

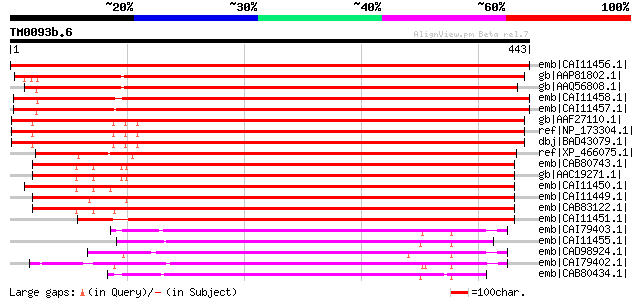
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093b.6
(443 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAI11456.1| putative glycosyltransferase [Lotus corniculatus... 924 0.0
gb|AAP81802.1| At1g74380 [Arabidopsis thaliana] gi|20260552|gb|A... 755 0.0
gb|AAQ56808.1| At5g07720 [Arabidopsis thaliana] gi|9759594|dbj|B... 747 0.0
emb|CAI11458.1| putative glycosyltransferase [Nicotiana benthami... 729 0.0
emb|CAI11457.1| putative glycosyltransferase [Solanum tuberosum] 725 0.0
gb|AAF27110.1| Similar to galactosyltransferase [Arabidopsis tha... 649 0.0
ref|NP_173304.1| galactosyl transferase GMA12/MNN10 family prote... 649 0.0
dbj|BAD43079.1| hypothetical protein [Arabidopsis thaliana] 649 0.0
ref|XP_466075.1| putative galactomannan galactosyltransferase [O... 647 0.0
emb|CAB80743.1| putative glycosyltransferase [Arabidopsis thalia... 555 e-156
gb|AAC19271.1| T14P8.23 [Arabidopsis thaliana] gi|30387559|gb|AA... 555 e-156
emb|CAI11450.1| beta-6-xylosyltransferase [Pinus taeda] 553 e-156
emb|CAI11449.1| beta-6-xylosyltransferase [Vitis vinifera] 540 e-152
emb|CAB83122.1| alpha galactosyltransferase-like protein [Arabid... 539 e-152
emb|CAI11451.1| beta-6-xylosyltransferase [Gossypium raimondii] 525 e-148
emb|CAI79403.1| galactomannan galactosyltransferase [Senna occid... 333 8e-90
emb|CAI11455.1| alpha-6-galactosyltransferase [Zea mays] 332 1e-89
emb|CAD98924.1| galactomannan galactosyltransferase [Lotus corni... 327 3e-88
emb|CAI79402.1| galactomannan galactosyltransferase [Cyamopsis t... 319 1e-85
emb|CAB80434.1| putative protein [Arabidopsis thaliana] gi|44689... 300 6e-80
>emb|CAI11456.1| putative glycosyltransferase [Lotus corniculatus var. japonicus]
Length = 443
Score = 924 bits (2388), Expect = 0.0
Identities = 443/443 (100%), Positives = 443/443 (100%)
Query: 1 MNGSAHKRTTTGLPTTTARGGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVNLSSSD 60
MNGSAHKRTTTGLPTTTARGGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVNLSSSD
Sbjct: 1 MNGSAHKRTTTGLPTTTARGGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVNLSSSD 60
Query: 61 ADAVNQNVIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAW 120
ADAVNQNVIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAW
Sbjct: 61 ADAVNQNVIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAW 120
Query: 121 LDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLA 180
LDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLA
Sbjct: 121 LDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLA 180
Query: 181 HLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGY 240
HLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGY
Sbjct: 181 HLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGY 240
Query: 241 PDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAF 300
PDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAF
Sbjct: 241 PDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAF 300
Query: 301 EADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDERWPF 360
EADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDERWPF
Sbjct: 301 EADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDERWPF 360
Query: 361 VTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNETV 420
VTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNETV
Sbjct: 361 VTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNETV 420
Query: 421 TPLEFVDQFDIRRHSSESRGSKS 443
TPLEFVDQFDIRRHSSESRGSKS
Sbjct: 421 TPLEFVDQFDIRRHSSESRGSKS 443
>gb|AAP81802.1| At1g74380 [Arabidopsis thaliana] gi|20260552|gb|AAM13174.1|
putative alpha galactosyltransferase [Arabidopsis
thaliana] gi|15221224|ref|NP_177578.1| galactosyl
transferase GMA12/MNN10 family protein [Arabidopsis
thaliana] gi|12324812|gb|AAG52374.1| putative alpha
galactosyltransferase; 16168-17541 [Arabidopsis
thaliana] gi|25349321|pir||E96772 hypothetical protein
F1M20.6 [imported] - Arabidopsis thaliana
gi|46576332|sp|Q9CA75|GT5_ARATH Putative
glycosyltransferase 5 (AtGT5)
Length = 457
Score = 755 bits (1950), Expect = 0.0
Identities = 362/449 (80%), Positives = 399/449 (88%), Gaps = 16/449 (3%)
Query: 5 AHKRTTT---GLPTTT-----ARGGR-----RGRQIQKTFNNVKITILCGFVTILVLRGT 51
AHKR + GLPTTT RGGR RGRQ+QKTFNN+KITILCGFVTILVLRGT
Sbjct: 8 AHKRPSGSGGGLPTTTLTNGGGRGGRGGLLPRGRQMQKTFNNIKITILCGFVTILVLRGT 67
Query: 52 IGV-NLSSSDADAVNQNVIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKI 110
IGV NL SS ADAVNQN+IEETNRILAEIRSD+DP+D D+ + +PNAT+ LGPKI
Sbjct: 68 IGVGNLGSSSADAVNQNIIEETNRILAEIRSDSDPTDLDEPQEGD--MNPNATYVLGPKI 125
Query: 111 TGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRL 170
T WD QRK WL+QNPE+P+ V GKARILLLTGSPPKPCDNPIGDHYLLKS+KNKIDYCRL
Sbjct: 126 TDWDSQRKVWLNQNPEFPSTVNGKARILLLTGSPPKPCDNPIGDHYLLKSVKNKIDYCRL 185
Query: 171 HGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKY 230
HGIEIVYN+AHLD ELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDA FTD++F++PL++Y
Sbjct: 186 HGIEIVYNMAHLDKELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDALFTDILFQIPLARY 245
Query: 231 DDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVL 290
+NLV+HGYPDLLF+QKSWIA+NTGSFL RNCQWSLDLLDAWAPMGPKGP+R+EAGKVL
Sbjct: 246 QKHNLVIHGYPDLLFDQKSWIALNTGSFLLRNCQWSLDLLDAWAPMGPKGPIRDEAGKVL 305
Query: 291 TANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYH 350
TA LKGRPAFEADDQSALIYLLLS+KD WM+K F+EN +YLHG+W GLVDRYEEMIEKYH
Sbjct: 306 TAYLKGRPAFEADDQSALIYLLLSQKDTWMEKVFVENQYYLHGFWEGLVDRYEEMIEKYH 365
Query: 351 PGLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSP 410
PGLGDERWPFVTHFVGCKPCGSY DY VE+CL SMERAFNFADNQVLKLYGF HRGLLSP
Sbjct: 366 PGLGDERWPFVTHFVGCKPCGSYADYAVERCLKSMERAFNFADNQVLKLYGFSHRGLLSP 425
Query: 411 KIKRIRNETVTPLEFVDQFDIRRHSSESR 439
KIKRIRNETV+PLEFVD+FDIRR E++
Sbjct: 426 KIKRIRNETVSPLEFVDKFDIRRTPVETK 454
>gb|AAQ56808.1| At5g07720 [Arabidopsis thaliana] gi|9759594|dbj|BAB11451.1| alpha
galactosyltransferase protein [Arabidopsis thaliana]
gi|9716848|emb|CAC01676.1| putative golgi
glycosyltransferase [Arabidopsis thaliana]
gi|15240848|ref|NP_196389.1| galactosyl transferase
GMA12/MNN10 family protein [Arabidopsis thaliana]
gi|46576342|sp|Q9LF80|GT3_ARATH Putative
glycosyltransferase 3 (AtGT3)
Length = 457
Score = 747 bits (1928), Expect = 0.0
Identities = 343/426 (80%), Positives = 388/426 (90%), Gaps = 7/426 (1%)
Query: 13 LPTTTARGG-----RRGRQIQKTFNNVKITILCGFVTILVLRGTIGVNLSSSDADAVNQN 67
LPTT A GG RGRQIQKTFNNVK+TILCGFVTILVLRGTIG+N +SDAD VNQN
Sbjct: 24 LPTTMASGGVRRPPPRGRQIQKTFNNVKMTILCGFVTILVLRGTIGINFGTSDADVVNQN 83
Query: 68 VIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAWLDQNPEY 127
+IEETNR+LAEIRSD+DP+D ++ ++ N T+TLGPKIT WD +RK WL QNP++
Sbjct: 84 IIEETNRLLAEIRSDSDPTDSNEPPDSD--LDLNMTYTLGPKITNWDQKRKLWLTQNPDF 141
Query: 128 PNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLAHLDVELA 187
P+F+ GKA++LLLTGSPPKPCDNPIGDHYLLKS+KNKIDYCR+HGIEIVYN+AHLD ELA
Sbjct: 142 PSFINGKAKVLLLTGSPPKPCDNPIGDHYLLKSVKNKIDYCRIHGIEIVYNMAHLDKELA 201
Query: 188 GYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQ 247
GYWAKLPMIRRLMLSHPE+EWIWWMDSDA FTDMVFE+PLS+Y+++NLV+HGYPDLLF+Q
Sbjct: 202 GYWAKLPMIRRLMLSHPEIEWIWWMDSDALFTDMVFEIPLSRYENHNLVIHGYPDLLFDQ 261
Query: 248 KSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSA 307
KSWIA+NTGSFLFRNCQWSLDLLDAWAPMGPKGP+REEAGK+LTANLKGRPAFEADDQSA
Sbjct: 262 KSWIALNTGSFLFRNCQWSLDLLDAWAPMGPKGPIREEAGKILTANLKGRPAFEADDQSA 321
Query: 308 LIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDERWPFVTHFVGC 367
LIYLLLS+K+ WM+K F+EN +YLHG+W GLVD+YEEM+EKYHPGLGDERWPF+THFVGC
Sbjct: 322 LIYLLLSQKETWMEKVFVENQYYLHGFWEGLVDKYEEMMEKYHPGLGDERWPFITHFVGC 381
Query: 368 KPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNETVTPLEFVD 427
KPCGSY DY VE+CL SMERAFNFADNQVLKLYGF HRGLLSPKIKRIRNET PL+FVD
Sbjct: 382 KPCGSYADYAVERCLKSMERAFNFADNQVLKLYGFGHRGLLSPKIKRIRNETTFPLKFVD 441
Query: 428 QFDIRR 433
+FDIRR
Sbjct: 442 RFDIRR 447
>emb|CAI11458.1| putative glycosyltransferase [Nicotiana benthamiana]
Length = 450
Score = 729 bits (1882), Expect = 0.0
Identities = 346/448 (77%), Positives = 386/448 (85%), Gaps = 13/448 (2%)
Query: 4 SAHKRTTTGLPTT-TARGGR------RGRQIQKTFNNVKITILCGFVTILVLRGTIGV-N 55
+A KR LPTT T GGR RGRQIQ+TFNN+KITILCGFVTILVLRGTIG +
Sbjct: 8 AAQKRGGGALPTTATPNGGRTSSVLPRGRQIQRTFNNIKITILCGFVTILVLRGTIGFGS 67
Query: 56 LSSSDADAVNQNVIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKITGWDL 115
L+SS +DA N N+IEETNRIL EIRSD D DP D TF N+T++LGPKIT WD
Sbjct: 68 LASSGSDAENANLIEETNRILDEIRSDTDLDDPSD-----TFSHLNSTYSLGPKITTWDA 122
Query: 116 QRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEI 175
RK WL +NP++PNF+ GK R+LL+TGSPP PCDNP GDHYLLK +KNKIDYCR+HGIEI
Sbjct: 123 DRKFWLQKNPDFPNFIHGKPRVLLVTGSPPNPCDNPTGDHYLLKVMKNKIDYCRIHGIEI 182
Query: 176 VYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNL 235
VYNLAHL+ E+AGYWAKLP+IR+LMLSHPEVEWIWWMDSDA FTDMVFE+P SKY+D+NL
Sbjct: 183 VYNLAHLEKEMAGYWAKLPLIRKLMLSHPEVEWIWWMDSDALFTDMVFEIPFSKYNDHNL 242
Query: 236 VLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLK 295
V+HGYPDLLFEQKSWIA+NTGSFL RNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLK
Sbjct: 243 VIHGYPDLLFEQKSWIALNTGSFLIRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLK 302
Query: 296 GRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGD 355
RPAFEADDQSALIYLL+S+KD+WMD+ F+ENS+YLHGYWAGLVDRYEEM+EKYHPGLGD
Sbjct: 303 SRPAFEADDQSALIYLLMSQKDQWMDQVFIENSYYLHGYWAGLVDRYEEMVEKYHPGLGD 362
Query: 356 ERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRI 415
ERW FVTHF GCKPCGSYGDYPVE+C+ SMERAFNFADNQVLKLYGFRH+GLLSP IKRI
Sbjct: 363 ERWAFVTHFAGCKPCGSYGDYPVERCMKSMERAFNFADNQVLKLYGFRHKGLLSPNIKRI 422
Query: 416 RNETVTPLEFVDQFDIRRHSSESRGSKS 443
RNET PL +VDQ D+R ES G +S
Sbjct: 423 RNETDNPLLYVDQLDLRHAKHESTGPQS 450
>emb|CAI11457.1| putative glycosyltransferase [Solanum tuberosum]
Length = 474
Score = 725 bits (1872), Expect = 0.0
Identities = 344/450 (76%), Positives = 385/450 (85%), Gaps = 11/450 (2%)
Query: 4 SAHKRTTTGLPTTTARGG---------RRGRQIQKTFNNVKITILCGFVTILVLRGTIGV 54
+A KR T TA GG RGRQI +TFNNVKITILCGFVTILVLRGTIG+
Sbjct: 8 TAQKRAGALPTTVTANGGVRGRSPNVLPRGRQINRTFNNVKITILCGFVTILVLRGTIGI 67
Query: 55 -NLSSSDADAVNQNVIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKITGW 113
N+SSS+A+A NQN+IEETNRIL++IRSD DP DP ETF S N T++LGPKI W
Sbjct: 68 GNVSSSEAEAENQNLIEETNRILSDIRSDKDPDDPV-GDQPETFMSLNDTYSLGPKIANW 126
Query: 114 DLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGI 173
D RK WL +NPE+PNFV GK RILL+TGSPP PCDN IGDHYLLK+IKNKIDYCR+HGI
Sbjct: 127 DKDRKMWLQKNPEFPNFVNGKPRILLVTGSPPNPCDNAIGDHYLLKAIKNKIDYCRIHGI 186
Query: 174 EIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDY 233
EI+YNLAHL+ E+AGYWAKLP+IRRLMLSHPEVEWIWWMDSDA FTDMVFE+PLSKY+ +
Sbjct: 187 EILYNLAHLEKEMAGYWAKLPLIRRLMLSHPEVEWIWWMDSDALFTDMVFEMPLSKYNRH 246
Query: 234 NLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTAN 293
NLV+HGYPDLLF+QKSWIA+NTGSFLFRNCQW LDLLDAWAPMGPKGP+REEAGK+LTA
Sbjct: 247 NLVIHGYPDLLFDQKSWIALNTGSFLFRNCQWFLDLLDAWAPMGPKGPIREEAGKILTAY 306
Query: 294 LKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGL 353
LKGRPAFEADDQSALIYLL S+KDKWM K +ENS+YLHGYWAGLVDRYEEMI+KYHPGL
Sbjct: 307 LKGRPAFEADDQSALIYLLTSQKDKWMHKVSIENSYYLHGYWAGLVDRYEEMIQKYHPGL 366
Query: 354 GDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIK 413
GDERWPFVTHFVGCKPCGSYGDYP E+CL +MERAFNFADNQVL LYGF+H+GLLSP IK
Sbjct: 367 GDERWPFVTHFVGCKPCGSYGDYPAERCLKNMERAFNFADNQVLNLYGFKHKGLLSPNIK 426
Query: 414 RIRNETVTPLEFVDQFDIRRHSSESRGSKS 443
RIRNET PL++VDQ D+R ES ++S
Sbjct: 427 RIRNETDNPLQYVDQLDVRHAKHESTETQS 456
>gb|AAF27110.1| Similar to galactosyltransferase [Arabidopsis thaliana]
gi|25349326|pir||F86320 hypothetical protein F6A14.20 -
Arabidopsis thaliana gi|46576343|sp|Q9M9U0|GT4_ARATH
Putative glycosyltransferase 4 (AtGT4)
Length = 513
Score = 649 bits (1675), Expect = 0.0
Identities = 326/509 (64%), Positives = 375/509 (73%), Gaps = 71/509 (13%)
Query: 2 NGSAHKRTTTGLPTTTA-------RGGRRGRQIQKT-FNNVKITILCGFVTILVLRGTIG 53
+GS + GL TT RG RG QIQ T FNN+K ILC FVTIL+L GTI
Sbjct: 4 DGSRSSGSGRGLSTTAVSNGGWRTRGFLRGWQIQNTLFNNIKFMILCCFVTILILLGTIR 63
Query: 54 V-NLSSSDADAVNQNVIEETNRILAEIRSDADPSD------------------------P 88
V NL SS+AD+VNQ+ I+ET ILAEI SD+ +D P
Sbjct: 64 VGNLGSSNADSVNQSFIKETIPILAEIPSDSHSTDLAEPPKADVSPNATYTLEPRIAEIP 123
Query: 89 DDAAAAETF------FSPNATFTLG--------------------------------PKI 110
D + + SPNAT+TLG PKI
Sbjct: 124 SDVHSTDLVELPKADISPNATYTLGPRIAEIPSDSHLTDLLEPPKADISPNATYTLGPKI 183
Query: 111 TGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRL 170
T WD QRK WL+QNPE+PN V GKARILLLTGS P PCD PIG++YLLK++KNKIDYCRL
Sbjct: 184 TNWDSQRKVWLNQNPEFPNIVNGKARILLLTGSSPGPCDKPIGNYYLLKAVKNKIDYCRL 243
Query: 171 HGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKY 230
HGIEIVYN+A+LD EL+GYW KLPMIR LMLSHPEVEWIWWMDSDA FTD++FE+PL +Y
Sbjct: 244 HGIEIVYNMANLDEELSGYWTKLPMIRTLMLSHPEVEWIWWMDSDALFTDILFEIPLPRY 303
Query: 231 DDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVL 290
+++NLV+HGYPDLLF QKSW+A+NTG FL RNCQWSLDLLDAWAPMGPKG +R+E GK+L
Sbjct: 304 ENHNLVIHGYPDLLFNQKSWVALNTGIFLLRNCQWSLDLLDAWAPMGPKGKIRDETGKIL 363
Query: 291 TANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYH 350
TA LKGRPAFEADDQSALIYLLLS+K+KW++K ++EN +YLHG+W GLVDRYEEMIEKYH
Sbjct: 364 TAYLKGRPAFEADDQSALIYLLLSQKEKWIEKVYVENQYYLHGFWEGLVDRYEEMIEKYH 423
Query: 351 PGLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSP 410
PGLGDERWPFVTHFVGCKPCGSY DY V++C SMERAFNFADNQVLKLYGF HRGLLSP
Sbjct: 424 PGLGDERWPFVTHFVGCKPCGSYADYAVDRCFKSMERAFNFADNQVLKLYGFSHRGLLSP 483
Query: 411 KIKRIRNETVTPLEFVDQFDIRRHSSESR 439
KIKRIRNETV+PLE VD+FDIRR E++
Sbjct: 484 KIKRIRNETVSPLESVDKFDIRRMHMETK 512
>ref|NP_173304.1| galactosyl transferase GMA12/MNN10 family protein [Arabidopsis
thaliana]
Length = 632
Score = 649 bits (1675), Expect = 0.0
Identities = 326/509 (64%), Positives = 375/509 (73%), Gaps = 71/509 (13%)
Query: 2 NGSAHKRTTTGLPTTTA-------RGGRRGRQIQKT-FNNVKITILCGFVTILVLRGTIG 53
+GS + GL TT RG RG QIQ T FNN+K ILC FVTIL+L GTI
Sbjct: 123 DGSRSSGSGRGLSTTAVSNGGWRTRGFLRGWQIQNTLFNNIKFMILCCFVTILILLGTIR 182
Query: 54 V-NLSSSDADAVNQNVIEETNRILAEIRSDADPSD------------------------P 88
V NL SS+AD+VNQ+ I+ET ILAEI SD+ +D P
Sbjct: 183 VGNLGSSNADSVNQSFIKETIPILAEIPSDSHSTDLAEPPKADVSPNATYTLEPRIAEIP 242
Query: 89 DDAAAAETF------FSPNATFTLG--------------------------------PKI 110
D + + SPNAT+TLG PKI
Sbjct: 243 SDVHSTDLVELPKADISPNATYTLGPRIAEIPSDSHLTDLLEPPKADISPNATYTLGPKI 302
Query: 111 TGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRL 170
T WD QRK WL+QNPE+PN V GKARILLLTGS P PCD PIG++YLLK++KNKIDYCRL
Sbjct: 303 TNWDSQRKVWLNQNPEFPNIVNGKARILLLTGSSPGPCDKPIGNYYLLKAVKNKIDYCRL 362
Query: 171 HGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKY 230
HGIEIVYN+A+LD EL+GYW KLPMIR LMLSHPEVEWIWWMDSDA FTD++FE+PL +Y
Sbjct: 363 HGIEIVYNMANLDEELSGYWTKLPMIRTLMLSHPEVEWIWWMDSDALFTDILFEIPLPRY 422
Query: 231 DDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVL 290
+++NLV+HGYPDLLF QKSW+A+NTG FL RNCQWSLDLLDAWAPMGPKG +R+E GK+L
Sbjct: 423 ENHNLVIHGYPDLLFNQKSWVALNTGIFLLRNCQWSLDLLDAWAPMGPKGKIRDETGKIL 482
Query: 291 TANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYH 350
TA LKGRPAFEADDQSALIYLLLS+K+KW++K ++EN +YLHG+W GLVDRYEEMIEKYH
Sbjct: 483 TAYLKGRPAFEADDQSALIYLLLSQKEKWIEKVYVENQYYLHGFWEGLVDRYEEMIEKYH 542
Query: 351 PGLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSP 410
PGLGDERWPFVTHFVGCKPCGSY DY V++C SMERAFNFADNQVLKLYGF HRGLLSP
Sbjct: 543 PGLGDERWPFVTHFVGCKPCGSYADYAVDRCFKSMERAFNFADNQVLKLYGFSHRGLLSP 602
Query: 411 KIKRIRNETVTPLEFVDQFDIRRHSSESR 439
KIKRIRNETV+PLE VD+FDIRR E++
Sbjct: 603 KIKRIRNETVSPLESVDKFDIRRMHMETK 631
>dbj|BAD43079.1| hypothetical protein [Arabidopsis thaliana]
Length = 513
Score = 649 bits (1673), Expect = 0.0
Identities = 326/509 (64%), Positives = 375/509 (73%), Gaps = 71/509 (13%)
Query: 2 NGSAHKRTTTGLPTTTA-------RGGRRGRQIQKT-FNNVKITILCGFVTILVLRGTIG 53
+GS + GL TT RG RG QIQ T FNN+K ILC FVTIL+L GTI
Sbjct: 4 DGSRSSGSGRGLSTTAVSNGGWRTRGFLRGWQIQNTLFNNIKFMILCCFVTILILLGTIR 63
Query: 54 V-NLSSSDADAVNQNVIEETNRILAEIRSDADPSD------------------------P 88
V NL SS+AD+VNQ+ I+ET ILAEI SD+ +D P
Sbjct: 64 VGNLGSSNADSVNQSFIKETIPILAEIPSDSYSTDLAEPPKADVSPNATYTLEPRIAEIP 123
Query: 89 DDAAAAETF------FSPNATFTLG--------------------------------PKI 110
D + + SPNAT+TLG PKI
Sbjct: 124 SDVHSTDLVELPKADISPNATYTLGPRIAEIPSDSHLTDLLEPPKADISPNATYTLGPKI 183
Query: 111 TGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRL 170
T WD QRK WL+QNPE+PN V GKARILLLTGS P PCD PIG++YLLK++KNKIDYCRL
Sbjct: 184 TNWDSQRKVWLNQNPEFPNIVNGKARILLLTGSSPGPCDKPIGNYYLLKAVKNKIDYCRL 243
Query: 171 HGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKY 230
HGIEIVYN+A+LD EL+GYW KLPMIR LMLSHPEVEWIWWMDSDA FTD++FE+PL +Y
Sbjct: 244 HGIEIVYNMANLDEELSGYWTKLPMIRTLMLSHPEVEWIWWMDSDALFTDILFEIPLPRY 303
Query: 231 DDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVL 290
+++NLV+HGYPDLLF QKSW+A+NTG FL RNCQWSLDLLDAWAPMGPKG +R+E GK+L
Sbjct: 304 ENHNLVIHGYPDLLFNQKSWVALNTGIFLLRNCQWSLDLLDAWAPMGPKGKIRDETGKIL 363
Query: 291 TANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYH 350
TA LKGRPAFEADDQSALIYLLLS+K+KW++K ++EN +YLHG+W GLVDRYEEMIEKYH
Sbjct: 364 TAYLKGRPAFEADDQSALIYLLLSQKEKWIEKVYVENQYYLHGFWEGLVDRYEEMIEKYH 423
Query: 351 PGLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSP 410
PGLGDERWPFVTHFVGCKPCGSY DY V++C SMERAFNFADNQVLKLYGF HRGLLSP
Sbjct: 424 PGLGDERWPFVTHFVGCKPCGSYADYAVDRCFKSMERAFNFADNQVLKLYGFSHRGLLSP 483
Query: 411 KIKRIRNETVTPLEFVDQFDIRRHSSESR 439
KIKRIRNETV+PLE VD+FDIRR E++
Sbjct: 484 KIKRIRNETVSPLESVDKFDIRRMHMETK 512
>ref|XP_466075.1| putative galactomannan galactosyltransferase [Oryza sativa
(japonica cultivar-group)] gi|49388322|dbj|BAD25434.1|
putative galactomannan galactosyltransferase [Oryza
sativa (japonica cultivar-group)]
Length = 480
Score = 647 bits (1669), Expect = 0.0
Identities = 310/433 (71%), Positives = 359/433 (82%), Gaps = 24/433 (5%)
Query: 23 RGRQIQKTFNNVKITILCGFVTILVLRGTIGVNLS----SSDADAV-NQNVIEETNRILA 77
R R+I +TFNNVKIT+LCG VTILVLRGTIG+NLS +DADA+ +E+ +RIL
Sbjct: 42 RSRKIHRTFNNVKITVLCGLVTILVLRGTIGLNLSLPNQPTDADALAGAKAVEDIDRILR 101
Query: 78 EIRSDADPSDPDDAAAAETFFSPNAT-----------------FTLGPKITGWDLQRKAW 120
EIRSD +D D AAA + S NAT + LGPKI+ WD QR+ W
Sbjct: 102 EIRSDGG-ADDDAAAAGDLAGSFNATALNATEAAAAYASAVERYALGPKISDWDGQRRRW 160
Query: 121 LDQNPEYPNFVRG-KARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNL 179
L QNP +P+ V G K RILL+TGS P PCDNP+GDHYLLK+ KNKIDYCRLHGIEIV+NL
Sbjct: 161 LRQNPGFPSTVAGGKPRILLVTGSQPGPCDNPLGDHYLLKTTKNKIDYCRLHGIEIVHNL 220
Query: 180 AHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHG 239
AHLD ELAGYWAKLP++RRLMLSHPEVEWIWWMDSDA FTDM FELPLS+Y D NL++HG
Sbjct: 221 AHLDTELAGYWAKLPLLRRLMLSHPEVEWIWWMDSDALFTDMAFELPLSRYQDRNLIIHG 280
Query: 240 YPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPA 299
Y DLLFE+ SWIA+NTGSFLFRNCQWSLDLLDAWAPMGPKG +R+EAGK+LTANLKGRPA
Sbjct: 281 YQDLLFEKHSWIALNTGSFLFRNCQWSLDLLDAWAPMGPKGFIRDEAGKILTANLKGRPA 340
Query: 300 FEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDERWP 359
FEADDQSALIYLLLS+K+KWM+K F+ENS+YLHG+WAGLVD+YEEM+E +HPGLGDERWP
Sbjct: 341 FEADDQSALIYLLLSQKEKWMNKVFIENSYYLHGFWAGLVDKYEEMMENHHPGLGDERWP 400
Query: 360 FVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNET 419
FVTHFVGCKPCGSYGDYPVE+CL SMERAFNFADNQVL+LYGF H+GL SPKIKR+RN+T
Sbjct: 401 FVTHFVGCKPCGSYGDYPVERCLRSMERAFNFADNQVLRLYGFAHKGLESPKIKRVRNQT 460
Query: 420 VTPLEFVDQFDIR 432
P++ + D++
Sbjct: 461 TKPIDDKENLDVK 473
>emb|CAB80743.1| putative glycosyltransferase [Arabidopsis thaliana]
gi|3892054|gb|AAC78266.1| putative glycosyltransferase
[Arabidopsis thaliana] gi|25349323|pir||H85031 probable
glycosyltransferase [imported] - Arabidopsis thaliana
Length = 459
Score = 555 bits (1430), Expect = e-156
Identities = 259/440 (58%), Positives = 323/440 (72%), Gaps = 28/440 (6%)
Query: 20 GGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVN---LSSSDADAVNQNVIE----ET 72
G R R+IQ+ +K+TILC +T++VLR TIG D D + Q+ E
Sbjct: 7 GAYRCRRIQRALRQLKVTILCLLLTVVVLRSTIGAGKFGTPEQDLDEIRQHFHARKRGEP 66
Query: 73 NRILAEIRSDADPSDPDDAAAA------ETFF--------------SPNATFTLGPKITG 112
+R+L EI++ D S D + ETF PN +TLGPKI+
Sbjct: 67 HRVLEEIQTGGDSSSGDGGGNSGGSNNYETFDINKIFVDEGEEEKPDPNKPYTLGPKISD 126
Query: 113 WDLQRKAWLDQNPEYPNFVR-GKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLH 171
WD QR WL +NP +PNF+ K R+LL+TGS PKPC+NP+GDHYLLKSIKNKIDYCRLH
Sbjct: 127 WDEQRSDWLAKNPSFPNFIGPNKPRVLLVTGSAPKPCENPVGDHYLLKSIKNKIDYCRLH 186
Query: 172 GIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYD 231
GIEI YN+A LD E+AG+WAKLP+IR+L+LSHPE+E++WWMDSDA FTDM FELP +Y
Sbjct: 187 GIEIFYNMALLDAEMAGFWAKLPLIRKLLLSHPEIEFLWWMDSDAMFTDMAFELPWERYK 246
Query: 232 DYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLT 291
DYNLV+HG+ +++++QK+WI +NTGSFL RN QW+LDLLD WAPMGPKG +REEAGKVLT
Sbjct: 247 DYNLVMHGWNEMVYDQKNWIGLNTGSFLLRNNQWALDLLDTWAPMGPKGKIREEAGKVLT 306
Query: 292 ANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHP 351
LK RP FEADDQSA++YLL +++D W +K +LE+ +YLHGYW LVDRYEEMIE YHP
Sbjct: 307 RELKDRPVFEADDQSAMVYLLATQRDAWGNKVYLESGYYLHGYWGILVDRYEEMIENYHP 366
Query: 352 GLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPK 411
GLGD RWP VTHFVGCKPCG +GDYPVE+CL M+RAFNF DNQ+L++YGF H+ L S K
Sbjct: 367 GLGDHRWPLVTHFVGCKPCGKFGDYPVERCLKQMDRAFNFGDNQILQIYGFTHKSLASRK 426
Query: 412 IKRIRNETVTPLEFVDQFDI 431
+KR+RNET PLE D+ +
Sbjct: 427 VKRVRNETSNPLEMKDELGL 446
>gb|AAC19271.1| T14P8.23 [Arabidopsis thaliana] gi|30387559|gb|AAP31945.1|
At4g02500 [Arabidopsis thaliana]
gi|22655160|gb|AAM98170.1| putative glycosyltransferase
[Arabidopsis thaliana] gi|9716844|emb|CAC01674.1|
putative golgi glycosyltransferase [Arabidopsis
thaliana] gi|16209669|gb|AAL14393.1| AT4g02500/T10P11_20
[Arabidopsis thaliana] gi|18411962|ref|NP_567241.1|
galactosyl transferase GMA12/MNN10 family protein
[Arabidopsis thaliana] gi|7487013|pir||T01300
hypothetical protein T14P8.23 - Arabidopsis thaliana
gi|46576207|sp|O22775|GT2_ARATH Putative
glycosyltransferase 2 (AtGT2)
Length = 461
Score = 555 bits (1430), Expect = e-156
Identities = 259/440 (58%), Positives = 323/440 (72%), Gaps = 28/440 (6%)
Query: 20 GGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVN---LSSSDADAVNQNVIE----ET 72
G R R+IQ+ +K+TILC +T++VLR TIG D D + Q+ E
Sbjct: 7 GAYRCRRIQRALRQLKVTILCLLLTVVVLRSTIGAGKFGTPEQDLDEIRQHFHARKRGEP 66
Query: 73 NRILAEIRSDADPSDPDDAAAA------ETFF--------------SPNATFTLGPKITG 112
+R+L EI++ D S D + ETF PN +TLGPKI+
Sbjct: 67 HRVLEEIQTGGDSSSGDGGGNSGGSNNYETFDINKIFVDEGEEEKPDPNKPYTLGPKISD 126
Query: 113 WDLQRKAWLDQNPEYPNFVR-GKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLH 171
WD QR WL +NP +PNF+ K R+LL+TGS PKPC+NP+GDHYLLKSIKNKIDYCRLH
Sbjct: 127 WDEQRSDWLAKNPSFPNFIGPNKPRVLLVTGSAPKPCENPVGDHYLLKSIKNKIDYCRLH 186
Query: 172 GIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYD 231
GIEI YN+A LD E+AG+WAKLP+IR+L+LSHPE+E++WWMDSDA FTDM FELP +Y
Sbjct: 187 GIEIFYNMALLDAEMAGFWAKLPLIRKLLLSHPEIEFLWWMDSDAMFTDMAFELPWERYK 246
Query: 232 DYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLT 291
DYNLV+HG+ +++++QK+WI +NTGSFL RN QW+LDLLD WAPMGPKG +REEAGKVLT
Sbjct: 247 DYNLVMHGWNEMVYDQKNWIGLNTGSFLLRNNQWALDLLDTWAPMGPKGKIREEAGKVLT 306
Query: 292 ANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHP 351
LK RP FEADDQSA++YLL +++D W +K +LE+ +YLHGYW LVDRYEEMIE YHP
Sbjct: 307 RELKDRPVFEADDQSAMVYLLATQRDAWGNKVYLESGYYLHGYWGILVDRYEEMIENYHP 366
Query: 352 GLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPK 411
GLGD RWP VTHFVGCKPCG +GDYPVE+CL M+RAFNF DNQ+L++YGF H+ L S K
Sbjct: 367 GLGDHRWPLVTHFVGCKPCGKFGDYPVERCLKQMDRAFNFGDNQILQIYGFTHKSLASRK 426
Query: 412 IKRIRNETVTPLEFVDQFDI 431
+KR+RNET PLE D+ +
Sbjct: 427 VKRVRNETSNPLEMKDELGL 446
>emb|CAI11450.1| beta-6-xylosyltransferase [Pinus taeda]
Length = 444
Score = 553 bits (1426), Expect = e-156
Identities = 253/432 (58%), Positives = 330/432 (75%), Gaps = 13/432 (3%)
Query: 13 LPTTTARGGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVN---LSSSDADAVNQNVI 69
+P A GGRR RQIQ+ N K+T+LC FVT+LVLRGTIG D + + +++I
Sbjct: 3 VPGFLALGGRRIRQIQRAMQNAKVTLLCLFVTVLVLRGTIGAGKFGTPEQDFNEIREHLI 62
Query: 70 E----ETNRILAEIRSDAD-----PSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAW 120
E +R+L E+ +++ P ++ + P+ ++ GPKI+ WD QR W
Sbjct: 63 VGRRGEPHRVLTEVVAESSNNKTPPVVEEEVEEPDVERDPSVPYSFGPKISDWDDQRSEW 122
Query: 121 LDQNPEYPNFVRG-KARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNL 179
L +NP +PNF+ G + RILL+TGS P PC++P+GDH LLKSIKNKIDYCRLHGIEI YN+
Sbjct: 123 LAENPAFPNFLPGGRPRILLVTGSAPAPCESPVGDHLLLKSIKNKIDYCRLHGIEIFYNM 182
Query: 180 AHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHG 239
AHLD E++G+WAKLP+IR+LML+HPEVE+IWWMDSDA FTDMVFELP +Y ++N ++HG
Sbjct: 183 AHLDHEMSGFWAKLPLIRKLMLTHPEVEFIWWMDSDAMFTDMVFELPFERYKNHNFIMHG 242
Query: 240 YPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPA 299
+ +L++ +K+WI +NTGSFL RNCQWSLD+LDAWAPMGPKG +R EAGK+LT +L GRP
Sbjct: 243 WEELVYNRKNWIGLNTGSFLLRNCQWSLDILDAWAPMGPKGKIRTEAGKLLTRSLVGRPE 302
Query: 300 FEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDERWP 359
FEADDQSAL+YLL+++++KW DK +LE+++YLHGYW LVD+YEEM+EK+HPGLGD RWP
Sbjct: 303 FEADDQSALVYLLITQREKWGDKVYLESAYYLHGYWGILVDKYEEMMEKHHPGLGDHRWP 362
Query: 360 FVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNET 419
VTHFVGCKPCG GDYPV +CL MERAFNF DNQ+L++YGF H+ L S +KR RN+T
Sbjct: 363 LVTHFVGCKPCGKVGDYPVAQCLRQMERAFNFGDNQILQIYGFTHKSLSSRGVKRTRNDT 422
Query: 420 VTPLEFVDQFDI 431
PLE D+ +
Sbjct: 423 DKPLEVKDELGL 434
>emb|CAI11449.1| beta-6-xylosyltransferase [Vitis vinifera]
Length = 450
Score = 540 bits (1392), Expect = e-152
Identities = 244/431 (56%), Positives = 318/431 (73%), Gaps = 19/431 (4%)
Query: 20 GGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVNLSSSDADAVNQ-------NVIEET 72
G RR RQ+Q+ F + KIT++C +T++VLRGTIG + N+ E
Sbjct: 7 GARRVRQMQRAFRHGKITLMCLLMTVVVLRGTIGAGKFGTPGQDFNEIREHFHNRKRVEP 66
Query: 73 NRILAEIRSDADPSDPDDAAAAETFF-----------SPNATFTLGPKITGWDLQRKAWL 121
+R+L E + S+ ++ A + PN ++LGPKI+ WD QR WL
Sbjct: 67 HRVLEEAEVSTESSESNNYATFDISKLTVDEGEDEKPDPNRPYSLGPKISDWDEQRAEWL 126
Query: 122 DQNPEYPNFVR-GKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLA 180
+NP +PNF+ K R+LL+TGS PKPC+NP GDHYLLK+IKNKIDYCRLHG+EI YN+A
Sbjct: 127 RKNPNFPNFIGPNKPRVLLVTGSSPKPCENPGGDHYLLKAIKNKIDYCRLHGVEIFYNMA 186
Query: 181 HLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGY 240
LD E+AG+WAKLP+IR+L+LSHPE+E++WWMDSDA FTDM FE+P +Y D+N V+HG+
Sbjct: 187 LLDAEMAGFWAKLPLIRKLLLSHPEIEFLWWMDSDAMFTDMAFEVPWERYKDHNFVMHGW 246
Query: 241 PDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAF 300
+++++QK+WI +NTGSFL RNCQW+LD+LDAWAPMGPKG +R EAGK+LT LK RP F
Sbjct: 247 NEMVYDQKNWIGLNTGSFLLRNCQWALDILDAWAPMGPKGKIRTEAGKILTRELKDRPVF 306
Query: 301 EADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDERWPF 360
EADDQSA++YLL +++D W DK +LE+++YLHGYW LVDRYEEMIE +HPGLGD RWP
Sbjct: 307 EADDQSAMVYLLATQRDNWGDKVYLESAYYLHGYWGILVDRYEEMIENHHPGLGDHRWPL 366
Query: 361 VTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNETV 420
VTHFVGCKPCG +GDYPVE+CL M+RAFNF DNQ+L++YGF H+ L S ++KR RN+
Sbjct: 367 VTHFVGCKPCGKFGDYPVERCLKQMDRAFNFGDNQILQIYGFTHKSLASRRVKRTRNDPS 426
Query: 421 TPLEFVDQFDI 431
PLE D +
Sbjct: 427 NPLEVKDDLGL 437
>emb|CAB83122.1| alpha galactosyltransferase-like protein [Arabidopsis thaliana]
gi|25141201|gb|AAN73295.1| At3g62720/F26K9_150
[Arabidopsis thaliana] gi|15983426|gb|AAL11581.1|
AT3g62720/F26K9_150 [Arabidopsis thaliana]
gi|15228843|ref|NP_191831.1| galactosyl transferase
GMA12/MNN10 family protein [Arabidopsis thaliana]
gi|11281898|pir||T48061 alpha galactosyltransferase-like
protein - Arabidopsis thaliana
gi|46577293|sp|Q9LZJ3|XT1_ARATH Xyloglucan
6-xylosyltransferase (AtXT1)
Length = 460
Score = 539 bits (1388), Expect = e-152
Identities = 250/439 (56%), Positives = 321/439 (72%), Gaps = 27/439 (6%)
Query: 20 GGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVN---LSSSDADAVNQNVIE-----E 71
G R R++Q+ K+TILC +T++VLRGTIG D + + ++ E
Sbjct: 7 GAHRFRRLQRFMRQGKVTILCLVLTVIVLRGTIGAGKFGTPEKDIEEIREHFFYTRKRGE 66
Query: 72 TNRILAEIRSDADPSDP------------------DDAAAAETFFSPNATFTLGPKITGW 113
+R+L E+ S S+ D+ ++ N ++LGPKI+ W
Sbjct: 67 PHRVLVEVSSKTTSSEDGGNGGNSYETFDINKLFVDEGDEEKSRDRTNKPYSLGPKISDW 126
Query: 114 DLQRKAWLDQNPEYPNFVR-GKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHG 172
D QR+ WL QNP +PNFV K R+LL+TGS PKPC+NP+GDHYLLKSIKNKIDYCR+HG
Sbjct: 127 DEQRRDWLKQNPSFPNFVAPNKPRVLLVTGSAPKPCENPVGDHYLLKSIKNKIDYCRIHG 186
Query: 173 IEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDD 232
IEI YN+A LD E+AG+WAKLP+IR+L+LSHPE+E++WWMDSDA FTDMVFELP +Y D
Sbjct: 187 IEIFYNMALLDAEMAGFWAKLPLIRKLLLSHPEIEFLWWMDSDAMFTDMVFELPWERYKD 246
Query: 233 YNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTA 292
YNLV+HG+ +++++QK+WI +NTGSFL RN QWSLDLLDAWAPMGPKG +REEAGKVLT
Sbjct: 247 YNLVMHGWNEMVYDQKNWIGLNTGSFLLRNSQWSLDLLDAWAPMGPKGKIREEAGKVLTR 306
Query: 293 NLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPG 352
LK RPAFEADDQSA++YLL ++++KW K +LE+ +YLHGYW LVDRYEEMIE + PG
Sbjct: 307 ELKDRPAFEADDQSAMVYLLATEREKWGGKVYLESGYYLHGYWGILVDRYEEMIENHKPG 366
Query: 353 LGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKI 412
GD RWP VTHFVGCKPCG +GDYPVE+CL M+RAFNF DNQ+L++YGF H+ L S ++
Sbjct: 367 FGDHRWPLVTHFVGCKPCGKFGDYPVERCLRQMDRAFNFGDNQILQMYGFTHKSLGSRRV 426
Query: 413 KRIRNETVTPLEFVDQFDI 431
K RN+T PL+ D+F +
Sbjct: 427 KPTRNQTDRPLDAKDEFGL 445
>emb|CAI11451.1| beta-6-xylosyltransferase [Gossypium raimondii]
Length = 413
Score = 525 bits (1353), Expect = e-148
Identities = 236/375 (62%), Positives = 293/375 (77%), Gaps = 14/375 (3%)
Query: 59 SDADAVNQNVIE-ETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPKITGWDLQR 117
+D +A N E + N+IL + D DP NA ++LGP+I+ WD QR
Sbjct: 38 ADTNAGTNNYHEFDINKILIDEEPDVPKRDP------------NAPYSLGPRISDWDEQR 85
Query: 118 KAWLDQNPEYPNFVR-GKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIV 176
WL +NP YPNF+ K R+LL+TGS PKPC+NP+GDHYLLKSIKNKIDYCRLHGIEI
Sbjct: 86 SRWLQENPNYPNFIGPNKPRVLLVTGSSPKPCENPVGDHYLLKSIKNKIDYCRLHGIEIF 145
Query: 177 YNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLV 236
YN+A LD E+AG+WAKLP+IR+L+LSHPEVE++WWMDSDA FTDM FE+P +Y D N V
Sbjct: 146 YNMALLDAEMAGFWAKLPLIRKLLLSHPEVEFLWWMDSDAMFTDMAFEVPWERYKDSNFV 205
Query: 237 LHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKG 296
+HG+ +++++QK+WI +NTGSFL RN QW+LD+LDAWAPMGPKG +REEAGKVLT LK
Sbjct: 206 MHGWNEMVYDQKNWIGLNTGSFLLRNGQWALDILDAWAPMGPKGKIREEAGKVLTRELKN 265
Query: 297 RPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHPGLGDE 356
RP FEADDQSA++YLL ++++KW DK +LENS+YLHGYW LVDRYEEMIE YHPGLGD
Sbjct: 266 RPVFEADDQSAMVYLLATQREKWGDKVYLENSYYLHGYWGILVDRYEEMIENYHPGLGDH 325
Query: 357 RWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSPKIKRIR 416
RWP VTHFVGCKPCG +GDY VE+CL M+RAFNF DNQ+L++YGF H+ L S ++KR+R
Sbjct: 326 RWPLVTHFVGCKPCGKFGDYSVERCLKQMDRAFNFGDNQILQIYGFTHKSLASRRVKRVR 385
Query: 417 NETVTPLEFVDQFDI 431
NET PLE D+ +
Sbjct: 386 NETSNPLEVKDELGL 400
>emb|CAI79403.1| galactomannan galactosyltransferase [Senna occidentalis]
Length = 449
Score = 333 bits (853), Expect = 8e-90
Identities = 161/375 (42%), Positives = 226/375 (59%), Gaps = 50/375 (13%)
Query: 87 DPDDAAAAETFFS-PNATFTLGPKITGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPP 145
DP +A TF+ P ++TL + WD +R+ WL+++P + K+RILL+TGS P
Sbjct: 85 DPPEA----TFYDDPETSYTLDKPMKNWDEKRQEWLNRHPSFS--AGAKSRILLVTGSQP 138
Query: 146 KPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPE 205
PC NPIGDH LL+ KNK+DYCRLHG +I YN A L ++ YWAK P++R M++HPE
Sbjct: 139 TPCKNPIGDHLLLRFFKNKVDYCRLHGYDIFYNNALLQPKMHTYWAKYPVVRAAMMAHPE 198
Query: 206 VEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQW 265
EWIWW+DSDA FTDM F+LPL++Y ++NL++HG+P L+ E KSW +N G FL RNCQW
Sbjct: 199 AEWIWWVDSDALFTDMEFKLPLNRYKNHNLIVHGWPTLIHEAKSWTGLNAGVFLIRNCQW 258
Query: 266 SLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFL 325
SLD +D WA MGP+ P E+ G+ L K + E+DDQ+ L YL+ +K+KW D+ +L
Sbjct: 259 SLDFMDVWASMGPQTPSYEKWGEKLRTTFKDKAFPESDDQTGLAYLIAVEKEKWADRIYL 318
Query: 326 ENSFYLHGYWAGLVDRYEEMIEKYHP---------------------------------G 352
E+ +Y GYW +V+ YE + +KYH G
Sbjct: 319 ESEYYFEGYWKEIVETYENITDKYHEVERKVRSLRRRHAEKVSESYGAVREPYVMVAGSG 378
Query: 353 LGDERWPFVTHFVGCKPCGSYGD--YPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSP 410
G R PF+THF GC+PC + Y + C + M +A FADNQVL+ +G+ H L
Sbjct: 379 RGSWRRPFITHFTGCQPCSGNHNAMYSPDACWNGMNKALIFADNQVLRKFGYVHPDL--- 435
Query: 411 KIKRIRNETVTPLEF 425
++ +V+P+ F
Sbjct: 436 -----QDNSVSPIPF 445
>emb|CAI11455.1| alpha-6-galactosyltransferase [Zea mays]
Length = 444
Score = 332 bits (851), Expect = 1e-89
Identities = 162/363 (44%), Positives = 216/363 (58%), Gaps = 42/363 (11%)
Query: 92 AAAETFFS-PNATFTLGPKITGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPPKPCDN 150
AAA TF+ P +T+ ITGWD +R WL +PE+ K R+L+++GS P PC +
Sbjct: 70 AAAHTFYDDPALAYTVDRPITGWDEKRAGWLRAHPEFAGGGE-KGRVLMVSGSQPTPCRS 128
Query: 151 PIGDHYLLKSIKNKIDYCRLHGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIW 210
P GDH L++ +KNK DYCRLHG++++YN+A L + YWAK+P+IR M++HPE EW+W
Sbjct: 129 PGGDHTLMRLLKNKADYCRLHGVQLLYNMALLRPSMDRYWAKIPVIRAAMVAHPEAEWVW 188
Query: 211 WMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLL 270
W+DSDA TDM F LPL +Y +NLV+HG+P L+FE SW ++N G FL RNCQWSLD +
Sbjct: 189 WVDSDAVLTDMDFRLPLRRYRAHNLVVHGWPSLVFEASSWTSLNAGVFLIRNCQWSLDFM 248
Query: 271 DAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFY 330
DAWA MGP P + G VL + K + E+DDQSAL+Y+LL + +W DK FLE+ +Y
Sbjct: 249 DAWAAMGPDSPDYQHWGAVLKSTFKDKVFDESDDQSALVYMLLQEGSRWRDKVFLESGYY 308
Query: 331 LHGYWAGLVDRYEEMIEKY--------------------------------------HPG 352
GYW +V R M E+Y G
Sbjct: 309 FEGYWMEIVGRLANMTERYEAMERRPGAAALRRRHAEREHAEYAVARNAALGGAGLAETG 368
Query: 353 LGDERWPFVTHFVGCKPCGSYGD--YPVEKCLSSMERAFNFADNQVLKLYGFRHRGLLSP 410
+ R PFVTHF GC+PC + Y + C M RA NFAD+QVL+ YGFRH LS
Sbjct: 369 VHGWRRPFVTHFTGCQPCSGQRNEHYSGDSCDQGMRRALNFADDQVLRAYGFRHASSLSD 428
Query: 411 KIK 413
++
Sbjct: 429 DVQ 431
>emb|CAD98924.1| galactomannan galactosyltransferase [Lotus corniculatus var.
japonicus]
Length = 437
Score = 327 bits (839), Expect = 3e-88
Identities = 163/401 (40%), Positives = 238/401 (58%), Gaps = 53/401 (13%)
Query: 67 NVIEETNRILAEIRSDADPSDPDDAAAAE---------TFFS-PNATFTLGPKITGWDLQ 116
N + N + A+++S P + ++A+ TF+ P ++T+ +T WD +
Sbjct: 43 NTDPKLNSVAAKLKSLNLPRNQITTSSAQDLLYDSPETTFYDDPEMSYTMDKPVTNWDEK 102
Query: 117 RKAWLDQNPEYPNFVRGKA-RILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEI 175
R+ WL +P+F G + RILL+TGS PK C NPIGDH LL+ KNK+DYCR+H I+I
Sbjct: 103 RRQWL---LHHPSFAAGASDRILLVTGSQPKRCHNPIGDHLLLRFFKNKVDYCRIHDIDI 159
Query: 176 VYNLAHLDVELAGYWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNL 235
+YN A L ++ YWAK P+++ M++HPEVEWIWW+DSDA TDM F+LPL++Y+++NL
Sbjct: 160 IYNNALLHPKMNSYWAKYPVVKAAMIAHPEVEWIWWVDSDAVITDMEFKLPLNRYNEFNL 219
Query: 236 VLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLK 295
++HG+ DL+ ++ SW +N G FL RNCQWSLD +D WA MGP P ++ G+ L A K
Sbjct: 220 IIHGWEDLVKKKHSWTGLNAGVFLMRNCQWSLDFMDVWAAMGPSSPDYKKWGEKLMATFK 279
Query: 296 GRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLV---------------- 339
+ ++DDQ+AL YL+ +DKW +K +LE +Y GYW L
Sbjct: 280 DKVIPDSDDQTALAYLIAMGEDKWTEKIYLEKDYYFEGYWVELAKMYENVSVRYDEVERR 339
Query: 340 -------------DRYEEMIEKYHPGLGDERWPFVTHFVGCKPCGSYGD--YPVEKCLSS 384
+RY EM E++ G R PF+THF GC+PC + + Y + C +
Sbjct: 340 VGGLRRRHAEKVSERYGEMREEHVKYFGQWRRPFITHFTGCQPCNGHHNPAYAADDCWNG 399
Query: 385 MERAFNFADNQVLKLYGFRHRGLLSPKIKRIRNETVTPLEF 425
M+RA NFADNQVL+ YG+ R L ++ VTP+ +
Sbjct: 400 MDRALNFADNQVLRTYGYVRRSL--------NDKAVTPIPY 432
>emb|CAI79402.1| galactomannan galactosyltransferase [Cyamopsis tetragonoloba]
Length = 435
Score = 319 bits (818), Expect = 1e-85
Identities = 173/447 (38%), Positives = 245/447 (54%), Gaps = 57/447 (12%)
Query: 18 ARGGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGVNLSSSDADAVNQNVIEETNRILA 77
A+ G R + K +N +L F +L+L G + + D + N +
Sbjct: 2 AKFGSRNKS-PKWISNGCCFLLGAFTALLLLWGLCSFIIPIPNTDP-------KLNSVAT 53
Query: 78 EIRSDADPSDP---------DDAAAAETFFSPNATFTLGPKITGWDLQRKAWLDQNPEYP 128
+RS P +P D + P ++T+ + WD +RK WL +P +
Sbjct: 54 SLRSLNFPKNPAATLPPNLQHDPPDTTFYDDPETSYTMDKPMKNWDEKRKEWLLHHPSFG 113
Query: 129 NFVRGKARILLLTGSPPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLAHLDVELAG 188
R K ILL+TGS PK C NPIGDH LL+ KNK+DYCRLH +I+YN A L ++
Sbjct: 114 AAARDK--ILLVTGSQPKRCHNPIGDHLLLRFFKNKVDYCRLHNYDIIYNNALLHPKMNS 171
Query: 189 YWAKLPMIRRLMLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQK 248
YWAK P+IR M++HPEVEW+WW+DSDA FTDM F+LPL +Y ++NLV+HG+ L+
Sbjct: 172 YWAKYPVIRAAMMAHPEVEWVWWVDSDAVFTDMEFKLPLKRYKNHNLVVHGWEGLVRLNH 231
Query: 249 SWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSAL 308
SW +N G FL RNCQWSL+ +D W MGP+ P E+ G+ L K + ++DDQ+AL
Sbjct: 232 SWTGLNAGVFLIRNCQWSLEFMDVWVSMGPQTPEYEKWGERLRETFKDKVLPDSDDQTAL 291
Query: 309 IYLLLS-KKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHP------GL-------- 353
YL+ + KD W +K FLE+ +Y GYW +V YE + E+Y GL
Sbjct: 292 AYLIATDNKDTWREKIFLESEYYFEGYWLEIVKTYENISERYDEVERKVEGLRRRHAEKV 351
Query: 354 -------------GDERWPFVTHFVGCKPCGSYGD--YPVEKCLSSMERAFNFADNQVLK 398
++R PF+THF GC+PC + + Y C + MERA NFADNQ+L+
Sbjct: 352 SEKYGAMREEYLKDNKRRPFITHFTGCQPCNGHHNPAYNANDCWNGMERALNFADNQILR 411
Query: 399 LYGFRHRGLLSPKIKRIRNETVTPLEF 425
YG+ + LL +++V+PL F
Sbjct: 412 TYGYHRQNLL--------DKSVSPLPF 430
>emb|CAB80434.1| putative protein [Arabidopsis thaliana] gi|4468994|emb|CAB38308.1|
putative protein [Arabidopsis thaliana]
gi|22329223|ref|NP_680773.1| galactosyl transferase
GMA12/MNN10 family protein [Arabidopsis thaliana]
gi|7485771|pir||T04726 hypothetical protein F19F18.180 -
Arabidopsis thaliana gi|46576348|sp|Q9SZG1|GT6_ARATH
Putative glycosyltransferase 6 (AtGT6)
Length = 432
Score = 300 bits (768), Expect = 6e-80
Identities = 147/362 (40%), Positives = 212/362 (57%), Gaps = 48/362 (13%)
Query: 84 DPSDPDDAAAAETFFSPNATFTLGPKITGWDLQRKAWLDQNPEYPNFVRGKARILLLTGS 143
DPS+P + P+ ++++ IT WD +R W + +P + + RI+++TGS
Sbjct: 63 DPSEPG------FYDDPDLSYSIEKPITKWDEKRNQWFESHPSFKP--GSENRIVMVTGS 114
Query: 144 PPKPCDNPIGDHYLLKSIKNKIDYCRLHGIEIVYNLAHLDVELAGYWAKLPMIRRLMLSH 203
PC NPIGDH LL+ KNK+DY R+HG +I Y+ + L ++ YWAKLP+++ ML+H
Sbjct: 115 QSSPCKNPIGDHLLLRCFKNKVDYARIHGHDIFYSNSLLHPKMNSYWAKLPVVKAAMLAH 174
Query: 204 PEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNC 263
PE EWIWW+DSDA FTDM F+ PL +Y +NLV+HG+P++++E++SW A+N G FL RNC
Sbjct: 175 PEAEWIWWVDSDAIFTDMEFKPPLHRYRQHNLVVHGWPNIIYEKQSWTALNAGVFLIRNC 234
Query: 264 QWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSALIYLLLSKKDKWMDKT 323
QWS+DL+D W MGP P ++ G + + K + E+DDQ+ALIYLL K+ + K
Sbjct: 235 QWSMDLIDTWKSMGPVSPDYKKWGPIQRSIFKDKLFPESDDQTALIYLLYKHKELYYPKI 294
Query: 324 FLENSFYLHGYWAGLVDRYEEMIEKY---------------------------------- 349
+LE +YL GYW G+ + + E+Y
Sbjct: 295 YLEAEYYLQGYWIGVFGDFANVTERYLEMEREDDTLRRRHAEKVSERYGAFREERFLKGE 354
Query: 350 HPGLGDERWPFVTHFVGCKPCGSYGD----YPVEKCLSSMERAFNFADNQVLKLYGFRHR 405
G G R F+THF GC+PC GD Y + C + M RA NFADNQV+++YG+ H
Sbjct: 355 FGGRGSRRRAFITHFTGCQPCS--GDHNPSYDGDTCWNEMIRALNFADNQVMRVYGYVHS 412
Query: 406 GL 407
L
Sbjct: 413 DL 414
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 826,158,334
Number of Sequences: 2540612
Number of extensions: 37077154
Number of successful extensions: 72174
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 72014
Number of HSP's gapped (non-prelim): 111
length of query: 443
length of database: 863,360,394
effective HSP length: 131
effective length of query: 312
effective length of database: 530,540,222
effective search space: 165528549264
effective search space used: 165528549264
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0093b.6


