

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093b.5
(495 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
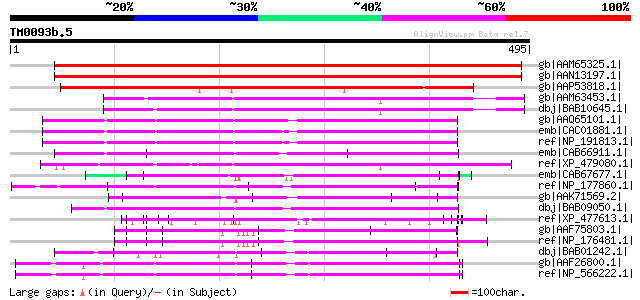
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM65325.1| unknown [Arabidopsis thaliana] 565 e-159
gb|AAN13197.1| unknown protein [Arabidopsis thaliana] gi|1752906... 564 e-159
gb|AAP53818.1| putative endopeptidase [Oryza sativa (japonica cu... 343 9e-93
gb|AAM63453.1| unknown [Arabidopsis thaliana] 166 2e-39
dbj|BAB10645.1| unnamed protein product [Arabidopsis thaliana] g... 164 8e-39
gb|AAQ65101.1| At3g62470 [Arabidopsis thaliana] gi|7340718|emb|C... 124 9e-27
emb|CAC01881.1| putative protein [Arabidopsis thaliana] gi|15241... 124 9e-27
ref|NP_191813.1| pentatricopeptide (PPR) repeat-containing prote... 122 2e-26
emb|CAB66911.1| putative protein [Arabidopsis thaliana] gi|15229... 122 3e-26
ref|XP_479080.1| unknown protein [Oryza sativa (japonica cultiva... 121 4e-26
emb|CAB67677.1| putative protein [Arabidopsis thaliana] gi|15982... 121 4e-26
ref|NP_177860.1| pentatricopeptide (PPR) repeat-containing prote... 120 1e-25
gb|AAK71569.2| putative reverse transcriptase [Oryza sativa (jap... 120 1e-25
dbj|BAB09050.1| unnamed protein product [Arabidopsis thaliana] g... 118 5e-25
ref|XP_477613.1| putative fertility restorer homologue [Oryza sa... 117 6e-25
gb|AAF75803.1| Contains weak similarity to leaf protein from Ipo... 115 4e-24
ref|NP_176481.1| pentatricopeptide (PPR) repeat-containing prote... 115 4e-24
dbj|BAB01242.1| unnamed protein product [Arabidopsis thaliana] g... 114 7e-24
gb|AAF26800.1| hypothetical protein [Arabidopsis thaliana] 114 9e-24
ref|NP_566222.1| pentatricopeptide (PPR) repeat-containing prote... 114 9e-24
>gb|AAM65325.1| unknown [Arabidopsis thaliana]
Length = 487
Score = 565 bits (1455), Expect = e-159
Identities = 278/447 (62%), Positives = 347/447 (77%), Gaps = 1/447 (0%)
Query: 43 LQELCGIVTSNVGGLDDLELSLNKFKGSLTSSLVAQAIESSKHEAQTRRLLRFFLWSSKN 102
L E+ IV+S VGGLDDLE +LN+ S +S+LV Q IES K+E RRLLRFF WS K+
Sbjct: 37 LHEVIRIVSSPVGGLDDLEENLNQVSVSPSSNLVTQVIESCKNETSPRRLLRFFSWSCKS 96
Query: 103 LRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDE 162
L L D ++NY LRV AEK+D+T+M IL+ DL+KE R MD QTF +VAE L+K+GKE++
Sbjct: 97 LGSSLHDKEFNYVLRVLAEKKDHTAMQILLSDLRKENRAMDKQTFSIVAETLVKIGKEED 156
Query: 163 ALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLY 222
A+GIFK LDK+ C + FTVTAII+ALCS+GH KRA GV+ HHKD I+G +YRSLL+
Sbjct: 157 AIGIFKILDKFSCPQDGFTVTAIISALCSRGHVKRALGVMHHHKDVISGNELSVYRSLLF 216
Query: 223 GWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEM 282
GWSV RNVKEARR+I++MKS G+ PDL C+N+ L CLCERN+ NPSGLVPE LN+M+EM
Sbjct: 217 GWSVQRNVKEARRVIQDMKSAGITPDLFCFNSLLTCLCERNVNRNPSGLVPEALNIMLEM 276
Query: 283 RSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRF 342
RS+K+ PTS+SYNILLSCLG+TRRV+ESCQILE MK SG PD SYY VVRVL+L+GRF
Sbjct: 277 RSYKIQPTSMSYNILLSCLGRTRRVRESCQILEQMKRSGCDPDTGSYYFVVRVLYLTGRF 336
Query: 343 GKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVL 402
GKG +IVD+MI +G P KFY+ LIG+LCGVERVN AL+LFEKMK SS+GGYG VYD+L
Sbjct: 337 GKGNQIVDEMIERGFRPERKFYYDLIGVLCGVERVNFALQLFEKMKRSSVGGYGQVYDLL 396
Query: 403 IPKLCRGGDFEKGRELWDEATSMGITLQCSKDILDPSITEVFKPVR-PEKISLADSPIAK 461
IPKLC+GG+FEKGRELW+EA S+ +TL CS +LDPS+TEVFKP++ E+ ++ D
Sbjct: 397 IPKLCKGGNFEKGRELWEEALSIDVTLSCSISLLDPSVTEVFKPMKMKEEAAMVDRRALN 456
Query: 462 SPKKAKKLLGKVKMRKKPTAVKKKKKR 488
A+ K K++ KP K KK+
Sbjct: 457 LKIHARMNKTKPKLKLKPKRRSKTKKK 483
>gb|AAN13197.1| unknown protein [Arabidopsis thaliana] gi|17529064|gb|AAL38742.1|
unknown protein [Arabidopsis thaliana]
gi|9757858|dbj|BAB08492.1| unnamed protein product
[Arabidopsis thaliana] gi|15240232|ref|NP_200945.1|
pentatricopeptide (PPR) repeat-containing protein
[Arabidopsis thaliana]
Length = 487
Score = 564 bits (1454), Expect = e-159
Identities = 278/447 (62%), Positives = 347/447 (77%), Gaps = 1/447 (0%)
Query: 43 LQELCGIVTSNVGGLDDLELSLNKFKGSLTSSLVAQAIESSKHEAQTRRLLRFFLWSSKN 102
L E+ IV+S VGGLDDLE +LN+ S +S+LV Q IES K+E RRLLRFF WS K+
Sbjct: 37 LHEVIRIVSSPVGGLDDLEENLNQVSVSPSSNLVTQVIESCKNETSPRRLLRFFSWSCKS 96
Query: 103 LRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDE 162
L L D ++NY LRV AEK+D+T+M IL+ DL+KE R MD QTF +VAE L+K+GKE++
Sbjct: 97 LGSSLHDKEFNYVLRVLAEKKDHTAMQILLSDLRKENRAMDKQTFSIVAETLVKVGKEED 156
Query: 163 ALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLY 222
A+GIFK LDK+ C + FTVTAII+ALCS+GH KRA GV+ HHKD I+G +YRSLL+
Sbjct: 157 AIGIFKILDKFSCPQDGFTVTAIISALCSRGHVKRALGVMHHHKDVISGNELSVYRSLLF 216
Query: 223 GWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEM 282
GWSV RNVKEARR+I++MKS G+ PDL C+N+ L CLCERN+ NPSGLVPE LN+M+EM
Sbjct: 217 GWSVQRNVKEARRVIQDMKSAGITPDLFCFNSLLTCLCERNVNRNPSGLVPEALNIMLEM 276
Query: 283 RSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRF 342
RS+K+ PTS+SYNILLSCLG+TRRV+ESCQILE MK SG PD SYY VVRVL+L+GRF
Sbjct: 277 RSYKIQPTSMSYNILLSCLGRTRRVRESCQILEQMKRSGCDPDTGSYYFVVRVLYLTGRF 336
Query: 343 GKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVL 402
GKG +IVD+MI +G P KFY+ LIG+LCGVERVN AL+LFEKMK SS+GGYG VYD+L
Sbjct: 337 GKGNQIVDEMIERGFRPERKFYYDLIGVLCGVERVNFALQLFEKMKRSSVGGYGQVYDLL 396
Query: 403 IPKLCRGGDFEKGRELWDEATSMGITLQCSKDILDPSITEVFKPVR-PEKISLADSPIAK 461
IPKLC+GG+FEKGRELW+EA S+ +TL CS +LDPS+TEVFKP++ E+ ++ D
Sbjct: 397 IPKLCKGGNFEKGRELWEEALSIDVTLSCSISLLDPSVTEVFKPMKMKEEAAMVDRRALN 456
Query: 462 SPKKAKKLLGKVKMRKKPTAVKKKKKR 488
A+ K K++ KP K KK+
Sbjct: 457 LKIHARMNKTKPKLKLKPKRRSKTKKK 483
>gb|AAP53818.1| putative endopeptidase [Oryza sativa (japonica cultivar-group)]
gi|37534458|ref|NP_921531.1| putative endopeptidase
[Oryza sativa (japonica cultivar-group)]
Length = 679
Score = 343 bits (879), Expect = 9e-93
Identities = 183/409 (44%), Positives = 260/409 (62%), Gaps = 16/409 (3%)
Query: 49 IVTSNVGGLDDLELSLNKFKGSLTSSLVAQAIES-SKHEAQTRRLLRFFLWSSKNLRHDL 107
+V S G LD++ +L++ +++ ++VA+ I++ S+ RRLLRF W +
Sbjct: 30 VVCSGAGSLDEVGGALDRLGVAVSPAMVARVIDACSERMGSGRRLLRFLSWCRSKDAGGI 89
Query: 108 EDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIF 167
D + A+ A D T+M I + D +K+GR M +TF +V E L+KLGKEDEA+ +F
Sbjct: 90 GDEALDSAIAALARMGDLTAMRIAVADAEKDGRRMSPETFTVVVEALVKLGKEDEAVRLF 149
Query: 168 KNLDKYKCSTNE----------FTVTAIINALCSKGHAKRAEGVVLHHKDKIT--GALPC 215
+ L++ + + A++ ALC KGHA+ A+GVV HHK +++ +
Sbjct: 150 RGLERQRLLPRRDAGDGGEGVWSSSLAMVQALCMKGHAREAQGVVWHHKSELSVEPMVSI 209
Query: 216 IYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPET 275
+ RSLL+GW VH N KEARR++ ++KS+ L +N +L CLC RNL+ NPS LV E
Sbjct: 210 VQRSLLHGWCVHGNAKEARRVLDDIKSSCTPLGLPSFNDYLHCLCHRNLKFNPSALVTEA 269
Query: 276 LNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMK--TSGVSPDWVSYYLVV 333
++V+ EMRS+ V P + S NILLSCLG+ RRVKES +IL M+ +G SPDWVSYYLVV
Sbjct: 270 MDVLSEMRSYGVTPDASSLNILLSCLGRARRVKESYRILYLMREGKAGCSPDWVSYYLVV 329
Query: 334 RVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLG 393
RVL+L+GR +GK +VD M+ G++P KF+ LIG+LCG E+V+H L++F MK L
Sbjct: 330 RVLYLTGRIIRGKRLVDDMLESGVLPTAKFFHGLIGVLCGTEKVDHGLDMFRLMKRCQLV 389
Query: 394 GYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGITLQCSKDILDPSITE 442
YD+LI KLCR G FE G+ELWD+A G L CS+D+LDP TE
Sbjct: 390 D-THTYDLLIEKLCRNGRFENGKELWDDAKKNGFMLGCSEDLLDPLKTE 437
>gb|AAM63453.1| unknown [Arabidopsis thaliana]
Length = 521
Score = 166 bits (420), Expect = 2e-39
Identities = 110/406 (27%), Positives = 191/406 (46%), Gaps = 29/406 (7%)
Query: 90 RRLLRFFLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGL 149
R L F W N D ++ + F ++D+ M +G + K + +T
Sbjct: 124 RAALGFNEWLDSNSNFSHTDETVSFFVDYFGRRKDFKGM---LGIISKYKGIAGGKTLES 180
Query: 150 VAENLIKLGKEDEALGIFKNLDK-YKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDK 208
+ L++ G+ + F+ ++ Y ++ ++T ++ LC KGHA AE +V + ++
Sbjct: 181 AIDRLVRAGRPKQVTDFFEKMENDYGLKRDKESLTLVVKKLCEKGHASIAEKMVKNTANE 240
Query: 209 ITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNP 268
I I L+ GW + + EA R+ EM G YN L C+C+ + +P
Sbjct: 241 IFPD-ENICDLLISGWCIAEKLDEATRLAGEMSRGGFEIGTKAYNMMLDCVCKLCRKKDP 299
Query: 269 SGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVS 328
L PE V++EM V + ++N+L++ L K RR +E+ + M G PD +
Sbjct: 300 FKLQPEVEKVLLEMEFRGVPRNTETFNVLINNLCKIRRTEEAMTLFGRMGEWGCQPDAET 359
Query: 329 YYLVVRVLFLSGRFGKGKEIVDQM--IGKGLVPNHKFYFSLIGILCGVERVNHALELFEK 386
Y +++R L+ + R G+G E++D+M G G + N K Y+ + ILCG+ER+ HA+ +F+
Sbjct: 360 YLVLIRSLYQAARIGEGDEMIDKMKSAGYGELLNKKEYYGFLKILCGIERLEHAMSVFKS 419
Query: 387 MKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGITLQCSKDILDPSITEVFKP 446
MK++ YD+L+ K+C + L+ EA GI + + +DP +
Sbjct: 420 MKANGCKPGIKTYDLLMGKMCANNQLTRANGLYKEAAKKGIAVSPKEYRVDPRFMK---- 475
Query: 447 VRPEKISLADSPIAKSPKKAKKLLGKVKMRKK-PTAVKKKKKRQKK 491
KK K++ VK R+ P +KKKR K+
Sbjct: 476 -----------------KKTKEVDSNVKKRETLPEKTARKKKRLKQ 504
>dbj|BAB10645.1| unnamed protein product [Arabidopsis thaliana]
gi|15239488|ref|NP_200904.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
Length = 521
Score = 164 bits (414), Expect = 8e-39
Identities = 110/406 (27%), Positives = 190/406 (46%), Gaps = 29/406 (7%)
Query: 90 RRLLRFFLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGL 149
R L F W N D ++ + F ++D+ M +I K + +T
Sbjct: 124 RAALGFNEWLDSNSNFSHTDETVSFFVDYFGRRKDFKGMLEIISKYKG---IAGGKTLES 180
Query: 150 VAENLIKLGKEDEALGIFKNLDK-YKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDK 208
+ L++ G+ + F+ ++ Y ++ ++T ++ LC KGHA AE +V + ++
Sbjct: 181 AIDRLVRAGRPKQVTDFFEKMENDYGLKRDKESLTLVVKKLCEKGHASIAEKMVKNTANE 240
Query: 209 ITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNP 268
I I L+ GW + + EA R+ EM G YN L C+C+ + +P
Sbjct: 241 IFPD-ENICDLLISGWCIAEKLDEATRLAGEMSRGGFEIGTKAYNMMLDCVCKLCRKKDP 299
Query: 269 SGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVS 328
L PE V++EM V + ++N+L++ L K RR +E+ + M G PD +
Sbjct: 300 FKLQPEVEKVLLEMEFRGVPRNTETFNVLINNLCKIRRTEEAMTLFGRMGEWGCQPDAET 359
Query: 329 YYLVVRVLFLSGRFGKGKEIVDQM--IGKGLVPNHKFYFSLIGILCGVERVNHALELFEK 386
Y +++R L+ + R G+G E++D+M G G + N K Y+ + ILCG+ER+ HA+ +F+
Sbjct: 360 YLVLIRSLYQAARIGEGDEMIDKMKSAGYGELLNKKEYYGFLKILCGIERLEHAMSVFKS 419
Query: 387 MKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGITLQCSKDILDPSITEVFKP 446
MK++ YD+L+ K+C + L+ EA GI + + +DP +
Sbjct: 420 MKANGCKPGIKTYDLLMGKMCANNQLTRANGLYKEAAKKGIAVSPKEYRVDPRFMK---- 475
Query: 447 VRPEKISLADSPIAKSPKKAKKLLGKVKMRKK-PTAVKKKKKRQKK 491
KK K++ VK R+ P +KKKR K+
Sbjct: 476 -----------------KKTKEVDSNVKKRETLPEKTARKKKRLKQ 504
>gb|AAQ65101.1| At3g62470 [Arabidopsis thaliana] gi|7340718|emb|CAB82961.1|
putative protein [Arabidopsis thaliana]
gi|15228767|ref|NP_191806.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
gi|11357975|pir||T48039 hypothetical protein T12C14.170
- Arabidopsis thaliana
Length = 599
Score = 124 bits (310), Expect = 9e-27
Identities = 86/396 (21%), Positives = 181/396 (44%), Gaps = 11/396 (2%)
Query: 32 STLQPISAPPHLQELCGIVTSNVGGLDDLELSLNKFKGSLTSSLVAQAIESSKHEAQTRR 91
S ++ + P ++ +C ++ ++E L++ K L+ L+ + +E +H +
Sbjct: 120 SCVESSTNPEEVERVCKVIDELFALDRNMEAVLDEMKLDLSHDLIVEVLERFRHARKPA- 178
Query: 92 LLRFFLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVA 151
RFF W+++ + YN + + A+ + + +M ++ ++ +G ++ +TF +
Sbjct: 179 -FRFFCWAAERQGFAHDSRTYNSMMSILAKTRQFETMVSVLEEMGTKG-LLTMETFTIAM 236
Query: 152 ENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITG 211
+ + +A+GIF+ + KYK T+ ++++L K A+ + K++ T
Sbjct: 237 KAFAAAKERKKAVGIFELMKKYKFKIGVETINCLLDSLGRAKLGKEAQVLFDKLKERFTP 296
Query: 212 ALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGL 271
+ Y LL GW RN+ EA RI +M G+ PD+V +N L L +
Sbjct: 297 NM-MTYTVLLNGWCRVRNLIEAARIWNDMIDQGLKPDIVAHNVMLEGLLRSRKKS----- 350
Query: 272 VPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYL 331
+ + + M+S P SY I++ K ++ + + + M SG+ PD Y
Sbjct: 351 --DAIKLFHVMKSKGPCPNVRSYTIMIRDFCKQSSMETAIEYFDDMVDSGLQPDAAVYTC 408
Query: 332 VVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSS 391
++ + E++ +M KG P+ K Y +LI ++ + HA ++ KM +
Sbjct: 409 LITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYNALIKLMANQKMPEHATRIYNKMIQNE 468
Query: 392 LGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
+ +++++ ++E GR +W+E GI
Sbjct: 469 IEPSIHTFNMIMKSYFMARNYEMGRAVWEEMIKKGI 504
>emb|CAC01881.1| putative protein [Arabidopsis thaliana]
gi|15241508|ref|NP_196986.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
gi|11358457|pir||T51427 hypothetical protein T9L3_120 -
Arabidopsis thaliana
Length = 598
Score = 124 bits (310), Expect = 9e-27
Identities = 87/396 (21%), Positives = 185/396 (45%), Gaps = 11/396 (2%)
Query: 32 STLQPISAPPHLQELCGIVTSNVGGLDDLELSLNKFKGSLTSSLVAQAIESSKHEAQTRR 91
S ++ + P ++ +C ++ ++E L++ K L+ L+ + +E +H +
Sbjct: 119 SCVESSTNPEEVERVCKVIDELFALDRNMEAVLDEMKLDLSHDLIVEVLERFRHARKPA- 177
Query: 92 LLRFFLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVA 151
RFF W+++ + YN + + A+ + + +M ++ ++ +G ++ +TF +
Sbjct: 178 -FRFFCWAAERQGFAHDSRTYNSMMSILAKTRQFETMVSVLEEMGTKG-LLTMETFTIAM 235
Query: 152 ENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITG 211
+ + +A+GIF+ + KYK T+ ++++L K A+ + K++ T
Sbjct: 236 KAFAAAKERKKAVGIFELMKKYKFKIGVETINCLLDSLGRAKLGKEAQVLFDKLKERFTP 295
Query: 212 ALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGL 271
+ Y LL GW RN+ EA RI +M +G+ PD+V +N L L R+++ +
Sbjct: 296 NM-MTYTVLLNGWCRVRNLIEAARIWNDMIDHGLKPDIVAHNVMLEGLL-RSMKKS---- 349
Query: 272 VPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYL 331
+ + + M+S P SY I++ K ++ + + + M SG+ PD Y
Sbjct: 350 --DAIKLFHVMKSKGPCPNVRSYTIMIRDFCKQSSMETAIEYFDDMVDSGLQPDAAVYTC 407
Query: 332 VVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSS 391
++ + E++ +M KG P+ K Y +LI ++ + H ++ KM +
Sbjct: 408 LITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYNALIKLMANQKMPEHGTRIYNKMIQNE 467
Query: 392 LGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
+ +++++ ++E GR +WDE GI
Sbjct: 468 IEPSIHTFNMIMKSYFVARNYEMGRAVWDEMIKKGI 503
>ref|NP_191813.1| pentatricopeptide (PPR) repeat-containing protein [Arabidopsis
thaliana]
Length = 599
Score = 122 bits (307), Expect = 2e-26
Identities = 87/396 (21%), Positives = 184/396 (45%), Gaps = 11/396 (2%)
Query: 32 STLQPISAPPHLQELCGIVTSNVGGLDDLELSLNKFKGSLTSSLVAQAIESSKHEAQTRR 91
S ++ + P ++ +C ++ ++E L++ K L+ L+ + +E +H +
Sbjct: 120 SCVESSTNPEEVERVCKVIDELFALDRNMEAVLDEMKLDLSHDLIVEVLERFRHARKPA- 178
Query: 92 LLRFFLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVA 151
RFF W+++ YN + + A+ + + +M ++ ++ +G ++ +TF +
Sbjct: 179 -FRFFCWAAERQGFAHASRTYNSMMSILAKTRQFETMVSVLEEMGTKG-LLTMETFTIAM 236
Query: 152 ENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITG 211
+ + +A+GIF+ + KYK T+ ++++L K A+ + K++ T
Sbjct: 237 KAFAAAKERKKAVGIFELMKKYKFKIGVETINCLLDSLGRAKLGKEAQVLFDKLKERFTP 296
Query: 212 ALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGL 271
+ Y LL GW RN+ EA RI +M +G+ PD+V +N L L R+++ +
Sbjct: 297 NM-MTYTVLLNGWCRVRNLIEAARIWNDMIDHGLKPDIVAHNVMLEGLL-RSMKKS---- 350
Query: 272 VPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYL 331
+ + + M+S P SY I++ K ++ + + + M SG+ PD Y
Sbjct: 351 --DAIKLFHVMKSKGPCPNVRSYTIMIRDFCKQSSMETAIEYFDDMVDSGLQPDAAVYTC 408
Query: 332 VVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSS 391
++ + E++ +M KG P+ K Y +LI ++ + H ++ KM +
Sbjct: 409 LITGFGTQKKLDTVYELLKEMQEKGHPPDGKTYNALIKLMANQKMPEHGTRIYNKMIQNE 468
Query: 392 LGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
+ +++++ ++E GR +WDE GI
Sbjct: 469 IEPSIHTFNMIMKSYFVARNYEMGRAVWDEMIKKGI 504
>emb|CAB66911.1| putative protein [Arabidopsis thaliana]
gi|15229604|ref|NP_190542.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
gi|11358047|pir||T46039 hypothetical protein T16K5.80 -
Arabidopsis thaliana
Length = 1184
Score = 122 bits (305), Expect = 3e-26
Identities = 88/388 (22%), Positives = 179/388 (45%), Gaps = 12/388 (3%)
Query: 43 LQELCGIVTSNVGGLDDLELSLNKFKGSLTSSLVAQAIESSKHEAQTRRLLRFFLWSSKN 102
++++ I+ ++ + LEL+LN+ L L+ + + RFFLW++K
Sbjct: 67 VEKIYRILRNHHSRVPKLELALNESGIDLRPGLIIRVLSRCGDAGNLG--YRFFLWATKQ 124
Query: 103 LRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEG-RVMDAQTFGLVAENLIKLGKED 161
+ + + ++ + + ++ LI +++K +++ + F ++
Sbjct: 125 PGYFHSYEVCKSMVMILSKMRQFGAVWGLIEEMRKTNPELIEPELFVVLMRRFASANMVK 184
Query: 162 EALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLL 221
+A+ + + KY +E+ +++ALC G K A V ++K L + SLL
Sbjct: 185 KAVEVLDEMPKYGLEPDEYVFGCLLDALCKNGSVKEASKVFEDMREKFPPNLR-YFTSLL 243
Query: 222 YGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMME 281
YGW + EA+ ++ +MK G+ PD+V + L + +G + + ++M +
Sbjct: 244 YGWCREGKLMEAKEVLVQMKEAGLEPDIVVFTNLLS-------GYAHAGKMADAYDLMND 296
Query: 282 MRSHKVLPTSISYNILLSCLGKT-RRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSG 340
MR P Y +L+ L +T +R+ E+ ++ M+ G D V+Y ++ G
Sbjct: 297 MRKRGFEPNVNCYTVLIQALCRTEKRMDEAMRVFVEMERYGCEADIVTYTALISGFCKWG 356
Query: 341 RFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYD 400
KG ++D M KG++P+ Y ++ E+ LEL EKMK +Y+
Sbjct: 357 MIDKGYSVLDDMRKKGVMPSQVTYMQIMVAHEKKEQFEECLELIEKMKRRGCHPDLLIYN 416
Query: 401 VLIPKLCRGGDFEKGRELWDEATSMGIT 428
V+I C+ G+ ++ LW+E + G++
Sbjct: 417 VVIRLACKLGEVKEAVRLWNEMEANGLS 444
Score = 67.8 bits (164), Expect = 8e-10
Identities = 44/193 (22%), Positives = 88/193 (44%), Gaps = 8/193 (4%)
Query: 131 LIGDLKKEGRVMDAQTFGLVAENLIKLGKE-DEALGIFKNLDKYKCSTNEFTVTAIINAL 189
L+ D++K G + + ++ + L + K DEA+ +F +++Y C + T TA+I+
Sbjct: 293 LMNDMRKRGFEPNVNCYTVLIQALCRTEKRMDEAMRVFVEMERYGCEADIVTYTALISGF 352
Query: 190 CSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDL 249
C G + V+ + K Y ++ +E +I++MK G PDL
Sbjct: 353 CKWGMIDKGYSVLDDMRKKGVMPSQVTYMQIMVAHEKKEQFEECLELIEKMKRRGCHPDL 412
Query: 250 VCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKE 309
+ YN +R C+ G V E + + EM ++ + P ++ I+++ + E
Sbjct: 413 LIYNVVIRLACK-------LGEVKEAVRLWNEMEANGLSPGVDTFVIMINGFTSQGFLIE 465
Query: 310 SCQILEAMKTSGV 322
+C + M + G+
Sbjct: 466 ACNHFKEMVSRGI 478
>ref|XP_479080.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|34394935|dbj|BAC84486.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
gi|34394565|dbj|BAC83868.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 486
Score = 121 bits (304), Expect = 4e-26
Identities = 111/459 (24%), Positives = 210/459 (45%), Gaps = 17/459 (3%)
Query: 30 LCSTLQPISAPPHL--QELCGIV-----TSNVGGLDDLELSLNKFKGSLTSSLVAQAIES 82
L ST P APP L G + T D SL+ T++L+A+A+
Sbjct: 30 LLSTASPDPAPPATLASSLAGALSALSSTPPPATSPDAYFSLHFSDVRPTNALLAEALAL 89
Query: 83 SKHEAQTRRLLRFFLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVM 142
+ A +R F + + D+ +R A ++D+ ++ L+ + +
Sbjct: 90 AP-PATSRAAAELFRFLVRRRSLHPSDSALAPVVRHLARRRDFPAVRSLVQEFPS---AL 145
Query: 143 DAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVV 202
T +L + G+ +A+ +F L + T + +T+++++L ++G AEG V
Sbjct: 146 GHDTLDAYLLSLARAGRATDAVKVFDELPP-QLRTRQ-ALTSLVSSLSAEGWPSHAEGAV 203
Query: 203 LHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCER 262
++I I L+ G++ + A R+I E + G P L YN L C+C
Sbjct: 204 KKVANEIFPD-DNICTLLVSGYANAGKLDHALRLIGETRRGGFQPGLDAYNAVLDCICRL 262
Query: 263 NLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGV 322
+ +P + E +++M ++ + + ++ +L++ L K R+ +++ + M G
Sbjct: 263 CRKKDPLRMPAEAEKFLVDMEANGIPRDAGTFRVLITNLCKIRKTEDAMNLFRRMGEWGC 322
Query: 323 SPDWVSYYLVVRVLFLSGRFGKGKEIVDQM--IGKGLVPNHKFYFSLIGILCGVERVNHA 380
SPD +Y ++++ L+ + R +G E++ M G G + K Y+ I ILCG+ERV HA
Sbjct: 323 SPDADTYLVLIKSLYQAARISEGDEMMTWMRSAGFGAKLDRKAYYGFIKILCGIERVEHA 382
Query: 381 LELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGITLQCSKDILDPSI 440
+++F MK Y +LI KL R ++ L+ EA + G+T+ +D
Sbjct: 383 VKVFRMMKGYGHAPGTKSYSLLIEKLTRHNLGDRANALFREAVARGVTVTPGVYKIDKKY 442
Query: 441 TEVFKPVRPEK-ISLADSPIAKSPKKAKKLLGKVKMRKK 478
+ K + +K ++L + KS + K + VK K+
Sbjct: 443 VKAKKEKKVKKRLTLPEKMRLKSKRLYKLRMSFVKKPKR 481
>emb|CAB67677.1| putative protein [Arabidopsis thaliana] gi|15982931|gb|AAL09812.1|
AT3g53700/F4P12_400 [Arabidopsis thaliana]
gi|15231863|ref|NP_190938.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
gi|11357800|pir||T45910 hypothetical protein F4P12.400 -
Arabidopsis thaliana
Length = 754
Score = 121 bits (304), Expect = 4e-26
Identities = 87/304 (28%), Positives = 140/304 (45%), Gaps = 15/304 (4%)
Query: 128 MDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIIN 187
MD+++ +EG D T+ V L KLG+ EA+ + + CS N T +I+
Sbjct: 318 MDVML----QEGYDPDVYTYNSVISGLCKLGEVKEAVEVLDQMITRDCSPNTVTYNTLIS 373
Query: 188 ALCSKGHAKRAEGVVLHHKDKITGALP--CIYRSLLYGWSVHRNVKEARRIIKEMKSNGV 245
LC + + A + K G LP C + SL+ G + RN + A + +EM+S G
Sbjct: 374 TLCKENQVEEATELARVLTSK--GILPDVCTFNSLIQGLCLTRNHRVAMELFEEMRSKGC 431
Query: 246 IPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTR 305
PD YN + LC + G + E LN++ +M + I+YN L+ K
Sbjct: 432 EPDEFTYNMLIDSLCSK-------GKLDEALNMLKQMELSGCARSVITYNTLIDGFCKAN 484
Query: 306 RVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYF 365
+ +E+ +I + M+ GVS + V+Y ++ L S R +++DQMI +G P+ Y
Sbjct: 485 KTREAEEIFDEMEVHGVSRNSVTYNTLIDGLCKSRRVEDAAQLMDQMIMEGQKPDKYTYN 544
Query: 366 SLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSM 425
SL+ C + A ++ + M S+ Y LI LC+ G E +L
Sbjct: 545 SLLTHFCRGGDIKKAADIVQAMTSNGCEPDIVTYGTLISGLCKAGRVEVASKLLRSIQMK 604
Query: 426 GITL 429
GI L
Sbjct: 605 GINL 608
Score = 120 bits (300), Expect = 1e-25
Identities = 79/319 (24%), Positives = 150/319 (46%), Gaps = 10/319 (3%)
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
+N ++ +++ D+ G V D +TF V + I+ G D AL I + +
Sbjct: 192 FNVLIKALCRAHQLRPAILMLEDMPSYGLVPDEKTFTTVMQGYIEEGDLDGALRIREQMV 251
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIY--RSLLYGWSVHRN 229
++ CS + +V I++ C +G + A + ++ G P Y +L+ G +
Sbjct: 252 EFGCSWSNVSVNVIVHGFCKEGRVEDALNFIQEMSNQ-DGFFPDQYTFNTLVNGLCKAGH 310
Query: 230 VKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLP 289
VK A I+ M G PD+ YN+ + LC+ G V E + V+ +M + P
Sbjct: 311 VKHAIEIMDVMLQEGYDPDVYTYNSVISGLCKL-------GEVKEAVEVLDQMITRDCSP 363
Query: 290 TSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIV 349
+++YN L+S L K +V+E+ ++ + + G+ PD ++ +++ L L+ E+
Sbjct: 364 NTVTYNTLISTLCKENQVEEATELARVLTSKGILPDVCTFNSLIQGLCLTRNHRVAMELF 423
Query: 350 DQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRG 409
++M KG P+ Y LI LC +++ AL + ++M+ S Y+ LI C+
Sbjct: 424 EEMRSKGCEPDEFTYNMLIDSLCSKGKLDEALNMLKQMELSGCARSVITYNTLIDGFCKA 483
Query: 410 GDFEKGRELWDEATSMGIT 428
+ E++DE G++
Sbjct: 484 NKTREAEEIFDEMEVHGVS 502
Score = 95.5 bits (236), Expect = 3e-18
Identities = 76/302 (25%), Positives = 136/302 (44%), Gaps = 12/302 (3%)
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
YN + ++ L L +G + D TF + + L A+ +F+ +
Sbjct: 368 YNTLISTLCKENQVEEATELARVLTSKGILPDVCTFNSLIQGLCLTRNHRVAMELFEEMR 427
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCI--YRSLLYGWSVHRN 229
C +EFT +I++LCSKG K E + + + +++G + Y +L+ G+
Sbjct: 428 SKGCEPDEFTYNMLIDSLCSKG--KLDEALNMLKQMELSGCARSVITYNTLIDGFCKANK 485
Query: 230 VKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLP 289
+EA I EM+ +GV + V YNT + LC+ S V + +M +M P
Sbjct: 486 TREAEEIFDEMEVHGVSRNSVTYNTLIDGLCK-------SRRVEDAAQLMDQMIMEGQKP 538
Query: 290 TSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIV 349
+YN LL+ + +K++ I++AM ++G PD V+Y ++ L +GR +++
Sbjct: 539 DKYTYNSLLTHFCRGGDIKKAADIVQAMTSNGCEPDIVTYGTLISGLCKAGRVEVASKLL 598
Query: 350 DQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPV-YDVLIPKLCR 408
+ KG+ Y +I L + A+ LF +M + V Y ++ LC
Sbjct: 599 RSIQMKGINLTPHAYNPVIQGLFRKRKTTEAINLFREMLEQNEAPPDAVSYRIVFRGLCN 658
Query: 409 GG 410
GG
Sbjct: 659 GG 660
Score = 92.8 bits (229), Expect = 2e-17
Identities = 95/411 (23%), Positives = 162/411 (39%), Gaps = 45/411 (10%)
Query: 73 SSLVAQAIESSKHEAQTRRLLRFFLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILI 132
SS + ++S + + LR F +SK E Y L + M ++
Sbjct: 47 SSTDVKLLDSLRSQPDDSAALRLFNLASKKPNFSPEPALYEEILLRLGRSGSFDDMKKIL 106
Query: 133 GDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFK-NLDKYKCSTNEFTVTAIINALCS 191
D+K M TF ++ E+ + +DE L + +D++ + ++N L
Sbjct: 107 EDMKSSRCEMGTSTFLILIESYAQFELQDEILSVVDWMIDEFGLKPDTHFYNRMLNLLVD 166
Query: 192 KGHAKRAEGVVLHHKDKITGALPCI--YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDL 249
K E + H K + G P + + L+ ++ A ++++M S G++PD
Sbjct: 167 GNSLKLVE--ISHAKMSVWGIKPDVSTFNVLIKALCRAHQLRPAILMLEDMPSYGLVPDE 224
Query: 250 VCYNTFLRCLCER---------------------NLRHN-------PSGLVPETLNVMME 281
+ T ++ E N+ N G V + LN + E
Sbjct: 225 KTFTTVMQGYIEEGDLDGALRIREQMVEFGCSWSNVSVNVIVHGFCKEGRVEDALNFIQE 284
Query: 282 MRSHK-VLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSG 340
M + P ++N L++ L K VK + +I++ M G PD +Y V+ L G
Sbjct: 285 MSNQDGFFPDQYTFNTLVNGLCKAGHVKHAIEIMDVMLQEGYDPDVYTYNSVISGLCKLG 344
Query: 341 RFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYD 400
+ E++DQMI + PN Y +LI LC +V A EL + S + ++
Sbjct: 345 EVKEAVEVLDQMITRDCSPNTVTYNTLISTLCKENQVEEATELARVLTSKGILPDVCTFN 404
Query: 401 VLIPKLCRGGDFEKGRELWDEATSMG-----------ITLQCSKDILDPSI 440
LI LC + EL++E S G I CSK LD ++
Sbjct: 405 SLIQGLCLTRNHRVAMELFEEMRSKGCEPDEFTYNMLIDSLCSKGKLDEAL 455
>ref|NP_177860.1| pentatricopeptide (PPR) repeat-containing protein [Arabidopsis
thaliana] gi|25406495|pir||F96802 hypothetical protein
F2P24.7 [imported] - Arabidopsis thaliana
gi|11079489|gb|AAG29201.1| hypothetical protein
[Arabidopsis thaliana]
Length = 481
Score = 120 bits (301), Expect = 1e-25
Identities = 82/334 (24%), Positives = 150/334 (44%), Gaps = 10/334 (2%)
Query: 94 RFFLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAEN 153
RFF WS K ++ Y+ + A+ + Y M LI ++K+ ++++ +TF +V
Sbjct: 83 RFFQWSEKQRHYEHSVRAYHMMIESTAKIRQYKLMWDLINAMRKK-KMLNVETFCIVMRK 141
Query: 154 LIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGAL 213
+ K DEA+ F ++KY N +++ALC + ++A+ V + +D+ T
Sbjct: 142 YARAQKVDEAIYAFNVMEKYDLPPNLVAFNGLLSALCKSKNVRKAQEVFENMRDRFTPDS 201
Query: 214 PCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVP 273
Y LL GW N+ +AR + +EM G PD+V Y+ + LC+ +G V
Sbjct: 202 K-TYSILLEGWGKEPNLPKAREVFREMIDAGCHPDIVTYSIMVDILCK-------AGRVD 253
Query: 274 ETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVV 333
E L ++ M PT+ Y++L+ G R++E+ M+ SG+ D + ++
Sbjct: 254 EALGIVRSMDPSICKPTTFIYSVLVHTYGTENRLEEAVDTFLEMERSGMKADVAVFNSLI 313
Query: 334 RVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLG 393
+ R ++ +M KG+ PN K ++ L + A ++F KM
Sbjct: 314 GAFCKANRMKNVYRVLKEMKSKGVTPNSKSCNIILRHLIERGEKDEAFDVFRKM-IKVCE 372
Query: 394 GYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
Y ++I C + E ++W G+
Sbjct: 373 PDADTYTMVIKMFCEKKEMETADKVWKYMRKKGV 406
Score = 78.2 bits (191), Expect = 6e-13
Identities = 83/388 (21%), Positives = 164/388 (41%), Gaps = 28/388 (7%)
Query: 2 KFKMLNSAWKLC-CLRKTRTRNFQLLSASLCSTLQPISAPPHLQELCGIVTSNVGGLDDL 60
K + W L +RK + N + + C ++ + + E I NV DL
Sbjct: 110 KIRQYKLMWDLINAMRKKKMLNVE----TFCIVMRKYARAQKVDE--AIYAFNVMEKYDL 163
Query: 61 ELSLNKFKGSLTSSLVAQAIESSKHEAQTRRLLRFFLWSSKNLRHDLEDNDYNYALRVFA 120
+L F G L++ ++ + ++ + R R + Y+ L +
Sbjct: 164 PPNLVAFNGLLSALCKSKNVRKAQEVFENMRD-----------RFTPDSKTYSILLEGWG 212
Query: 121 EKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEF 180
++ + + ++ G D T+ ++ + L K G+ DEALGI +++D C F
Sbjct: 213 KEPNLPKAREVFREMIDAGCHPDIVTYSIMVDILCKAGRVDEALGIVRSMDPSICKPTTF 272
Query: 181 TVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEM 240
+ +++ ++ + A L + A ++ SL+ + +K R++KEM
Sbjct: 273 IYSVLVHTYGTENRLEEAVDTFLEMERSGMKADVAVFNSLIGAFCKANRMKNVYRVLKEM 332
Query: 241 KSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVL-PTSISYNILLS 299
KS GV P+ N LR L ER G E +V +M KV P + +Y +++
Sbjct: 333 KSKGVTPNSKSCNIILRHLIER-------GEKDEAFDVFRKM--IKVCEPDADTYTMVIK 383
Query: 300 CLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVP 359
+ + ++ + ++ + M+ GV P ++ +++ L K ++++MI G+ P
Sbjct: 384 MFCEKKEMETADKVWKYMRKKGVFPSMHTFSVLINGLCEERTTQKACVLLEEMIEMGIRP 443
Query: 360 NHKFYFSLIGILCGVERVNHALELFEKM 387
+ + L +L ER + L EKM
Sbjct: 444 SGVTFGRLRQLLIKEEREDVLKFLNEKM 471
Score = 64.3 bits (155), Expect = 8e-09
Identities = 53/201 (26%), Positives = 88/201 (43%), Gaps = 8/201 (3%)
Query: 228 RNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKV 287
+ V EA M+ + P+LV +N L LC+ S V + V MR +
Sbjct: 146 QKVDEAIYAFNVMEKYDLPPNLVAFNGLLSALCK-------SKNVRKAQEVFENMRD-RF 197
Query: 288 LPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKE 347
P S +Y+ILL GK + ++ ++ M +G PD V+Y ++V +L +GR +
Sbjct: 198 TPDSKTYSILLEGWGKEPNLPKAREVFREMIDAGCHPDIVTYSIMVDILCKAGRVDEALG 257
Query: 348 IVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLC 407
IV M P Y L+ R+ A++ F +M+ S + V++ LI C
Sbjct: 258 IVRSMDPSICKPTTFIYSVLVHTYGTENRLEEAVDTFLEMERSGMKADVAVFNSLIGAFC 317
Query: 408 RGGDFEKGRELWDEATSMGIT 428
+ + + E S G+T
Sbjct: 318 KANRMKNVYRVLKEMKSKGVT 338
>gb|AAK71569.2| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|50918647|ref|XP_469720.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1833
Score = 120 bits (300), Expect = 1e-25
Identities = 79/318 (24%), Positives = 146/318 (45%), Gaps = 16/318 (5%)
Query: 95 FFLWSSKNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENL 154
FF+W + + Y+ + + + + + M LI + + G +M T V L
Sbjct: 1431 FFMWVGTQGGYCHCADSYDLMVDILGKFKQFDLMWGLINQMVEVGGLMSLMTMTKVMRRL 1490
Query: 155 IKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALP 214
+ EA+ F +D++ + + +++ LC + KRA GV ++ G +P
Sbjct: 1491 AGASRWTEAIDAFHKMDRFGVVKDTKAMNVLLDTLCKERSVKRARGVF----QELRGTIP 1546
Query: 215 C---IYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLC-ERNLRHNPSG 270
+ +L++GW R +KEA ++EMK +G P +V Y + + C E++ +
Sbjct: 1547 PDENSFNTLVHGWCKARMLKEALDTMEEMKQHGFSPSVVTYTSLVEAYCMEKDFQ----- 1601
Query: 271 LVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYY 330
++ EMR + P ++Y IL+ LGK R +E+ + +K GV+PD Y
Sbjct: 1602 ---TVYALLDEMRKRRCPPNVVTYTILMHALGKAGRTREALDTFDKLKEDGVAPDASFYN 1658
Query: 331 LVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSS 390
++ +L +GR +V++M G+ PN + +LI C + +AL+L KM+
Sbjct: 1659 SLIYILGRAGRLEDAYSVVEEMRTTGIAPNVTTFNTLISAACDHSQAENALKLLVKMEEQ 1718
Query: 391 SLGGYGPVYDVLIPKLCR 408
S Y L+ C+
Sbjct: 1719 SCNPDIKTYTPLLKLCCK 1736
Score = 72.4 bits (176), Expect = 3e-11
Identities = 54/259 (20%), Positives = 112/259 (42%), Gaps = 11/259 (4%)
Query: 108 EDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIF 167
++N +N + + + + + ++K+ G T+ + E +
Sbjct: 1548 DENSFNTLVHGWCKARMLKEALDTMEEMKQHGFSPSVVTYTSLVEAYCMEKDFQTVYALL 1607
Query: 168 KNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALP--CIYRSLLYGWS 225
+ K +C N T T +++AL G + E + K K G P Y SL+Y
Sbjct: 1608 DEMRKRRCPPNVVTYTILMHALGKAGRTR--EALDTFDKLKEDGVAPDASFYNSLIYILG 1665
Query: 226 VHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSH 285
+++A +++EM++ G+ P++ +NT + C+ + N L ++++M
Sbjct: 1666 RAGRLEDAYSVVEEMRTTGIAPNVTTFNTLISAACDHSQAEN-------ALKLLVKMEEQ 1718
Query: 286 KVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKG 345
P +Y LL K + VK ++ M +SPD+ +Y L+V L +G+ +
Sbjct: 1719 SCNPDIKTYTPLLKLCCKRQWVKILLFLVCHMFRKDISPDFSTYTLLVSWLCRNGKVAQS 1778
Query: 346 KEIVDQMIGKGLVPNHKFY 364
+++M+ KG P + +
Sbjct: 1779 CLFLEEMVSKGFAPKQETF 1797
Score = 66.6 bits (161), Expect = 2e-09
Identities = 55/196 (28%), Positives = 81/196 (41%), Gaps = 8/196 (4%)
Query: 232 EARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTS 291
EA +M GV+ D N L LC+ V V E+R + P
Sbjct: 1498 EAIDAFHKMDRFGVVKDTKAMNVLLDTLCKERS-------VKRARGVFQELRG-TIPPDE 1549
Query: 292 ISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQ 351
S+N L+ K R +KE+ +E MK G SP V+Y +V + F ++D+
Sbjct: 1550 NSFNTLVHGWCKARMLKEALDTMEEMKQHGFSPSVVTYTSLVEAYCMEKDFQTVYALLDE 1609
Query: 352 MIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGD 411
M + PN Y L+ L R AL+ F+K+K + Y+ LI L R G
Sbjct: 1610 MRKRRCPPNVVTYTILMHALGKAGRTREALDTFDKLKEDGVAPDASFYNSLIYILGRAGR 1669
Query: 412 FEKGRELWDEATSMGI 427
E + +E + GI
Sbjct: 1670 LEDAYSVVEEMRTTGI 1685
>dbj|BAB09050.1| unnamed protein product [Arabidopsis thaliana]
gi|15239161|ref|NP_201383.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
Length = 637
Score = 118 bits (295), Expect = 5e-25
Identities = 85/370 (22%), Positives = 170/370 (44%), Gaps = 11/370 (2%)
Query: 60 LELSLNKFKGSLTSSLVAQAIESSKHEAQTRRLLRFFLWSSKNLRHDLEDNDYNYALRVF 119
LEL+LN+ L L+ + + RFF+W++K R+ Y +++
Sbjct: 100 LELALNESGVELRPGLIERVLNRCGDAGNLG--YRFFVWAAKQPRYCHSIEVYKSMVKIL 157
Query: 120 AEKQDYTSMDILIGDLKKEG-RVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTN 178
++ + + ++ LI +++KE ++++ + F ++ + +A+ + + K+ +
Sbjct: 158 SKMRQFGAVWGLIEEMRKENPQLIEPELFVVLVQRFASADMVKKAIEVLDEMPKFGFEPD 217
Query: 179 EFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIK 238
E+ +++ALC G K A + + + L + SLLYGW + EA+ ++
Sbjct: 218 EYVFGCLLDALCKHGSVKDAAKLFEDMRMRFPVNLR-YFTSLLYGWCRVGKMMEAKYVLV 276
Query: 239 EMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILL 298
+M G PD+V Y L +G + + +++ +MR P + Y +L+
Sbjct: 277 QMNEAGFEPDIVDYTNLLSGYAN-------AGKMADAYDLLRDMRRRGFEPNANCYTVLI 329
Query: 299 SCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLV 358
L K R++E+ ++ M+ D V+Y +V G+ K ++D MI KGL+
Sbjct: 330 QALCKVDRMEEAMKVFVEMERYECEADVVTYTALVSGFCKWGKIDKCYIVLDDMIKKGLM 389
Query: 359 PNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGREL 418
P+ Y ++ E LEL EKM+ +Y+V+I C+ G+ ++ L
Sbjct: 390 PSELTYMHIMVAHEKKESFEECLELMEKMRQIEYHPDIGIYNVVIRLACKLGEVKEAVRL 449
Query: 419 WDEATSMGIT 428
W+E G++
Sbjct: 450 WNEMEENGLS 459
>ref|XP_477613.1| putative fertility restorer homologue [Oryza sativa (japonica
cultivar-group)] gi|50509027|dbj|BAD31989.1| putative
fertility restorer [Oryza sativa (japonica
cultivar-group)] gi|34394343|dbj|BAC84898.1| putative
fertility restorer homologue [Oryza sativa (japonica
cultivar-group)]
Length = 1013
Score = 117 bits (294), Expect = 6e-25
Identities = 78/316 (24%), Positives = 158/316 (49%), Gaps = 7/316 (2%)
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
Y+ + + + +++ + ++ ++++ G ++ T+ ++ L + G +EA G K+++
Sbjct: 247 YSTLIEAYCKVREFDTAKKVLVEMRERGCGLNTVTYNVLIAGLCRSGAVEEAFGFKKDME 306
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVK 231
Y + FT A+IN LC + A+ ++ +Y +L+ G+ N
Sbjct: 307 DYGLVPDGFTYGALINGLCKSRRSNEAKALLDEMSCAELKPNVVVYANLIDGFMREGNAD 366
Query: 232 EARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTS 291
EA ++IKEM + GV P+ + Y+ +R LC+ S L+ + M SH+ P +
Sbjct: 367 EAFKMIKEMVAAGVQPNKITYDNLVRGLCKMGQMDRASLLLKQ-----MVRDSHR--PDT 419
Query: 292 ISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQ 351
I+YN+++ + K++ ++L M+ +G+SP+ +Y +++ L SG K +++++
Sbjct: 420 ITYNLIIEGHFRHHSKKDAFRLLSEMENAGISPNVYTYSIMIHGLCQSGEPEKASDLLEE 479
Query: 352 MIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGD 411
M KGL PN Y LI C V+ A E+F+KM ++ Y+ LI L + G
Sbjct: 480 MTTKGLKPNAFVYAPLISGYCREGNVSLACEIFDKMTKVNVLPDLYCYNSLIFGLSKVGR 539
Query: 412 FEKGRELWDEATSMGI 427
E+ + + + G+
Sbjct: 540 VEESTKYFAQMQERGL 555
Score = 92.0 bits (227), Expect = 4e-17
Identities = 75/281 (26%), Positives = 125/281 (43%), Gaps = 11/281 (3%)
Query: 143 DAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVV 202
D + + L K+G+ +E+ F + + NEFT + +I+ G + AE +V
Sbjct: 523 DLYCYNSLIFGLSKVGRVEESTKYFAQMQERGLLPNEFTYSGLIHGYLKNGDLESAEQLV 582
Query: 203 LHHKDKITGALP--CIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLC 260
D TG P IY LL + ++++ K M GV+ D Y + L
Sbjct: 583 QRMLD--TGLKPNDVIYIDLLESYFKSDDIEKVSSTFKSMLDQGVMLDNRIYGILIHNL- 639
Query: 261 ERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTS 320
+ SG + V+ + + +P Y+ L+S L KT +++ IL+ M
Sbjct: 640 ------SSSGNMEAAFRVLSGIEKNGSVPDVHVYSSLISGLCKTADREKAFGILDEMSKK 693
Query: 321 GVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHA 380
GV P+ V Y ++ L SG + + + ++ KGLVPN Y SLI C V +++A
Sbjct: 694 GVDPNIVCYNALIDGLCKSGDISYARNVFNSILAKGLVPNCVTYTSLIDGSCKVGDISNA 753
Query: 381 LELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDE 421
L+ +M ++ + VY VL GD E+ L +E
Sbjct: 754 FYLYNEMLATGITPDAFVYSVLTTGCSSAGDLEQAMFLIEE 794
Score = 89.0 bits (219), Expect = 3e-16
Identities = 73/355 (20%), Positives = 138/355 (38%), Gaps = 42/355 (11%)
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
Y+ +R + +L+ + ++ D T+ L+ E + + +A + ++
Sbjct: 387 YDNLVRGLCKMGQMDRASLLLKQMVRDSHRPDTITYNLIIEGHFRHHSKKDAFRLLSEME 446
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVK 231
S N +T + +I+ LC G ++A ++ K +Y L+ G+ NV
Sbjct: 447 NAGISPNVYTYSIMIHGLCQSGEPEKASDLLEEMTTKGLKPNAFVYAPLISGYCREGNVS 506
Query: 232 EARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTS 291
A I +M V+PDL CYN+ + L + G V E+ +M+ +LP
Sbjct: 507 LACEIFDKMTKVNVLPDLYCYNSLIFGLSK-------VGRVEESTKYFAQMQERGLLPNE 559
Query: 292 ISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQ 351
+Y+ L+ K ++ + Q+++ M +G+ P+ V Y ++ F S K
Sbjct: 560 FTYSGLIHGYLKNGDLESAEQLVQRMLDTGLKPNDVIYIDLLESYFKSDDIEKVSSTFKS 619
Query: 352 MIGKGL-----------------------------------VPNHKFYFSLIGILCGVER 376
M+ +G+ VP+ Y SLI LC
Sbjct: 620 MLDQGVMLDNRIYGILIHNLSSSGNMEAAFRVLSGIEKNGSVPDVHVYSSLISGLCKTAD 679
Query: 377 VNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGITLQC 431
A + ++M + Y+ LI LC+ GD R +++ + G+ C
Sbjct: 680 REKAFGILDEMSKKGVDPNIVCYNALIDGLCKSGDISYARNVFNSILAKGLVPNC 734
Score = 84.3 bits (207), Expect = 8e-15
Identities = 66/296 (22%), Positives = 116/296 (38%), Gaps = 40/296 (13%)
Query: 152 ENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVL------HH 205
+ ++ G DEA + K + N+ T ++ LC G RA ++ H
Sbjct: 357 DGFMREGNADEAFKMIKEMVAAGVQPNKITYDNLVRGLCKMGQMDRASLLLKQMVRDSHR 416
Query: 206 KDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLR 265
D IT Y ++ G H + K+A R++ EM++ G+ P++ Y+ + LC+
Sbjct: 417 PDTIT------YNLIIEGHFRHHSKKDAFRLLSEMENAGISPNVYTYSIMIHGLCQSGEP 470
Query: 266 HNPSGLVPE----------------------------TLNVMMEMRSHKVLPTSISYNIL 297
S L+ E + +M VLP YN L
Sbjct: 471 EKASDLLEEMTTKGLKPNAFVYAPLISGYCREGNVSLACEIFDKMTKVNVLPDLYCYNSL 530
Query: 298 LSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGL 357
+ L K RV+ES + M+ G+ P+ +Y ++ +G +++V +M+ GL
Sbjct: 531 IFGLSKVGRVEESTKYFAQMQERGLLPNEFTYSGLIHGYLKNGDLESAEQLVQRMLDTGL 590
Query: 358 VPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFE 413
PN Y L+ + + F+ M + +Y +LI L G+ E
Sbjct: 591 KPNDVIYIDLLESYFKSDDIEKVSSTFKSMLDQGVMLDNRIYGILIHNLSSSGNME 646
Score = 84.0 bits (206), Expect = 1e-14
Identities = 72/316 (22%), Positives = 140/316 (43%), Gaps = 23/316 (7%)
Query: 128 MDILIGDLKKEGRVMDAQTFGLVAEN------------LIKLGKEDEALGIFKNLDKYKC 175
+D+L+ KK GRV DA L+ + L+K +A+ + + ++
Sbjct: 177 LDVLVDTYKKSGRVQDAAEVVLMMRDRGMAPSIRCCNALLKDLLRADAMALLWKVREFMV 236
Query: 176 ----STNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVK 231
S + +T + +I A C A+ V++ +++ G Y L+ G V+
Sbjct: 237 GAGISPDVYTYSTLIEAYCKVREFDTAKKVLVEMRERGCGLNTVTYNVLIAGLCRSGAVE 296
Query: 232 EARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTS 291
EA K+M+ G++PD Y + LC ++ R N E ++ EM ++ P
Sbjct: 297 EAFGFKKDMEDYGLVPDGFTYGALINGLC-KSRRSN------EAKALLDEMSCAELKPNV 349
Query: 292 ISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQ 351
+ Y L+ + E+ ++++ M +GV P+ ++Y +VR L G+ + ++ Q
Sbjct: 350 VVYANLIDGFMREGNADEAFKMIKEMVAAGVQPNKITYDNLVRGLCKMGQMDRASLLLKQ 409
Query: 352 MIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGD 411
M+ P+ Y +I A L +M+++ + Y ++I LC+ G+
Sbjct: 410 MVRDSHRPDTITYNLIIEGHFRHHSKKDAFRLLSEMENAGISPNVYTYSIMIHGLCQSGE 469
Query: 412 FEKGRELWDEATSMGI 427
EK +L +E T+ G+
Sbjct: 470 PEKASDLLEEMTTKGL 485
Score = 76.6 bits (187), Expect = 2e-12
Identities = 70/330 (21%), Positives = 138/330 (41%), Gaps = 36/330 (10%)
Query: 131 LIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALC 190
L+ +++ G + T+ ++ L + G+ ++A + + + N F +I+ C
Sbjct: 441 LLSEMENAGISPNVYTYSIMIHGLCQSGEPEKASDLLEEMTTKGLKPNAFVYAPLISGYC 500
Query: 191 SKGHAKRAEGVVLHHKDKIT--GALPCIY--RSLLYGWSVHRNVKEARRIIKEMKSNGVI 246
+G+ A + DK+T LP +Y SL++G S V+E+ + +M+ G++
Sbjct: 501 REGNVSLACEIF----DKMTKVNVLPDLYCYNSLIFGLSKVGRVEESTKYFAQMQERGLL 556
Query: 247 PDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMME------------------------- 281
P+ Y+ + + + LV L+ ++
Sbjct: 557 PNEFTYSGLIHGYLKNGDLESAEQLVQRMLDTGLKPNDVIYIDLLESYFKSDDIEKVSST 616
Query: 282 ---MRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFL 338
M V+ + Y IL+ L + ++ + ++L ++ +G PD Y ++ L
Sbjct: 617 FKSMLDQGVMLDNRIYGILIHNLSSSGNMEAAFRVLSGIEKNGSVPDVHVYSSLISGLCK 676
Query: 339 SGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPV 398
+ K I+D+M KG+ PN Y +LI LC +++A +F + + L
Sbjct: 677 TADREKAFGILDEMSKKGVDPNIVCYNALIDGLCKSGDISYARNVFNSILAKGLVPNCVT 736
Query: 399 YDVLIPKLCRGGDFEKGRELWDEATSMGIT 428
Y LI C+ GD L++E + GIT
Sbjct: 737 YTSLIDGSCKVGDISNAFYLYNEMLATGIT 766
Score = 67.4 bits (163), Expect = 1e-09
Identities = 63/346 (18%), Positives = 143/346 (41%), Gaps = 28/346 (8%)
Query: 107 LEDNDYNYA--LRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEAL 164
L+ ND Y L + + D + + +G ++D + +G++ NL G + A
Sbjct: 590 LKPNDVIYIDLLESYFKSDDIEKVSSTFKSMLDQGVMLDNRIYGILIHNLSSSGNMEAAF 649
Query: 165 GIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGW 224
+ ++K + +++I+ LC ++A G++ K Y +L+ G
Sbjct: 650 RVLSGIEKNGSVPDVHVYSSLISGLCKTADREKAFGILDEMSKKGVDPNIVCYNALIDGL 709
Query: 225 SVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRS 284
++ AR + + + G++P+ V Y + + C+ G + + EM +
Sbjct: 710 CKSGDISYARNVFNSILAKGLVPNCVTYTSLIDGSCK-------VGDISNAFYLYNEMLA 762
Query: 285 HKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGK 344
+ P + Y++L + ++++ ++E M G + S+ +V G+ +
Sbjct: 763 TGITPDAFVYSVLTTGCSSAGDLEQAMFLIEEMFLRGHA-SISSFNNLVDGFCKRGKMQE 821
Query: 345 GKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHA----LELFEKMKSSSLGGYGPVY- 399
+++ ++G+GLVPN ++I L +++ +EL +K S+ + ++
Sbjct: 822 TLKLLHVIMGRGLVPNALTIENIISGLSEAGKLSEVHTIFVELQQKTSESAARHFSSLFM 881
Query: 400 -------------DVLIPKLCRGGDFEKGRELWDEATSMGITLQCS 432
D +I C+ G+ +K L D + + CS
Sbjct: 882 DMINQGKIPLDVVDDMIRDHCKEGNLDKALMLRDVIVAKSAPMGCS 927
Score = 62.0 bits (149), Expect = 4e-08
Identities = 58/241 (24%), Positives = 105/241 (43%), Gaps = 9/241 (3%)
Query: 214 PCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVP 273
P + L+ + V++A ++ M+ G+ P + C N L+ L LR + L+
Sbjct: 174 PAVLDVLVDTYKKSGRVQDAAEVVLMMRDRGMAPSIRCCNALLKDL----LRADAMALLW 229
Query: 274 ETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVV 333
+ M+ + P +Y+ L+ K R + ++L M+ G + V+Y +++
Sbjct: 230 KVREFMV---GAGISPDVYTYSTLIEAYCKVREFDTAKKVLVEMRERGCGLNTVTYNVLI 286
Query: 334 RVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLG 393
L SG + M GLVP+ Y +LI LC R N A L ++M + L
Sbjct: 287 AGLCRSGAVEEAFGFKKDMEDYGLVPDGFTYGALINGLCKSRRSNEAKALLDEMSCAELK 346
Query: 394 GYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGITLQCSKDILDPSITEVFKPVRPEKIS 453
VY LI R G+ ++ ++ E + G+ Q +K D + + K + ++ S
Sbjct: 347 PNVVVYANLIDGFMREGNADEAFKMIKEMVAAGV--QPNKITYDNLVRGLCKMGQMDRAS 404
Query: 454 L 454
L
Sbjct: 405 L 405
>gb|AAF75803.1| Contains weak similarity to leaf protein from Ipomea nil gb|D85101
and contains a RepB PF|01051 protein and multiple PPR
PF|01535 repeats. [Arabidopsis thaliana]
gi|25404380|pir||H96653 hypothetical protein F16P17.7
[imported] - Arabidopsis thaliana
Length = 613
Score = 115 bits (287), Expect = 4e-24
Identities = 69/282 (24%), Positives = 130/282 (45%), Gaps = 7/282 (2%)
Query: 146 TFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHH 205
TF + L K EA+ + + C + FT ++N LC +G A ++
Sbjct: 171 TFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLKKM 230
Query: 206 KDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLR 265
+ A IY +++ ++NV +A + EM + G+ P++V YN+ +RCLC
Sbjct: 231 EKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNY--- 287
Query: 266 HNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPD 325
G + ++ +M K+ P ++++ L+ K ++ E+ ++ + M + PD
Sbjct: 288 ----GRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPD 343
Query: 326 WVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFE 385
+Y ++ + R + K + + MI K PN Y +LI C +RV +ELF
Sbjct: 344 IFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFR 403
Query: 386 KMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
+M L G Y+ LI L + GD + ++++ + S G+
Sbjct: 404 EMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGV 445
Score = 102 bits (254), Expect = 3e-20
Identities = 78/329 (23%), Positives = 147/329 (43%), Gaps = 11/329 (3%)
Query: 101 KNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKE 160
+NLR + YN + F + ++G + K G D T + +
Sbjct: 91 QNLRISYDLYSYNILINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRI 150
Query: 161 DEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCI--YR 218
EA+ + + + N T +I+ L H K +E V L + G P + Y
Sbjct: 151 SEAVALVDQMFVMEYQPNTVTFNTLIHGLFL--HNKASEAVALIDRMVARGCQPDLFTYG 208
Query: 219 SLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNV 278
+++ G ++ A ++K+M+ + D+V Y T + LC N ++ V + LN+
Sbjct: 209 TVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYTTIIDALC--NYKN-----VNDALNL 261
Query: 279 MMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFL 338
EM + + P ++YN L+ CL R ++ ++L M ++P+ V++ ++
Sbjct: 262 FTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVK 321
Query: 339 SGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPV 398
G+ + +++ D+MI + + P+ Y SLI C +R++ A +FE M S
Sbjct: 322 EGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVT 381
Query: 399 YDVLIPKLCRGGDFEKGRELWDEATSMGI 427
Y+ LI C+ E+G EL+ E + G+
Sbjct: 382 YNTLIKGFCKAKRVEEGMELFREMSQRGL 410
Score = 96.3 bits (238), Expect = 2e-18
Identities = 76/317 (23%), Positives = 140/317 (43%), Gaps = 11/317 (3%)
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
YN +R ++ L+ D+ + + TF + + +K GK EA ++ +
Sbjct: 277 YNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMI 336
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGV--VLHHKDKITGALPCIYRSLLYGWSVHRN 229
K + FT +++IN C A+ + ++ KD + Y +L+ G+ +
Sbjct: 337 KRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVT--YNTLIKGFCKAKR 394
Query: 230 VKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLP 289
V+E + +EM G++ + V YNT ++ L + +G + +M S V P
Sbjct: 395 VEEGMELFREMSQRGLVGNTVTYNTLIQGLFQ-------AGDCDMAQKIFKKMVSDGVPP 447
Query: 290 TSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIV 349
I+Y+ILL L K +++++ + E ++ S + PD +Y +++ + +G+ G ++
Sbjct: 448 DIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWDLF 507
Query: 350 DQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRG 409
+ KG+ PN Y ++I C A LF +MK Y+ LI R
Sbjct: 508 CSLSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIRARLRD 567
Query: 410 GDFEKGRELWDEATSMG 426
GD EL E S G
Sbjct: 568 GDKAASAELIKEMRSCG 584
Score = 95.5 bits (236), Expect = 3e-18
Identities = 79/332 (23%), Positives = 140/332 (41%), Gaps = 42/332 (12%)
Query: 131 LIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALC 190
LI + G D T+G V L K G D AL + K ++K K + T II+ALC
Sbjct: 191 LIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYTTIIDALC 250
Query: 191 SKGHAKRAEGVVLHHKDKITGALPCIYRSLL-----YG-WS---------VHRNVK---- 231
+ + A + +K Y SL+ YG WS + R +
Sbjct: 251 NYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVV 310
Query: 232 ----------------EARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPET 275
EA ++ EM + PD+ Y++ + C + + E
Sbjct: 311 TFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDR-------LDEA 363
Query: 276 LNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRV 335
++ M S P ++YN L+ K +RV+E ++ M G+ + V+Y +++
Sbjct: 364 KHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNTLIQG 423
Query: 336 LFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGY 395
LF +G ++I +M+ G+ P+ Y L+ LC ++ AL +FE ++ S +
Sbjct: 424 LFQAGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPD 483
Query: 396 GPVYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
Y+++I +C+ G E G +L+ + G+
Sbjct: 484 IYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGV 515
Score = 67.4 bits (163), Expect = 1e-09
Identities = 43/190 (22%), Positives = 88/190 (45%), Gaps = 7/190 (3%)
Query: 238 KEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNIL 297
+ M++ + DL YN + C C R+ +P L V+ +M P ++ + L
Sbjct: 88 ERMQNLRISYDLYSYNILINCFCRRS-------QLPLALAVLGKMMKLGYEPDIVTLSSL 140
Query: 298 LSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGL 357
L+ +R+ E+ +++ M P+ V++ ++ LFL + + ++D+M+ +G
Sbjct: 141 LNGYCHGKRISEAVALVDQMFVMEYQPNTVTFNTLIHGLFLHNKASEAVALIDRMVARGC 200
Query: 358 VPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRE 417
P+ Y +++ LC ++ AL L +KM+ + +Y +I LC +
Sbjct: 201 QPDLFTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALN 260
Query: 418 LWDEATSMGI 427
L+ E + GI
Sbjct: 261 LFTEMDNKGI 270
Score = 57.8 bits (138), Expect = 8e-07
Identities = 50/234 (21%), Positives = 99/234 (41%), Gaps = 10/234 (4%)
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
YN ++ F + + L ++ + G V + T+ + + L + G D A IFK +
Sbjct: 382 YNTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMV 441
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLH-HKDKITGALPCIYRSLLYGWSVHRNV 230
+ T + +++ LC G ++A V + K K+ + Y ++ G V
Sbjct: 442 SDGVPPDIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDI-YTYNIMIEGMCKAGKV 500
Query: 231 KEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPT 290
++ + + GV P+++ Y T + C + L+ L EM+ LP
Sbjct: 501 EDGWDLFCSLSLKGVKPNVIIYTTMISGFCRKGLKEEADAL-------FREMKEDGTLPN 553
Query: 291 SISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGK 344
S +YN L+ + S ++++ M++ G D + +V+ +L GR K
Sbjct: 554 SGTYNTLIRARLRDGDKAASAELIKEMRSCGFVGDASTISMVINMLH-DGRLEK 606
Score = 40.8 bits (94), Expect = 0.099
Identities = 25/114 (21%), Positives = 54/114 (46%)
Query: 274 ETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVV 333
+ +++ EM + LP+ + +N LLS + K + + E M+ +S D SY +++
Sbjct: 47 DAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYNILI 106
Query: 334 RVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKM 387
+ ++ +M+ G P+ SL+ C +R++ A+ L ++M
Sbjct: 107 NCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQM 160
>ref|NP_176481.1| pentatricopeptide (PPR) repeat-containing protein [Arabidopsis
thaliana]
Length = 659
Score = 115 bits (287), Expect = 4e-24
Identities = 69/282 (24%), Positives = 130/282 (45%), Gaps = 7/282 (2%)
Query: 146 TFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHH 205
TF + L K EA+ + + C + FT ++N LC +G A ++
Sbjct: 187 TFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLKKM 246
Query: 206 KDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLR 265
+ A IY +++ ++NV +A + EM + G+ P++V YN+ +RCLC
Sbjct: 247 EKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNY--- 303
Query: 266 HNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPD 325
G + ++ +M K+ P ++++ L+ K ++ E+ ++ + M + PD
Sbjct: 304 ----GRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPD 359
Query: 326 WVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFE 385
+Y ++ + R + K + + MI K PN Y +LI C +RV +ELF
Sbjct: 360 IFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFR 419
Query: 386 KMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
+M L G Y+ LI L + GD + ++++ + S G+
Sbjct: 420 EMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGV 461
Score = 102 bits (254), Expect = 3e-20
Identities = 78/329 (23%), Positives = 147/329 (43%), Gaps = 11/329 (3%)
Query: 101 KNLRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKE 160
+NLR + YN + F + ++G + K G D T + +
Sbjct: 107 QNLRISYDLYSYNILINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRI 166
Query: 161 DEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCI--YR 218
EA+ + + + N T +I+ L H K +E V L + G P + Y
Sbjct: 167 SEAVALVDQMFVMEYQPNTVTFNTLIHGLFL--HNKASEAVALIDRMVARGCQPDLFTYG 224
Query: 219 SLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNV 278
+++ G ++ A ++K+M+ + D+V Y T + LC N ++ V + LN+
Sbjct: 225 TVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYTTIIDALC--NYKN-----VNDALNL 277
Query: 279 MMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFL 338
EM + + P ++YN L+ CL R ++ ++L M ++P+ V++ ++
Sbjct: 278 FTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVK 337
Query: 339 SGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPV 398
G+ + +++ D+MI + + P+ Y SLI C +R++ A +FE M S
Sbjct: 338 EGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVT 397
Query: 399 YDVLIPKLCRGGDFEKGRELWDEATSMGI 427
Y+ LI C+ E+G EL+ E + G+
Sbjct: 398 YNTLIKGFCKAKRVEEGMELFREMSQRGL 426
Score = 97.4 bits (241), Expect = 9e-19
Identities = 83/353 (23%), Positives = 153/353 (42%), Gaps = 18/353 (5%)
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
YN +R ++ L+ D+ + + TF + + +K GK EA ++ +
Sbjct: 293 YNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMI 352
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGV--VLHHKDKITGALPCIYRSLLYGWSVHRN 229
K + FT +++IN C A+ + ++ KD + Y +L+ G+ +
Sbjct: 353 KRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVT--YNTLIKGFCKAKR 410
Query: 230 VKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLP 289
V+E + +EM G++ + V YNT ++ L + +G + +M S V P
Sbjct: 411 VEEGMELFREMSQRGLVGNTVTYNTLIQGLFQ-------AGDCDMAQKIFKKMVSDGVPP 463
Query: 290 TSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIV 349
I+Y+ILL L K +++++ + E ++ S + PD +Y +++ + +G+ G ++
Sbjct: 464 DIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWDLF 523
Query: 350 DQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRG 409
+ KG+ PN Y ++I C A LF +MK Y+ LI R
Sbjct: 524 CSLSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIRARLRD 583
Query: 410 GDFEKGRELWDEATSMG-------ITLQCSKDILDPSITEVFKPVRPEKISLA 455
GD EL E S G I++ +L SI E +P + +A
Sbjct: 584 GDKAASAELIKEMRSCGFVGDASTISMLSGNAVLKQSILEKRIGEQPSSLVIA 636
Score = 95.5 bits (236), Expect = 3e-18
Identities = 79/332 (23%), Positives = 140/332 (41%), Gaps = 42/332 (12%)
Query: 131 LIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALC 190
LI + G D T+G V L K G D AL + K ++K K + T II+ALC
Sbjct: 207 LIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYTTIIDALC 266
Query: 191 SKGHAKRAEGVVLHHKDKITGALPCIYRSLL-----YG-WS---------VHRNVK---- 231
+ + A + +K Y SL+ YG WS + R +
Sbjct: 267 NYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVV 326
Query: 232 ----------------EARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPET 275
EA ++ EM + PD+ Y++ + C + + E
Sbjct: 327 TFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDR-------LDEA 379
Query: 276 LNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRV 335
++ M S P ++YN L+ K +RV+E ++ M G+ + V+Y +++
Sbjct: 380 KHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNTLIQG 439
Query: 336 LFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGY 395
LF +G ++I +M+ G+ P+ Y L+ LC ++ AL +FE ++ S +
Sbjct: 440 LFQAGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPD 499
Query: 396 GPVYDVLIPKLCRGGDFEKGRELWDEATSMGI 427
Y+++I +C+ G E G +L+ + G+
Sbjct: 500 IYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGV 531
Score = 67.4 bits (163), Expect = 1e-09
Identities = 43/190 (22%), Positives = 88/190 (45%), Gaps = 7/190 (3%)
Query: 238 KEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNIL 297
+ M++ + DL YN + C C R+ +P L V+ +M P ++ + L
Sbjct: 104 ERMQNLRISYDLYSYNILINCFCRRS-------QLPLALAVLGKMMKLGYEPDIVTLSSL 156
Query: 298 LSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGL 357
L+ +R+ E+ +++ M P+ V++ ++ LFL + + ++D+M+ +G
Sbjct: 157 LNGYCHGKRISEAVALVDQMFVMEYQPNTVTFNTLIHGLFLHNKASEAVALIDRMVARGC 216
Query: 358 VPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRE 417
P+ Y +++ LC ++ AL L +KM+ + +Y +I LC +
Sbjct: 217 QPDLFTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALN 276
Query: 418 LWDEATSMGI 427
L+ E + GI
Sbjct: 277 LFTEMDNKGI 286
Score = 40.8 bits (94), Expect = 0.099
Identities = 25/114 (21%), Positives = 54/114 (46%)
Query: 274 ETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVV 333
+ +++ EM + LP+ + +N LLS + K + + E M+ +S D SY +++
Sbjct: 63 DAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYNILI 122
Query: 334 RVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKM 387
+ ++ +M+ G P+ SL+ C +R++ A+ L ++M
Sbjct: 123 NCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQM 176
>dbj|BAB01242.1| unnamed protein product [Arabidopsis thaliana]
gi|15228825|ref|NP_188906.1| pentatricopeptide (PPR)
repeat-containing protein [Arabidopsis thaliana]
Length = 562
Score = 114 bits (285), Expect = 7e-24
Identities = 88/370 (23%), Positives = 166/370 (44%), Gaps = 15/370 (4%)
Query: 43 LQELCGIVTSNVGGLDDLELSLNKFKGSLTSSLVAQAIESSKHEAQTRRLLRFFLWSSKN 102
+ ++C + +D+ L+K +T SLV Q + + + FF+W++
Sbjct: 102 IDKVCDFLNKKDTSHEDVVKELSKCDVVVTESLVLQVLR--RFSNGWNQAYGFFIWANSQ 159
Query: 103 LRHDLEDNDYNYALRVFAEKQDYTSMDILIGDLKK--EGRVMDAQTFGLVAENLIKLGKE 160
+ + YN + V + +++ M L+ ++ K E +++ T V L K GK
Sbjct: 160 TGYVHSGHTYNAMVDVLGKCRNFDLMWELVNEMNKNEESKLVTLDTMSKVMRRLAKSGKY 219
Query: 161 DEALGIFKNLDK-YKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRS 219
++A+ F ++K Y T+ + ++++AL + + A V L D I +
Sbjct: 220 NKAVDAFLEMEKSYGVKTDTIAMNSLMDALVKENSIEHAHEVFLKLFDTIKPDART-FNI 278
Query: 220 LLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVM 279
L++G+ R +AR ++ MK PD+V Y +F+ C+ G ++
Sbjct: 279 LIHGFCKARKFDDARAMMDLMKVTEFTPDVVTYTSFVEAYCKE-------GDFRRVNEML 331
Query: 280 MEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLS 339
EMR + P ++Y I++ LGK+++V E+ + E MK G PD Y ++ +L +
Sbjct: 332 EEMRENGCNPNVVTYTIVMHSLGKSKQVAEALGVYEKMKEDGCVPDAKFYSSLIHILSKT 391
Query: 340 GRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVY 399
GRF EI + M +G+ + Y ++I R AL L ++M+ P
Sbjct: 392 GRFKDAAEIFEDMTNQGVRRDVLVYNTMISAALHHSRDEMALRLLKRMEDEEGESCSPNV 451
Query: 400 DVLIP--KLC 407
+ P K+C
Sbjct: 452 ETYAPLLKMC 461
Score = 69.3 bits (168), Expect = 3e-10
Identities = 73/317 (23%), Positives = 125/317 (39%), Gaps = 61/317 (19%)
Query: 100 SKNLRHDLEDNDYNYALRVFAE-------KQDYTSMDILIGDLKKEGRVM---------- 142
SK +R + YN A+ F E K D +M+ L+ L KE +
Sbjct: 207 SKVMRRLAKSGKYNKAVDAFLEMEKSYGVKTDTIAMNSLMDALVKENSIEHAHEVFLKLF 266
Query: 143 -----DAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKR 197
DA+TF ++ K K D+A + + + + + T T+ + A C +G +R
Sbjct: 267 DTIKPDARTFNILIHGFCKARKFDDARAMMDLMKVTEFTPDVVTYTSFVEAYCKEGDFRR 326
Query: 198 ---------------------------------AEGVVLHHKDKITGALPC--IYRSLLY 222
AE + ++ K K G +P Y SL++
Sbjct: 327 VNEMLEEMRENGCNPNVVTYTIVMHSLGKSKQVAEALGVYEKMKEDGCVPDAKFYSSLIH 386
Query: 223 GWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEM 282
S K+A I ++M + GV D++ YNT + L H+ + L M +
Sbjct: 387 ILSKTGRFKDAAEIFEDMTNQGVRRDVLVYNTMISAA----LHHSRDEMALRLLKRMEDE 442
Query: 283 RSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRF 342
P +Y LL +++K +L M + VS D +Y L++R L +SG+
Sbjct: 443 EGESCSPNVETYAPLLKMCCHKKKMKLLGILLHHMVKNDVSIDVSTYILLIRGLCMSGKV 502
Query: 343 GKGKEIVDQMIGKGLVP 359
+ ++ + KG+VP
Sbjct: 503 EEACLFFEEAVRKGMVP 519
Score = 65.5 bits (158), Expect = 4e-09
Identities = 47/187 (25%), Positives = 88/187 (46%), Gaps = 8/187 (4%)
Query: 241 KSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSC 300
KS GV D + N+ + L + N + V +++ + P + ++NIL+
Sbjct: 231 KSYGVKTDTIAMNSLMDALVKENS-------IEHAHEVFLKLFD-TIKPDARTFNILIHG 282
Query: 301 LGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPN 360
K R+ ++ +++ MK + +PD V+Y V G F + E++++M G PN
Sbjct: 283 FCKARKFDDARAMMDLMKVTEFTPDVVTYTSFVEAYCKEGDFRRVNEMLEEMRENGCNPN 342
Query: 361 HKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWD 420
Y ++ L ++V AL ++EKMK Y LI L + G F+ E+++
Sbjct: 343 VVTYTIVMHSLGKSKQVAEALGVYEKMKEDGCVPDAKFYSSLIHILSKTGRFKDAAEIFE 402
Query: 421 EATSMGI 427
+ T+ G+
Sbjct: 403 DMTNQGV 409
>gb|AAF26800.1| hypothetical protein [Arabidopsis thaliana]
Length = 572
Score = 114 bits (284), Expect = 9e-24
Identities = 97/434 (22%), Positives = 190/434 (43%), Gaps = 24/434 (5%)
Query: 6 LNSAWKLCCLRKTRTRNFQLLSASLCSTLQPISAPPHLQELCGIVTSNVGGLDDLELSLN 65
LN + + T +N S L TL S +E+ ++ G D +
Sbjct: 80 LNPSSSSIAIFSTFIKNLSTASEQLPETLDEYSQS---EEIWNVIVGRDGDRDSEDDVFK 136
Query: 66 KFKG-------SLTSSLVAQAIESSKHEAQTRRLLRFFLWSSKNLRHDLEDNDYNYALRV 118
+ +L+ LV + + + + R L W+ H + Y+ A+ +
Sbjct: 137 RLSSDEICKRVNLSDGLVHKLLHRFRDD--WRSALGILKWAESCKGHKHSSDAYDMAVDI 194
Query: 119 FAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTN 178
+ + + M + ++ + +++ T + G+ +EA+GIF L ++ N
Sbjct: 195 LGKAKKWDRMKEFVERMRGD-KLVTLNTVAKIMRRFAGAGEWEEAVGIFDRLGEFGLEKN 253
Query: 179 EFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIK 238
++ +++ LC + ++A V+L K IT + ++GW V+EA I+
Sbjct: 254 TESMNLLLDTLCKEKRVEQARVVLLQLKSHITPNAHT-FNIFIHGWCKANRVEEALWTIQ 312
Query: 239 EMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILL 298
EMK +G P ++ Y T +RC C++ + ++ EM ++ P SI+Y ++
Sbjct: 313 EMKGHGFRPCVISYTTIIRCYCQQ-------FEFIKVYEMLSEMEANGSPPNSITYTTIM 365
Query: 299 SCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVD-QMIGKGL 357
S L + +E+ ++ MK SG PD + Y ++ L +GR + + + +M G+
Sbjct: 366 SSLNAQKEFEEALRVATRMKRSGCKPDSLFYNCLIHTLARAGRLEEAERVFRVEMPELGV 425
Query: 358 VPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYG-PVYDVLIPKLCRGGD-FEKG 415
N Y S+I + C + + A+EL ++M+SS+L Y L+ + GD E G
Sbjct: 426 SINTSTYNSMIAMYCHHDEEDKAIELLKEMESSNLCNPDVHTYQPLLRSCFKRGDVVEVG 485
Query: 416 RELWDEATSMGITL 429
+ L + T ++L
Sbjct: 486 KLLKEMVTKHHLSL 499
Score = 63.5 bits (153), Expect = 1e-08
Identities = 50/203 (24%), Positives = 90/203 (43%), Gaps = 9/203 (4%)
Query: 231 KEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPT 290
+EA I + G+ + N L LC+ V + V+++++SH + P
Sbjct: 236 EEAVGIFDRLGEFGLEKNTESMNLLLDTLCKEKR-------VEQARVVLLQLKSH-ITPN 287
Query: 291 SISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVD 350
+ ++NI + K RV+E+ ++ MK G P +SY ++R F K E++
Sbjct: 288 AHTFNIFIHGWCKANRVEEALWTIQEMKGHGFRPCVISYTTIIRCYCQQFEFIKVYEMLS 347
Query: 351 QMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGG 410
+M G PN Y +++ L + AL + +MK S Y+ LI L R G
Sbjct: 348 EMEANGSPPNSITYTTIMSSLNAQKEFEEALRVATRMKRSGCKPDSLFYNCLIHTLARAG 407
Query: 411 DFEKGRELWD-EATSMGITLQCS 432
E+ ++ E +G+++ S
Sbjct: 408 RLEEAERVFRVEMPELGVSINTS 430
>ref|NP_566222.1| pentatricopeptide (PPR) repeat-containing protein [Arabidopsis
thaliana]
Length = 508
Score = 114 bits (284), Expect = 9e-24
Identities = 97/434 (22%), Positives = 190/434 (43%), Gaps = 24/434 (5%)
Query: 6 LNSAWKLCCLRKTRTRNFQLLSASLCSTLQPISAPPHLQELCGIVTSNVGGLDDLELSLN 65
LN + + T +N S L TL S +E+ ++ G D +
Sbjct: 16 LNPSSSSIAIFSTFIKNLSTASEQLPETLDEYSQS---EEIWNVIVGRDGDRDSEDDVFK 72
Query: 66 KFKG-------SLTSSLVAQAIESSKHEAQTRRLLRFFLWSSKNLRHDLEDNDYNYALRV 118
+ +L+ LV + + + + R L W+ H + Y+ A+ +
Sbjct: 73 RLSSDEICKRVNLSDGLVHKLLHRFRDD--WRSALGILKWAESCKGHKHSSDAYDMAVDI 130
Query: 119 FAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLDKYKCSTN 178
+ + + M + ++ + +++ T + G+ +EA+GIF L ++ N
Sbjct: 131 LGKAKKWDRMKEFVERMRGD-KLVTLNTVAKIMRRFAGAGEWEEAVGIFDRLGEFGLEKN 189
Query: 179 EFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIK 238
++ +++ LC + ++A V+L K IT + ++GW V+EA I+
Sbjct: 190 TESMNLLLDTLCKEKRVEQARVVLLQLKSHITPNAHT-FNIFIHGWCKANRVEEALWTIQ 248
Query: 239 EMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILL 298
EMK +G P ++ Y T +RC C++ + ++ EM ++ P SI+Y ++
Sbjct: 249 EMKGHGFRPCVISYTTIIRCYCQQ-------FEFIKVYEMLSEMEANGSPPNSITYTTIM 301
Query: 299 SCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVD-QMIGKGL 357
S L + +E+ ++ MK SG PD + Y ++ L +GR + + + +M G+
Sbjct: 302 SSLNAQKEFEEALRVATRMKRSGCKPDSLFYNCLIHTLARAGRLEEAERVFRVEMPELGV 361
Query: 358 VPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYG-PVYDVLIPKLCRGGD-FEKG 415
N Y S+I + C + + A+EL ++M+SS+L Y L+ + GD E G
Sbjct: 362 SINTSTYNSMIAMYCHHDEEDKAIELLKEMESSNLCNPDVHTYQPLLRSCFKRGDVVEVG 421
Query: 416 RELWDEATSMGITL 429
+ L + T ++L
Sbjct: 422 KLLKEMVTKHHLSL 435
Score = 63.5 bits (153), Expect = 1e-08
Identities = 50/203 (24%), Positives = 90/203 (43%), Gaps = 9/203 (4%)
Query: 231 KEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPT 290
+EA I + G+ + N L LC+ V + V+++++SH + P
Sbjct: 172 EEAVGIFDRLGEFGLEKNTESMNLLLDTLCKEKR-------VEQARVVLLQLKSH-ITPN 223
Query: 291 SISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVD 350
+ ++NI + K RV+E+ ++ MK G P +SY ++R F K E++
Sbjct: 224 AHTFNIFIHGWCKANRVEEALWTIQEMKGHGFRPCVISYTTIIRCYCQQFEFIKVYEMLS 283
Query: 351 QMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGG 410
+M G PN Y +++ L + AL + +MK S Y+ LI L R G
Sbjct: 284 EMEANGSPPNSITYTTIMSSLNAQKEFEEALRVATRMKRSGCKPDSLFYNCLIHTLARAG 343
Query: 411 DFEKGRELWD-EATSMGITLQCS 432
E+ ++ E +G+++ S
Sbjct: 344 RLEEAERVFRVEMPELGVSINTS 366
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 835,321,971
Number of Sequences: 2540612
Number of extensions: 35198496
Number of successful extensions: 111347
Number of sequences better than 10.0: 963
Number of HSP's better than 10.0 without gapping: 607
Number of HSP's successfully gapped in prelim test: 368
Number of HSP's that attempted gapping in prelim test: 99308
Number of HSP's gapped (non-prelim): 4967
length of query: 495
length of database: 863,360,394
effective HSP length: 132
effective length of query: 363
effective length of database: 527,999,610
effective search space: 191663858430
effective search space used: 191663858430
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0093b.5


