

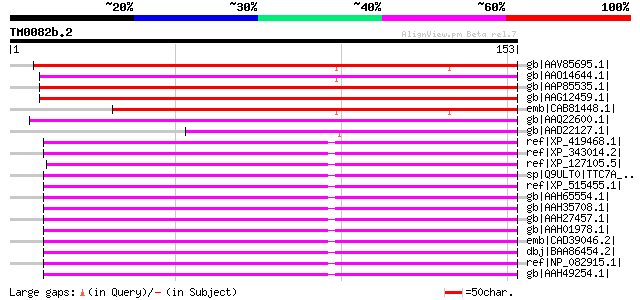
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.2
(153 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAV85695.1| At4g28600 [Arabidopsis thaliana] gi|51536508|gb|A... 164 5e-40
gb|AAO14644.1| pollen-specific calmodulin-binding protein [Arabi... 137 7e-32
gb|AAP85535.1| calmodulin-binding protein [Oryza sativa (indica ... 127 4e-29
gb|AAG12459.1| calmodulin-binding protein MPCBP [Zea mays] 127 5e-29
emb|CAB81448.1| putative protein [Arabidopsis thaliana] gi|74876... 117 7e-26
gb|AAQ22600.1| At1g27460/F17L21_26 [Arabidopsis thaliana] gi|183... 115 2e-25
gb|AAD22127.1| hypothetical protein [Arabidopsis thaliana] gi|25... 86 2e-16
ref|XP_419468.1| PREDICTED: similar to hypothetical protein [Gal... 67 9e-11
ref|XP_343014.2| PREDICTED: similar to tetratricopeptide repeat ... 67 1e-10
ref|XP_127105.5| PREDICTED: tetratricopeptide repeat domain 7B [... 67 1e-10
sp|Q9ULT0|TTC7A_HUMAN Tetratricopeptide repeat protein 7A (TPR r... 66 1e-10
ref|XP_515455.1| PREDICTED: hypothetical protein XP_515455 [Pan ... 66 1e-10
gb|AAH65554.1| TTC7A protein [Homo sapiens] 66 1e-10
gb|AAH35708.1| TTC7A protein [Homo sapiens] 66 1e-10
gb|AAH27457.1| TTC7A protein [Homo sapiens] 66 1e-10
gb|AAH01978.1| TTC7A protein [Homo sapiens] 66 1e-10
emb|CAD39046.2| hypothetical protein [Homo sapiens] gi|55741819|... 66 1e-10
dbj|BAA86454.2| KIAA1140 protein [Homo sapiens] 66 1e-10
ref|NP_082915.1| tetratricopeptide repeat domain 7 [Mus musculus... 66 2e-10
gb|AAH49254.1| Ttc7 protein [Mus musculus] 66 2e-10
>gb|AAV85695.1| At4g28600 [Arabidopsis thaliana] gi|51536508|gb|AAU05492.1|
At4g28600 [Arabidopsis thaliana]
gi|22329002|ref|NP_194589.2| calmodulin-binding protein
[Arabidopsis thaliana]
Length = 739
Score = 164 bits (414), Expect = 5e-40
Identities = 87/150 (58%), Positives = 107/150 (71%), Gaps = 4/150 (2%)
Query: 8 NLEEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKA 67
+LE WHDLA+ YI+LSQW DAE CLS+S+ Y++ R H+ G ++ +G +EA++A
Sbjct: 589 SLELGTWHDLAHIYINLSQWRDAESCLSRSRLIAPYSSVRYHIEGVLYNRRGQLEEAMEA 648
Query: 68 FRDALNIDPRHVPSLISTAVALRRWSNQSD-PVVRSFLMDALRYDRLNASAWYNLGILHK 126
F AL+IDP HVPSL S A L N+S VVRSFLM+ALR DRLN SAWYNLG + K
Sbjct: 649 FTTALDIDPMHVPSLTSKAEILLEVGNRSGIAVVRSFLMEALRIDRLNHSAWYNLGKMFK 708
Query: 127 AEGRV---LEAVECFQAANSLEETAPIEPF 153
AEG V EAVECFQAA +LEET P+EPF
Sbjct: 709 AEGSVSSMQEAVECFQAAVTLEETMPVEPF 738
>gb|AAO14644.1| pollen-specific calmodulin-binding protein [Arabidopsis thaliana]
gi|30689234|ref|NP_850382.1| calmodulin-binding protein
[Arabidopsis thaliana]
Length = 704
Score = 137 bits (344), Expect = 7e-32
Identities = 69/147 (46%), Positives = 89/147 (59%), Gaps = 3/147 (2%)
Query: 10 EEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFR 69
E E+WH LAY Y SLS W+D EVCL K+ +QY+AS H G M E + +K A+ AF
Sbjct: 554 EFEVWHGLAYLYSSLSHWNDVEVCLKKAGELKQYSASMLHTEGRMWEGRKEFKPALAAFL 613
Query: 70 DALNIDPRHVPSLISTAVALRRWSNQSD---PVVRSFLMDALRYDRLNASAWYNLGILHK 126
D L +D VP ++ L PV RS L DALR D N AWY LG++HK
Sbjct: 614 DGLLLDGSSVPCKVAVGALLSERGKDHQPTLPVARSLLSDALRIDPTNRKAWYYLGMVHK 673
Query: 127 AEGRVLEAVECFQAANSLEETAPIEPF 153
++GR+ +A +CFQAA+ LEE+ PIE F
Sbjct: 674 SDGRIADATDCFQAASMLEESDPIESF 700
>gb|AAP85535.1| calmodulin-binding protein [Oryza sativa (indica cultivar-group)]
Length = 697
Score = 127 bits (320), Expect = 4e-29
Identities = 64/144 (44%), Positives = 91/144 (62%)
Query: 10 EEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFR 69
E EIW LA Y SLS W DAE+CL K++A + Y+A+ H G M EA+ KEA+ A+
Sbjct: 550 EFEIWQGLANLYSSLSIWRDAEICLRKARALKSYSAATMHAEGYMLEARDQNKEALAAYV 609
Query: 70 DALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEG 129
+A +I+ HVPS ++ L + ++ P R FL DALR + N AW +LG +H+ +G
Sbjct: 610 NAFSIELEHVPSKVAIGALLCKQGSRYLPAARCFLSDALRIEPTNRMAWLHLGKVHRNDG 669
Query: 130 RVLEAVECFQAANSLEETAPIEPF 153
R+ +A +CFQAA LEE+ P+E F
Sbjct: 670 RINDAADCFQAAVMLEESDPVESF 693
>gb|AAG12459.1| calmodulin-binding protein MPCBP [Zea mays]
Length = 659
Score = 127 bits (319), Expect = 5e-29
Identities = 62/144 (43%), Positives = 90/144 (62%)
Query: 10 EEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFR 69
E EIW LA Y SLS W DAE+CL+K++A + Y+A+ H G MHEA+ +A+ A+
Sbjct: 512 EFEIWQGLANLYSSLSYWRDAEICLNKARALKLYSAATLHAEGYMHEARDQTTDALAAYV 571
Query: 70 DALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEG 129
+A + + HVPS ++ L + + P R FL DALR + N AW LG +H+++G
Sbjct: 572 NAFSTELEHVPSKVAIGAMLSKQGPRFLPAARCFLSDALRVEPTNRMAWLYLGKVHRSDG 631
Query: 130 RVLEAVECFQAANSLEETAPIEPF 153
R+ +A +CFQAA LEE+ P+E F
Sbjct: 632 RISDAADCFQAAVMLEESDPVESF 655
>emb|CAB81448.1| putative protein [Arabidopsis thaliana] gi|7487684|pir||T10654
hypothetical protein T5F17.50 - Arabidopsis thaliana
Length = 710
Score = 117 bits (292), Expect = 7e-26
Identities = 68/126 (53%), Positives = 83/126 (64%), Gaps = 4/126 (3%)
Query: 32 VCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRR 91
VCL ++ A + G ++ +G +EA++AF AL+IDP HVPSL S A L
Sbjct: 584 VCLFETSKAVILIAYSYRLAGVLYNRRGQLEEAMEAFTTALDIDPMHVPSLTSKAEILLE 643
Query: 92 WSNQSD-PVVRSFLMDALRYDRLNASAWYNLGILHKAEGRV---LEAVECFQAANSLEET 147
N+S VVRSFLM+ALR DRLN SAWYNLG + KAEG V EAVECFQAA +LEET
Sbjct: 644 VGNRSGIAVVRSFLMEALRIDRLNHSAWYNLGKMFKAEGSVSSMQEAVECFQAAVTLEET 703
Query: 148 APIEPF 153
P+EPF
Sbjct: 704 MPVEPF 709
>gb|AAQ22600.1| At1g27460/F17L21_26 [Arabidopsis thaliana]
gi|18396347|ref|NP_564285.1| calmodulin-binding protein
[Arabidopsis thaliana] gi|16226498|gb|AAL16183.1|
At1g27460/F17L21_26 [Arabidopsis thaliana]
gi|9802536|gb|AAF99738.1| F17L21.25 [Arabidopsis
thaliana]
Length = 694
Score = 115 bits (288), Expect = 2e-25
Identities = 60/147 (40%), Positives = 88/147 (59%)
Query: 7 RNLEEEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVK 66
+ E E W DLA Y L W DAE CL K+++ Y+ + G EAK L++EA+
Sbjct: 547 QKFETEAWQDLASVYGKLGSWSDAETCLEKARSMCYYSPRGWNETGLCLEAKSLHEEALI 606
Query: 67 AFRDALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHK 126
+F +L+I+P HVPS++S A + + ++S P +SFLM+ALR D N AW LG + K
Sbjct: 607 SFFLSLSIEPDHVPSIVSIAEVMMKSGDESLPTAKSFLMNALRLDPRNHDAWMKLGHVAK 666
Query: 127 AEGRVLEAVECFQAANSLEETAPIEPF 153
+G +A E +QAA LE +AP++ F
Sbjct: 667 KQGLSQQAAEFYQAAYELELSAPVQSF 693
>gb|AAD22127.1| hypothetical protein [Arabidopsis thaliana] gi|25408831|pir||C84861
hypothetical protein At2g43040 [imported] - Arabidopsis
thaliana
Length = 666
Score = 85.9 bits (211), Expect = 2e-16
Identities = 45/103 (43%), Positives = 60/103 (57%), Gaps = 3/103 (2%)
Query: 54 MHEAKGLYKEAVKAFRDALNIDPRHVPSLISTAVALRRWSNQSDP---VVRSFLMDALRY 110
M E + +K A+ AF D L +D VP ++ L P V RS L DALR
Sbjct: 560 MWEGRKEFKPALAAFLDGLLLDGSSVPCKVAVGALLSERGKDHQPTLPVARSLLSDALRI 619
Query: 111 DRLNASAWYNLGILHKAEGRVLEAVECFQAANSLEETAPIEPF 153
D N AWY LG++HK++GR+ +A +CFQAA+ LEE+ PIE F
Sbjct: 620 DPTNRKAWYYLGMVHKSDGRIADATDCFQAASMLEESDPIESF 662
>ref|XP_419468.1| PREDICTED: similar to hypothetical protein [Gallus gallus]
Length = 779
Score = 67.0 bits (162), Expect = 9e-11
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + + + ++ G + E KG + A + + +
Sbjct: 632 EQIWLQAAELFMEQQHLKEAGFCIQEAASLFPTSHAVLYMRGRLAEMKGNLEVARQLYDE 691
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R + + + L DA+R + AW +LG + +A+GR
Sbjct: 692 ALTVNPDGVEIMHSLGLVLNRLERRE--LAQKVLRDAIRIQNTSHRAWNSLGEVLQAQGR 749
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AVECF A LE ++P+ PF
Sbjct: 750 NEAAVECFLTALDLESSSPVIPF 772
>ref|XP_343014.2| PREDICTED: similar to tetratricopeptide repeat domain 7 [Rattus
norvegicus]
Length = 1107
Score = 66.6 bits (161), Expect = 1e-10
Identities = 39/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ Q +A C+ ++ + S ++ G + E KG ++EA + +++
Sbjct: 960 EQIWLQAAELFMEQRQLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGSFEEAKQLYKE 1019
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ + AW LG + + +G
Sbjct: 1020 ALTVNPDGVCIMHSLGLILSRLGHKS--LAQKVLRDAVERQSTHHEAWQGLGEVLQDQGH 1077
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
A +CF A LE ++P+ PF
Sbjct: 1078 NEAAADCFLTALELEASSPVLPF 1100
>ref|XP_127105.5| PREDICTED: tetratricopeptide repeat domain 7B [Mus musculus]
Length = 843
Score = 66.6 bits (161), Expect = 1e-10
Identities = 40/142 (28%), Positives = 69/142 (48%), Gaps = 2/142 (1%)
Query: 12 EIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRDA 71
+IW A YI + + +A C ++ + + ++ G + E +G + EA + + +A
Sbjct: 697 QIWLHAAEVYIGIGKPAEATACTQEAANLFPMSHNVLYMRGQVAELRGHFDEARRWYEEA 756
Query: 72 LNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGRV 131
L+I P HV S+ A+ L + S + L DA++ + W LG + +A+G
Sbjct: 757 LSISPTHVKSMQRLALVLHQLGRYS--LAEKILRDAVQVNSTAHEVWNGLGEVLQAQGND 814
Query: 132 LEAVECFQAANSLEETAPIEPF 153
A ECF A LE ++P PF
Sbjct: 815 AAATECFLTALELEASSPAVPF 836
>sp|Q9ULT0|TTC7A_HUMAN Tetratricopeptide repeat protein 7A (TPR repeat protein 7)
Length = 858
Score = 66.2 bits (160), Expect = 1e-10
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + S ++ G + E KG +EA + +++
Sbjct: 711 EQIWLQAAELFMEQQHLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGNLEEAKQLYKE 770
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ AW LG + +A+G+
Sbjct: 771 ALTVNPDGVRIMHSLGLMLSRLGHKS--LAQKVLRDAVERQSTCHEAWQGLGEVLQAQGQ 828
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 829 NEAAVDCFLTALELEASSPVLPF 851
>ref|XP_515455.1| PREDICTED: hypothetical protein XP_515455 [Pan troglodytes]
Length = 1903
Score = 66.2 bits (160), Expect = 1e-10
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + S ++ G + E KG +EA + +++
Sbjct: 1756 EQIWLQAAELFMEQQHLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGNLEEAKQLYKE 1815
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ AW LG + +A+G+
Sbjct: 1816 ALTVNPDGVRIMHSLGLMLSRLGHKS--LAQKVLRDAVERQSTCHEAWQGLGEVLQAQGQ 1873
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 1874 NEAAVDCFLTALELEASSPVLPF 1896
>gb|AAH65554.1| TTC7A protein [Homo sapiens]
Length = 686
Score = 66.2 bits (160), Expect = 1e-10
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + S ++ G + E KG +EA + +++
Sbjct: 539 EQIWLQAAELFMEQQHLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGNLEEAKQLYKE 598
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ AW LG + +A+G+
Sbjct: 599 ALTVNPDGVRIMHSLGLMLSRLGHKS--LAQKVLRDAVERQSTCHEAWQGLGEVLQAQGQ 656
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 657 NEAAVDCFLTALELEASSPVLPF 679
>gb|AAH35708.1| TTC7A protein [Homo sapiens]
Length = 663
Score = 66.2 bits (160), Expect = 1e-10
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + S ++ G + E KG +EA + +++
Sbjct: 516 EQIWLQAAELFMEQQHLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGNLEEAKQLYKE 575
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ AW LG + +A+G+
Sbjct: 576 ALTVNPDGVRIMHSLGLMLSRLGHKS--LAQKVLRDAVERQSTCHEAWQGLGEVLQAQGQ 633
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 634 NEAAVDCFLTALELEASSPVLPF 656
>gb|AAH27457.1| TTC7A protein [Homo sapiens]
Length = 168
Score = 66.2 bits (160), Expect = 1e-10
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + S ++ G + E KG +EA + +++
Sbjct: 21 EQIWLQAAELFMEQQHLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGNLEEAKQLYKE 80
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ AW LG + +A+G+
Sbjct: 81 ALTVNPDGVRIMHSLGLMLSRLGHKS--LAQKVLRDAVERQSTCHEAWQGLGEVLQAQGQ 138
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 139 NEAAVDCFLTALELEASSPVLPF 161
>gb|AAH01978.1| TTC7A protein [Homo sapiens]
Length = 450
Score = 66.2 bits (160), Expect = 1e-10
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + S ++ G + E KG +EA + +++
Sbjct: 303 EQIWLQAAELFMEQQHLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGNLEEAKQLYKE 362
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ AW LG + +A+G+
Sbjct: 363 ALTVNPDGVRIMHSLGLMLSRLGHKS--LAQKVLRDAVERQSTCHEAWQGLGEVLQAQGQ 420
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 421 NEAAVDCFLTALELEASSPVLPF 443
>emb|CAD39046.2| hypothetical protein [Homo sapiens] gi|55741819|ref|NP_065191.1|
tetratricopeptide repeat domain 7A [Homo sapiens]
Length = 858
Score = 66.2 bits (160), Expect = 1e-10
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + S ++ G + E KG +EA + +++
Sbjct: 711 EQIWLQAAELFMEQQHLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGNLEEAKQLYKE 770
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ AW LG + +A+G+
Sbjct: 771 ALTVNPDGVRIMHSLGLMLSRLGHKS--LAQKVLRDAVERQSTCHEAWQGLGEVLQAQGQ 828
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 829 NEAAVDCFLTALELEASSPVLPF 851
>dbj|BAA86454.2| KIAA1140 protein [Homo sapiens]
Length = 752
Score = 66.2 bits (160), Expect = 1e-10
Identities = 40/143 (27%), Positives = 73/143 (50%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ +A C+ ++ + S ++ G + E KG +EA + +++
Sbjct: 605 EQIWLQAAELFMEQQHLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGNLEEAKQLYKE 664
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L R ++S + + L DA+ AW LG + +A+G+
Sbjct: 665 ALTVNPDGVRIMHSLGLMLSRLGHKS--LAQKVLRDAVERQSTCHEAWQGLGEVLQAQGQ 722
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 723 NEAAVDCFLTALELEASSPVLPF 745
>ref|NP_082915.1| tetratricopeptide repeat domain 7 [Mus musculus]
gi|37515285|gb|AAH42512.2| Tetratricopeptide repeat
domain 7 [Mus musculus]
gi|34222845|sp|Q8BGB2|TTC7A_MOUSE Tetratricopeptide
repeat protein 7A (TPR repeat protein 7)
gi|26348451|dbj|BAC37865.1| unnamed protein product [Mus
musculus] gi|26340982|dbj|BAC34153.1| unnamed protein
product [Mus musculus] gi|26333833|dbj|BAC30634.1|
unnamed protein product [Mus musculus]
Length = 858
Score = 65.9 bits (159), Expect = 2e-10
Identities = 39/143 (27%), Positives = 74/143 (51%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ Q +A C+ ++ + S ++ G + E KG ++EA + +++
Sbjct: 711 EQIWLQAAELFMEQRQLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGSFEEAKQLYKE 770
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L + ++S + + L DA+ AW LG + + +G+
Sbjct: 771 ALTVNPDGVRIMHSLGLMLSQLGHKS--LAQKVLRDAVERQSTFHEAWQGLGEVLQDQGQ 828
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 829 NEAAVDCFLTALELEASSPVLPF 851
>gb|AAH49254.1| Ttc7 protein [Mus musculus]
Length = 211
Score = 65.9 bits (159), Expect = 2e-10
Identities = 39/143 (27%), Positives = 74/143 (51%), Gaps = 2/143 (1%)
Query: 11 EEIWHDLAYAYISLSQWHDAEVCLSKSKAFRQYTASRCHVIGTMHEAKGLYKEAVKAFRD 70
E+IW A ++ Q +A C+ ++ + S ++ G + E KG ++EA + +++
Sbjct: 64 EQIWLQAAELFMEQRQLKEAGFCIQEAAGLFPTSHSVLYMRGRLAEVKGSFEEAKQLYKE 123
Query: 71 ALNIDPRHVPSLISTAVALRRWSNQSDPVVRSFLMDALRYDRLNASAWYNLGILHKAEGR 130
AL ++P V + S + L + ++S + + L DA+ AW LG + + +G+
Sbjct: 124 ALTVNPDGVRIMHSLGLMLSQLGHKS--LAQKVLRDAVERQSTFHEAWQGLGEVLQDQGQ 181
Query: 131 VLEAVECFQAANSLEETAPIEPF 153
AV+CF A LE ++P+ PF
Sbjct: 182 NEAAVDCFLTALELEASSPVLPF 204
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 242,616,232
Number of Sequences: 2540612
Number of extensions: 8354527
Number of successful extensions: 26240
Number of sequences better than 10.0: 754
Number of HSP's better than 10.0 without gapping: 260
Number of HSP's successfully gapped in prelim test: 497
Number of HSP's that attempted gapping in prelim test: 24418
Number of HSP's gapped (non-prelim): 1982
length of query: 153
length of database: 863,360,394
effective HSP length: 129
effective length of query: 24
effective length of database: 535,621,446
effective search space: 12854914704
effective search space used: 12854914704
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0082b.2


