTGVIPDLQIAVAGTVESAKRK 125
>ref|NP_915934.1| similar to SET1 [Oryza sativa (japonica cultivar-group)]
gi|20160708|dbj|BAB89651.1| putative SET domain protein
113 [Oryza sativa (japonica cultivar-group)]
gi|18844764|dbj|BAB85235.1| putative SET domain protein
113 [Oryza sativa (japonica cultivar-group)]
Length = 736
Score = 552 bits (1422), Expect = e-155
Identities = 323/734 (44%), Positives = 421/734 (57%), Gaps = 72/734 (9%)
Query: 21 VLNVKPLRTLVPVFPSPSNPSSSATPQGGAPFVCVSPSGPFPSGVA----PFYPFFVSPE 76
VL VKPLRTL P+FP+P P V V+P+G FP G P F +
Sbjct: 15 VLEVKPLRTLAPMFPAPLG-IDVLNRLTAPPLVFVAPAGQFPGGFGSLNIPAVRSFAAFG 73
Query: 77 SQRLSEQNAQTPTAQRAA--PISAAVPINSFRTPTGATNG--------------DVGSSR 120
Q S Q A+ +A ++ R T A G +VG+S
Sbjct: 74 GQDASGGKTAGGGDQDASGGKTAAGGDQDAGRGETAAFGGQETVRGEFVANGTPNVGASA 133
Query: 121 RKSRGQLP--------------EDDNFVDLSEVDGEGGTGDGKRRKPQKRIREKRCSSDV 166
P +DD+ D G + G++ K ++ S +
Sbjct: 134 TGPIDATPISACKSTQPSVISLDDDDNDDDEPYGGNQTSASGRKIKRPSHLKGYNVSDGL 193
Query: 167 DPDA---------VANEILKTINPGVFEILNQPDGS--RDAVAYTLMIYEVMRRKLGQID 215
D+ +N T N EI P S R+ V LM +E +RR+ Q+D
Sbjct: 194 GTDSSNGTKKRPKTSNRKAATDN----EISLMPPSSDPREVVEVLLMTFEALRRRHLQLD 249
Query: 216 EKAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFFRFELCLVGLHAP 275
E + S KR DLKAG +M +RAN KRIG VPGVEVGDIF+FR ELC++GLHAP
Sbjct: 250 ETQETS----KRADLKAGAIMLASNLRANIGKRIGAVPGVEVGDIFYFRMELCIIGLHAP 305
Query: 276 SMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTSRE-KGASDQKLE 334
SM GIDY+ K E++ +A+ IV++G YE++ +D D L+YSG GG SR + DQKLE
Sbjct: 306 SMGGIDYMN-KFGDEDDSVAICIVAAGVYENDDDDTDTLVYSGSGGISRNSEEKQDQKLE 364
Query: 335 RGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKL 394
RGNLALERSL R N +RV+RG +D A TGKVY+YDGLYKI SW E+ K+G N FKYKL
Sbjct: 365 RGNLALERSLSRKNVIRVVRGYKDPACLTGKVYIYDGLYKIHESWKERTKTGINCFKYKL 424
Query: 395 VRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDNEKGPAYFTY 454
R PGQP A IWK +W + A+R V+ PDL+SGAE LPVCL+NDV++EKGP +F Y
Sbjct: 425 QREPGQPDAVAIWKMCQRWVENPAARGKVLHPDLSSGAENLPVCLINDVNSEKGPGHFNY 484
Query: 455 SPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYEC 514
+K L L ++ +GC C C PG C C Q NGG LPYS++GLL K +VYEC
Sbjct: 485 ITQVKYLKPLRSMKPFQGCRCTSVCLPGDTSCDCAQHNGGDLPYSSSGLLVCRKLMVYEC 544
Query: 515 GPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARV 574
G SC C +CRNRV+Q G+++ LEVFRT +GWGLRSWDPIRAG+FICEY GEV+D+ +V
Sbjct: 545 GESCRCSINCRNRVAQKGVRIHLEVFRTTNRGWGLRSWDPIRAGSFICEYVGEVVDDTKV 604
Query: 575 EELSGENEDDYIF------DSTRIY----QQLEVFSSDVEAPKI-PSPLYITARNEGNVA 623
+ + EDDY+F + T + + + S ++ A P P+ I+A GNVA
Sbjct: 605 ---NLDGEDDYLFRTVCPGEKTLKWNYGPELIGEHSINISADTFEPLPIKISAMKMGNVA 661
Query: 624 RFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIV--LPLKVGQKK 681
RFMNHSC PN W+PV ++ + H+ F+A++HIPPM ELTYDYG + VG +
Sbjct: 662 RFMNHSCNPNTFWQPVQFDHGEDGYPHIMFFALKHIPPMTELTYDYGDIGCESRGVGSRA 721
Query: 682 KKCLCGSVKCRGYF 695
K CLCGS CRG+F
Sbjct: 722 KNCLCGSSNCRGFF 735
>gb|AAN41253.1| SET domain protein 113 [Zea mays]
Length = 766
Score = 536 bits (1382), Expect = e-151
Identities = 318/761 (41%), Positives = 434/761 (56%), Gaps = 96/761 (12%)
Query: 21 VLNVKPLRTLVPVFPSPSNPSS---SATPQGGAPFVCVSPS-------GPFPSGVAPFYP 70
VL+VKPLRTL P+FP+P ++ S TP P + V+PS G + + A +
Sbjct: 15 VLDVKPLRTLAPMFPAPLGVNTFNHSTTP----PLIFVTPSGQFQGGFGDWNNSAAKSFF 70
Query: 71 FFVSPESQ-----RLSEQNAQTPTAQRAAPISAAVPINSFR----TPTG--ATNGD---V 116
F S +++ S+QN T A A N F T TG AT GD
Sbjct: 71 AFSSKDARGGKADTFSDQNTGTGKAATFGDQDAFGSQNVFASDHSTGTGKAATFGDQDAF 130
Query: 117 GSS------RRKSRGQLPED------------------------DNFVDLSEVDGEGGTG 146
GS + ++GQ D N + L + D +
Sbjct: 131 GSQNVATGVQDAAKGQTATDWTDVSANPNGPIDAIAISAYRSTQPNVISLDDDDDDDDDE 190
Query: 147 DGKRRKPQKRIREKRCSSDVDPDAVANEIL-----------------KTINPGVFEILNQ 189
K R+ + S + V++ ++ K +L
Sbjct: 191 PYAANKTSASGRKIKRPSHLSGYNVSDALVSDSSNSMKVKRPKSSHGKAAADNEHALLPP 250
Query: 190 PDGSRDAVAYTLMIYEVMRRKLGQIDEKAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRI 249
+ R+ V LM +E +RR+ Q+DE + S KR DLKA ++ + IRAN KRI
Sbjct: 251 SEDPREIVEAVLMTFEALRRRHLQLDEAQETS----KRADLKASAILMSSNIRANPGKRI 306
Query: 250 GVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVE 309
GVVPGVE+GDIF+FR ELC++GLHAPSMAGIDY+ K E++ +A+ IV++GGY++N +
Sbjct: 307 GVVPGVEIGDIFYFRMELCIIGLHAPSMAGIDYMTAKFGDEDDSVAICIVAAGGYDNNDD 366
Query: 310 DGDVLIYSGQGGTSREKGAS-DQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYV 368
D DVL+YSG GG S+ DQKLERGNLALERSL R N +RV+RG +D TGKVY+
Sbjct: 367 DTDVLVYSGSGGNSKNSEEKHDQKLERGNLALERSLSRKNVIRVVRGYKDPGCLTGKVYI 426
Query: 369 YDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDL 428
YDGLY+I SW EK KSG FKYKL+R PGQP IWK +W +R V+ PDL
Sbjct: 427 YDGLYRIHESWKEKTKSGIFCFKYKLLREPGQPDGVAIWKMSQKWVQNPLTRGSVLNPDL 486
Query: 429 TSGAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGC 488
+SGAE LPVCLVND+D+++ P +FTY+ +++L L+ V+ +GC C C PG C C
Sbjct: 487 SSGAENLPVCLVNDIDSDEVPHHFTYTTQVEHLKPLSSVKPLQGCRCLSVCLPGDANCCC 546
Query: 489 TQKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWG 548
Q+NGG LPYS++GLL K++VYECG SC C +CRNRV+Q G+++ EVF+T +GWG
Sbjct: 547 AQRNGGSLPYSSSGLLVCRKTMVYECGESCRCSFNCRNRVTQKGVRIHFEVFKTGNRGWG 606
Query: 549 LRSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQ----------LEV 598
LRSWD IRAG+FICEY GEVID+A + + EDDYIF + ++ +
Sbjct: 607 LRSWDAIRAGSFICEYVGEVIDDANIN--LNDIEDDYIFQMSCPGERTLKWNFGPELIGE 664
Query: 599 FSSDVEAPKIPS-PLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIR 657
S++V A + P+ I+A+ GN++RFMNHSC PNV W+PV +++++ H+ F+A++
Sbjct: 665 QSTNVSADTFETLPIKISAKRIGNISRFMNHSCAPNVFWQPVQFDHEDDHRPHIMFFALK 724
Query: 658 HIPPMMELTYDYGIVLPLKVG---QKKKKCLCGSVKCRGYF 695
HIPPM ELTYDYG V G + K CLC S CRG+F
Sbjct: 725 HIPPMTELTYDYGDVGADPSGVRSPRAKNCLCESSNCRGFF 765
>gb|AAT47547.1| SET domain protein [Triticum aestivum]
Length = 428
Score = 447 bits (1149), Expect = e-124
Identities = 218/431 (50%), Positives = 286/431 (65%), Gaps = 15/431 (3%)
Query: 277 MAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTSRE-KGASDQKLER 335
M GIDY+ K +E+ +A+ IV++GGYE+ +D D L+YSG GG SR + DQKLER
Sbjct: 1 MGGIDYMSAKFGADEDSVAICIVAAGGYENEDDDTDTLVYSGSGGNSRNTEERHDQKLER 60
Query: 336 GNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLV 395
GNLALERS+HR N++RV+RG +D A GK+Y+YDGLYKIQ SW E+ K G N FKY+L
Sbjct: 61 GNLALERSMHRKNEIRVVRGFKDPAMVAGKIYIYDGLYKIQESWTERTKFGVNCFKYRLQ 120
Query: 396 RLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDNEKGPAYFTYS 455
R PGQ IWK +W ++R VIL DL+SG E +PVCLVN+VD+EKGP FTY+
Sbjct: 121 REPGQRDGAAIWKMTQRWIQDPSTRGRVILRDLSSGIESIPVCLVNEVDHEKGPGQFTYT 180
Query: 456 PTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNGGYLPYSAAGLLADLKSVVYECG 515
+K L ++ + +GC C C PG C C Q NGG LPYS++G+L K +VYECG
Sbjct: 181 NQVKYLRPVSSMTPMQGCGCQSVCLPGDANCACGQHNGGDLPYSSSGVLVCRKPIVYECG 240
Query: 516 PSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVE 575
+CHC +CRNRVSQ G++ EVFRT +GWGLR W+PIRAG FICEY GEVID +V
Sbjct: 241 EACHCTLNCRNRVSQKGIRFHFEVFRTANRGWGLRCWEPIRAGAFICEYTGEVIDELKVN 300
Query: 576 ELSGENEDDYIFDSTRIYQQ----------LEVFSSDVEAPKI-PSPLYITARNEGNVAR 624
++EDDYIF + ++ + S+ V A + P P+ I+A+ GNV+R
Sbjct: 301 --LDDSEDDYIFQTVCPGEKTLKWNFGPELIGEQSTYVSAEEFQPLPIKISAKKMGNVSR 358
Query: 625 FMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQKKKKC 684
FMNHSC+PNV W+PV + ++ H+ F+A+ HI PM ELTYDYG+V + + K C
Sbjct: 359 FMNHSCSPNVFWQPVQYNHGDDKHPHIMFFALNHIAPMTELTYDYGVV-GEETSHRAKTC 417
Query: 685 LCGSVKCRGYF 695
LCGS+ CRG F
Sbjct: 418 LCGSLTCRGLF 428
>gb|AAO32934.1| SET domain protein SDG111 [Zea mays]
Length = 486
Score = 424 bits (1090), Expect = e-117
Identities = 209/399 (52%), Positives = 272/399 (67%), Gaps = 17/399 (4%)
Query: 312 DVLIYSGQGGTSRE-KGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYD 370
DVL+YSGQGG SR + DQKLERGNLALERSLHR N++RV+RG +D TGK+Y+YD
Sbjct: 90 DVLVYSGQGGNSRNTEERQDQKLERGNLALERSLHRKNEIRVVRGFKDPFCLTGKIYIYD 149
Query: 371 GLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTS 430
GLYKI SW EK +SG N FKYKL+R PGQ +WK +W D A+R V+L DL+S
Sbjct: 150 GLYKIHESWKEKTRSGINCFKYKLLREPGQRDGAALWKMTQKWIDDPATRGRVLLADLSS 209
Query: 431 GAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQ 490
AE +PV LVN+VD+EKGPA+FTY+ +K + L+ ++ +GC C C PG C C Q
Sbjct: 210 KAETIPVSLVNEVDHEKGPAHFTYTNQVKYVRPLSSMKKLQGCGCQSVCLPGDASCACGQ 269
Query: 491 KNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLR 550
NGG LPYS+ GLL+ K ++YECG SC+C +CRNRV+Q G +L EVFRT +GWGLR
Sbjct: 270 HNGGDLPYSSLGLLSCRKPMIYECGESCNCSTNCRNRVTQKGPRLHFEVFRTTNRGWGLR 329
Query: 551 SWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQ----------LEVFS 600
W+P+RAG+FICEYAGEVID +V + EDDYIF + ++ L S
Sbjct: 330 CWEPVRAGSFICEYAGEVIDELKVN--LNDTEDDYIFQTVCPGEKTLKWNCGPELLGEAS 387
Query: 601 SDVEAPKI-PSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHI 659
+ V A + P P+ I+A+N GNV+RFMNHSC+PNV W+PV ++ ++ H+ F+A++HI
Sbjct: 388 TYVSADEFEPLPIKISAKNMGNVSRFMNHSCSPNVFWQPVQYDHGDDGHPHIMFFALKHI 447
Query: 660 PPMMELTYDYGIVLPLKVG---QKKKKCLCGSVKCRGYF 695
PPM ELTYDYG+ G ++ K C+CGS CRG F
Sbjct: 448 PPMTELTYDYGVAGAESSGSGSRRTKNCMCGSQNCRGLF 486
Score = 43.5 bits (101), Expect = 0.023
Identities = 22/46 (47%), Positives = 31/46 (66%), Gaps = 6/46 (13%)
Query: 21 VLNVKPLRTLVPVFPSPS--NPSSSATPQGGAPFVCVSPSGPFPSG 64
+L+ KPLR+L P+FP+P N + S+TP P VCV+P G F +G
Sbjct: 15 LLDAKPLRSLAPMFPAPMGVNVNQSSTP----PLVCVTPVGQFSAG 56
>ref|NP_564036.1| SET domain-containing protein (SUVH7) [Arabidopsis thaliana]
gi|13517755|gb|AAK28972.1| SUVH7 [Arabidopsis thaliana]
gi|30580524|sp|Q9C5P1|SUVH7_ARATH Histone-lysine
N-methyltransferase, H3 lysine-9 specific 7 (Histone
H3-K9 methyltransferase 7) (H3-K9-HMTase 7) (Suppressor
of variegation 3-9 homolog 7) (Su(var)3-9 homolog 7)
Length = 693
Score = 416 bits (1069), Expect = e-114
Identities = 264/699 (37%), Positives = 367/699 (51%), Gaps = 75/699 (10%)
Query: 35 PSPSNPSSSATPQGGAPFVCVSPSGPFPSGVAPFY-----PFFVSPESQRLSEQNAQTPT 89
P+ +P + P +P + V P +Y P P + S+ T
Sbjct: 31 PTMVSPVLTNMPSATSPLLMVPPLRTIWPSNKEWYDGDAGPSSTGPIKREASDNTNDTAH 90
Query: 90 AQRAAPISAAVPINSFRTPTGATNGDVGSSRRKSRGQLPEDDNFVDLSEVDGEGGTGDGK 149
A P +P+ + R ++N + S G + G G K
Sbjct: 91 NTFAPPPEMVIPLITIRPSDDSSNYSCDAGAGPSTGPVKR-----------GRGRPKGSK 139
Query: 150 RRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEI----LNQPDGSRDAVAYTLMIYE 205
P + + K DP++ LK + G F+ G+++ V +M ++
Sbjct: 140 NSTPTEPKKPKV----YDPNS-----LKVTSRGNFDSEITEAETETGNQEIVDSVMMRFD 190
Query: 206 VMRRKLGQID--EKAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFF 263
+RR+L QI+ E + SG G++ N+R+RIG VPG+ VGDIF++
Sbjct: 191 AVRRRLCQINHPEDILTTASGN----------CTKMGVKTNTRRRIGAVPGIHVGDIFYY 240
Query: 264 RFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTS 323
E+CLVGLH + GID+ S E A+ +V++G Y+ E D LIYSGQGGT
Sbjct: 241 WGEMCLVGLHKSNYGGIDFFTAAESAVEGHAAMCVVTAGQYDGETEGLDTLIYSGQGGTD 300
Query: 324 REKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKA 383
A DQ+++ GNLALE S+ +GNDVRV+RG+ K+Y+YDG+Y + W
Sbjct: 301 VYGNARDQEMKGGNLALEASVSKGNDVRVVRGVIHPHENNQKIYIYDGMYLVSKFWTVTG 360
Query: 384 KSGFNVFKYKLVRLPGQPQAYMIWKSI--LQWTDKSASRVGVILPDLTSGAEKLPVCLVN 441
KSGF F++KLVR P QP AY IWK++ L+ D SR G IL DL+ GAE L V LVN
Sbjct: 361 KSGFKEFRFKLVRKPNQPPAYAIWKTVENLRNHDLIDSRQGFILEDLSFGAELLRVPLVN 420
Query: 442 DVDNEKG--PAYFTYSPTLKNLNRLAPV----ESSEGCTCNGGCQPGSHK-CGCTQKNGG 494
+VD + P F Y P+ + + S GC N QP H+ C C Q+NG
Sbjct: 421 EVDEDDKTIPEDFDYIPSQCHSGMMTHEFHFDRQSLGCQ-NCRHQPCMHQNCTCVQRNGD 479
Query: 495 YLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDP 554
LPY +L K ++YECG SC CP C R+ Q GLKL LEVF+T+ GWGLRSWDP
Sbjct: 480 LLPYH-NNILVCRKPLIYECGGSCPCPDHCPTRLVQTGLKLHLEVFKTRNCGWGLRSWDP 538
Query: 555 IRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL------EVFSSD-----V 603
IRAGTFICE+AG VEE +DDY+FD+++IYQ+ E+ D
Sbjct: 539 IRAGTFICEFAGLRKTKEEVEE-----DDDYLFDTSKIYQRFRWNYEPELLLEDSWEQVS 593
Query: 604 EAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMM 663
E +P+ + I+A+ +GNV RFMNHSC+PNV W+P+ EN+ + L + +A++HIPPM
Sbjct: 594 EFINLPTQVLISAKEKGNVGRFMNHSCSPNVFWQPIEYENRGDVYLLIGLFAMKHIPPMT 653
Query: 664 ELTYDYGIVL-------PLKVGQKKKKCLCGSVKCRGYF 695
ELTYDYG+ + + + KK CLCGSVKCRG F
Sbjct: 654 ELTYDYGVSCVERSEEDEVLLYKGKKTCLCGSVKCRGSF 692
>pir||G86312 hypothetical protein F2H15.1 - Arabidopsis thaliana
gi|9665056|gb|AAF97258.1| Contains a DNA binding domain
with preference for A/T rich regions PF|02178, a domain
of unknown function PF|02182 and a SET domain PF|00856.
[Arabidopsis thaliana]
Length = 954
Score = 397 bits (1021), Expect = e-109
Identities = 251/668 (37%), Positives = 351/668 (51%), Gaps = 68/668 (10%)
Query: 35 PSPSNPSSSATPQGGAPFVCVSPSGPFPSGVAPFY-----PFFVSPESQRLSEQNAQTPT 89
P+ +P + P +P + V P +Y P P + S+ T
Sbjct: 31 PTMVSPVLTNMPSATSPLLMVPPLRTIWPSNKEWYDGDAGPSSTGPIKREASDNTNDTAH 90
Query: 90 AQRAAPISAAVPINSFRTPTGATNGDVGSSRRKSRGQLPEDDNFVDLSEVDGEGGTGDGK 149
A P +P+ + R ++N + S G + G G K
Sbjct: 91 NTFAPPPEMVIPLITIRPSDDSSNYSCDAGAGPSTGPVKR-----------GRGRPKGSK 139
Query: 150 RRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEI----LNQPDGSRDAVAYTLMIYE 205
P + + K DP++ LK + G F+ G+++ V +M ++
Sbjct: 140 NSTPTEPKKPKV----YDPNS-----LKVTSRGNFDSEITEAETETGNQEIVDSVMMRFD 190
Query: 206 VMRRKLGQID--EKAKGSHSGAKRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFF 263
+RR+L QI+ E + SG G++ N+R+RIG VPG+ VGDIF++
Sbjct: 191 AVRRRLCQINHPEDILTTASGN----------CTKMGVKTNTRRRIGAVPGIHVGDIFYY 240
Query: 264 RFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTS 323
E+CLVGLH + GID+ S E A+ +V++G Y+ E D LIYSGQGGT
Sbjct: 241 WGEMCLVGLHKSNYGGIDFFTAAESAVEGHAAMCVVTAGQYDGETEGLDTLIYSGQGGTD 300
Query: 324 REKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKA 383
A DQ+++ GNLALE S+ +GNDVRV+RG+ K+Y+YDG+Y + W
Sbjct: 301 VYGNARDQEMKGGNLALEASVSKGNDVRVVRGVIHPHENNQKIYIYDGMYLVSKFWTVTG 360
Query: 384 KSGFNVFKYKLVRLPGQPQAYMIWKSI--LQWTDKSASRVGVILPDLTSGAEKLPVCLVN 441
KSGF F++KLVR P QP AY IWK++ L+ D SR G IL DL+ GAE L V LVN
Sbjct: 361 KSGFKEFRFKLVRKPNQPPAYAIWKTVENLRNHDLIDSRQGFILEDLSFGAELLRVPLVN 420
Query: 442 DVDNEKG--PAYFTYSPTLKNLNRLAPV----ESSEGCTCNGGCQPGSHK-CGCTQKNGG 494
+VD + P F Y P+ + + S GC N QP H+ C C Q+NG
Sbjct: 421 EVDEDDKTIPEDFDYIPSQCHSGMMTHEFHFDRQSLGCQ-NCRHQPCMHQNCTCVQRNGD 479
Query: 495 YLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDP 554
LPY +L K ++YECG SC CP C R+ Q GLKL LEVF+T+ GWGLRSWDP
Sbjct: 480 LLPYH-NNILVCRKPLIYECGGSCPCPDHCPTRLVQTGLKLHLEVFKTRNCGWGLRSWDP 538
Query: 555 IRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQL------EVFSSD-----V 603
IRAGTFICE+AG VEE +DDY+FD+++IYQ+ E+ D
Sbjct: 539 IRAGTFICEFAGLRKTKEEVEE-----DDDYLFDTSKIYQRFRWNYEPELLLEDSWEQVS 593
Query: 604 EAPKIPSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMM 663
E +P+ + I+A+ +GNV RFMNHSC+PNV W+P+ EN+ + L + +A++HIPPM
Sbjct: 594 EFINLPTQVLISAKEKGNVGRFMNHSCSPNVFWQPIEYENRGDVYLLIGLFAMKHIPPMT 653
Query: 664 ELTYDYGI 671
ELTYDYG+
Sbjct: 654 ELTYDYGV 661
>gb|AAK28973.1| SUVH8 [Arabidopsis thaliana] gi|30580523|sp|Q9C5P0|SUVH8_ARATH
Histone-lysine N-methyltransferase, H3 lysine-9 specific
8 (Histone H3-K9 methyltransferase 8) (H3-K9-HMTase 8)
(Suppressor of variegation 3-9 homolog 8) (Su(var)3-9
homolog 8) gi|30682537|ref|NP_180049.2| SET
domain-containing protein (SUVH8) [Arabidopsis thaliana]
Length = 755
Score = 377 bits (968), Expect = e-103
Identities = 247/615 (40%), Positives = 331/615 (53%), Gaps = 67/615 (10%)
Query: 108 PTGATNGDVGSSRRKSRGQLPEDDNFVDLSE--VDGEGGTGDGKRRKPQKRIREKRCSSD 165
P G+ NG RK + D+N D S G G G+ + + R R+ +
Sbjct: 180 PKGSKNGS-----RKPKKPKAYDNNSTDASAGPSSGLGKRRCGRPKGLKNRSRKPKKPKA 234
Query: 166 VDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLMIYEVMRRKLGQIDEKAKGSHSGA 225
DP++ + + E + G+++ V LM ++ +RR+L Q++ +
Sbjct: 235 DDPNSKMVISCPDFDSRITEA-ERESGNQEIVDSILMRFDAVRRRLCQLNYR-------- 285
Query: 226 KRPDLKAGTLMNTKGIRANSRKRIGVVPGVEVGDIFFFRFELCLVGLHAPSMAGIDYLGT 285
K L A T G+R N +RIG +PGV+VGDIF++ E+CLVGLH + GID L
Sbjct: 286 KDKILTASTNCMNLGVRTNMTRRIGPIPGVQVGDIFYYWCEMCLVGLHRNTAGGIDSLLA 345
Query: 286 KVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGTSREKGASDQKLERGNLALERSLH 345
K S + P A S+V+SG Y++ ED + LIYSG GG DQ L+RGN ALE S+
Sbjct: 346 KESGVDGPAATSVVTSGKYDNETEDLETLIYSGHGGK-----PCDQVLQRGNRALEASVR 400
Query: 346 RGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYM 405
R N+VRVIRG E + KVY+YDGLY + + W KSGF +++KL+R PGQP Y
Sbjct: 401 RRNEVRVIRG---ELYNNEKVYIYDGLYLVSDCWQVTGKSGFKEYRFKLLRKPGQPPGYA 457
Query: 406 IWKSI--LQWTDKSASRVGVILPDLTSGAEKLPVCLVNDVDNEKG--PAYFTY--SPTLK 459
IWK + L+ + R G IL DL+ G E L V LVN+VD E P F Y S
Sbjct: 458 IWKLVENLRNHELIDPRQGFILGDLSFGEEGLRVPLVNEVDEEDKTIPDDFDYIRSQCYS 517
Query: 460 NLNRLAPVESSEGCTCNGGCQPGSHK-CGCTQKNGGYLPYSAAGLLADLKSVVYECGPSC 518
+ V+S Q H+ C C KN G LPY +L K ++YECG
Sbjct: 518 GMTNDVNVDS------QSLVQSYIHQNCTCILKNCGQLPYH-DNILVCRKPLIYECG--- 567
Query: 519 HCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIRAGTFICEYAGEVIDNARVEELS 578
SC R+ + GLKL LEVF+T GWGLRSWDPIRAGTFICE+ G VEE
Sbjct: 568 ---GSCPTRMVETGLKLHLEVFKTSNCGWGLRSWDPIRAGTFICEFTGVSKTKEEVEE-- 622
Query: 579 GENEDDYIFDSTRIYQQL------EVFSSDV-----EAPKIPSPLYITARNEGNVARFMN 627
+DDY+FD++RIY E+ D E +P+ + I+A+ +GNV RFMN
Sbjct: 623 ---DDDYLFDTSRIYHSFRWNYEPELLCEDACEQVSEDANLPTQVLISAKEKGNVGRFMN 679
Query: 628 HSCTPNVLWRPV-VRENKNEADLHVAFYAIRHIPPMMELTYDYGIVLPLKVGQK------ 680
H+C PNV W+P+ +N + + +A++HIPPM ELTYDYGI K G+
Sbjct: 680 HNCWPNVFWQPIEYDDNNGHIYVRIGLFAMKHIPPMTELTYDYGISCVEKTGEDEVIYKG 739
Query: 681 KKKCLCGSVKCRGYF 695
KK CLCGSVKCRG F
Sbjct: 740 KKICLCGSVKCRGSF 754
>gb|AAO30037.1| putative SET-domain protein [Arabidopsis thaliana]
gi|3668088|gb|AAC61820.1| similar to mammalian MHC III
region protein G9a [Arabidopsis thaliana]
gi|17065318|gb|AAL32813.1| putative SET-domain protein
[Arabidopsis thaliana] gi|13517751|gb|AAK28970.1| SUVH5
[Arabidopsis thaliana] gi|25408359|pir||D84765 similar
to mammalian MHC III region protein G9a [imported] -
Arabidopsis thaliana gi|15226918|ref|NP_181061.1| SET
domain-containing protein (SUVH5) [Arabidopsis thaliana]
gi|30580519|sp|O82175|SUV5_ARATH Histone-lysine
N-methyltransferase, H3 lysine-9 specific 5 (Histone
H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Suppressor
of variegation 3-9 homolog 5) (Su(var)3-9 homolog 5)
Length = 794
Score = 372 bits (954), Expect = e-101
Identities = 220/564 (39%), Positives = 327/564 (57%), Gaps = 49/564 (8%)
Query: 143 GGTGDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLM 202
G G+G RK +R+ +D +A++ + N G+ + D +R V T+
Sbjct: 264 GDYGEGSMRKNSERVA-------LDKKRLASKF-RLSNGGLPSCSSSGDSARYKVKETMR 315
Query: 203 IYEVMRRKLGQIDEKAKGSHSGAK-RPDLKAGTLMNTKGIRANSRKRI-GVVPGVEVGDI 260
++ +K+ Q +E G + +A ++ +KG S +I G VPGVEVGD
Sbjct: 316 LFHETCKKIMQEEEARPRKRDGGNFKVVCEASKILKSKGKNLYSGTQIIGTVPGVEVGDE 375
Query: 261 FFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQG 320
F +R EL L+G+H PS +GIDY+ E +A SIVSSGGY D +++ DVLIY+GQG
Sbjct: 376 FQYRMELNLLGIHRPSQSGIDYMK---DDGGELVATSIVSSGGYNDVLDNSDVLIYTGQG 432
Query: 321 GTSREKGAS----DQKLERGNLALERSLHRGNDVRVIRGMRD---EAHPTGKVYVYDGLY 373
G +K + DQ+L GNLAL+ S+++ N VRVIRG+++ ++ K YVYDGLY
Sbjct: 433 GNVGKKKNNEPPKDQQLVTGNLALKNSINKKNPVRVIRGIKNTTLQSSVVAKNYVYDGLY 492
Query: 374 KIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTSGAE 433
++ W E G VFK+KL R+PGQP+ + WK + + + KS R G+ D+T G E
Sbjct: 493 LVEEYWEETGSHGKLVFKFKLRRIPGQPE--LPWKEVAK-SKKSEFRDGLCNVDITEGKE 549
Query: 434 KLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHKCGCTQKNG 493
LP+C VN++D+EK P F Y+ + + P+ + C C GC S C C KNG
Sbjct: 550 TLPICAVNNLDDEKPPP-FIYTAKMIYPDWCRPIPP-KSCGCTNGCSK-SKNCACIVKNG 606
Query: 494 GYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWD 553
G +PY G + ++K +VYECGP C CPPSC RVSQ G+K++LE+F+T+ +GWG+RS +
Sbjct: 607 GKIPYYD-GAIVEIKPLVYECGPHCKCPPSCNMRVSQHGIKIKLEIFKTESRGWGVRSLE 665
Query: 554 PIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIYQQLEVFSSDVEAPKIPSPLY 613
I G+FICEYAGE++++ + E L+G +D+Y+FD + D P
Sbjct: 666 SIPIGSFICEYAGELLEDKQAESLTG--KDEYLFD---------LGDED-------DPFT 707
Query: 614 ITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELTYDYGI-- 671
I A +GN+ RF+NHSC+PN+ + V+ +++ H+ F+A+ +IPP+ EL+YDY
Sbjct: 708 INAAQKGNIGRFINHSCSPNLYAQDVLYDHEEIRIPHIMFFALDNIPPLQELSYDYNYKI 767
Query: 672 --VLPLKVGQKKKKCLCGSVKCRG 693
V KKK C CGS +C G
Sbjct: 768 DQVYDSNGNIKKKFCYCGSAECSG 791
>gb|AAL38815.1| putative mammalian MHC III region protein G9a [Arabidopsis
thaliana]
Length = 790
Score = 359 bits (921), Expect = 2e-97
Identities = 223/578 (38%), Positives = 315/578 (53%), Gaps = 48/578 (8%)
Query: 146 GDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLMIYE 205
G+G R+K K+ R +D + E L+ + G + D SR+ V TL ++
Sbjct: 226 GEGSRKKKSKKNLYWRDRESLD----SPEQLRILGVGTSSGSSSGDSSRNKVKETLRLFH 281
Query: 206 VMRRKLGQIDEKAKGSHSGAK----RPDLKAGTLMNTKGIRANSRKRI-GVVPGVEVGDI 260
+ RK+ Q DE AK K R D +A T++ G NS I G VPGVEVGD
Sbjct: 282 GVCRKILQEDE-AKPEDQRRKGKGLRIDFEASTILKRNGKFLNSGVHILGEVPGVEVGDE 340
Query: 261 FFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQG 320
F +R EL ++G+H PS AGIDY+ + +A SIV+SGGY+D+++D DVL Y+GQG
Sbjct: 341 FQYRMELNILGIHKPSQAGIDYM----KYGKAKVATSIVASGGYDDHLDDSDVLTYTGQG 396
Query: 321 GTSRE--------KGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKV--YVYD 370
G + K DQKL GNLAL S+ + VRVIRG H K YVYD
Sbjct: 397 GNVMQVKKKGEELKEPEDQKLITGNLALATSIEKQTPVRVIRGKHKSTHDKSKGGNYVYD 456
Query: 371 GLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTS 430
GLY ++ W + G NVFK++L R+PGQP+ + W + + KS R G+ D++
Sbjct: 457 GLYLVEKYWQQVGSHGMNVFKFQLRRIPGQPE--LSWVEVKK--SKSKYREGLCKLDISE 512
Query: 431 GAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHK-CGCT 489
G E+ P+ VN++D+EK P FTY+ L + PV + C C C + C C
Sbjct: 513 GKEQSPISAVNEIDDEK-PPLFTYTVKLIYPDWCRPVPP-KSCCCTTRCTEAEARVCACV 570
Query: 490 QKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGL 549
+KNGG +PY+ G + K +YECGP C CP SC RV+Q G+KL LE+F+TK +GWG+
Sbjct: 571 EKNGGEIPYNFDGAIVGAKPTIYECGPLCKCPSSCYLRVTQHGIKLPLEIFKTKSRGWGV 630
Query: 550 RSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIY-----QQLEVFSSDVE 604
R I G+FICEY GE+++++ E G D+Y+FD Y Q + +
Sbjct: 631 RCLKSIPIGSFICEYVGELLEDSEAERRIG--NDEYLFDIGNRYDNSLAQGMSELMLGTQ 688
Query: 605 APKI------PSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRH 658
A + S I A ++GNV RF+NHSC+PN+ + V+ ++++ HV F+A +
Sbjct: 689 AGRSMAEGDESSGFTIDAASKGNVGRFINHSCSPNLYAQNVLYDHEDSRIPHVMFFAQDN 748
Query: 659 IPPMMELTYDYGIVL----PLKVGQKKKKCLCGSVKCR 692
IPP+ EL YDY L K K+K C CG+ CR
Sbjct: 749 IPPLQELCYDYNYALDQVRDSKGNIKQKPCFCGAAVCR 786
>gb|AAO22580.1| putative mammalian MHC III region protein G9a [Arabidopsis
thaliana] gi|13517753|gb|AAK28971.1| SUVH6 [Arabidopsis
thaliana] gi|30681803|ref|NP_850030.1| SET
domain-containing protein (SUVH6) [Arabidopsis thaliana]
gi|42570881|ref|NP_973514.1| SET domain-containing
protein (SUVH6) [Arabidopsis thaliana]
gi|30580521|sp|Q8VZ17|SUV6_ARATH Histone-lysine
N-methyltransferase, H3 lysine-9 specific 6 (Histone
H3-K9 methyltransferase 6) (H3-K9-HMTase 6) (Suppressor
of variegation 3-9 homolog 6) (Su(var)3-9 homolog 6)
Length = 790
Score = 357 bits (916), Expect = 7e-97
Identities = 222/578 (38%), Positives = 315/578 (54%), Gaps = 48/578 (8%)
Query: 146 GDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLMIYE 205
G+G R+K K+ R +D + E L+ + G + D SR+ V TL ++
Sbjct: 226 GEGSRKKKSKKNLYWRDRESLD----SPEQLRILGVGTSSGSSSGDSSRNKVKETLRLFH 281
Query: 206 VMRRKLGQIDEKAKGSHSGAK----RPDLKAGTLMNTKGIRANSRKRI-GVVPGVEVGDI 260
+ RK+ Q DE AK K R D +A T++ G NS I G VPGVEVGD
Sbjct: 282 GVCRKILQEDE-AKPEDQRRKGKGLRIDFEASTILKRNGKFLNSGVHILGEVPGVEVGDE 340
Query: 261 FFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQG 320
F +R EL ++G+H PS AGIDY+ + +A SIV+SGGY+D++++ DVL Y+GQG
Sbjct: 341 FQYRMELNILGIHKPSQAGIDYM----KYGKAKVATSIVASGGYDDHLDNSDVLTYTGQG 396
Query: 321 GTSRE--------KGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKV--YVYD 370
G + K DQKL GNLAL S+ + VRVIRG H K YVYD
Sbjct: 397 GNVMQVKKKGEELKEPEDQKLITGNLALATSIEKQTPVRVIRGKHKSTHDKSKGGNYVYD 456
Query: 371 GLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTS 430
GLY ++ W + G NVFK++L R+PGQP+ + W + + KS R G+ D++
Sbjct: 457 GLYLVEKYWQQVGSHGMNVFKFQLRRIPGQPE--LSWVEVKK--SKSKYREGLCKLDISE 512
Query: 431 GAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHK-CGCT 489
G E+ P+ VN++D+EK P FTY+ L + PV + C C C + C C
Sbjct: 513 GKEQSPISAVNEIDDEK-PPLFTYTVKLIYPDWCRPVPP-KSCCCTTRCTEAEARVCACV 570
Query: 490 QKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGL 549
+KNGG +PY+ G + K +YECGP C CP SC RV+Q G+KL LE+F+TK +GWG+
Sbjct: 571 EKNGGEIPYNFDGAIVGAKPTIYECGPLCKCPSSCYLRVTQHGIKLPLEIFKTKSRGWGV 630
Query: 550 RSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIY-----QQLEVFSSDVE 604
R I G+FICEY GE+++++ E G D+Y+FD Y Q + +
Sbjct: 631 RCLKSIPIGSFICEYVGELLEDSEAERRIG--NDEYLFDIGNRYDNSLAQGMSELMLGTQ 688
Query: 605 APKI------PSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRH 658
A + S I A ++GNV RF+NHSC+PN+ + V+ ++++ HV F+A +
Sbjct: 689 AGRSMAEGDESSGFTIDAASKGNVGRFINHSCSPNLYAQNVLYDHEDSRIPHVMFFAQDN 748
Query: 659 IPPMMELTYDYGIVL----PLKVGQKKKKCLCGSVKCR 692
IPP+ EL YDY L K K+K C CG+ CR
Sbjct: 749 IPPLQELCYDYNYALDQVRDSKGNIKQKPCFCGAAVCR 786
>gb|AAD15582.1| similar to mammalian MHC III region protein G9a [Arabidopsis
thaliana] gi|25412134|pir||C84616 similar to mammalian
MHC III region protein G9a [imported] - Arabidopsis
thaliana
Length = 788
Score = 357 bits (916), Expect = 7e-97
Identities = 222/578 (38%), Positives = 315/578 (54%), Gaps = 48/578 (8%)
Query: 146 GDGKRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLMIYE 205
G+G R+K K+ R +D + E L+ + G + D SR+ V TL ++
Sbjct: 224 GEGSRKKKSKKNLYWRDRESLD----SPEQLRILGVGTSSGSSSGDSSRNKVKETLRLFH 279
Query: 206 VMRRKLGQIDEKAKGSHSGAK----RPDLKAGTLMNTKGIRANSRKRI-GVVPGVEVGDI 260
+ RK+ Q DE AK K R D +A T++ G NS I G VPGVEVGD
Sbjct: 280 GVCRKILQEDE-AKPEDQRRKGKGLRIDFEASTILKRNGKFLNSGVHILGEVPGVEVGDE 338
Query: 261 FFFRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQG 320
F +R EL ++G+H PS AGIDY+ + +A SIV+SGGY+D++++ DVL Y+GQG
Sbjct: 339 FQYRMELNILGIHKPSQAGIDYM----KYGKAKVATSIVASGGYDDHLDNSDVLTYTGQG 394
Query: 321 GTSRE--------KGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKV--YVYD 370
G + K DQKL GNLAL S+ + VRVIRG H K YVYD
Sbjct: 395 GNVMQVKKKGEELKEPEDQKLITGNLALATSIEKQTPVRVIRGKHKSTHDKSKGGNYVYD 454
Query: 371 GLYKIQNSWVEKAKSGFNVFKYKLVRLPGQPQAYMIWKSILQWTDKSASRVGVILPDLTS 430
GLY ++ W + G NVFK++L R+PGQP+ + W + + KS R G+ D++
Sbjct: 455 GLYLVEKYWQQVGSHGMNVFKFQLRRIPGQPE--LSWVEVKK--SKSKYREGLCKLDISE 510
Query: 431 GAEKLPVCLVNDVDNEKGPAYFTYSPTLKNLNRLAPVESSEGCTCNGGCQPGSHK-CGCT 489
G E+ P+ VN++D+EK P FTY+ L + PV + C C C + C C
Sbjct: 511 GKEQSPISAVNEIDDEK-PPLFTYTVKLIYPDWCRPVPP-KSCCCTTRCTEAEARVCACV 568
Query: 490 QKNGGYLPYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGL 549
+KNGG +PY+ G + K +YECGP C CP SC RV+Q G+KL LE+F+TK +GWG+
Sbjct: 569 EKNGGEIPYNFDGAIVGAKPTIYECGPLCKCPSSCYLRVTQHGIKLPLEIFKTKSRGWGV 628
Query: 550 RSWDPIRAGTFICEYAGEVIDNARVEELSGENEDDYIFDSTRIY-----QQLEVFSSDVE 604
R I G+FICEY GE+++++ E G D+Y+FD Y Q + +
Sbjct: 629 RCLKSIPIGSFICEYVGELLEDSEAERRIG--NDEYLFDIGNRYDNSLAQGMSELMLGTQ 686
Query: 605 APKI------PSPLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRH 658
A + S I A ++GNV RF+NHSC+PN+ + V+ ++++ HV F+A +
Sbjct: 687 AGRSMAEGDESSGFTIDAASKGNVGRFINHSCSPNLYAQNVLYDHEDSRIPHVMFFAQDN 746
Query: 659 IPPMMELTYDYGIVL----PLKVGQKKKKCLCGSVKCR 692
IPP+ EL YDY L K K+K C CG+ CR
Sbjct: 747 IPPLQELCYDYNYALDQVRDSKGNIKQKPCFCGAAVCR 784
>emb|CAB78388.1| putative protein [Arabidopsis thaliana] gi|4678372|emb|CAB41104.1|
putative protein [Arabidopsis thaliana]
gi|13517759|gb|AAK28974.1| SUVH9 [Arabidopsis thaliana]
gi|15236295|ref|NP_193082.1| SET domain-containing
protein (SUVH9) [Arabidopsis thaliana]
gi|7487739|pir||T06648 hypothetical protein T6G15.10 -
Arabidopsis thaliana gi|30580529|sp|Q9T0G7|SUV9_ARATH
Probable histone-lysine N-methyltransferase, H3 lysine-9
specific 9 (Histone H3-K9 methyltransferase 9)
(H3-K9-HMTase 9) (Suppressor of variegation 3-9 homolog
9) (Su(var)3-9 homolog 9)
Length = 650
Score = 351 bits (901), Expect = 4e-95
Identities = 242/666 (36%), Positives = 344/666 (51%), Gaps = 70/666 (10%)
Query: 49 GAPFVCVSPS-GPFPSGVAPFYPFFVSPESQRLSEQNAQTPTAQRAAPISAAVPINSFRT 107
G+ + + PS P PS + P V+ +Q L+ Q P A IS+AV + T
Sbjct: 2 GSSHIPLDPSLNPSPSLIPKLEP--VTESTQNLA---FQLPNTNPQALISSAVSDFNEAT 56
Query: 108 PTGATNGDVGSSRRKSRGQ-LPEDDNFVDLSEVDGE--------------GGTGDG---- 148
+ V S R + Q L D+ L + G T D
Sbjct: 57 DFSSDYNTVAESARSAFAQRLQRHDDVAVLDSLTGAIVPVEENPEPEPNPYSTSDSSPSV 116
Query: 149 --KRRKPQKRIREKRCSSDVDPDAVANEILKTINPGVFEILNQPDGSRDAVAYTLMIYEV 206
+R +PQ R E +DV P++ R+ V T MIY+
Sbjct: 117 ATQRPRPQPRSSELVRITDVGPESERQ-------------------FREHVRKTRMIYDS 157
Query: 207 MRRKLGQIDEKAKGSHSGAKRPDLKAG---TLMNTKGIRANSRKRI-GVVPGVEVGDIFF 262
+R L + K G R D KAG ++M + N KRI G +PGV+VGDIFF
Sbjct: 158 LRMFLMMEEAKRNGVGGRRARADGKAGKAGSMMRDCMLWMNRDKRIVGSIPGVQVGDIFF 217
Query: 263 FRFELCLVGLHAPSMAGIDYLGTKVSQEEEPLAVSIVSSGGYEDNVEDGDVLIYSGQGGT 322
FRFELC++GLH +GID+L +S EP+A S++ SGGYED+ + GDV++Y+GQGG
Sbjct: 218 FRFELCVMGLHGHPQSGIDFLTGSLSSNGEPIATSVIVSGGYEDDDDQGDVIMYTGQGGQ 277
Query: 323 SR-EKGASDQKLERGNLALERSLHRGNDVRVIRGMRDEAHPTGKVYVYDGLYKIQNSWVE 381
R + A Q+LE GNLA+ERS++ G +VRVIRG++ E + +VYVYDGL++I +SW +
Sbjct: 278 DRLGRQAEHQRLEGGNLAMERSMYYGIEVRVIRGLKYENEVSSRVYVYDGLFRIVDSWFD 337
Query: 382 KAKSGFNVFKYKLVRLPGQPQ---AYMIWKSILQWTDKSASRVGVILPDLTSGAEKLPVC 438
KSGF VFKY+L R+ GQ + + + + L+ S G I D+++G E +PV
Sbjct: 338 VGKSGFGVFKYRLERIEGQAEMGSSVLKFARTLKTNPLSVRPRGYINFDISNGKENVPVY 397
Query: 439 LVNDVDNEKGPAYFTYSPTLKNLNRLAPVES--SEGCTCNGGCQPGSHKCGCTQKNGGYL 496
L ND+D+++ P Y+ Y L +S + GC C GC G C C KN G +
Sbjct: 398 LFNDIDSDQEPLYYEYLAQTSFPPGLFVQQSGNASGCDCVNGCGSG---CLCEAKNSGEI 454
Query: 497 PYSAAGLLADLKSVVYECGPSCHCPPSCRNRVSQGGLKLRLEVFRTKGKGWGLRSWDPIR 556
Y G L K +++ECG +C CPPSCRNRV+Q GL+ RLEVFR+ GWG+RS D +
Sbjct: 455 AYDYNGTLIRQKPLIHECGSACQCPPSCRNRVTQKGLRNRLEVFRSLETGWGVRSLDVLH 514
Query: 557 AGTFICEYAGEVIDNARVEELSGENEDDYI----FDSTR--IYQQLEVFSSDVEAPKIPS 610
AG FICEYAG + + L+ N D + F S R + L +D E P P
Sbjct: 515 AGAFICEYAGVALTREQANILT-MNGDTLVYPARFSSARWEDWGDLSQVLADFERPSYPD 573
Query: 611 ----PLYITARNEGNVARFMNHSCTPNVLWRPVVRENKNEADLHVAFYAIRHIPPMMELT 666
+ NVA +++HS PNV+ + V+ ++ + V +A +IPPM EL+
Sbjct: 574 IPPVDFAMDVSKMRNVACYISHSTDPNVIVQFVLHDHNSLMFPRVMLFAAENIPPMTELS 633
Query: 667 YDYGIV 672
DYG+V
Sbjct: 634 LDYGVV 639
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,317,051,137
Number of Sequences: 2540612
Number of extensions: 62295734
Number of successful extensions: 164174
Number of sequences better than 10.0: 934
Number of HSP's better than 10.0 without gapping: 453
Number of HSP's successfully gapped in prelim test: 482
Number of HSP's that attempted gapping in prelim test: 161190
Number of HSP's gapped (non-prelim): 1618
length of query: 696
length of database: 863,360,394
effective HSP length: 135
effective length of query: 561
effective length of database: 520,377,774
effective search space: 291931931214
effective search space used: 291931931214
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0098b.1


