

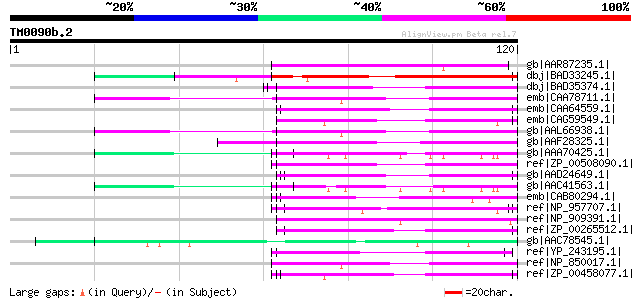
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0090b.2
(120 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAR87235.1| hypothetical protein [Oryza sativa (japonica cult... 65 4e-10
dbj|BAD33245.1| putative glycine-rich cell wall structural prote... 64 8e-10
dbj|BAD35374.1| putative glycine-rich cell wall structural prote... 59 3e-08
emb|CAA78711.1| glycine rich protein [Arabidopsis thaliana] gi|4... 57 8e-08
emb|CAA64559.1| Tfm5 [Lycopersicon esculentum] gi|7489011|pir||T... 57 8e-08
emb|CAG59549.1| unnamed protein product [Candida glabrata CBS138... 57 8e-08
gb|AAL66938.1| glycine-rich RNA binding protein [Arabidopsis tha... 57 8e-08
gb|AAF28325.1| EsV-1-144 [Ectocarpus siliculosus virus] gi|13242... 57 8e-08
gb|AAA70425.1| unknown protein 57 8e-08
ref|ZP_00508090.1| hypothetical protein BproDRAFT_1716 [Polaromo... 57 1e-07
gb|AAD24649.1| unknown protein [Arabidopsis thaliana] gi|2529569... 57 1e-07
gb|AAC41563.1| RNA binding protein gi|532788|gb|AAA86955.1| RNA ... 57 1e-07
emb|CAB80294.1| putative glycine-rich cell wall protein [Arabido... 56 2e-07
ref|NP_957707.1| keratin complex 1, acidic, gene 9 [Mus musculus... 56 2e-07
ref|NP_909391.1| B1189A09.32 [Oryza sativa (japonica cultivar-gr... 56 2e-07
ref|ZP_00265512.1| hypothetical protein Pflu02001988 [Pseudomona... 56 2e-07
gb|AAC78545.1| late embryogenesis abundant M17 protein [Arabidop... 56 2e-07
ref|YP_243195.1| adsorption protein [Xanthomonas campestris pv. ... 56 2e-07
ref|NP_850017.1| glycine-rich RNA-binding protein (GRP7) [Arabid... 56 2e-07
ref|ZP_00458077.1| putative lipoprotein [Burkholderia cenocepaci... 56 2e-07
>gb|AAR87235.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|50582695|gb|AAT78765.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 273
Score = 65.1 bits (157), Expect = 4e-10
Identities = 32/60 (53%), Positives = 33/60 (54%), Gaps = 4/60 (6%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDG----GGSGSDDGGGSGGGGGG 118
DGGGG G G GG G G W GRRR+WW DG GG G DGGG G GGGG
Sbjct: 77 DGGGGGLGDGGGGRLGDGGGRWLGRRRRWWARATAAVDGLGDSGGGGLGDGGGGGLGGGG 136
>dbj|BAD33245.1| putative glycine-rich cell wall structural protein [Oryza sativa
(japonica cultivar-group)]
Length = 239
Score = 63.9 bits (154), Expect = 8e-10
Identities = 32/58 (55%), Positives = 35/58 (60%), Gaps = 8/58 (13%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
DGGGG GGG GGSSGG + ++ W W D GGG S GGG GGGGGGGG
Sbjct: 74 DGGGG--GGGGGGSSGGSSWSYG------WGWGWGTDSGGGGSSGGGGGGGGGGGGGG 123
Score = 60.8 bits (146), Expect = 7e-09
Identities = 40/108 (37%), Positives = 46/108 (42%), Gaps = 32/108 (29%)
Query: 40 VVVVAVTVVVIVV------------EAVVVEAVVVDGGGGND---------------GGG 72
VVV+A V++VV + +E V GGGGN GGG
Sbjct: 20 VVVLAFMAVMVVVPVFGADGGDGRVQVQSLERPVGGGGGGNGTSYNATSVAGRKDGGGGG 79
Query: 73 SGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG G +W W W W D GG GS GGG GGGGGGGG
Sbjct: 80 GGGGGSSGGSSW----SYGWGW-GWGTDSGGGGSSGGGGGGGGGGGGG 122
Score = 55.8 bits (133), Expect = 2e-07
Identities = 35/99 (35%), Positives = 39/99 (39%), Gaps = 18/99 (18%)
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG SGG W + W ++ + GGGG GGG GG GGG
Sbjct: 82 GGGSSGGSSWSYGWGWGWGT------------DSGGGGSSGGGGGGGGGGGGGGGGGGGG 129
Query: 81 ARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G RR WW G G GG GGGGG G
Sbjct: 130 GGGGGGGRRCWW------GCGNGRRRHKGGKEGGGGGEG 162
Score = 37.7 bits (86), Expect = 0.065
Identities = 28/86 (32%), Positives = 33/86 (37%), Gaps = 28/86 (32%)
Query: 44 AVTVVVIVVEAVVVEAVVVDGGGGNDG-----------GGSGGSSGGGARAWWGRRRQWW 92
A VVV++ V+ V V G G DG GG GG +G A
Sbjct: 16 AAAVVVVLAFMAVMVVVPVFGADGGDGRVQVQSLERPVGGGGGGNGTSYNAT-------- 67
Query: 93 RWRQWCDDGGGSGSDDGGGSGGGGGG 118
+G DGGG GGGGGG
Sbjct: 68 ---------SVAGRKDGGGGGGGGGG 84
>dbj|BAD35374.1| putative glycine-rich cell wall structural protein [Oryza sativa
(japonica cultivar-group)]
Length = 321
Score = 58.9 bits (141), Expect = 3e-08
Identities = 29/57 (50%), Positives = 29/57 (50%), Gaps = 12/57 (21%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG R G GGG G GGG GGGGGGGG
Sbjct: 103 GGGGGGGGGGGGGGGGGGRGGGG------------GGGGGGGGGGGGGGGGGGGGGG 147
Score = 58.5 bits (140), Expect = 4e-08
Identities = 29/57 (50%), Positives = 29/57 (50%), Gaps = 13/57 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GR GGG G GGG GGGGGGGG
Sbjct: 98 GGGGGGGGGGGGGGGGGGGGGGGR-------------GGGGGGGGGGGGGGGGGGGG 141
Score = 54.7 bits (130), Expect = 5e-07
Identities = 28/60 (46%), Positives = 28/60 (46%), Gaps = 18/60 (30%)
Query: 61 VVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
V GGGG GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 79 VWSGGGGGGGGGGGGGGGGGG------------------GGGGGGGGGGGGGGGGGGGGG 120
Score = 50.4 bits (119), Expect = 1e-05
Identities = 26/57 (45%), Positives = 26/57 (45%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G R GGG G GGG GG GG G
Sbjct: 97 GGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGGGGGGGGNGGDDG 153
Score = 48.5 bits (114), Expect = 4e-05
Identities = 26/59 (44%), Positives = 28/59 (47%), Gaps = 9/59 (15%)
Query: 62 VDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
V GGG GGG GGS + G W GGG G D G+GGGGGGGG
Sbjct: 216 VIGGGNKGGGGDGGSDNAQSGDGGGG---------WESSGGGGGRGDVSGAGGGGGGGG 265
Score = 46.2 bits (108), Expect = 2e-04
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 20/57 (35%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 76 GWGVWSGGGGGGGGGGG--------------------GGGGGGGGGGGGGGGGGGGG 112
Score = 45.8 bits (107), Expect = 2e-04
Identities = 24/57 (42%), Positives = 25/57 (43%), Gaps = 12/57 (21%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GGG G GGG+GG G G
Sbjct: 112 GGGGGGGGGRGGGGGGGGGGGGG------------GGGGGGGGGGGGGNGGDDGDNG 156
Score = 43.1 bits (100), Expect = 0.002
Identities = 27/70 (38%), Positives = 28/70 (39%), Gaps = 27/70 (38%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDG-------------G 110
GGGG GGG GG GGG G GGG+G DDG G
Sbjct: 122 GGGGGGGGGGGGGGGGGGGGGGG--------------GGGNGGDDGDNGDDGEDGDGDAG 167
Query: 111 GSGGGGGGGG 120
G GG G G G
Sbjct: 168 GRGGKGRGRG 177
Score = 42.4 bits (98), Expect = 0.003
Identities = 18/32 (56%), Positives = 18/32 (56%)
Query: 89 RQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
R W W GGG G GGG GGGGGGGG
Sbjct: 75 RGWGVWSGGGGGGGGGGGGGGGGGGGGGGGGG 106
Score = 35.0 bits (79), Expect = 0.42
Identities = 32/95 (33%), Positives = 35/95 (36%), Gaps = 38/95 (40%)
Query: 64 GGGGNDGGGSGGSSGG----------------GARAWWGRRRQWWRWR---------QWC 98
GGGG GGG GG +GG G R GR R R Q
Sbjct: 135 GGGGGGGGGGGGGNGGDDGDNGDDGEDGDGDAGGRGGKGRGRGRGRGMGSGIGRGNGQNG 194
Query: 99 DDG-------------GGSGSDDGGGSGGGGGGGG 120
D G GGS + GGG+ GGGG GG
Sbjct: 195 DRGINSGGRGKIKGSNGGSRNVIGGGNKGGGGDGG 229
>emb|CAA78711.1| glycine rich protein [Arabidopsis thaliana]
gi|4567224|gb|AAD23639.1| glycine-rich RNA binding
protein 7 [Arabidopsis thaliana]
gi|16226372|gb|AAL16149.1| At2g22292/F2G1.7_
[Arabidopsis thaliana] gi|15810032|gb|AAL06943.1|
At2g21660/F2G1.7 [Arabidopsis thaliana]
gi|544424|sp|Q03250|GRP7_ARATH Glycine-rich RNA-binding
protein 7 gi|15226605|ref|NP_179760.1| glycine-rich
RNA-binding protein (GRP7) [Arabidopsis thaliana]
gi|166837|gb|AAA32853.1| RNA-binding protein
Length = 176
Score = 57.4 bits (137), Expect = 8e-08
Identities = 40/101 (39%), Positives = 45/101 (43%), Gaps = 34/101 (33%)
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSS-GG 79
GGG SGGGG RR +GGGG GGG G SS GG
Sbjct: 108 GGGYSGGGGSYGGGGGRR------------------------EGGGGYSGGGGGYSSRGG 143
Query: 80 GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G ++ G RR+ GGG G +GGG GG GGGGG
Sbjct: 144 GGGSYGGGRRE---------GGGGYGGGEGGGYGGSGGGGG 175
Score = 46.6 bits (109), Expect = 1e-04
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 10/57 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG G S GGG+ G RR +GGG S GGG GGGGG
Sbjct: 100 GGGYRSGGGGGYSGGGGSYGGGGGRR----------EGGGGYSGGGGGYSSRGGGGG 146
Score = 39.7 bits (91), Expect = 0.017
Identities = 25/65 (38%), Positives = 28/65 (42%), Gaps = 19/65 (29%)
Query: 54 AVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG 113
++ V G GG GGG G GGG R+ GGG G GGGS
Sbjct: 78 SITVNEAQSRGSGG--GGGHRGGGGGGYRS-----------------GGGGGYSGGGGSY 118
Query: 114 GGGGG 118
GGGGG
Sbjct: 119 GGGGG 123
Score = 31.2 bits (69), Expect = 6.1
Identities = 18/52 (34%), Positives = 19/52 (35%), Gaps = 17/52 (32%)
Query: 68 NDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
N+ G GGG R GGG G GGG G GGGG
Sbjct: 82 NEAQSRGSGGGGGHRG-----------------GGGGGYRSGGGGGYSGGGG 116
>emb|CAA64559.1| Tfm5 [Lycopersicon esculentum] gi|7489011|pir||T07381 glycine-rich
protein Tfm5 - tomato
Length = 207
Score = 57.4 bits (137), Expect = 8e-08
Identities = 29/57 (50%), Positives = 32/57 (55%), Gaps = 9/57 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GS GGG+ + G GGGSG+ GGGSGGGGGGGG
Sbjct: 62 GGGSGSGGGGSGSGGGGSGSGGGGSGS---------GGGGSGTGGGGGSGGGGGGGG 109
Score = 55.8 bits (133), Expect = 2e-07
Identities = 28/57 (49%), Positives = 29/57 (50%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG+ GG G SGGG G GGGSG GGG GGGGGGGG
Sbjct: 61 GGGGSGSGGGGSGSGGGGSGSGGGGSGSGGGGSGTGGGGGSGGGGGGGGGGGGGGGG 117
Score = 53.5 bits (127), Expect = 1e-06
Identities = 27/57 (47%), Positives = 28/57 (48%), Gaps = 10/57 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GS GGG+ G GGG G GGG GGGGGGGG
Sbjct: 76 GGGSGSGGGGSGSGGGGSGTGGGGG----------SGGGGGGGGGGGGGGGGGGGGG 122
Score = 45.1 bits (105), Expect = 4e-04
Identities = 23/55 (41%), Positives = 25/55 (44%), Gaps = 16/55 (29%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G G GGG GS GGG+ + GGG GS GGG G GGGG
Sbjct: 40 GSGGSGGGGSGSGGGGSGS----------------GGGGGGSGSGGGGSGSGGGG 78
Score = 39.3 bits (90), Expect = 0.022
Identities = 21/49 (42%), Positives = 23/49 (46%), Gaps = 18/49 (36%)
Query: 72 GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GSGGS GGG+ + GG GS GGG GG G GGG
Sbjct: 40 GSGGSGGGGSGS------------------GGGGSGSGGGGGGSGSGGG 70
Score = 32.3 bits (72), Expect = 2.7
Identities = 12/19 (63%), Positives = 13/19 (68%)
Query: 64 GGGGNDGGGSGGSSGGGAR 82
GGGG GGG GG GGG +
Sbjct: 106 GGGGGGGGGGGGGGGGGGQ 124
Score = 30.8 bits (68), Expect = 7.9
Identities = 12/19 (63%), Positives = 12/19 (63%)
Query: 64 GGGGNDGGGSGGSSGGGAR 82
GGGG GGG GG GG R
Sbjct: 108 GGGGGGGGGGGGGGGGQVR 126
Score = 30.8 bits (68), Expect = 7.9
Identities = 13/25 (52%), Positives = 14/25 (56%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRR 89
GGG GGG GG GGG G+ R
Sbjct: 102 GGGGGGGGGGGGGGGGGGGGGGQVR 126
>emb|CAG59549.1| unnamed protein product [Candida glabrata CBS138]
gi|50288385|ref|XP_446622.1| unnamed protein product
[Candida glabrata]
Length = 419
Score = 57.4 bits (137), Expect = 8e-08
Identities = 29/56 (51%), Positives = 30/56 (52%), Gaps = 15/56 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGGSGG SGGG+ GGGSG GGGSGGG GGG
Sbjct: 58 GGGGGSGGGSGGGSGGGSGG---------------GSGGGSGGGSGGGSGGGSGGG 98
Score = 56.2 bits (134), Expect = 2e-07
Identities = 29/56 (51%), Positives = 30/56 (52%), Gaps = 11/56 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGGSGG SGGG+ G GGGSG GGGSGGG GGG
Sbjct: 70 GSGGGSGGGSGGGSGGGSGGGSGG-----------GSGGGSGGGSGGGSGGGSGGG 114
Score = 53.5 bits (127), Expect = 1e-06
Identities = 30/59 (50%), Positives = 31/59 (51%), Gaps = 13/59 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
G GG GGGSGG SGGG+ G GGGSG GGGSGG GGG GG
Sbjct: 66 GSGGGSGGGSGGGSGGGSGGGSGG-----------GSGGGSGGGSGGGSGGGSGGGSGG 113
Score = 50.8 bits (120), Expect = 7e-06
Identities = 28/59 (47%), Positives = 30/59 (50%), Gaps = 22/59 (37%)
Query: 64 GGGGNDGGGS---GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG +GGG GG SGGG+ GGGSG GGGSGGG GGG
Sbjct: 47 GGGGQEGGGGQGGGGGSGGGS-------------------GGGSGGGSGGGSGGGSGGG 86
Score = 41.6 bits (96), Expect = 0.004
Identities = 22/70 (31%), Positives = 26/70 (36%), Gaps = 18/70 (25%)
Query: 51 VVEAVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGG 110
+ + + G N GG GG GGG + GGG GS G
Sbjct: 26 IAPPLFTNTTIPHNNGTNGTGGGGGQEGGGGQ------------------GGGGGSGGGS 67
Query: 111 GSGGGGGGGG 120
G G GGG GG
Sbjct: 68 GGGSGGGSGG 77
>gb|AAL66938.1| glycine-rich RNA binding protein [Arabidopsis thaliana]
gi|14596077|gb|AAK68766.1| glycine-rich RNA binding
protein [Arabidopsis thaliana]
Length = 115
Score = 57.4 bits (137), Expect = 8e-08
Identities = 40/101 (39%), Positives = 45/101 (43%), Gaps = 34/101 (33%)
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSS-GG 79
GGG SGGGG RR +GGGG GGG G SS GG
Sbjct: 47 GGGYSGGGGSYGGGGGRR------------------------EGGGGYSGGGGGYSSRGG 82
Query: 80 GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G ++ G RR+ GGG G +GGG GG GGGGG
Sbjct: 83 GGGSYGGGRRE---------GGGGYGGGEGGGYGGSGGGGG 114
Score = 46.6 bits (109), Expect = 1e-04
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 10/57 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG G S GGG+ G RR +GGG S GGG GGGGG
Sbjct: 39 GGGYRSGGGGGYSGGGGSYGGGGGRR----------EGGGGYSGGGGGYSSRGGGGG 85
Score = 39.7 bits (91), Expect = 0.017
Identities = 25/65 (38%), Positives = 28/65 (42%), Gaps = 19/65 (29%)
Query: 54 AVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG 113
++ V G GG GGG G GGG R+ GGG G GGGS
Sbjct: 17 SITVNEAQSRGSGG--GGGHRGGGGGGYRS-----------------GGGGGYSGGGGSY 57
Query: 114 GGGGG 118
GGGGG
Sbjct: 58 GGGGG 62
Score = 31.2 bits (69), Expect = 6.1
Identities = 18/52 (34%), Positives = 19/52 (35%), Gaps = 17/52 (32%)
Query: 68 NDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
N+ G GGG R GGG G GGG G GGGG
Sbjct: 21 NEAQSRGSGGGGGHRG-----------------GGGGGYRSGGGGGYSGGGG 55
>gb|AAF28325.1| EsV-1-144 [Ectocarpus siliculosus virus]
gi|13242615|ref|NP_077629.1| EsV-1-144 [Ectocarpus
siliculosus virus]
Length = 698
Score = 57.4 bits (137), Expect = 8e-08
Identities = 29/57 (50%), Positives = 30/57 (51%), Gaps = 10/57 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG SGG+SGGG G GGG G GGG GGGGGGGG
Sbjct: 456 GGGGGTGGASGGASGGGGGGGTGG----------AGGGGGGGGGGGGGGGGGGGGGG 502
Score = 52.4 bits (124), Expect = 3e-06
Identities = 28/71 (39%), Positives = 34/71 (47%)
Query: 50 IVVEAVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDG 109
+ +++++ A G GG GG SGG GGG G GGG G G
Sbjct: 430 VKLDSLIQNAGTGGGTGGASGGASGGGGGGGTGGASGGASGGGGGGGTGGAGGGGGGGGG 489
Query: 110 GGSGGGGGGGG 120
GG GGGGGGGG
Sbjct: 490 GGGGGGGGGGG 500
>gb|AAA70425.1| unknown protein
Length = 365
Score = 57.4 bits (137), Expect = 8e-08
Identities = 43/124 (34%), Positives = 46/124 (36%), Gaps = 47/124 (37%)
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSG------ 74
GGG GGGG R+ R GGGGN GGG G
Sbjct: 192 GGGGGGGGGGGGGGRFDRG-----------------------GGGGGNGGGGGGRYDRGG 228
Query: 75 -GSSGGGARAWWGRRRQW---------WRWRQWC--------DDGGGSGSDDGGGSGGGG 116
G GGG R +W + WR C DD G SG GGG GGGG
Sbjct: 229 GGGGGGGGGNVQPRDGEWKCNSCNNTNFAWRNECNRCKTPKGDDEGSSGGGGGGGYGGGG 288
Query: 117 GGGG 120
GGGG
Sbjct: 289 GGGG 292
Score = 48.1 bits (113), Expect = 5e-05
Identities = 30/63 (47%), Positives = 32/63 (50%), Gaps = 10/63 (15%)
Query: 64 GGGGNDGGGSGGSSG--GGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG----GGGG 117
GGGG GGG GG G GG G R+ D GGG G + GGG G GGGG
Sbjct: 175 GGGGGGGGGRGGFGGRRGGGGGGGGGGGGGGRF----DRGGGGGGNGGGGGGRYDRGGGG 230
Query: 118 GGG 120
GGG
Sbjct: 231 GGG 233
Score = 47.0 bits (110), Expect = 1e-04
Identities = 27/61 (44%), Positives = 29/61 (47%), Gaps = 14/61 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGG----GSGGGGGGG 119
G GG GGG GG GGG + R GGG+G GG G GGGGGGG
Sbjct: 186 GFGGRRGGGGGGGGGGGGGGRFDRG----------GGGGGNGGGGGGRYDRGGGGGGGGG 235
Query: 120 G 120
G
Sbjct: 236 G 236
Score = 46.6 bits (109), Expect = 1e-04
Identities = 25/60 (41%), Positives = 29/60 (47%), Gaps = 2/60 (3%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDG--GGSGSDDGGGSGGGGGGGG 120
D G G+ G G G+ GGG +W R + D G G GGG GGGGGGGG
Sbjct: 11 DSGSGHRGSGGSGNGGGGGGSWNDRGGNSYGNGGASKDSYNKGPGGYSGGGGGGGGGGGG 70
Score = 46.6 bits (109), Expect = 1e-04
Identities = 28/57 (49%), Positives = 29/57 (50%), Gaps = 17/57 (29%)
Query: 68 NDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG----GGGGGG 120
N GGG GG GGG R +G RR GGG G GGG GG GGGGGG
Sbjct: 172 NKGGGGGG--GGGGRGGFGGRR-----------GGGGGGGGGGGGGGRFDRGGGGGG 215
Score = 43.9 bits (102), Expect = 0.001
Identities = 24/58 (41%), Positives = 27/58 (46%), Gaps = 5/58 (8%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
D G + GGG GG GGG + R + GG D GG S GGGGGGG
Sbjct: 271 DDEGSSGGGGGGGYGGGGGGGGYDRGND-----RGSGGGGYHNRDRGGNSQGGGGGGG 323
Score = 42.4 bits (98), Expect = 0.003
Identities = 26/63 (41%), Positives = 31/63 (48%), Gaps = 10/63 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGS------DDGGGSGGGGG 117
GGGG D G GS GGG + R + + GGG G ++GGG GG GG
Sbjct: 289 GGGGYDRGNDRGSGGGG----YHNRDRGGNSQGGGGGGGGGGGYSRFNDNNGGGRGGRGG 344
Query: 118 GGG 120
GGG
Sbjct: 345 GGG 347
Score = 32.7 bits (73), Expect = 2.1
Identities = 16/33 (48%), Positives = 16/33 (48%), Gaps = 5/33 (15%)
Query: 93 RWRQWCDDGGGSGSDDGGGS-----GGGGGGGG 120
R W GGG G G G GGGGGGGG
Sbjct: 167 RQNNWNKGGGGGGGGGGRGGFGGRRGGGGGGGG 199
>ref|ZP_00508090.1| hypothetical protein BproDRAFT_1716 [Polaromonas sp. JS666]
gi|67778285|gb|EAM37908.1| hypothetical protein
BproDRAFT_1716 [Polaromonas sp. JS666]
Length = 261
Score = 56.6 bits (135), Expect = 1e-07
Identities = 28/57 (49%), Positives = 28/57 (49%), Gaps = 16/57 (28%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGGN GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 219 GGGGNGGGGGGGGGGGGGGG----------------GGGGGGGGGGGGGGGGGGGGG 259
Score = 55.5 bits (132), Expect = 3e-07
Identities = 27/58 (46%), Positives = 29/58 (49%), Gaps = 19/58 (32%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGGG +GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 217 NGGGGGNGGGGGGGGGGGG-------------------GGGGGGGGGGGGGGGGGGGG 255
Score = 55.1 bits (131), Expect = 4e-07
Identities = 27/57 (47%), Positives = 29/57 (50%), Gaps = 18/57 (31%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGGN GG GG+ GGG +GGG G GGG GGGGGGGG
Sbjct: 205 GGGGNAGGNGGGNGGGGG------------------NGGGGGGGGGGGGGGGGGGGG 243
Score = 54.7 bits (130), Expect = 5e-07
Identities = 27/58 (46%), Positives = 29/58 (49%), Gaps = 14/58 (24%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGG GGG+GG GGG G GGG G GGG GGGGGGGG
Sbjct: 213 NGGGNGGGGGNGGGGGGGGGGGGG--------------GGGGGGGGGGGGGGGGGGGG 256
Score = 52.8 bits (125), Expect = 2e-06
Identities = 27/57 (47%), Positives = 29/57 (50%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGGSG GGG G +GGG G+ GGG GGGGGGGG
Sbjct: 192 GSGGGKGGGSGAGGGGGNAGGNGG-----------GNGGGGGNGGGGGGGGGGGGGG 237
Score = 47.4 bits (111), Expect = 8e-05
Identities = 24/57 (42%), Positives = 25/57 (43%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GGG GG SG G + GGG G GGG GGGGGGGG
Sbjct: 188 GNAAGSGGGKGGGSGAGGGGGNAGGNGGGNGGGGGNGGGGGGGGGGGGGGGGGGGGG 244
Score = 32.7 bits (73), Expect = 2.1
Identities = 13/25 (52%), Positives = 14/25 (56%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRR 88
GGGG GGG GG GGG G +
Sbjct: 237 GGGGGGGGGGGGGGGGGGGGGGGNK 261
>gb|AAD24649.1| unknown protein [Arabidopsis thaliana] gi|25295693|pir||C84470
hypothetical protein At2g05580 [imported] - Arabidopsis
thaliana
Length = 302
Score = 56.6 bits (135), Expect = 1e-07
Identities = 30/57 (52%), Positives = 31/57 (53%), Gaps = 10/57 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG+ GGG GG GGG R G R GGGSG GGGSG GGGGGG
Sbjct: 254 GQGGHKGGGGGGHVGGGGRGGGGGGR----------GGGGSGGGGGGGSGRGGGGGG 300
Score = 48.9 bits (115), Expect = 3e-05
Identities = 25/54 (46%), Positives = 26/54 (47%)
Query: 66 GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GG GGG GG GGG G + Q GGG G GGG GGGGGG
Sbjct: 225 GGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGHKGGGGGGHVGGGGRGGGGGG 278
Score = 48.5 bits (114), Expect = 4e-05
Identities = 26/55 (47%), Positives = 27/55 (48%), Gaps = 1/55 (1%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GG GGG GG GGG R G + Q GGG G GG GGGGGGG
Sbjct: 178 GGQKGGGGQGGQKGGGGRGQGGMKGGGGGG-QGGHKGGGGGGGQGGHKGGGGGGG 231
Score = 46.6 bits (109), Expect = 1e-04
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG GGG G GGG G GG GGGGG GG
Sbjct: 215 GGGGGQGGHKGGGGGGGQGGHKG--------------GGGGGQGGGGHKGGGGGQGG 257
Score = 46.6 bits (109), Expect = 1e-04
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 15/57 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG GGG Q GGG G GG GGGGGG G
Sbjct: 203 GGGGGQGGHKGGGGGGG---------------QGGHKGGGGGGGQGGHKGGGGGGQG 244
Score = 46.2 bits (108), Expect = 2e-04
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 1/57 (1%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG GG + G+ Q Q GGG G GG GGGGGG G
Sbjct: 154 GGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGG-GRGQGGMKGGGGGGQG 209
Score = 45.8 bits (107), Expect = 2e-04
Identities = 24/56 (42%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG GGG GG GGG + G++ R + GGG G G GGGGG GG
Sbjct: 169 GGQKGGGGQGGQKGGGGQG--GQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGG 222
Score = 45.4 bits (106), Expect = 3e-04
Identities = 25/57 (43%), Positives = 27/57 (46%), Gaps = 2/57 (3%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG G GGG + G + Q GGG G GGG GGGGG G
Sbjct: 202 GGGGGGQGGHKGGGGGGGQG--GHKGGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQG 256
Score = 43.9 bits (102), Expect = 0.001
Identities = 27/65 (41%), Positives = 29/65 (44%), Gaps = 8/65 (12%)
Query: 64 GGGGNDGG---GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG-----G 115
G GG GG G GG GGG G++ GGG G GGG GG G
Sbjct: 83 GQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGGGGGQGGQKGGGGGGQGGQKG 142
Query: 116 GGGGG 120
GGGGG
Sbjct: 143 GGGGG 147
Score = 43.9 bits (102), Expect = 0.001
Identities = 24/57 (42%), Positives = 27/57 (47%), Gaps = 2/57 (3%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G GGG GG GGG G++ + Q GGG G GGG GG GGG
Sbjct: 139 GQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQ--KGGGGQGGQKGGGGQGGQKGGG 193
Score = 41.2 bits (95), Expect = 0.006
Identities = 26/63 (41%), Positives = 28/63 (44%), Gaps = 15/63 (23%)
Query: 64 GGGGNDGGGSG-GSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG-----GGG 117
G GG GGG G G GG G++ GGG G GGG GG GGG
Sbjct: 105 GQGGQKGGGGGQGGQKGGGGGQGGQKG---------GGGGGQGGQKGGGGGGQGGQKGGG 155
Query: 118 GGG 120
GGG
Sbjct: 156 GGG 158
Score = 40.8 bits (94), Expect = 0.008
Identities = 24/59 (40%), Positives = 26/59 (43%), Gaps = 18/59 (30%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG--GGGGGGG 120
G GG+ GGG GG GGG + GGG G GGG G GGGG G
Sbjct: 231 GQGGHKGGGGGGQGGGGHKG----------------GGGGQGGHKGGGGGGHVGGGGRG 273
Score = 40.8 bits (94), Expect = 0.008
Identities = 25/59 (42%), Positives = 28/59 (47%), Gaps = 3/59 (5%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG--GGGG 120
G G GGG GG GGG G++ + GGG G GGG G GG GGGG
Sbjct: 128 GQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGG-GQGGQKGGGG 185
Score = 38.9 bits (89), Expect = 0.029
Identities = 21/54 (38%), Positives = 23/54 (41%), Gaps = 13/54 (24%)
Query: 67 GNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G G GG GGG G++ GG G GG GGGGG GG
Sbjct: 78 GGIGRGQGGQKGGGGGGQGGQK-------------GGGGGGQGGQKGGGGGQGG 118
Score = 38.9 bits (89), Expect = 0.029
Identities = 25/60 (41%), Positives = 27/60 (44%), Gaps = 11/60 (18%)
Query: 64 GGGGNDGGGSG-GSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG--GGG 120
G GG GGG G G GG G ++ GGG G GGG GG GG GGG
Sbjct: 115 GQGGQKGGGGGQGGQKGGGGGGQGGQKG--------GGGGGQGGQKGGGGGGQGGQKGGG 166
Score = 37.7 bits (86), Expect = 0.065
Identities = 24/59 (40%), Positives = 24/59 (40%), Gaps = 14/59 (23%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG--GGGG 120
GG G GG G GGG G GGG G GGG G GG GGGG
Sbjct: 78 GGIGRGQGGQKGGGGGGQGGQKG------------GGGGGQGGQKGGGGGQGGQKGGGG 124
Score = 37.4 bits (85), Expect = 0.085
Identities = 26/63 (41%), Positives = 30/63 (47%), Gaps = 11/63 (17%)
Query: 65 GGGNDG--GGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG-----GGG 117
GG G GG GG GGG + G++ + Q GGG G GGG G GGG
Sbjct: 149 GGQKGGGGGGQGGQKGGGGQG--GQKGGGGQGGQ--KGGGGQGGQKGGGGRGQGGMKGGG 204
Query: 118 GGG 120
GGG
Sbjct: 205 GGG 207
>gb|AAC41563.1| RNA binding protein gi|532788|gb|AAA86955.1| RNA binding protein
Length = 404
Score = 56.6 bits (135), Expect = 1e-07
Identities = 43/124 (34%), Positives = 45/124 (35%), Gaps = 47/124 (37%)
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSG------ 74
GGG GGGG R+ R GGGGN GGG G
Sbjct: 230 GGGGGGGGGGGGGGRFDRG-----------------------GGGGGNGGGGGGRYDRGG 266
Query: 75 -GSSGGGARAWWGRRRQW---------WRWRQWC--------DDGGGSGSDDGGGSGGGG 116
G GGG R W + WR C DD G SG GGG GGGG
Sbjct: 267 GGGGGGGGGNVQPRDGDWKCNSCNNTNFAWRNECNRCKTPKGDDEGSSGGGGGGGYGGGG 326
Query: 117 GGGG 120
GGGG
Sbjct: 327 GGGG 330
Score = 48.1 bits (113), Expect = 5e-05
Identities = 30/63 (47%), Positives = 32/63 (50%), Gaps = 10/63 (15%)
Query: 64 GGGGNDGGGSGGSSG--GGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG----GGGG 117
GGGG GGG GG G GG G R+ D GGG G + GGG G GGGG
Sbjct: 213 GGGGGGGGGRGGFGGRRGGGGGGGGGGGGGGRF----DRGGGGGGNGGGGGGRYDRGGGG 268
Query: 118 GGG 120
GGG
Sbjct: 269 GGG 271
Score = 47.0 bits (110), Expect = 1e-04
Identities = 27/61 (44%), Positives = 29/61 (47%), Gaps = 14/61 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGG----GSGGGGGGG 119
G GG GGG GG GGG + R GGG+G GG G GGGGGGG
Sbjct: 224 GFGGRRGGGGGGGGGGGGGGRFDRG----------GGGGGNGGGGGGRYDRGGGGGGGGG 273
Query: 120 G 120
G
Sbjct: 274 G 274
Score = 46.6 bits (109), Expect = 1e-04
Identities = 28/57 (49%), Positives = 29/57 (50%), Gaps = 17/57 (29%)
Query: 68 NDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG----GGGGGG 120
N GGG GG GGG R +G RR GGG G GGG GG GGGGGG
Sbjct: 210 NKGGGGGG--GGGGRGGFGGRR-----------GGGGGGGGGGGGGGRFDRGGGGGG 253
Score = 46.6 bits (109), Expect = 1e-04
Identities = 25/60 (41%), Positives = 29/60 (47%), Gaps = 2/60 (3%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDG--GGSGSDDGGGSGGGGGGGG 120
D G G+ G G G+ GGG +W R + D G G GGG GGGGGGGG
Sbjct: 49 DSGSGHRGSGGSGNGGGGGGSWNDRGGNSYGNGGASKDSYNKGPGGYSGGGGGGGGGGGG 108
Score = 43.9 bits (102), Expect = 0.001
Identities = 24/58 (41%), Positives = 27/58 (46%), Gaps = 5/58 (8%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
D G + GGG GG GGG + R + GG D GG S GGGGGGG
Sbjct: 309 DDEGSSGGGGGGGYGGGGGGGGYDRGND-----RGSGGGGYHNRDRGGNSQGGGGGGG 361
Score = 42.4 bits (98), Expect = 0.003
Identities = 26/63 (41%), Positives = 31/63 (48%), Gaps = 10/63 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGS------DDGGGSGGGGG 117
GGGG D G GS GGG + R + + GGG G ++GGG GG GG
Sbjct: 327 GGGGYDRGNDRGSGGGG----YHNRDRGGNSQGGGGGGGGGGGYSRFNDNNGGGRGGRGG 382
Query: 118 GGG 120
GGG
Sbjct: 383 GGG 385
Score = 37.7 bits (86), Expect = 0.065
Identities = 20/57 (35%), Positives = 25/57 (43%), Gaps = 13/57 (22%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
D GG + GGG GG GGG + ++ D+ GG GGG G GG
Sbjct: 348 DRGGNSQGGGGGGGGGGG-------------YSRFNDNNGGGRGGRGGGGGNRRDGG 391
Score = 32.7 bits (73), Expect = 2.1
Identities = 16/33 (48%), Positives = 16/33 (48%), Gaps = 5/33 (15%)
Query: 93 RWRQWCDDGGGSGSDDGGGS-----GGGGGGGG 120
R W GGG G G G GGGGGGGG
Sbjct: 205 RQNNWNKGGGGGGGGGGRGGFGGRRGGGGGGGG 237
>emb|CAB80294.1| putative glycine-rich cell wall protein [Arabidopsis thaliana]
gi|2961382|emb|CAA18129.1| putative glycine-rich cell
wall protein [Arabidopsis thaliana]
gi|7484986|pir||T04592 glycine-rich cell wall structural
protein homolog F23E13.120 - Arabidopsis thaliana
Length = 221
Score = 56.2 bits (134), Expect = 2e-07
Identities = 29/57 (50%), Positives = 30/57 (51%), Gaps = 10/57 (17%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG SG G+ R R GGG G GGG GGGGGGGG
Sbjct: 131 GGGGGGGGGGGGGSGNGSG----------RGRGGGGGGGGGGGGGGGGGGGGGGGGG 177
Score = 52.8 bits (125), Expect = 2e-06
Identities = 29/66 (43%), Positives = 30/66 (44%), Gaps = 9/66 (13%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGS---------GG 114
GGGG GGG GS GGG G + R GGG G GGG GG
Sbjct: 95 GGGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGG 154
Query: 115 GGGGGG 120
GGGGGG
Sbjct: 155 GGGGGG 160
Score = 48.5 bits (114), Expect = 4e-05
Identities = 27/64 (42%), Positives = 31/64 (48%), Gaps = 18/64 (28%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDD--------GGGSGGGG 116
GGG GGG GG GGG + G + GG+G D+ GGG GGGG
Sbjct: 89 GGGGGGGGGGGGGGGGQGSGGGGG----------EGNGGNGKDNSHKRNKSSGGGGGGGG 138
Query: 117 GGGG 120
GGGG
Sbjct: 139 GGGG 142
Score = 47.4 bits (111), Expect = 8e-05
Identities = 26/57 (45%), Positives = 26/57 (45%), Gaps = 4/57 (7%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GGS G R G GGG G GGG GGGGG G
Sbjct: 132 GGGGGGGGGGGGSGNGSGRGRGGGGGGGGGG----GGGGGGGGGGGGGGGGGGGSDG 184
Score = 44.3 bits (103), Expect = 7e-04
Identities = 25/58 (43%), Positives = 26/58 (44%), Gaps = 22/58 (37%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+G G GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 146 NGSGRGRGGGGGGGGGGGG-------------------GGGGG---GGGGGGGGGGGG 181
Score = 42.7 bits (99), Expect = 0.002
Identities = 26/58 (44%), Positives = 26/58 (44%), Gaps = 20/58 (34%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSD-DGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG GSD GGG G G G GG
Sbjct: 159 GGGGGGGGGGGGGGGGGG-------------------GGGGGSDGKGGGWGFGFGWGG 197
Score = 40.8 bits (94), Expect = 0.008
Identities = 23/57 (40%), Positives = 23/57 (40%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G GG G G G G G GG G
Sbjct: 157 GGGGGGGGGGGGGGGGGGGGGGG--------------GGSDGKGGGWGFGFGWGGDG 199
Score = 39.7 bits (91), Expect = 0.017
Identities = 23/56 (41%), Positives = 23/56 (41%), Gaps = 15/56 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGG DG G G G G GG G GG
Sbjct: 162 GGGGGGGGGGGGGGGGGGGG---------------SDGKGGGWGFGFGWGGDGPGG 202
Score = 34.3 bits (77), Expect = 0.72
Identities = 13/23 (56%), Positives = 14/23 (60%)
Query: 98 CDDGGGSGSDDGGGSGGGGGGGG 120
C+ G G GGG GGGGGG G
Sbjct: 84 CESRAGGGGGGGGGGGGGGGGQG 106
>ref|NP_957707.1| keratin complex 1, acidic, gene 9 [Mus musculus]
gi|40744571|gb|AAR89518.1| cytokeratin 9 [Mus musculus]
Length = 743
Score = 56.2 bits (134), Expect = 2e-07
Identities = 29/57 (50%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG + GGGSGGS GGG+ G + + GGGSG GGGSG GGG GG
Sbjct: 656 GGGSSSGGGSGGSYGGGSGGSHGGK-SGGSYGGGSSSGGGSGGSYGGGSGSGGGSGG 711
Score = 50.1 bits (118), Expect = 1e-05
Identities = 28/59 (47%), Positives = 32/59 (53%), Gaps = 11/59 (18%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
G GG+ GG SGG GGG+ + G + GGGSG GGGSGG GGG GG
Sbjct: 542 GSGGSHGGKSGGGYGGGSSSGGGSGGSY---------GGGSGGSHGGGSGGSYGGGSGG 591
Score = 50.1 bits (118), Expect = 1e-05
Identities = 29/59 (49%), Positives = 32/59 (54%), Gaps = 17/59 (28%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
G GG+ GGGSGGS GGG+ + GGGSG GGGSGG GGG GG
Sbjct: 462 GSGGSYGGGSGGSYGGGSGGSY---------------GGGSGGSYGGGSGGSYGGGSGG 505
Score = 49.7 bits (117), Expect = 2e-05
Identities = 27/56 (48%), Positives = 30/56 (53%), Gaps = 15/56 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG+ GGGSGGS GGG+ +G GGSG GG SGGG GGG
Sbjct: 564 GSGGSYGGGSGGSHGGGSGGSYG---------------GGSGGSHGGKSGGGYGGG 604
Score = 49.7 bits (117), Expect = 2e-05
Identities = 28/57 (49%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query: 64 GGGGNDGGGSGGSSGGGAR-AWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG + GGGSGGS GGG+ + G+ + GGGSG GG SGGG GGG
Sbjct: 602 GGGTSSGGGSGGSYGGGSGGSHGGKSGGSYGGGSGGSYGGGSGGSHGGKSGGGYGGG 658
Score = 49.3 bits (116), Expect = 2e-05
Identities = 28/59 (47%), Positives = 32/59 (53%), Gaps = 11/59 (18%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
G GG+ GGGSGGS GGG+ G + G G G+ GGGSGG GGG GG
Sbjct: 572 GSGGSHGGGSGGSYGGGSGGSHGGKS---------GGGYGGGTSSGGGSGGSYGGGSGG 621
Score = 48.5 bits (114), Expect = 4e-05
Identities = 29/59 (49%), Positives = 32/59 (54%), Gaps = 9/59 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
G GG+ GGGSGGS GGG+ +G GGGSG GG SGG GGG GG
Sbjct: 470 GSGGSYGGGSGGSYGGGSGGSYGGGSGG-------SYGGGSGGSHGGKSGGSHGGGSGG 521
Score = 48.5 bits (114), Expect = 4e-05
Identities = 29/59 (49%), Positives = 32/59 (54%), Gaps = 9/59 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
G GG+ GGGSGGS GGG+ G + GGGSG GG SGG GGG GG
Sbjct: 486 GSGGSYGGGSGGSYGGGSGGSHGGKSGG-------SHGGGSGGSYGGESGGSHGGGSGG 537
Score = 47.8 bits (112), Expect = 6e-05
Identities = 26/55 (47%), Positives = 29/55 (52%), Gaps = 7/55 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
G GG+ GG SGGS GGG+ +G GGGSG GGGSGG GG
Sbjct: 502 GSGGSHGGKSGGSHGGGSGGSYG-------GESGGSHGGGSGGSYGGGSGGSHGG 549
Score = 47.4 bits (111), Expect = 8e-05
Identities = 28/57 (49%), Positives = 31/57 (54%), Gaps = 11/57 (19%)
Query: 66 GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
GG+ GGGSGGS GGG+ G + G G GS GGGSGG GGG GG
Sbjct: 628 GGSYGGGSGGSYGGGSGGSHGGKS---------GGGYGGGSSSGGGSGGSYGGGSGG 675
Score = 46.6 bits (109), Expect = 1e-04
Identities = 26/56 (46%), Positives = 29/56 (51%), Gaps = 15/56 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG+ GG SGGS GGG+ +G GGSG GG SGGG GGG
Sbjct: 518 GSGGSYGGESGGSHGGGSGGSYG---------------GGSGGSHGGKSGGGYGGG 558
Score = 46.2 bits (108), Expect = 2e-04
Identities = 26/57 (45%), Positives = 29/57 (50%), Gaps = 7/57 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG+ GG SGGS GGG+ +G GG SG GGGS GGG GG
Sbjct: 618 GSGGSHGGKSGGSYGGGSGGSYGGGSGG-------SHGGKSGGGYGGGSSSGGGSGG 667
Score = 44.7 bits (104), Expect = 5e-04
Identities = 25/53 (47%), Positives = 25/53 (47%), Gaps = 19/53 (35%)
Query: 66 GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
GG GGG GG SGGG GGGSG GGGSGGG GG
Sbjct: 92 GGGSGGGFGGGSGGGF-------------------GGGSGGGFGGGSGGGEGG 125
Score = 44.3 bits (103), Expect = 7e-04
Identities = 28/60 (46%), Positives = 33/60 (54%), Gaps = 4/60 (6%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGGGG 120
+ GG + GGSGGS GGG+ G + + GGGSG GGGSGG GGG GG
Sbjct: 526 ESGGSHG-GGSGGSYGGGSGGSHG-GKSGGGYGGGSSSGGGSGGSYGGGSGGSHGGGSGG 583
Score = 43.9 bits (102), Expect = 0.001
Identities = 26/56 (46%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG+ G GG GGG G + GGGSG GGGSGGG GGG
Sbjct: 65 GGGGSFGSSYGGGYGGGFST--GSYSGMFGGGSGGGFGGGSGGGFGGGSGGGFGGG 118
Score = 41.2 bits (95), Expect = 0.006
Identities = 23/53 (43%), Positives = 24/53 (44%), Gaps = 19/53 (35%)
Query: 67 GNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GGGSGG GGG+ GGG G GGG GGG GGG
Sbjct: 89 GMFGGGSGGGFGGGS-------------------GGGFGGGSGGGFGGGSGGG 122
Score = 38.9 bits (89), Expect = 0.029
Identities = 21/49 (42%), Positives = 25/49 (50%), Gaps = 17/49 (34%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGS 112
GGG + GGGSGGS GGG+ + GGGSG GGG+
Sbjct: 686 GGGSSSGGGSGGSYGGGSGS-----------------GGGSGGSYGGGN 717
Score = 35.0 bits (79), Expect = 0.42
Identities = 22/56 (39%), Positives = 22/56 (39%), Gaps = 16/56 (28%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG S G SGGG A GGG G G GGG GGG
Sbjct: 42 GGGGGRFNSSSGFSGGGFSAC----------------GGGGGGSFGSSYGGGYGGG 81
Score = 33.9 bits (76), Expect = 0.94
Identities = 22/55 (40%), Positives = 26/55 (47%), Gaps = 10/55 (18%)
Query: 70 GGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGG----GGGGG 120
GGG G SGG R+ + R + GGG + G SGGG GGGGG
Sbjct: 19 GGGGRGGSGGSMRSSFSRSSRAG------GGGGGRFNSSSGFSGGGFSACGGGGG 67
Score = 33.5 bits (75), Expect = 1.2
Identities = 25/63 (39%), Positives = 28/63 (43%), Gaps = 18/63 (28%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGG------SGSDDGGGSGGGGG 117
GGGG GG SGG R+ + R + GGG SG GG S GGG
Sbjct: 18 GGGGGRGG-----SGGSMRSSFSRSSR-------AGGGGGGRFNSSSGFSGGGFSACGGG 65
Query: 118 GGG 120
GGG
Sbjct: 66 GGG 68
Score = 33.5 bits (75), Expect = 1.2
Identities = 14/19 (73%), Positives = 14/19 (73%)
Query: 64 GGGGNDGGGSGGSSGGGAR 82
GGG GGGSGGS GGG R
Sbjct: 700 GGGSGSGGGSGGSYGGGNR 718
Score = 32.0 bits (71), Expect = 3.6
Identities = 26/59 (44%), Positives = 29/59 (49%), Gaps = 11/59 (18%)
Query: 65 GGGNDGGGSG-GSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG---GGGGG 119
GG D SG G G G+ G+ RQ + GGGSG GGGSGG GG GG
Sbjct: 438 GGQQDFESSGAGQIGFGS----GKGRQRGSGGSY---GGGSGGSYGGGSGGSYGGGSGG 489
>ref|NP_909391.1| B1189A09.32 [Oryza sativa (japonica cultivar-group)]
Length = 535
Score = 56.2 bits (134), Expect = 2e-07
Identities = 28/57 (49%), Positives = 31/57 (54%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG + G + + GGG G GGG GGGGGGGG
Sbjct: 58 GGGGYGGGGVGGGYGGGGGGYGGGGGGYGGGGRGGGGGGGYGGGGGGGRGGGGGGGG 114
Score = 50.1 bits (118), Expect = 1e-05
Identities = 27/57 (47%), Positives = 27/57 (47%), Gaps = 9/57 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GG GG GGG R G GGG G GGG GGG GGGG
Sbjct: 72 GGGGGGYGGGGGGYGGGGRGGGGGGGY---------GGGGGGGRGGGGGGGGRGGGG 119
Score = 48.1 bits (113), Expect = 5e-05
Identities = 30/72 (41%), Positives = 32/72 (43%), Gaps = 16/72 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQW-----------WRWRQWCDDGGGSGSDDGGGS 112
GGGG GGG GG GGG R GR W + R C+ G GGG
Sbjct: 102 GGGGRGGGGGGGGRGGGGRG-GGRDGDWVCPDPSCGNVNFARRTECNKCGAPSPAGGGGG 160
Query: 113 GGGGG----GGG 120
GGGGG GGG
Sbjct: 161 GGGGGYNKSGGG 172
Score = 41.2 bits (95), Expect = 0.006
Identities = 29/72 (40%), Positives = 30/72 (41%), Gaps = 17/72 (23%)
Query: 64 GGGGNDGGGSGG-SSGGGARAWWGRRRQWWRWRQW-CDDGG---------------GSGS 106
GGGG GGG GG GGG G R R W C D G+ S
Sbjct: 94 GGGGYGGGGGGGRGGGGGGGGRGGGGRGGGRDGDWVCPDPSCGNVNFARRTECNKCGAPS 153
Query: 107 DDGGGSGGGGGG 118
GGG GGGGGG
Sbjct: 154 PAGGGGGGGGGG 165
Score = 40.4 bits (93), Expect = 0.010
Identities = 26/63 (41%), Positives = 28/63 (44%), Gaps = 10/63 (15%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDD------GGGSGGGGG 117
GGGG GGG S GGG G R + G G D GGG+GGGG
Sbjct: 157 GGGGGGGGGYNKSGGGGG----GYNRGGGDFSSGGGGGYNRGGGDYNSGGRGGGTGGGGR 212
Query: 118 GGG 120
GGG
Sbjct: 213 GGG 215
Score = 37.7 bits (86), Expect = 0.065
Identities = 24/59 (40%), Positives = 27/59 (45%), Gaps = 2/59 (3%)
Query: 64 GGGG--NDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG + GG G + R + GG G GGG GGG GG
Sbjct: 162 GGGGYNKSGGGGGGYNRGGGDFSSGGGGGYNRGGGDYNSGGRGGGTGGGGRGGGYNRGG 220
Score = 37.0 bits (84), Expect = 0.11
Identities = 21/51 (41%), Positives = 21/51 (41%), Gaps = 19/51 (37%)
Query: 70 GGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GG GG GGG G GGG GGG GGGG
Sbjct: 44 GGGGGGRGRGGG-------------------GGGGGGYGGGGVGGGYGGGG 75
Score = 35.8 bits (81), Expect = 0.25
Identities = 26/74 (35%), Positives = 30/74 (40%), Gaps = 22/74 (29%)
Query: 47 VVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGS 106
VV ++ + +VV GG GGG G GGG GGG G
Sbjct: 25 VVCVLASGSAPDWLVVFFVGG--GGGGRGRGGGG--------------------GGGGGY 62
Query: 107 DDGGGSGGGGGGGG 120
GG GG GGGGG
Sbjct: 63 GGGGVGGGYGGGGG 76
Score = 35.4 bits (80), Expect = 0.32
Identities = 22/57 (38%), Positives = 24/57 (41%), Gaps = 8/57 (14%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG N GGG S GGG G + G G G+ GG GG GGG
Sbjct: 173 GGGYNRGGGDFSSGGGGGYNRGGG--------DYNSGGRGGGTGGGGRGGGYNRGGG 221
Score = 35.4 bits (80), Expect = 0.32
Identities = 23/57 (40%), Positives = 25/57 (43%), Gaps = 14/57 (24%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGS--GSDDGGGSGGGGGG 118
GGG + GG GG +GGG R R GGG G DD G GG GG
Sbjct: 194 GGGDYNSGGRGGGTGGGGRGGGYNR------------GGGDDRGFDDHRGGRGGYGG 238
Score = 32.3 bits (72), Expect = 2.7
Identities = 15/25 (60%), Positives = 16/25 (64%), Gaps = 5/25 (20%)
Query: 101 GGGSGSDD-----GGGSGGGGGGGG 120
GGG G+ GGG GGGGGGGG
Sbjct: 347 GGGPGAPPPYHGGGGGGGGGGGGGG 371
>ref|ZP_00265512.1| hypothetical protein Pflu02001988 [Pseudomonas fluorescens PfO-1]
Length = 220
Score = 56.2 bits (134), Expect = 2e-07
Identities = 28/57 (49%), Positives = 31/57 (54%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG+ GGG GG +GGG G GGG G + GGGSGGGGGG G
Sbjct: 41 GGGGSGGGGGGGGNGGGGGGNGGG-----------GSGGGGGGNGGGGSGGGGGGNG 86
Score = 53.9 bits (128), Expect = 9e-07
Identities = 27/57 (47%), Positives = 31/57 (54%), Gaps = 7/57 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG +GGG GG+ GGG+ G GGG G + GGGSGG GGG G
Sbjct: 49 GGGGGNGGGGGGNGGGGSGGGGGGNGGG-------GSGGGGGGNGGGGSGGNGGGHG 98
Score = 52.4 bits (124), Expect = 3e-06
Identities = 26/55 (47%), Positives = 29/55 (52%), Gaps = 18/55 (32%)
Query: 66 GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG+ GGGSGG GGG +GGG G + GGGSGGGGGG G
Sbjct: 38 GGSGGGGSGGGGGGGG------------------NGGGGGGNGGGGSGGGGGGNG 74
Score = 50.4 bits (119), Expect = 1e-05
Identities = 26/57 (45%), Positives = 28/57 (48%), Gaps = 12/57 (21%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGGN GGGSGG GG G GGG+G GG+GGG GG G
Sbjct: 57 GGGGNGGGGSGGGGGGNGGGGSG------------GGGGGNGGGGSGGNGGGHGGSG 101
Score = 49.3 bits (116), Expect = 2e-05
Identities = 25/56 (44%), Positives = 27/56 (47%), Gaps = 12/56 (21%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGGN GGGSGG GG G +GGG G GG+ G GGGG
Sbjct: 69 GGGGNGGGGSGGGGGGNGGGGSG------------GNGGGHGGSGSGGNSGSGGGG 112
Score = 42.7 bits (99), Expect = 0.002
Identities = 24/57 (42%), Positives = 26/57 (45%), Gaps = 15/57 (26%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGGN GGGSGG+ GG + GG SGS GG SG G G
Sbjct: 81 GGGGNGGGGSGGNGGGHGGS---------------GSGGNSGSGGGGNSGHSGSDHG 122
Score = 35.0 bits (79), Expect = 0.42
Identities = 13/21 (61%), Positives = 14/21 (65%)
Query: 100 DGGGSGSDDGGGSGGGGGGGG 120
+GG G GGG GGGG GGG
Sbjct: 37 EGGSGGGGSGGGGGGGGNGGG 57
Score = 34.7 bits (78), Expect = 0.55
Identities = 13/22 (59%), Positives = 14/22 (63%)
Query: 99 DDGGGSGSDDGGGSGGGGGGGG 120
+ G G G GGG GGG GGGG
Sbjct: 37 EGGSGGGGSGGGGGGGGNGGGG 58
Score = 33.5 bits (75), Expect = 1.2
Identities = 22/60 (36%), Positives = 25/60 (41%), Gaps = 13/60 (21%)
Query: 63 DGGGGND--GGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGGG+ GGG GGS GG G + G SGSD G G G G
Sbjct: 85 NGGGGSGGNGGGHGGSGSGGNSGSGGG-----------GNSGHSGSDHGSGHSGNTASSG 133
>gb|AAC78545.1| late embryogenesis abundant M17 protein [Arabidopsis thaliana]
gi|3342551|gb|AAC27641.1| late-embryogenesis abundant
M17 protein [Arabidopsis thaliana]
gi|25408738|pir||G84839 late embryogenesis abundant M17
protein [imported] - Arabidopsis thaliana
gi|42571165|ref|NP_973656.1| glycine-rich protein / late
embryogenesis abundant protein (M17) [Arabidopsis
thaliana]
Length = 280
Score = 56.2 bits (134), Expect = 2e-07
Identities = 45/123 (36%), Positives = 49/123 (39%), Gaps = 29/123 (23%)
Query: 21 GGGDSGGGGWC-------WWRRWRRSVV--VVAVTVVVIVVEAVVVEAVVVDGGGGNDGG 71
GGG GG G C WWR R + VV VE VE GG GG
Sbjct: 123 GGGGGGGRGGCRWGCCGGWWRGRCRYCCRSQAEASEVVETVEPNDVEPQQ----GGRGGG 178
Query: 72 GSGGSSGGGARAWWGRRRQWWRWR-QWCDDGGGSGSD-------------DGGGSGGGGG 117
G GG GG R WG WWR R ++C S+ GG GGGGG
Sbjct: 179 GGGGGGRGGCR--WGCCGGWWRGRCRYCCRSQAEASEVVETVEPNDVEPQQGGRGGGGGG 236
Query: 118 GGG 120
GGG
Sbjct: 237 GGG 239
Score = 52.0 bits (123), Expect = 3e-06
Identities = 48/137 (35%), Positives = 53/137 (38%), Gaps = 30/137 (21%)
Query: 7 VAAAVVVAMLAVVEGGGDSGGGGWC-------WWR---RWRRSVVVVAVTVVVIVVEAVV 56
+A A V A VVE D GG G C WWR R+ A VV V V
Sbjct: 54 IAEAQVEANDPVVEPQQDWGGRGGCRWGCCGGWWRGRCRYCCRSQAEASEVVETVEPNDV 113
Query: 57 VEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWR-QWCDDGGGSGSD-------- 107
GGGG GGG GG WG WWR R ++C S+
Sbjct: 114 EPQQGGRGGGGGGGGGRGGCR-------WGCCGGWWRGRCRYCCRSQAEASEVVETVEPN 166
Query: 108 ----DGGGSGGGGGGGG 120
GG GGGGGGGG
Sbjct: 167 DVEPQQGGRGGGGGGGG 183
Score = 43.5 bits (101), Expect = 0.001
Identities = 33/84 (39%), Positives = 34/84 (40%), Gaps = 15/84 (17%)
Query: 21 GGGDSGGGGWC-------WWRRWRRSVV--VVAVTVVVIVVEAVVVEAVVVDGGGGNDGG 71
GGG GG G C WWR R + VV VE VE GG GG
Sbjct: 178 GGGGGGGRGGCRWGCCGGWWRGRCRYCCRSQAEASEVVETVEPNDVEPQQ----GGRGGG 233
Query: 72 GSGGSSGGGARAWWGRRRQWWRWR 95
G GG GG R WG WWR R
Sbjct: 234 GGGGGGRGGCR--WGCCGGWWRGR 255
>ref|YP_243195.1| adsorption protein [Xanthomonas campestris pv. campestris str.
8004] gi|66573765|gb|AAY49175.1| adsorption protein
[Xanthomonas campestris pv. campestris str. 8004]
Length = 356
Score = 56.2 bits (134), Expect = 2e-07
Identities = 28/57 (49%), Positives = 28/57 (49%), Gaps = 15/57 (26%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
DG GG DGGG GG GGG DGGG G DGGG GGG GGG
Sbjct: 191 DGDGGGDGGGDGGGDGGGDGG---------------GDGGGDGGGDGGGDGGGDGGG 232
Score = 48.5 bits (114), Expect = 4e-05
Identities = 27/57 (47%), Positives = 28/57 (48%), Gaps = 13/57 (22%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
DGG G+DG G GG GG G DGGG G DGGG GGG GGG
Sbjct: 185 DGGDGSDGDG-GGDGGGDGGGDGG------------GDGGGDGGGDGGGDGGGDGGG 228
Score = 44.7 bits (104), Expect = 5e-04
Identities = 24/54 (44%), Positives = 24/54 (44%), Gaps = 11/54 (20%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG 117
G GG DGGG GG GGG G DGGG G DG G G G G
Sbjct: 200 GDGGGDGGGDGGGDGGGDGGGDGG-----------GDGGGDGGGDGDGDGDGDG 242
Score = 43.1 bits (100), Expect = 0.002
Identities = 24/56 (42%), Positives = 24/56 (42%), Gaps = 11/56 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG DGGG GG GGG G DGGG G DG G G G G
Sbjct: 204 GDGGGDGGGDGGGDGGGDGGGDGG-----------GDGGGDGDGDGDGDGDTPGDG 248
Score = 38.9 bits (89), Expect = 0.029
Identities = 23/57 (40%), Positives = 24/57 (41%), Gaps = 13/57 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG DGGG GG GGG G DG G DG G+ G G GG
Sbjct: 216 GDGGGDGGGDGGGDGGGDGDGDG-------------DGDGDTPGDGDGTTPGDGEGG 259
Score = 35.8 bits (81), Expect = 0.25
Identities = 14/21 (66%), Positives = 14/21 (66%)
Query: 99 DDGGGSGSDDGGGSGGGGGGG 119
DDGG DGGG GGG GGG
Sbjct: 184 DDGGDGSDGDGGGDGGGDGGG 204
Score = 34.3 bits (77), Expect = 0.72
Identities = 14/21 (66%), Positives = 14/21 (66%)
Query: 100 DGGGSGSDDGGGSGGGGGGGG 120
D GG GSD GG GGG GGG
Sbjct: 184 DDGGDGSDGDGGGDGGGDGGG 204
>ref|NP_850017.1| glycine-rich RNA-binding protein (GRP7) [Arabidopsis thaliana]
Length = 159
Score = 56.2 bits (134), Expect = 2e-07
Identities = 30/59 (50%), Positives = 35/59 (58%), Gaps = 10/59 (16%)
Query: 63 DGGGGNDGGGSGGSS-GGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGGG GGG G SS GGG ++ G RR+ GGG G +GGG GG GGGGG
Sbjct: 109 EGGGGYSGGGGGYSSRGGGGGSYGGGRRE---------GGGGYGGGEGGGYGGSGGGGG 158
Score = 40.8 bits (94), Expect = 0.008
Identities = 26/67 (38%), Positives = 29/67 (42%), Gaps = 15/67 (22%)
Query: 54 AVVVEAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSG 113
++ V G GG G GGS GGG GRR +GGG S GGG
Sbjct: 78 SITVNEAQSRGSGGGGGHRGGGSYGGGG----GRR-----------EGGGGYSGGGGGYS 122
Query: 114 GGGGGGG 120
GGGGG
Sbjct: 123 SRGGGGG 129
>ref|ZP_00458077.1| putative lipoprotein [Burkholderia cenocepacia AU 1054]
gi|67091651|gb|EAM09221.1| putative lipoprotein
[Burkholderia cenocepacia AU 1054]
Length = 518
Score = 55.8 bits (133), Expect = 2e-07
Identities = 27/58 (46%), Positives = 31/58 (52%), Gaps = 13/58 (22%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGGG GGG GG GGG G GGG+G +GGG+GGGG GGG
Sbjct: 321 NGGGGGGGGGGGGGGGGGGGGGGG-------------GGGGNGGGNGGGNGGGGNGGG 365
Score = 53.5 bits (127), Expect = 1e-06
Identities = 26/57 (45%), Positives = 29/57 (50%), Gaps = 13/57 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G +GGG+G +GGG GGG GGG
Sbjct: 326 GGGGGGGGGGGGGGGGGGGGGGG-------------NGGGNGGGNGGGGNGGGNGGG 369
Score = 53.5 bits (127), Expect = 1e-06
Identities = 27/58 (46%), Positives = 30/58 (51%), Gaps = 11/58 (18%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
DGGGG+ GG G+ GGG G +GGG G GGG GGGGGGGG
Sbjct: 295 DGGGGHGNGGGHGNGGGGNGNGGGH-----------GNGGGGGGGGGGGGGGGGGGGG 341
Score = 53.5 bits (127), Expect = 1e-06
Identities = 26/58 (44%), Positives = 28/58 (47%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGG DGGG G+ GG G GGG G GGG GGGGGGGG
Sbjct: 289 NGGGHGDGGGGHGNGGGHGNGGGGNGNGGGHGNGGGGGGGGGGGGGGGGGGGGGGGGG 346
Score = 52.0 bits (123), Expect = 3e-06
Identities = 26/58 (44%), Positives = 28/58 (47%), Gaps = 17/58 (29%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGGGN GG G+ GGG GGG G GGG GGGGGGGG
Sbjct: 308 NGGGGNGNGGGHGNGGGGGGG-----------------GGGGGGGGGGGGGGGGGGGG 348
Score = 52.0 bits (123), Expect = 3e-06
Identities = 26/55 (47%), Positives = 32/55 (57%), Gaps = 7/55 (12%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGN GGG+GG +GGG G +GGG+G +GGG+GGG GGG
Sbjct: 354 GGGNGGGGNGGGNGGGNGGGNGGGNGG-------GNGGGNGGGNGGGNGGGNGGG 401
Score = 51.6 bits (122), Expect = 4e-06
Identities = 25/58 (43%), Positives = 29/58 (49%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGG DGGG G+ GG G +G G G +GGG GGGGGGGG
Sbjct: 276 NGGGHGDGGGGHGNGGGHGDGGGGHGNGGGHGNGGGGNGNGGGHGNGGGGGGGGGGGG 333
Score = 49.7 bits (117), Expect = 2e-05
Identities = 25/57 (43%), Positives = 31/57 (53%), Gaps = 11/57 (19%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG +GGG+GG +GGG G +GGG+G +GGG G GGGG G
Sbjct: 365 GNGGGNGGGNGGGNGGGNGGGNGG-----------GNGGGNGGGNGGGHGNGGGGHG 410
Score = 47.8 bits (112), Expect = 6e-05
Identities = 24/58 (41%), Positives = 30/58 (51%), Gaps = 4/58 (6%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGG +G GSGG+ GGA R + GGS + GGG+G GGG GG
Sbjct: 411 NGGGNGNGNGSGGAGNGGANGVGNGRGNGGN----SGNAGGSNGNGGGGAGNGGGSGG 464
Score = 47.4 bits (111), Expect = 8e-05
Identities = 24/58 (41%), Positives = 28/58 (47%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGGGN GG GG+ GG G +GGG+G G G GG G GGG
Sbjct: 357 NGGGGNGGGNGGGNGGGNGGGNGGGNGGGNGGGNGGGNGGGNGGGHGNGGGGHGNGGG 414
Score = 46.6 bits (109), Expect = 1e-04
Identities = 23/57 (40%), Positives = 29/57 (50%), Gaps = 12/57 (21%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG +GGG+GG +GGG G + GGG G+ G G+G G GG G
Sbjct: 381 GNGGGNGGGNGGGNGGGNGGGHG------------NGGGGHGNGGGNGNGNGSGGAG 425
Score = 46.6 bits (109), Expect = 1e-04
Identities = 23/56 (41%), Positives = 28/56 (49%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG +GGG+GG +GGG G G G G+ +G GSGG G GG
Sbjct: 373 GNGGGNGGGNGGGNGGGNGGGNGGGNGGGHGNGGGGHGNGGGNGNGNGSGGAGNGG 428
Score = 46.2 bits (108), Expect = 2e-04
Identities = 26/58 (44%), Positives = 29/58 (49%), Gaps = 13/58 (22%)
Query: 65 GGGNDGGGS--GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG+ GG+SGGG G DGGG G+ G G GGGG G G
Sbjct: 244 GGGTSGGGTSGGGTSGGGGHGGGGD-----------GDGGGHGNGGGHGDGGGGHGNG 290
Score = 44.7 bits (104), Expect = 5e-04
Identities = 22/58 (37%), Positives = 29/58 (49%), Gaps = 14/58 (24%)
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGG +G GSGG+ GGA +G G+G+ +GGG G GG GG
Sbjct: 473 NGGGNGNGNGSGGAGNGGANG--------------VGNGHGTGNGNGGGHGNGGSSGG 516
Score = 44.3 bits (103), Expect = 7e-04
Identities = 24/57 (42%), Positives = 26/57 (45%), Gaps = 12/57 (21%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG+ GGG G G G G D GGG G+ G G GGGG G G
Sbjct: 259 GGGGHGGGGDGDGGGHGNGGGHG------------DGGGGHGNGGGHGDGGGGHGNG 303
Score = 43.9 bits (102), Expect = 0.001
Identities = 24/57 (42%), Positives = 28/57 (49%), Gaps = 7/57 (12%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG + GG SGG + GG + G DG G G +GGG G GGGG G
Sbjct: 239 GGGTSGGGTSGGGTSGGGTSGGGGHGGG-------GDGDGGGHGNGGGHGDGGGGHG 288
Score = 43.5 bits (101), Expect = 0.001
Identities = 22/56 (39%), Positives = 27/56 (47%), Gaps = 18/56 (32%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG +GGGSGG++G G G G G+ +G GSGG G GG
Sbjct: 453 GGGAGNGGGSGGANGTGGH------------------GNGGGNGNGNGSGGAGNGG 490
Score = 42.7 bits (99), Expect = 0.002
Identities = 25/59 (42%), Positives = 28/59 (47%), Gaps = 19/59 (32%)
Query: 64 GGGGNDG--GGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GGN G GGS G+ GGGA +GGGSG +G G G GGG G
Sbjct: 437 GNGGNSGNAGGSNGNGGGGA-----------------GNGGGSGGANGTGGHGNGGGNG 478
Score = 41.2 bits (95), Expect = 0.006
Identities = 23/56 (41%), Positives = 24/56 (42%), Gaps = 12/56 (21%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG DG G G +GGG G G G G DGGG G GGG G
Sbjct: 264 GGGGDGDGGGHGNGGGHGDGGGGH------------GNGGGHGDGGGGHGNGGGHG 307
Score = 40.0 bits (92), Expect = 0.013
Identities = 24/60 (40%), Positives = 29/60 (48%), Gaps = 15/60 (25%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGS---GSDDGGGSGGGGGGGG 120
GGG + GG SGG + GG + G GGG+ G+ GG SGGG GGG
Sbjct: 184 GGGTSGGGTSGGGTSGGGTSGGGT------------SGGGTSGGGTSGGGTSGGGTSGGG 231
Score = 40.0 bits (92), Expect = 0.013
Identities = 24/60 (40%), Positives = 29/60 (48%), Gaps = 15/60 (25%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGS---GSDDGGGSGGGGGGGG 120
GGG + GG SGG + GG + G GGG+ G+ GG SGGG GGG
Sbjct: 189 GGGTSGGGTSGGGTSGGGTSGGGT------------SGGGTSGGGTSGGGTSGGGTSGGG 236
Score = 40.0 bits (92), Expect = 0.013
Identities = 24/60 (40%), Positives = 29/60 (48%), Gaps = 15/60 (25%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGS---GSDDGGGSGGGGGGGG 120
GGG + GG SGG + GG + G GGG+ G+ GG SGGG GGG
Sbjct: 174 GGGTSGGGTSGGGTSGGGTSGGGT------------SGGGTSGGGTSGGGTSGGGTSGGG 221
Score = 40.0 bits (92), Expect = 0.013
Identities = 24/60 (40%), Positives = 29/60 (48%), Gaps = 15/60 (25%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGS---GSDDGGGSGGGGGGGG 120
GGG + GG SGG + GG + G GGG+ G+ GG SGGG GGG
Sbjct: 159 GGGTSGGGTSGGGTSGGGTSGGGT------------SGGGTSGGGTSGGGTSGGGTSGGG 206
Score = 40.0 bits (92), Expect = 0.013
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 8/66 (12%)
Query: 63 DGGGGNDGGGS------GGSSGGGARAWWGRRRQWWRWRQWCDDGG--GSGSDDGGGSGG 114
+G GGN+G G+ GG++GG + + G R GG GSG+ GG SGG
Sbjct: 106 NGSGGNNGNGAVGPAGVGGTTGGTSGSTSGTGRGGNASGNGGSGGGTSGSGTSGGGTSGG 165
Query: 115 GGGGGG 120
G GGG
Sbjct: 166 GTSGGG 171
Score = 40.0 bits (92), Expect = 0.013
Identities = 24/60 (40%), Positives = 29/60 (48%), Gaps = 15/60 (25%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGS---GSDDGGGSGGGGGGGG 120
GGG + GG SGG + GG + G GGG+ G+ GG SGGG GGG
Sbjct: 209 GGGTSGGGTSGGGTSGGGTSGGGT------------SGGGTSGGGTSGGGTSGGGTSGGG 256
Score = 39.3 bits (90), Expect = 0.022
Identities = 26/62 (41%), Positives = 29/62 (45%), Gaps = 14/62 (22%)
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG-----GGGG 118
G GG +G GSGG++G GA G GG SGS G G GG GG G
Sbjct: 99 GNGGGNGNGSGGNNGNGAVGPAG---------VGGTTGGTSGSTSGTGRGGNASGNGGSG 149
Query: 119 GG 120
GG
Sbjct: 150 GG 151
Score = 35.4 bits (80), Expect = 0.32
Identities = 19/55 (34%), Positives = 23/55 (41%), Gaps = 18/55 (32%)
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GN G G+GGS G +G G+G +G GSGG G G
Sbjct: 79 GVGNGGAGAGGSGSGNG------------------NGSGNGGGNGNGSGGNNGNG 115
Score = 33.9 bits (76), Expect = 0.94
Identities = 19/58 (32%), Positives = 25/58 (42%), Gaps = 19/58 (32%)
Query: 61 VVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
V +GG G G GSG +G G +GGG+G+ GG +G G G
Sbjct: 80 VGNGGAGAGGSGSGNGNGSG-------------------NGGGNGNGSGGNNGNGAVG 118
Score = 32.3 bits (72), Expect = 2.7
Identities = 19/59 (32%), Positives = 23/59 (38%), Gaps = 19/59 (32%)
Query: 61 VVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
V G G +GG G SG G +G GSG+ G G+G GG G
Sbjct: 74 VYAGRGVGNGGAGAGGSGSG-------------------NGNGSGNGGGNGNGSGGNNG 113
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.142 0.499
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 274,276,414
Number of Sequences: 2540612
Number of extensions: 16264323
Number of successful extensions: 655825
Number of sequences better than 10.0: 8018
Number of HSP's better than 10.0 without gapping: 4228
Number of HSP's successfully gapped in prelim test: 3895
Number of HSP's that attempted gapping in prelim test: 258780
Number of HSP's gapped (non-prelim): 98266
length of query: 120
length of database: 863,360,394
effective HSP length: 96
effective length of query: 24
effective length of database: 619,461,642
effective search space: 14867079408
effective search space used: 14867079408
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0090b.2


