

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0097b.12
(292 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
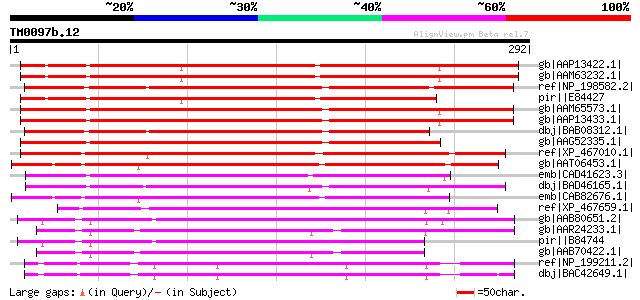
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP13422.1| At2g01660 [Arabidopsis thaliana] gi|20197542|gb|A... 336 5e-91
gb|AAM63232.1| unknown [Arabidopsis thaliana] 330 4e-89
ref|NP_198582.2| receptor-like protein kinase-related [Arabidops... 286 6e-76
pir||E84427 hypothetical protein At2g01660 [imported] - Arabidop... 283 4e-75
gb|AAM65573.1| unknown [Arabidopsis thaliana] 263 3e-69
gb|AAP13433.1| At1g70690 [Arabidopsis thaliana] gi|27311729|gb|A... 262 7e-69
dbj|BAB08312.1| unnamed protein product [Arabidopsis thaliana] 258 2e-67
gb|AAG52335.1| unknown protein; 52490-51678 [Arabidopsis thalian... 229 7e-59
ref|XP_467010.1| receptor-like protein kinase-like [Oryza sativa... 208 2e-52
gb|AAT06453.1| At3g60720 [Arabidopsis thaliana] gi|42566097|ref|... 199 6e-50
emb|CAD41623.3| OSJNBa0091D06.12 [Oryza sativa (japonica cultiva... 192 7e-48
dbj|BAD46165.1| 33 kDa secretory protein-like [Oryza sativa (jap... 179 6e-44
emb|CAB82676.1| secretory protein-like [Arabidopsis thaliana] gi... 175 1e-42
ref|XP_467659.1| 33 kDa secretory protein-like [Oryza sativa (ja... 166 7e-40
gb|AAB80651.2| expressed protein [Arabidopsis thaliana] gi|20148... 137 4e-31
gb|AAR24233.1| At1g04520 [Arabidopsis thaliana] gi|42561693|ref|... 133 7e-30
pir||B84744 hypothetical protein At2g33330 [imported] - Arabidop... 123 5e-27
gb|AAB70422.1| F19P19.1 [Arabidopsis thaliana] gi|25406792|pir||... 119 8e-26
ref|NP_199211.2| receptor-like protein kinase-related [Arabidops... 113 7e-24
dbj|BAC42649.1| unknown protein [Arabidopsis thaliana] 112 1e-23
>gb|AAP13422.1| At2g01660 [Arabidopsis thaliana] gi|20197542|gb|AAD12705.2|
expressed protein [Arabidopsis thaliana]
gi|17065482|gb|AAL32895.1| Unknown protein [Arabidopsis
thaliana] gi|18379273|ref|NP_565272.1| 33 kDa secretory
protein-related [Arabidopsis thaliana]
Length = 288
Score = 336 bits (861), Expect = 5e-91
Identities = 168/285 (58%), Positives = 216/285 (74%), Gaps = 10/285 (3%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNY 66
+LFI +S + T S S+++DTFI+GGCSQ K P S YE+ VNSLLTS V+SAS Y
Sbjct: 7 VLFIAVVSLLGTFS-SAAVDTFIYGGCSQEKYFPGSP--YESNVNSLLTSFVSSASLYTY 63
Query: 67 NNFTVPATAGSGDTLYGLYQCRGDLNNDQ--CYRCVARAVSQLGTLCSAVRGGALQLEGC 124
NNFT +G ++YGLYQCRGDL++ C RCVARAVS+LG+LC+ GGALQLEGC
Sbjct: 64 NNFTTNGISGDSSSVYGLYQCRGDLSSGSGDCARCVARAVSRLGSLCAMASGGALQLEGC 123
Query: 125 FVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGD 184
FVKYDN FLGVEDKT+VV++CGP +G SD +TRRD+V+ L S G ++R+ G+
Sbjct: 124 FVKYDNTTFLGVEDKTVVVRRCGPPVGYNSDEMTRRDSVVGSLAASSG--GSYRVGVSGE 181
Query: 185 FQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSR---H 241
QG AQCTGDLS +ECQDCL E+I RL+T+CG + WG++YLAKCYARYS G HSR +
Sbjct: 182 LQGVAQCTGDLSATECQDCLMEAIGRLRTDCGGAAWGDVYLAKCYARYSARGGHSRANGY 241
Query: 242 SDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEKQRG 286
+ +N++DDEIEKTLAI++GLIAGV L++VFLSF++K CE+ +G
Sbjct: 242 GGNRNNNDDDEIEKTLAIIVGLIAGVTLLVVFLSFMAKSCERGKG 286
>gb|AAM63232.1| unknown [Arabidopsis thaliana]
Length = 289
Score = 330 bits (845), Expect = 4e-89
Identities = 168/286 (58%), Positives = 216/286 (74%), Gaps = 11/286 (3%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNY 66
+LFI +S + T S S+++DTFI+GGCSQ K S YE+ VNSLLTS V+SAS Y
Sbjct: 7 VLFIAVVSLLGTFS-SAAVDTFIYGGCSQAKYFQGSP--YESNVNSLLTSFVSSASLYTY 63
Query: 67 NNFTVPATAG-SGDTLYGLYQCRGDLNNDQ--CYRCVARAVSQLGTLCSAVRGGALQLEG 123
NNFT +G S ++YGLYQCRGDL++ C RCVARAVS+LG+LC+ GGALQLEG
Sbjct: 64 NNFTANGISGDSSSSVYGLYQCRGDLSSGSGDCARCVARAVSRLGSLCAMASGGALQLEG 123
Query: 124 CFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYG 183
CFVKYDN FLGVEDKT+VV++CGP +G SD +TRRD+V+ L S G ++R+ G
Sbjct: 124 CFVKYDNTTFLGVEDKTVVVRRCGPPVGYNSDEMTRRDSVVGSLAASSG--GSYRVGVSG 181
Query: 184 DFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSR--- 240
+ QG AQCTGDLS +ECQDCL E+I RL+T+CG + WG++YLAKCYARYS G HSR
Sbjct: 182 ELQGVAQCTGDLSATECQDCLMEAIGRLRTDCGGAAWGDVYLAKCYARYSARGGHSRANG 241
Query: 241 HSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEKQRG 286
+ + +N++DDEIEKTLAI++GLIAGV L++VFLSF++K CE+ +G
Sbjct: 242 YGGNRNNNDDDEIEKTLAIIVGLIAGVTLLVVFLSFMAKSCERGKG 287
>ref|NP_198582.2| receptor-like protein kinase-related [Arabidopsis thaliana]
gi|63025162|gb|AAY27054.1| At5g37660 [Arabidopsis
thaliana]
Length = 288
Score = 286 bits (731), Expect = 6e-76
Identities = 147/276 (53%), Positives = 192/276 (69%), Gaps = 9/276 (3%)
Query: 9 FILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNN 68
F++ +A + S +S+ DTF+FGGCSQ K +P S YE+ +NSLLTSLVNSA++++YNN
Sbjct: 18 FLIAATAPSLSSATSATDTFVFGGCSQQKFSPAS--AYESNLNSLLTSLVNSATYSSYNN 75
Query: 69 FTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKY 128
FT+ ++ S DT GL+QCRGDL+ C CVARAVSQ+G LC GGALQL GC++KY
Sbjct: 76 FTIMGSSSS-DTARGLFQCRGDLSMPDCATCVARAVSQVGPLCPFTCGGALQLAGCYIKY 134
Query: 129 DNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGDFQGA 188
DN+ FLG EDKT+V+KKCG S G +D ++RRDAVL L G +R GD QG
Sbjct: 135 DNISFLGQEDKTVVLKKCGSSEGYNTDGISRRDAVLTELVNGGGYFRA---GGSGDVQGM 191
Query: 189 AQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSRHSDDDSNH 248
QC GDL+ SECQDCL +I RLK +CG + +G+M+LAKCYARYS G ++H N+
Sbjct: 192 GQCVGDLTVSECQDCLGTAIGRLKNDCGTAVFGDMFLAKCYARYSTDG--AQHYAKSHNY 249
Query: 249 NDD-EIEKTLAILIGLIAGVALIIVFLSFLSKVCEK 283
+ EKT AI+IGL+A V L+I+FL FL VC +
Sbjct: 250 KTNYGGEKTFAIIIGLLAAVVLLIIFLLFLRGVCSR 285
>pir||E84427 hypothetical protein At2g01660 [imported] - Arabidopsis thaliana
gi|42570641|ref|NP_973394.1| 33 kDa secretory
protein-related [Arabidopsis thaliana]
Length = 259
Score = 283 bits (724), Expect = 4e-75
Identities = 141/236 (59%), Positives = 176/236 (73%), Gaps = 7/236 (2%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNY 66
+LFI +S + T S S+++DTFI+GGCSQ K P S YE+ VNSLLTS V+SAS Y
Sbjct: 7 VLFIAVVSLLGTFS-SAAVDTFIYGGCSQEKYFPGSP--YESNVNSLLTSFVSSASLYTY 63
Query: 67 NNFTVPATAGSGDTLYGLYQCRGDLNNDQ--CYRCVARAVSQLGTLCSAVRGGALQLEGC 124
NNFT +G ++YGLYQCRGDL++ C RCVARAVS+LG+LC+ GGALQLEGC
Sbjct: 64 NNFTTNGISGDSSSVYGLYQCRGDLSSGSGDCARCVARAVSRLGSLCAMASGGALQLEGC 123
Query: 125 FVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGD 184
FVKYDN FLGVEDKT+VV++CGP +G SD +TRRD+V+ L S G ++R+ G+
Sbjct: 124 FVKYDNTTFLGVEDKTVVVRRCGPPVGYNSDEMTRRDSVVGSLAASSG--GSYRVGVSGE 181
Query: 185 FQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSR 240
QG AQCTGDLS +ECQDCL E+I RL+T+CG + WG++YLAKCYARYS G HSR
Sbjct: 182 LQGVAQCTGDLSATECQDCLMEAIGRLRTDCGGAAWGDVYLAKCYARYSARGGHSR 237
>gb|AAM65573.1| unknown [Arabidopsis thaliana]
Length = 299
Score = 263 bits (673), Expect = 3e-69
Identities = 137/287 (47%), Positives = 185/287 (63%), Gaps = 15/287 (5%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNY 66
L F+LT + PS+SS D +I+ CS K +P S YET +NSLL+S V S + T Y
Sbjct: 10 LCFLLTAVILMNPSSSSPTDNYIYAVCSPAKFSPSSG--YETNLNSLLSSFVTSTAQTRY 67
Query: 67 NNFTVPATAGSGD-TLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCF 125
NFTVP T+YG+YQCRGDL+ C CV+ AV+Q+G LCS G LQ+E C
Sbjct: 68 ANFTVPTGKSEPTVTVYGIYQCRGDLDPTACSTCVSSAVAQVGALCSNSYSGFLQMENCL 127
Query: 126 VKYDNVKFLGVEDKTMVVKKCGPSIGLT-SDALTRRDAVLAYLQTSDGVYRTWRISAYGD 184
++YDN FLGV+DKT+++ KCG + DALT+ V+ L T DG YRT G+
Sbjct: 128 IRYDNKSFLGVQDKTLILNKCGQPMEFNDQDALTKASDVIGSLGTGDGSYRT---GGNGN 184
Query: 185 FQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSR---- 240
QG AQC+GDLS S+CQDCLS++I RLK++CG + G +YL+KCYAR+S GG H+R
Sbjct: 185 VQGVAQCSGDLSTSQCQDCLSDAIGRLKSDCGMAQGGYVYLSKCYARFSVGGSHARQTPG 244
Query: 241 ----HSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEK 283
H + N +D+ + KTLAI+IG+I + L++VFL+F+ K C K
Sbjct: 245 PNFGHEGEKGNKDDNGVGKTLAIIIGMITLIILLVVFLAFVGKCCRK 291
>gb|AAP13433.1| At1g70690 [Arabidopsis thaliana] gi|27311729|gb|AAO00830.1| Unknown
protein [Arabidopsis thaliana]
gi|18409653|ref|NP_564997.1| kinase-related [Arabidopsis
thaliana]
Length = 299
Score = 262 bits (670), Expect = 7e-69
Identities = 136/287 (47%), Positives = 185/287 (64%), Gaps = 15/287 (5%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNY 66
L F+LT + PS+SS D +I+ CS K +P S YET +NSLL+S V S + T Y
Sbjct: 10 LCFLLTAVILMNPSSSSPTDNYIYAVCSPAKFSPSSG--YETNLNSLLSSFVTSTAQTRY 67
Query: 67 NNFTVPATAGSGD-TLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCF 125
NFTVP T+YG+YQCRGDL+ C CV+ AV+Q+G LCS G LQ+E C
Sbjct: 68 ANFTVPTGKPEPTVTVYGIYQCRGDLDPTACSTCVSSAVAQVGALCSNSYSGFLQMENCL 127
Query: 126 VKYDNVKFLGVEDKTMVVKKCGPSIGLT-SDALTRRDAVLAYLQTSDGVYRTWRISAYGD 184
++YDN FLGV+DKT+++ KCG + DALT+ V+ L T DG YRT G+
Sbjct: 128 IRYDNKSFLGVQDKTLILNKCGQPMEFNDQDALTKASDVIGSLGTGDGSYRT---GGNGN 184
Query: 185 FQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSR---- 240
QG AQC+GDLS S+CQDCLS++I RLK++CG + G +YL+KCYAR+S GG H+R
Sbjct: 185 VQGVAQCSGDLSTSQCQDCLSDAIGRLKSDCGMAQGGYVYLSKCYARFSVGGSHARQTPG 244
Query: 241 ----HSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEK 283
H + N +D+ + KTLAI+IG++ + L++VFL+F+ K C K
Sbjct: 245 PNFGHEGEKGNKDDNGVGKTLAIIIGIVTLIILLVVFLAFVGKCCRK 291
>dbj|BAB08312.1| unnamed protein product [Arabidopsis thaliana]
Length = 270
Score = 258 bits (658), Expect = 2e-67
Identities = 127/228 (55%), Positives = 163/228 (70%), Gaps = 6/228 (2%)
Query: 9 FILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNN 68
F++ +A + S +S+ DTF+FGGCSQ K +P S YE+ +NSLLTSLVNSA++++YNN
Sbjct: 18 FLIAATAPSLSSATSATDTFVFGGCSQQKFSPAS--AYESNLNSLLTSLVNSATYSSYNN 75
Query: 69 FTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKY 128
FT+ ++ S DT GL+QCRGDL+ C CVARAVSQ+G LC GGALQL GC++KY
Sbjct: 76 FTIMGSSSS-DTARGLFQCRGDLSMPDCATCVARAVSQVGPLCPFTCGGALQLAGCYIKY 134
Query: 129 DNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGDFQGA 188
DN+ FLG EDKT+V+KKCG S G +D ++RRDAVL L G +R GD QG
Sbjct: 135 DNISFLGQEDKTVVLKKCGSSEGYNTDGISRRDAVLTELVNGGGYFRA---GGSGDVQGM 191
Query: 189 AQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGG 236
QC GDL+ SECQDCL +I RLK +CG + +G+M+LAKCYARYS G
Sbjct: 192 GQCVGDLTVSECQDCLGTAIGRLKNDCGTAVFGDMFLAKCYARYSTDG 239
>gb|AAG52335.1| unknown protein; 52490-51678 [Arabidopsis thaliana]
gi|25406066|pir||C96731 unknown protein F5A18.13
[imported] - Arabidopsis thaliana
Length = 270
Score = 229 bits (584), Expect = 7e-59
Identities = 118/238 (49%), Positives = 156/238 (64%), Gaps = 7/238 (2%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNY 66
L F+LT + PS+SS D +I+ CS K +P S YET +NSLL+S V S + T Y
Sbjct: 10 LCFLLTAVILMNPSSSSPTDNYIYAVCSPAKFSPSSG--YETNLNSLLSSFVTSTAQTRY 67
Query: 67 NNFTVPATAGSGD-TLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCF 125
NFTVP T+YG+YQCRGDL+ C CV+ AV+Q+G LCS G LQ+E C
Sbjct: 68 ANFTVPTGKPEPTVTVYGIYQCRGDLDPTACSTCVSSAVAQVGALCSNSYSGFLQMENCL 127
Query: 126 VKYDNVKFLGVEDKTMVVKKCGPSIGLT-SDALTRRDAVLAYLQTSDGVYRTWRISAYGD 184
++YDN FLGV+DKT+++ KCG + DALT+ V+ L T DG YRT G+
Sbjct: 128 IRYDNKSFLGVQDKTLILNKCGQPMEFNDQDALTKASDVIGSLGTGDGSYRT---GGNGN 184
Query: 185 FQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSRHS 242
QG AQC+GDLS S+CQDCLS++I RLK++CG + G +YL+KCYAR+S GG H+R +
Sbjct: 185 VQGVAQCSGDLSTSQCQDCLSDAIGRLKSDCGMAQGGYVYLSKCYARFSVGGSHARQT 242
>ref|XP_467010.1| receptor-like protein kinase-like [Oryza sativa (japonica
cultivar-group)] gi|49388651|dbj|BAD25786.1|
receptor-like protein kinase-like [Oryza sativa
(japonica cultivar-group)]
Length = 297
Score = 208 bits (529), Expect = 2e-52
Identities = 108/278 (38%), Positives = 170/278 (60%), Gaps = 14/278 (5%)
Query: 7 LLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASF-TN 65
+L +++L A + ++ TFI+ GCS K P + +++ +NSLL+S+ ++AS
Sbjct: 14 VLVLISLGAAPPAAVDAATATFIYAGCSPSKYEP--NTAFQSNLNSLLSSIASTASSGAA 71
Query: 66 YNNFTVPATAG----SGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQL 121
YN+FT AG +G YGLYQCRGDL+ C CV + V++LG +C+ +LQ+
Sbjct: 72 YNSFTAGGGAGPDPAAGTAAYGLYQCRGDLSPGDCVACVRQTVARLGAVCANAYAASLQV 131
Query: 122 EGCFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISA 181
+GC+V+YD F+G D T +KC S L+ RD VL LQ + G +++S
Sbjct: 132 DGCYVRYDAADFIGRADTTTAYRKCSSSTSRDGAFLSSRDGVLGELQAAAG----YKLST 187
Query: 182 YGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSRH 241
G QG AQC GD+ ++C CL+E++ +LK CG + ++YLA+CY RY G + R
Sbjct: 188 SGTVQGVAQCLGDVPANDCTACLAEAVGQLKGACGTALAADVYLAQCYVRYWANGYYFRP 247
Query: 242 SDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSK 279
+ D+S D++ +T+AI+IG++AG+AL++VF+SFL K
Sbjct: 248 NSDNSG---DDVGRTVAIIIGILAGLALLVVFISFLRK 282
>gb|AAT06453.1| At3g60720 [Arabidopsis thaliana] gi|42566097|ref|NP_191631.2|
receptor-like protein kinase-related [Arabidopsis
thaliana]
Length = 279
Score = 199 bits (507), Expect = 6e-50
Identities = 111/278 (39%), Positives = 172/278 (60%), Gaps = 13/278 (4%)
Query: 2 VLLLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSA 61
+ L LLF+ S+ ++ S+S S FI+GGCS K TP + +E+ ++ L+S+V S+
Sbjct: 4 LFLFSLLFLFFYSSSSSRSSSES-HIFIYGGCSPEKYTP--NTPFESNRDTFLSSVVTSS 60
Query: 62 SFTNYNNFTV---PATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGA 118
S ++N+F V +++ S ++GLYQCR DL + C +C+ +V Q+ +C G +
Sbjct: 61 SDASFNSFAVGNDSSSSSSSSAVFGLYQCRDDLRSSDCSKCIQTSVDQITLICPYSYGAS 120
Query: 119 LQLEGCFVKYDNVKFLGVEDKTMVVKKC-GPSIGLTSDALTRRDAVLAYLQTSDGVYRTW 177
LQLEGCF++Y+ FLG D ++ KKC S+ D RRD VL+ L+++ Y
Sbjct: 121 LQLEGCFLRYETNDFLGKPDTSLRYKKCSSKSVENDYDFFKRRDDVLSDLESTQLGY--- 177
Query: 178 RISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGD 237
++S G +G AQC GDLSPS+C CL+ES+ +LK CG++ E+YLA+CYARY G
Sbjct: 178 KVSRSGLVEGYAQCVGDLSPSDCTACLAESVGKLKNLCGSAVAAEVYLAQCYARYWGSGY 237
Query: 238 HSRHSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLS 275
+ SD N D + K++AI++G+IAG A+++V LS
Sbjct: 238 YDFSSDPT---NGDHVGKSIAIIVGVIAGFAILVVLLS 272
>emb|CAD41623.3| OSJNBa0091D06.12 [Oryza sativa (japonica cultivar-group)]
gi|50926855|ref|XP_473334.1| OSJNBa0091D06.12 [Oryza
sativa (japonica cultivar-group)]
Length = 273
Score = 192 bits (489), Expect = 7e-48
Identities = 103/245 (42%), Positives = 142/245 (57%), Gaps = 10/245 (4%)
Query: 10 ILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNNF 69
++ L I ++ TFI+ GCS K P + +E +NSLL S+ N+A YN+F
Sbjct: 11 VVVLVVIGATVAEAATGTFIYAGCSPSKYQPGTP--FEGNLNSLLASIANAAPNGGYNSF 68
Query: 70 TVPATA-GSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKY 128
T + G G YGLYQCRGDL N C CV AV QL +C+A +LQLEGC+V+Y
Sbjct: 69 TAGSNGTGDGAAAYGLYQCRGDLGNADCAACVRDAVGQLNEVCAAAYAASLQLEGCYVRY 128
Query: 129 DNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGDFQGA 188
D+ F+G D MV +KC S D L RDAVLA LQ G+ +++S+ G+ QG
Sbjct: 129 DSSNFVGQPDNAMVYRKCSTSTSGDGDFLKNRDAVLAALQ--GGLANGYKVSSSGNVQGV 186
Query: 189 AQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGDHSRHSD----- 243
+QC GDL+ +C CL++++ +LK CG S ++YLA+CY RY G + R S
Sbjct: 187 SQCLGDLAAGDCTTCLAQAVGQLKGTCGTSLAADVYLAQCYVRYWANGFYFRPSQACSAK 246
Query: 244 DDSNH 248
D+SNH
Sbjct: 247 DESNH 251
>dbj|BAD46165.1| 33 kDa secretory protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 346
Score = 179 bits (455), Expect = 6e-44
Identities = 115/311 (36%), Positives = 159/311 (50%), Gaps = 47/311 (15%)
Query: 10 ILTLSAITTPSTSSSIDT-FIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNN 68
+L L A T + + T F++ GCSQ + + Y V++ L++L NSA +T Y N
Sbjct: 9 LLVLCACATRARGADDYTAFVYAGCSQARYDAGTQ--YAADVDTALSALTNSAGYTAYAN 66
Query: 69 FTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKY 128
+T P+ A SG L G+YQCR DL C CV A ++L +LC++ G +QL CFV+Y
Sbjct: 67 YTSPSAA-SGTGLVGVYQCRSDLPAAICGGCVRSAATKLASLCNSAAGAGVQLRACFVRY 125
Query: 129 DNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYL-----QTSDGVYRTWRISAYG 183
N FLG +D T++ KKCG G + + RDA L L DG YR A G
Sbjct: 126 GNDSFLGRQDTTVLFKKCGGEGGSDTGVVAMRDAALGALVAAAAPAGDGSYRA---GAAG 182
Query: 184 DFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSE--------- 234
Q +QC GDL C DC+S + +LK CG ++ GE+YL KCYAR+
Sbjct: 183 YVQAMSQCVGDLGAKACTDCVSAASSQLKAGCGYASAGEVYLGKCYARFWSNAGTGDNNG 242
Query: 235 -------------------------GGDHSRHSDDDSNHND-DEIEKTLAILIGLIAGVA 268
GG+ + ++D DE KTLAI+IGL+A VA
Sbjct: 243 GGISGGGGGIGGGGNGISGGGGAVGGGNGYAYGFVPHTYSDHDESGKTLAIIIGLVAAVA 302
Query: 269 LIIVFLSFLSK 279
L+IVFLSF+ +
Sbjct: 303 LVIVFLSFVRR 313
>emb|CAB82676.1| secretory protein-like [Arabidopsis thaliana]
gi|11358832|pir||T47883 secretory protein-like -
Arabidopsis thaliana
Length = 247
Score = 175 bits (444), Expect = 1e-42
Identities = 98/250 (39%), Positives = 150/250 (59%), Gaps = 10/250 (4%)
Query: 2 VLLLRLLFILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSA 61
+ L LLF+ S+ ++ S+S S FI+GGCS K TP + +E+ ++ L+S+V S+
Sbjct: 4 LFLFSLLFLFFYSSSSSRSSSES-HIFIYGGCSPEKYTP--NTPFESNRDTFLSSVVTSS 60
Query: 62 SFTNYNNFTV---PATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGA 118
S ++N+F V +++ S ++GLYQCR DL + C +C+ +V Q+ +C G +
Sbjct: 61 SDASFNSFAVGNDSSSSSSSSAVFGLYQCRDDLRSSDCSKCIQTSVDQITLICPYSYGAS 120
Query: 119 LQLEGCFVKYDNVKFLGVEDKTMVVKKC-GPSIGLTSDALTRRDAVLAYLQTSDGVYRTW 177
LQLEGCF++Y+ FLG D ++ KKC S+ D RRD VL+ L+++ Y
Sbjct: 121 LQLEGCFLRYETNDFLGKPDTSLRYKKCSSKSVENDYDFFKRRDDVLSDLESTQLGY--- 177
Query: 178 RISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYSEGGD 237
++S G +G AQC GDLSPS+C CL+ES+ +LK CG++ E+YLA+CYARY G
Sbjct: 178 KVSRSGLVEGYAQCVGDLSPSDCTACLAESVGKLKNLCGSAVAAEVYLAQCYARYWGSGY 237
Query: 238 HSRHSDDDSN 247
+ S N
Sbjct: 238 YDFSSGQSIN 247
>ref|XP_467659.1| 33 kDa secretory protein-like [Oryza sativa (japonica
cultivar-group)] gi|46390426|dbj|BAD15888.1| 33 kDa
secretory protein-like [Oryza sativa (japonica
cultivar-group)] gi|46390682|dbj|BAD16164.1| 33 kDa
secretory protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 278
Score = 166 bits (420), Expect = 7e-40
Identities = 96/252 (38%), Positives = 136/252 (53%), Gaps = 11/252 (4%)
Query: 28 FIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNNFTVPATAGSGDTLYGLYQC 87
F++ GCSQ + S Y + V+S+LTS+ NSA ++ Y NFT P + +++ G+YQC
Sbjct: 33 FVYAGCSQGRYA--SGTQYASDVDSVLTSVANSAPYSPYANFTSPTS----NSVVGVYQC 86
Query: 88 RGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKFLGVEDKTMVVKKCG 147
R DL C CV A+S+L +LC+ GGA+QL CFV+Y N FLG +D ++ KKCG
Sbjct: 87 RSDLPASVCTGCVRSAISRLSSLCAWATGGAVQLRACFVRYGNDTFLGKQDTAVLFKKCG 146
Query: 148 PSIGLTSDALTRRDAVLAYLQTSDGVYRTWRISAYGDFQGAAQCTGDLSPSECQDCLSES 207
S G A R A+ A + + +R G Q +QC GDL C DC+S +
Sbjct: 147 GSPGDAGGAAMRDSALGALVAAAAPAGGGYRAGGSGGVQAMSQCVGDLGAKACSDCVSAA 206
Query: 208 IQRLKTECGASTWGEMYLAKCYARY--SEGGDHSRHSDDD---SNHNDDEIEKTLAILIG 262
+LK CG +T GE+YL KCYAR+ + GG S + D S H L++ G
Sbjct: 207 AGQLKAGCGYATAGEVYLGKCYARFWGNGGGGFSSGAAGDAYGSGHRVSGNRFVLSVAGG 266
Query: 263 LIAGVALIIVFL 274
+A I V +
Sbjct: 267 FFTSLAYIFVLM 278
>gb|AAB80651.2| expressed protein [Arabidopsis thaliana] gi|20148723|gb|AAM10252.1|
unknown protein [Arabidopsis thaliana]
gi|14596051|gb|AAK68753.1| Unknown protein [Arabidopsis
thaliana] gi|18403243|ref|NP_565764.1| 33 kDa secretory
protein-related [Arabidopsis thaliana]
Length = 304
Score = 137 bits (345), Expect = 4e-31
Identities = 85/297 (28%), Positives = 144/297 (47%), Gaps = 22/297 (7%)
Query: 5 LRLLFILTLSAIT----TPSTSSSIDTFIFGGCSQPKNTPVSDP--VYETGVNSLLTSLV 58
L LL+I+ ++ + S+S I+ GC++ + +SDP +Y ++++ LV
Sbjct: 9 LLLLYIIIMALFSDLKLAKSSSPEYTNLIYKGCARQR---LSDPSGLYSQALSAMYGLLV 65
Query: 59 NSASFTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGA 118
++ T + T T+ + T GL+QCRGDL+N+ CY CV+R G LC
Sbjct: 66 TQSTKTRFYKTTTGTTSQTSVT--GLFQCRGDLSNNDCYNCVSRLPVLSGKLCGKTIAAR 123
Query: 119 LQLEGCFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWR 178
+QL GC++ Y+ F + ++ K CG + + RRD +Q +
Sbjct: 124 VQLSGCYLLYEISGFAQISGMELLFKTCGKNNVAGTGFEQRRDTAFGVMQNGVVQGHGFY 183
Query: 179 ISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS--EGG 236
+ Y QC GD+ S+C C+ ++QR + ECG+S G++YL KC+ YS G
Sbjct: 184 ATTYESVYVLGQCEGDIGDSDCSGCIKNALQRAQVECGSSISGQIYLHKCFVGYSFYPNG 243
Query: 237 DHSRHS---------DDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEKQ 284
R S S+ + KT+AI++G AGV +++ L F+ + +K+
Sbjct: 244 VPKRSSPYPSSGSSGSSSSSSSSGTTGKTVAIIVGGTAGVGFLVICLLFVKNLMKKK 300
>gb|AAR24233.1| At1g04520 [Arabidopsis thaliana] gi|42561693|ref|NP_171946.2| 33
kDa secretory protein-related [Arabidopsis thaliana]
gi|45592918|gb|AAS68113.1| At1g04520 [Arabidopsis
thaliana]
Length = 307
Score = 133 bits (334), Expect = 7e-30
Identities = 83/291 (28%), Positives = 137/291 (46%), Gaps = 32/291 (10%)
Query: 16 ITTPSTSSSIDTFIFGGCSQPKNTPVSDP--VYETGVNSLLTSLVNSASFTNYNNFTVPA 73
+ S ++ T I+ GC++ + SDP +Y ++++ SLV+ ++ T + T
Sbjct: 23 VVVKSATTEYTTLIYKGCARQQ---FSDPSGLYSQALSAMFGSLVSQSTKTRFYKTT--- 76
Query: 74 TAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKF 133
T S T+ GL+QCRGDL+N CY CV+R LC +QL GC++ Y+ F
Sbjct: 77 TGTSTTTITGLFQCRGDLSNHDCYNCVSRLPVLSDKLCGKTIASRVQLSGCYLLYEVSGF 136
Query: 134 LGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQ----TSDGVYRTWRISAYGDFQGAA 189
+ M+ K CG + + RRD +Q + G Y T S Y
Sbjct: 137 SQISGMEMLFKTCGKNNIAGTGFEERRDTAFGVMQNGVVSGHGFYATTYESVY----VLG 192
Query: 190 QCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY----------------S 233
QC GD+ ++C C+ ++++ + ECG+S G++YL KC+ Y S
Sbjct: 193 QCEGDVGDTDCSGCVKNALEKAQVECGSSISGQIYLHKCFIAYSYYPNGVPRRSSSSSSS 252
Query: 234 EGGDHSRHSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKVCEKQ 284
S S+ D + + KT+AI++G AGV +++ L F + K+
Sbjct: 253 SSSSSSGSSNSDPSTSTGATGKTVAIIVGGAAGVGFLVICLLFAKNLMRKK 303
>pir||B84744 hypothetical protein At2g33330 [imported] - Arabidopsis thaliana
Length = 303
Score = 123 bits (309), Expect = 5e-27
Identities = 70/235 (29%), Positives = 117/235 (49%), Gaps = 11/235 (4%)
Query: 5 LRLLFILTLSAIT----TPSTSSSIDTFIFGGCSQPKNTPVSDP--VYETGVNSLLTSLV 58
L LL+I+ ++ + S+S I+ GC++ + +SDP +Y ++++ LV
Sbjct: 9 LLLLYIIIMALFSDLKLAKSSSPEYTNLIYKGCARQR---LSDPSGLYSQALSAMYGLLV 65
Query: 59 NSASFTNYNNFTVPATAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGA 118
++ T + T T+ + T GL+QCRGDL+N+ CY CV+R G LC
Sbjct: 66 TQSTKTRFYKTTTGTTSQTSVT--GLFQCRGDLSNNDCYNCVSRLPVLSGKLCGKTIAAR 123
Query: 119 LQLEGCFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWR 178
+QL GC++ Y+ F + ++ K CG + + RRD +Q +
Sbjct: 124 VQLSGCYLLYEISGFAQISGMELLFKTCGKNNVAGTGFEQRRDTAFGVMQNGVVQGHGFY 183
Query: 179 ISAYGDFQGAAQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
+ Y QC GD+ S+C C+ ++QR + ECG+S G++YL KC+ YS
Sbjct: 184 ATTYESVYVLGQCEGDIGDSDCSGCIKNALQRAQVECGSSISGQIYLHKCFVGYS 238
>gb|AAB70422.1| F19P19.1 [Arabidopsis thaliana] gi|25406792|pir||F86177 protein
F19P19.1 [imported] - Arabidopsis thaliana
Length = 270
Score = 119 bits (299), Expect = 8e-26
Identities = 69/224 (30%), Positives = 112/224 (49%), Gaps = 16/224 (7%)
Query: 16 ITTPSTSSSIDTFIFGGCSQPKNTPVSDP--VYETGVNSLLTSLVNSASFTNYNNFTVPA 73
+ S ++ T I+ GC++ + SDP +Y ++++ SLV+ ++ T + T
Sbjct: 23 VVVKSATTEYTTLIYKGCARQQ---FSDPSGLYSQALSAMFGSLVSQSTKTRFYKTT--- 76
Query: 74 TAGSGDTLYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVRGGALQLEGCFVKYDNVKF 133
T S T+ GL+QCRGDL+N CY CV+R LC +QL GC++ Y+ F
Sbjct: 77 TGTSTTTITGLFQCRGDLSNHDCYNCVSRLPVLSDKLCGKTIASRVQLSGCYLLYEVSGF 136
Query: 134 LGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQ----TSDGVYRTWRISAYGDFQGAA 189
+ M+ K CG + + RRD +Q + G Y T S Y
Sbjct: 137 SQISGMEMLFKTCGKNNIAGTGFEERRDTAFGVMQNGVVSGHGFYATTYESVY----VLG 192
Query: 190 QCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARYS 233
QC GD+ ++C C+ ++++ + ECG+S G++YL KC+ YS
Sbjct: 193 QCEGDVGDTDCSGCVKNALEKAQVECGSSISGQIYLHKCFIAYS 236
>ref|NP_199211.2| receptor-like protein kinase-related [Arabidopsis thaliana]
Length = 303
Score = 113 bits (282), Expect = 7e-24
Identities = 79/304 (25%), Positives = 134/304 (43%), Gaps = 47/304 (15%)
Query: 9 FILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNN 68
F L AI S IF GC+ K+ P V+ + +L TSLV+ +S +++ +
Sbjct: 12 FFLPFFAI---SGDDDYKNLIFKGCANQKS-PDPTGVFSQNLKNLFTSLVSQSSQSSFAS 67
Query: 69 FTVPATAGSGDT--LYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVR------GGALQ 120
T+G+ +T + G++QCRGDL N QCY CV++ + LC R +
Sbjct: 68 ----VTSGTDNTTAVIGVFQCRGDLQNAQCYDCVSKIPKLVSKLCGGGRDDGNVVAARVH 123
Query: 121 LEGCFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRIS 180
L GC+++Y++ F M+ + CG + +R+ + +
Sbjct: 124 LAGCYIRYESSGFRQTSGTEMLFRVCGKKDSNDPGFVGKRETAFGMAENGVKTGSSGGGG 183
Query: 181 AYGDFQGA--------AQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
G F QC G L S+C +C+ + ++ K+ECG S G++YL KC+ Y
Sbjct: 184 GGGGFYAGQYESVYVLGQCEGSLGNSDCGECVKDGFEKAKSECGESNSGQVYLQKCFVSY 243
Query: 233 S------------EGGDHSRHSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKV 280
S GG+ +H+ E+T+A+ +G + + +IV L L
Sbjct: 244 SYYSHGVPNIEPLSGGEKRQHT-----------ERTIALAVGGVFVLGFVIVCLLVLRSA 292
Query: 281 CEKQ 284
+K+
Sbjct: 293 MKKK 296
>dbj|BAC42649.1| unknown protein [Arabidopsis thaliana]
Length = 297
Score = 112 bits (280), Expect = 1e-23
Identities = 79/304 (25%), Positives = 136/304 (43%), Gaps = 48/304 (15%)
Query: 9 FILTLSAITTPSTSSSIDTFIFGGCSQPKNTPVSDPVYETGVNSLLTSLVNSASFTNYNN 68
F L AI S IF GC+ K+ P V+ + +L TSLV+ +S +++ +
Sbjct: 12 FFLPFFAI---SGDDDYKNLIFKGCANQKS-PDPTGVFSQNLKNLFTSLVSQSSQSSFAS 67
Query: 69 FTVPATAGSGDT--LYGLYQCRGDLNNDQCYRCVARAVSQLGTLCSAVR------GGALQ 120
T+G+ +T + G++QCRGDL N QCY CV++ + LC R +
Sbjct: 68 ----VTSGTDNTTAVIGVFQCRGDLQNAQCYDCVSKIPKLVSKLCGGGRDDGNVVAARVH 123
Query: 121 LEGCFVKYDNVKFLGVEDKTMVVKKCGPSIGLTSDALTRRDAVLAYLQTSDGVYRTWRIS 180
L GC+++Y++ F M+ + CG + +R+ + +
Sbjct: 124 LAGCYIRYESSGFRQTSGTEMLFRVCGKKDSNDPGFVGKRETAFGMAENGVKTGSSGGGG 183
Query: 181 AYGDFQGA--------AQCTGDLSPSECQDCLSESIQRLKTECGASTWGEMYLAKCYARY 232
G F QC G L S+C +C+ + ++ K+ECG S G++YL KC+ Y
Sbjct: 184 GGGGFYAGQYESVYVLGQCEGSLGNSDCGECVKDGFEKAKSECGESNSGQVYLQKCFVSY 243
Query: 233 S------------EGGDHSRHSDDDSNHNDDEIEKTLAILIGLIAGVALIIVFLSFLSKV 280
S GG+ +H+ E+T+A+ +G + + +IV L + +V
Sbjct: 244 SYYSHGVPNIEPLSGGEKRQHT-----------ERTIALAVGGVFVLGFVIVCL-LVFEV 291
Query: 281 CEKQ 284
C ++
Sbjct: 292 CHEE 295
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 495,690,365
Number of Sequences: 2540612
Number of extensions: 20414730
Number of successful extensions: 60800
Number of sequences better than 10.0: 248
Number of HSP's better than 10.0 without gapping: 206
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 59878
Number of HSP's gapped (non-prelim): 572
length of query: 292
length of database: 863,360,394
effective HSP length: 127
effective length of query: 165
effective length of database: 540,702,670
effective search space: 89215940550
effective search space used: 89215940550
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0097b.12


