

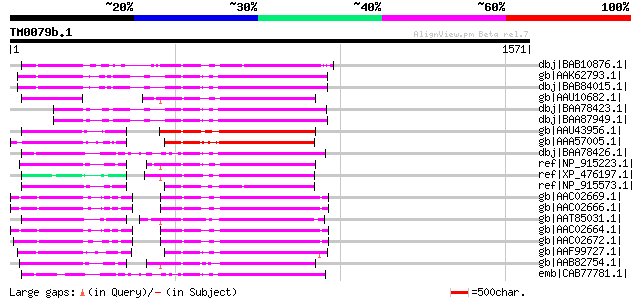
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0079b.1
(1571 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB10876.1| polyprotein [Arabidopsis thaliana] 422 e-116
gb|AAK62793.1| polyprotein, putative [Arabidopsis thaliana] 416 e-114
dbj|BAB84015.1| polyprotein [Arabidopsis thaliana] gi|14475941|g... 416 e-114
gb|AAU10682.1| putative polyprotein [Oryza sativa (japonica cult... 380 e-103
dbj|BAA78423.1| polyprotein [Arabidopsis thaliana] 372 e-101
dbj|BAA87949.1| retrotransposon homolog [Arabidopsis thaliana] g... 372 e-101
gb|AAU43956.1| unknown protein [Oryza sativa (japonica cultivar-... 372 e-101
gb|AAA57005.1| copia-like retrotransposon Hopscotch polyprotein ... 371 e-101
dbj|BAA78426.1| polyprotein [Arabidopsis thaliana] 369 e-100
ref|NP_915223.1| putative rice retrotransposon retrofit gag/pol ... 369 e-100
ref|XP_476197.1| putative polyprotein [Oryza sativa (japonica cu... 365 5e-99
ref|NP_915573.1| putative gag/pol polyprotein [Oryza sativa (jap... 365 7e-99
gb|AAC02669.1| polyprotein [Arabidopsis thaliana] 364 1e-98
gb|AAC02666.1| polyprotein [Arabidopsis thaliana] 364 1e-98
gb|AAT85031.1| putative polyprotein [Oryza sativa (japonica cult... 364 1e-98
gb|AAC02664.1| polyprotein [Arabidopsis thaliana] 362 4e-98
gb|AAC02672.1| polyprotein [Arabidopsis arenosa] gi|7522104|pir|... 359 5e-97
gb|AAF99727.1| F17L21.7 [Arabidopsis thaliana] 348 6e-94
gb|AAB82754.1| retrofit [Oryza longistaminata] gi|7444451|pir||T... 346 3e-93
emb|CAB77781.1| putative polyprotein of LTR transposon [Arabidop... 342 5e-92
>dbj|BAB10876.1| polyprotein [Arabidopsis thaliana]
Length = 1429
Score = 422 bits (1084), Expect = e-116
Identities = 299/946 (31%), Positives = 446/946 (46%), Gaps = 170/946 (17%)
Query: 35 KLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRANSVENPAFLVWEQHDS 94
KL +NFL W++ V ++ + L ++ E P ++ + NP + +W++ D
Sbjct: 6 KLTSTNFLMWRRQVHALLDGYDLAGYVDGSIEEPHTTVTVHGVTSP--NPEYKLWKRQDK 63
Query: 95 ALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSITKGSKNT 154
+++ L+ ++S +V P + S QIW ++D + S Q+R +++ KGS++
Sbjct: 64 LIYSGLIGAISVAVQPLLSQATTSAQIWRKLVDTYANPSRGHKQQIREQIKQWKKGSRSI 123
Query: 155 SDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTSREFPCSISQAEAM 214
DY+ + + + L + + + + + + I GL +DY ++ + R+ SI++
Sbjct: 124 DDYVLGLTTRFDQLALLEEAIPHEDQIAYILGGLSDDYRRVIDQIEGRDISPSITELHEK 183
Query: 215 VIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQAPPPVAIAP 274
+I E +L QA P STP +
Sbjct: 184 LINFELKL------------------QAMVPDSSTPVTANAASYNNNN------------ 213
Query: 275 PSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGRGSYNNRGRGGDRSSV 334
+ N S G+ + ++ +SRS NNRG G+G Y R
Sbjct: 214 -NGRNNRSSSRGNQNNQWQQNQTQQSRS---------NNRGSQGKG-YQGR--------- 253
Query: 335 QCQICHKYGHDASVCYYRGNSTSPATNSQPNMTNQAPAPVAPTFGFGSSFGMMPQFGYSP 394
CQIC +GH A C SQ
Sbjct: 254 -CQICGVHGHSARRC------------SQ------------------------------- 269
Query: 395 FRGFQPYGAPSPFGMSFPNSGFGSGSPYASGGMFTPPGYGFGSGYGFGLPPRAPARHIRA 454
FQPYG GSG G P GY G+ P AP +
Sbjct: 270 ---FQPYG--------------GSG-----GSQSVPSGYPTN---GYSPSPMAPWQ---- 300
Query: 455 PQTMLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSA 514
P+ +A+AP W DSGATHH+T D + +S TG ++V + +G GLPI GSA
Sbjct: 301 PRANIATAPP-FNPWVLDSGATHHLTSDLANLSMHQPYTGGEEVTIADGSGLPISHTGSA 359
Query: 515 SFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLL 574
+P +L LK++L VPN++KNL+SV + N V EF P H V +T LL
Sbjct: 360 LLPTP---SRSLALKDILYVPNVSKNLISVYRLCNANQVSVEFFPAHFQVKDLNTGARLL 416
Query: 575 RGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYD 634
+G ++ LY + PV+ ASP+ T +S+
Sbjct: 417 QGRTRNE-LYEW---------------PVNQKSITILTASPSPKTDLSS----------- 449
Query: 635 LWHSRLGHPHYETLQSALTTCKISVPHK-SKYTICHSCCVAKSHRLPSSASSTLYTAPLE 693
WH RLGHP L+ ++ + + + K C C + KSH+LP ++ + + PLE
Sbjct: 450 -WHQRLGHPALPILKDVVSHFHLPLSNTIPKQLPCSDCSINKSHKLPFFTNTIVSSQPLE 508
Query: 694 LIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGL 753
++ D+W I S Y Y+L VD F+R+TW+Y LK+KS+ VF+ F+A+VE +F
Sbjct: 509 YLYTDVWTSPCI-SVDNYKYYLVIVDHFTRYTWMYPLKQKSQVKDVFVAFKALVENRFQS 567
Query: 754 KIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+I+++ +D G EF L P GI+H + PHT NG ERKHRHIVETGL+LL A+
Sbjct: 568 RIRTLYSDNGGEFIGLRPFLAAHGISHLTSPPHTPEHNGLAERKHRHIVETGLALLTHAS 627
Query: 814 MPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNS 873
+P ++W +AF TA +LINR+PT VL SPY KLF++ +Y LRVFG CYP LRPYN+
Sbjct: 628 LPKTFWTYAFATAVYLINRMPTEVLQGTSPYVKLFQMSPNYLKLRVFGCLCYPWLRPYNT 687
Query: 874 SKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTLFSSEFPASNP 932
+KL RS CVFLGYS + Y CL + RIY S+ VQF E FP+++ +SE ++
Sbjct: 688 NKLEARSTMCVFLGYSLTQSAYLCLDIATNRIYTSRHVQFVESSFPFASPRTSETDSTQT 747
Query: 933 SSVSSVIPVISYVPSLQQPIPSPHHSHPTASHLLIQLIPCSHCPIH 978
S + VI P LQ+P PH + PTA + L P H P H
Sbjct: 748 MSQPTTTNVI---PLLQRP---PHIAPPTA----LPLCPIFHSPPH 783
>gb|AAK62793.1| polyprotein, putative [Arabidopsis thaliana]
Length = 1466
Score = 416 bits (1069), Expect = e-114
Identities = 290/947 (30%), Positives = 449/947 (46%), Gaps = 172/947 (18%)
Query: 23 LSSTSLVH---KLLLKLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRAN 79
L++TS+++ + KL +N+L W + V + + L FL +PP + D A
Sbjct: 10 LNNTSILNVNMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGT-DAAP 68
Query: 80 SVENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQ 139
V NP + W++ D +++ +L ++S SV P V + QIWE + + S TQ
Sbjct: 69 RV-NPDYTRWKRQDKLIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQ 127
Query: 140 LRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIV 199
LR++L+ TKG+K DY++ + + + L +G P+ + E +E + + L E+Y ++ +
Sbjct: 128 LRTQLKQWTKGTKTIDDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQI 187
Query: 200 TSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQ 259
+++ P ++++ ++ HE+++ LA ++A
Sbjct: 188 AAKDTPPTLTEIHERLLNHESKI----------------LAVSSA--------------- 216
Query: 260 SPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGR 319
I ++ V++ + T +++ + + +R+ DNR+ NN + +
Sbjct: 217 ----------TVIPITANAVSHRNTTTTNN----NNNGNRNNRYDNRNN---NNNSKPWQ 259
Query: 320 GSYNNRGRGGDRSSV---QCQICHKYGHDASVCYYRGNSTSPATNSQPNMTNQAPAPVAP 376
S N ++S +CQIC GH A C + S + QP
Sbjct: 260 QSSTNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQP------------ 307
Query: 377 TFGFGSSFGMMPQFGYSPFRGFQPYGAPSPFGMSFPNSGFGSGSPYASGGMFTPPGYGFG 436
PSPF P + GSPY+S
Sbjct: 308 ---------------------------PSPFTPWQPRANLALGSPYSSN----------- 329
Query: 437 SGYGFGLPPRAPARHIRAPQTMLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGND 496
NW DSGATHH+T D + +S TG D
Sbjct: 330 -------------------------------NWLLDSGATHHITSDFNNLSLHQPYTGGD 358
Query: 497 QVYMGNGQGLPIQSIGSASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFE 556
V + +G +PI GS S S+ L L N+L VPNI KNL+SV + NGV E
Sbjct: 359 DVMVADGSTIPISHTGSTSLSTK---SRPLNLHNILYVPNIHKNLISVYRLCNANGVSVE 415
Query: 557 FHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPA 616
F P V +T LL+G D LY + P++S+ +S ASP+
Sbjct: 416 FFPASFQVKDLNTGVPLLQGKT-KDELYEW---------------PIASSQPVSLFASPS 459
Query: 617 VNTIVSTSNNVPSPTAYDLWHSRLGHPHYETLQSALTTCKISVPHKS-KYTICHSCCVAK 675
S + WH+RLGHP L S ++ +SV + S K+ C C + K
Sbjct: 460 ------------SKATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLSCSDCLINK 507
Query: 676 SHRLPSSASSTLYTAPLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSE 735
S+++P S S+ T PLE I++D+W + I S Y Y++ VD F+R+TW+Y LK+KS+
Sbjct: 508 SNKVPFSQSTINSTRPLEYIYSDVWS-SPILSHDNYRYYVIFVDHFTRYTWLYPLKQKSQ 566
Query: 736 TSTVFIQFQAMVELKFGLKIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVE 795
FI F+ ++E +F +I + +D G EF L +F + GI+H + PHT NG E
Sbjct: 567 VKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFSQHGISHLTSPPHTPEHNGLSE 626
Query: 796 RKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYK 855
RKHRHIVETGL+LL+ A++P +YW +AF A +LINRLPTP+L SP+ KLF +Y
Sbjct: 627 RKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLFGTSPNYD 686
Query: 856 ILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNE 914
LRVFG ACYP LRPYN KL +S++CVFLGYS + Y CL R+YIS+ V+F+E
Sbjct: 687 KLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDE 746
Query: 915 FKFPYSTLFSSEFPASNPSSVSSVI-PVISYVPSLQQPIPSPHHSHP 960
FP+S ++ P SS + + +P+ +P+P S P
Sbjct: 747 NCFPFSNYLATLSPVQEQRRESSCVWSPHTTLPTRTPVLPAPSCSDP 793
>dbj|BAB84015.1| polyprotein [Arabidopsis thaliana] gi|14475941|gb|AAK62788.1|
polyprotein, putative [Arabidopsis thaliana]
Length = 1466
Score = 416 bits (1069), Expect = e-114
Identities = 290/947 (30%), Positives = 449/947 (46%), Gaps = 172/947 (18%)
Query: 23 LSSTSLVH---KLLLKLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRAN 79
L++TS+++ + KL +N+L W + V + + L FL +PP + D A
Sbjct: 10 LNNTSILNVNMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGT-DAAP 68
Query: 80 SVENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQ 139
V NP + W++ D +++ +L ++S SV P V + QIWE + + S TQ
Sbjct: 69 RV-NPDYTRWKRQDKLIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQ 127
Query: 140 LRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIV 199
LR++L+ TKG+K DY++ + + + L +G P+ + E +E + + L E+Y ++ +
Sbjct: 128 LRTQLKQWTKGTKTIDDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQI 187
Query: 200 TSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQ 259
+++ P ++++ ++ HE+++ LA ++A
Sbjct: 188 AAKDTPPTLTEIHERLLNHESKI----------------LAVSSA--------------- 216
Query: 260 SPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGR 319
I ++ V++ + T +++ + + +R+ DNR+ NN + +
Sbjct: 217 ----------TVIPITANAVSHRNTTTTNN----NNNGNRNNRYDNRNN---NNNSKPWQ 259
Query: 320 GSYNNRGRGGDRSSV---QCQICHKYGHDASVCYYRGNSTSPATNSQPNMTNQAPAPVAP 376
S N ++S +CQIC GH A C + S + QP
Sbjct: 260 QSSTNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQP------------ 307
Query: 377 TFGFGSSFGMMPQFGYSPFRGFQPYGAPSPFGMSFPNSGFGSGSPYASGGMFTPPGYGFG 436
PSPF P + GSPY+S
Sbjct: 308 ---------------------------PSPFTPWQPRANLALGSPYSSN----------- 329
Query: 437 SGYGFGLPPRAPARHIRAPQTMLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGND 496
NW DSGATHH+T D + +S TG D
Sbjct: 330 -------------------------------NWLLDSGATHHITSDFNNLSLHQPYTGGD 358
Query: 497 QVYMGNGQGLPIQSIGSASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFE 556
V + +G +PI GS S S+ L L N+L VPNI KNL+SV + NGV E
Sbjct: 359 DVMVADGSTIPISHTGSTSLSTK---SRPLNLHNILYVPNIHKNLISVYRLCNANGVSVE 415
Query: 557 FHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPA 616
F P V +T LL+G D LY + P++S+ +S ASP+
Sbjct: 416 FFPASFQVKDLNTGVPLLQGKT-KDELYEW---------------PIASSQPVSLFASPS 459
Query: 617 VNTIVSTSNNVPSPTAYDLWHSRLGHPHYETLQSALTTCKISVPHKS-KYTICHSCCVAK 675
S + WH+RLGHP L S ++ +SV + S K+ C C + K
Sbjct: 460 ------------SKATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLSCSDCLINK 507
Query: 676 SHRLPSSASSTLYTAPLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSE 735
S+++P S S+ T PLE I++D+W + I S Y Y++ VD F+R+TW+Y LK+KS+
Sbjct: 508 SNKVPFSQSTINSTRPLEYIYSDVWS-SPILSHDNYRYYVIFVDHFTRYTWLYPLKQKSQ 566
Query: 736 TSTVFIQFQAMVELKFGLKIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVE 795
FI F+ ++E +F +I + +D G EF L +F + GI+H + PHT NG E
Sbjct: 567 VKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFSQHGISHLTSPPHTPEHNGLSE 626
Query: 796 RKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYK 855
RKHRHIVETGL+LL+ A++P +YW +AF A +LINRLPTP+L SP+ KLF +Y
Sbjct: 627 RKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLFGTSPNYD 686
Query: 856 ILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNE 914
LRVFG ACYP LRPYN KL +S++CVFLGYS + Y CL R+YIS+ V+F+E
Sbjct: 687 KLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDE 746
Query: 915 FKFPYSTLFSSEFPASNPSSVSSVI-PVISYVPSLQQPIPSPHHSHP 960
FP+S ++ P SS + + +P+ +P+P S P
Sbjct: 747 NCFPFSNYLATLSPVQEQRRESSCVWSPHTTLPTRTPVLPAPSCSDP 793
>gb|AAU10682.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1210
Score = 380 bits (975), Expect = e-103
Identities = 221/546 (40%), Positives = 312/546 (56%), Gaps = 56/546 (10%)
Query: 401 YGAPSPFGMSFPNSGFGSGSPYASGGMFTPPGYGFGSGYGFGLPPRAPAR------HIRA 454
+G + G +F N G + GG G G G+G G PR + H+ A
Sbjct: 229 HGRNNGGGGNFNNGGGYFSNRGRGGGRGDRGGCGRGNG-GRNFKPRPTCQLCSKVGHVVA 287
Query: 455 -----------PQTMLASAPS--TLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMG 501
P +A+A S NW+ D+GAT H+T++ ++ G +Q++
Sbjct: 288 DCWHCYDDSFVPDARVAAAASYGVDSNWYVDTGATDHITNELEKLTTRDRYNGKEQIHTA 347
Query: 502 NGQGLPIQSIGSASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFH 561
+G G+ I+ IG ++ +P L L+N+L VP KNL+S + A DN + E HP
Sbjct: 348 SGSGMDIKHIGQSTIRTPTRN---LYLRNILHVPRTKKNLISAHRLAVDNHAFVEVHPRF 404
Query: 562 CTVNSQDTCKTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIV 621
+ Q T TLLRG +GLY P+ ++ S SK A+ V
Sbjct: 405 FAIKDQITKNTLLRGPC-CNGLY-----------------PLPASTSSSKQVLGAIKPTV 446
Query: 622 STSNNVPSPTAYDLWHSRLGHPHYETLQSALTTCKISVPHKSKYT-ICHSCCVAKSHRLP 680
S +WHSRLGHP ++ +Q ++ + +SK +C +C KSH+LP
Sbjct: 447 S------------IWHSRLGHPSWQIVQKVVSHNNLPCLGESKELGVCDACQKGKSHQLP 494
Query: 681 SSASSTLYTAPLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVF 740
S++ PLEL+F+D+WGPA +S G Y+++ +D FS+FTWIYLLK+KSE F
Sbjct: 495 FLKSTSASNFPLELVFSDVWGPAP-DSVGGRKYYVSFIDDFSKFTWIYLLKQKSEVFHKF 553
Query: 741 IQFQAMVELKFGLKIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRH 800
+FQ +VE F KI ++QTD G E++ L F K+GI+H ++CPHTH QNGS ERKHRH
Sbjct: 554 HEFQQLVERIFYRKIIAMQTDWGGEYEKLHDFFTKIGISHHVSCPHTHQQNGSAERKHRH 613
Query: 801 IVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVF 860
IVE GLSLLA A+MP +WD AF+ AT+LINR P+ V+ +P LF Y LRVF
Sbjct: 614 IVEVGLSLLAHASMPLKFWDEAFIAATYLINRTPSKVIQYETPLEHLFHQKPSYTSLRVF 673
Query: 861 GSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPY 919
G AC+P+LRPYN+ KL FRS++CVFLGYS+ HKG+KCL + GR+YIS+DV F+E FP+
Sbjct: 674 GCACWPNLRPYNARKLQFRSKQCVFLGYSNLHKGFKCLDVAAGRVYISRDVVFDENVFPF 733
Query: 920 STLFSS 925
++L S+
Sbjct: 734 ASLHSN 739
Score = 89.4 bits (220), Expect = 9e-16
Identities = 48/190 (25%), Positives = 91/190 (47%), Gaps = 5/190 (2%)
Query: 35 KLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDR-----ANSVENPAFLVW 89
KL +NF WK H+ ++ R++ +L ++P + ++ V NPA+ W
Sbjct: 20 KLTKNNFSLWKTHILPVICGARMEGYLTGATQVPSAEIEVKEGEKGEITKKVSNPAYEAW 79
Query: 90 EQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSITK 149
D + +LLSS+S +L V N + W+ ++ S A++ R L + K
Sbjct: 80 IAADQQVLGFLLSSISKEILIQVANVDTAAHAWKMIVGLLSTQSRARALNTRIALATTQK 139
Query: 150 GSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTSREFPCSIS 209
G + SDY+ ++K++ + + S G P+ E I GL DY +++ + R +IS
Sbjct: 140 GESSVSDYISKMKTLADEMASAGKPLDDEEFTSYILAGLDSDYEQVVSSIVGRSEGVTIS 199
Query: 210 QAEAMVIAHE 219
+ + +++ E
Sbjct: 200 EVYSQLLSFE 209
Score = 37.4 bits (85), Expect = 4.1
Identities = 21/66 (31%), Positives = 30/66 (44%), Gaps = 17/66 (25%)
Query: 302 SDDNRDRGGYNNRGRGGRGSYNNRGRGGDRSS----------------VQCQICHKYGHD 345
++ R+ GG N GG G ++NRGRGG R CQ+C K GH
Sbjct: 227 ANHGRNNGGGGNFNNGG-GYFSNRGRGGGRGDRGGCGRGNGGRNFKPRPTCQLCSKVGHV 285
Query: 346 ASVCYY 351
+ C++
Sbjct: 286 VADCWH 291
>dbj|BAA78423.1| polyprotein [Arabidopsis thaliana]
Length = 1431
Score = 372 bits (956), Expect = e-101
Identities = 262/837 (31%), Positives = 387/837 (45%), Gaps = 172/837 (20%)
Query: 133 SLAQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDY 192
S TQLR++L+ TKG+K DY++ + + + L +G P+ + E +E + + L E+Y
Sbjct: 86 SYGHVTQLRTQLKQWTKGTKTIDDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEY 145
Query: 193 SHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQA 252
++ + +++ P ++++ ++ HE+++ A SS + A A
Sbjct: 146 KPVIDQIAAKDTPPTLTEIHERLLNHESKI-------LAVSSATVIPITANAV------- 191
Query: 253 PPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYN 312
S N + T +++ +R D+R+ N
Sbjct: 192 ------------------------SHRNTTTTTNNNNGNRNNRYDNRNN----------N 217
Query: 313 NRGRGGRGSYNNRGRGGDRSSV---QCQICHKYGHDASVCYYRGNSTSPATNSQPNMTNQ 369
N + + S N ++S +CQIC GH A C + S + QP
Sbjct: 218 NNSKPWQQSSTNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQP----- 272
Query: 370 APAPVAPTFGFGSSFGMMPQFGYSPFRGFQPYGAPSPFGMSFPNSGFGSGSPYASGGMFT 429
PSPF P + GSPY+S
Sbjct: 273 ----------------------------------PSPFTPWQPRANLALGSPYSSN---- 294
Query: 430 PPGYGFGSGYGFGLPPRAPARHIRAPQTMLASAPSTLGNWHPDSGATHHVTHDASAISDG 489
NW DSGATHH+T D + +S
Sbjct: 295 --------------------------------------NWLLDSGATHHITSDFNNLSLH 316
Query: 490 MSVTGNDQVYMGNGQGLPIQSIGSASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQ 549
TG D V + +G +PI GS S S+ L L N+L VPNI KNL+SV +
Sbjct: 317 QPYTGGDDVMVADGSTIPISHTGSTSLSTK---SRPLNLHNILYVPNIHKNLISVYRLCN 373
Query: 550 DNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSL 609
NGV EF P V +T LL+G D LY + P++S+ +
Sbjct: 374 ANGVSVEFFPASFQVKDLNTGVPLLQGKT-KDELYEW---------------PIASSQPV 417
Query: 610 SKPASPAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETLQSALTTCKISVPHKS-KYTIC 668
S ASP+ S + WH+RLGHP L S ++ +SV + S K+ C
Sbjct: 418 SLFASPS------------SKATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLSC 465
Query: 669 HSCCVAKSHRLPSSASSTLYTAPLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIY 728
C + KS+++P S S+ T PLE I++D+W + I S Y Y++ VD F+R+TW+Y
Sbjct: 466 SDCLINKSNKVPFSQSTINSTRPLEYIYSDVWS-SPILSHDNYRYYVIFVDHFTRYTWLY 524
Query: 729 LLKRKSETSTVFIQFQAMVELKFGLKIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTH 788
LK+KS+ FI F+ ++E +F +I + +D G EF L +F + GI+H + PHT
Sbjct: 525 PLKQKSQVKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFSQHGISHLTSPPHTP 584
Query: 789 HQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLF 848
NG ERKHRHIVETGL+LL+ A++P +YW +AF A +LINRLPTP+L SP+ KLF
Sbjct: 585 EHNGLSERKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLF 644
Query: 849 KVPADYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYIS 907
+Y LRVFG ACYP LRPYN KL +S++CVFLGYS + Y CL R+YIS
Sbjct: 645 GTSPNYDKLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYIS 704
Query: 908 KDVQFNEFKFPYSTLFSSEFPASNPSSVSSVI-----PVISYVPSLQQPIPS-PHHS 958
+ V+F+E FP+S ++ P SS + + + P L P S PHH+
Sbjct: 705 RHVRFDENCFPFSNYLATLSPVQEQRRESSCVWSPHTTLPTRTPGLPAPSGSDPHHA 761
>dbj|BAA87949.1| retrotransposon homolog [Arabidopsis thaliana]
gi|25301703|pir||T52436 RF28 protein - Arabidopsis
thaliana retrotransposon (fragment)
Length = 847
Score = 372 bits (955), Expect = e-101
Identities = 260/834 (31%), Positives = 387/834 (46%), Gaps = 167/834 (20%)
Query: 133 SLAQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDY 192
S TQLR++L+ TKG+K DY++ + + + L +G P+ + E +E + + L E+Y
Sbjct: 86 SYGHVTQLRTQLKQWTKGTKTIDDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEY 145
Query: 193 SHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQA 252
++ + +++ P ++++ ++ HE+++ A SS + A A
Sbjct: 146 KPVIDQIAAKDTPPTLTEIHERLLNHESKI-------LAVSSATVIPITANAV------- 191
Query: 253 PPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYN 312
S N + T +++ +R D+R+ N
Sbjct: 192 ------------------------SHRNTTTTTNNNNGNRNNRYDNRNN----------N 217
Query: 313 NRGRGGRGSYNNRGRGGDRSSV---QCQICHKYGHDASVCYYRGNSTSPATNSQPNMTNQ 369
N + + S N ++S +CQIC GH A C + S + QP
Sbjct: 218 NNSKPWQQSSTNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQP----- 272
Query: 370 APAPVAPTFGFGSSFGMMPQFGYSPFRGFQPYGAPSPFGMSFPNSGFGSGSPYASGGMFT 429
PSPF P + GSPY+S
Sbjct: 273 ----------------------------------PSPFTPWQPRANLALGSPYSSN---- 294
Query: 430 PPGYGFGSGYGFGLPPRAPARHIRAPQTMLASAPSTLGNWHPDSGATHHVTHDASAISDG 489
NW DSGATHH+T D + +S
Sbjct: 295 --------------------------------------NWLLDSGATHHITSDFNNLSLH 316
Query: 490 MSVTGNDQVYMGNGQGLPIQSIGSASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQ 549
TG D V + +G +PI GS S S+ L L N+L VPNI KNL+SV +
Sbjct: 317 QPYTGGDDVMVADGSTIPISHTGSTSLST---KSRPLNLHNILYVPNIHKNLISVYRLCN 373
Query: 550 DNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSL 609
NGV EF P V +T LL+G D LY + P++S+ +
Sbjct: 374 ANGVSVEFFPASFQVKDLNTGVPLLQGKT-KDELYEW---------------PIASSQPV 417
Query: 610 SKPASPAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETLQSALTTCKISVPHKS-KYTIC 668
S ASP+ S + WH+RLGHP L S ++ +SV + S K+ C
Sbjct: 418 SLFASPS------------SKATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLSC 465
Query: 669 HSCCVAKSHRLPSSASSTLYTAPLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIY 728
C + KS+++P S S+ T PLE I++D+W + I S Y Y++ VD F+R+TW+Y
Sbjct: 466 SDCLINKSNKVPFSQSTINSTRPLEYIYSDVWS-SPILSHDNYRYYVIFVDHFTRYTWLY 524
Query: 729 LLKRKSETSTVFIQFQAMVELKFGLKIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTH 788
LK+KS+ FI F+ ++E +F +I + +D G EF L +F + GI+H + PHT
Sbjct: 525 PLKQKSQVKETFITFKNLLENRFQTRIGTFYSDNGGEFVALWEYFSQHGISHLTSPPHTP 584
Query: 789 HQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLF 848
NG ERKHRHIVETGL+LL+ A++P +YW +AF A +LINRLPTP+L SP+ KLF
Sbjct: 585 EHNGLSERKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLF 644
Query: 849 KVPADYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYIS 907
+Y LRVFG ACYP LRPYN KL +S++CVFLGYS + Y CL R+YIS
Sbjct: 645 GTSPNYDKLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYIS 704
Query: 908 KDVQFNEFKFPYSTLFSSEFPASNPSSVSSVI-PVISYVPSLQQPIPSPHHSHP 960
+ V+F+E FP+S ++ P SS + + +P+ +P+P S P
Sbjct: 705 RHVRFDENCFPFSNYLATLSPVQEQRRESSCVWSPHTTLPTRTPVLPAPSCSDP 758
>gb|AAU43956.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|52353503|gb|AAU44069.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1447
Score = 372 bits (954), Expect = e-101
Identities = 201/476 (42%), Positives = 288/476 (60%), Gaps = 34/476 (7%)
Query: 455 PQTMLASAPSTLG---NWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSI 511
P A + ++ G NW+ D+ AT HVT + ++ G DQV+ +G G+ I I
Sbjct: 302 PDEKYAGSATSYGIDTNWYVDTSATDHVTGELDKLTVRDRYKGQDQVHTASGAGMEISHI 361
Query: 512 GSASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCK 571
G ++ +P + L+N+L VPN KNLVS ++ DN Y E + + + T K
Sbjct: 362 GHSTVRTP---NRDIHLRNILYVPNANKNLVSANRLVSDNSAYMELYSKYFNLKDLATKK 418
Query: 572 TLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPT 631
L RG LY+ + SS+ H + P+ P+ AF KP+
Sbjct: 419 LLFRGPCRGR-LYA---LPSSSPH-ERPR-PLKEAFGAIKPS------------------ 454
Query: 632 AYDLWHSRLGHPHYETLQSALTTCKIS-VPHKSKYTICHSCCVAKSHRLPSSASSTLYTA 690
++ WHSRLGHP ++ ++ + + +K ++C +C KSH+LP S SS++ +
Sbjct: 455 -FERWHSRLGHPASPIVEKVISKNNLPCLAESNKQSVCDACQQGKSHQLPYSRSSSMSSH 513
Query: 691 PLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELK 750
PLELI++D+WGPA + S G Y+++ + +S+FTW+YL+K KSE F +FQA+VE
Sbjct: 514 PLELIYSDVWGPA-LTSVGGKQYYVSFIGDYSKFTWLYLIKHKSEVIQKFHEFQALVERL 572
Query: 751 FGLKIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLA 810
F KI ++Q+D G E++ L F K+GITH ++CPHTH QNGS ERKHRHIVE GL+LLA
Sbjct: 573 FNRKIIAMQSDWGGEYEKLHSFFTKIGITHHVSCPHTHQQNGSAERKHRHIVEVGLTLLA 632
Query: 811 CANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRP 870
++MP +WD AF A +LINR PT +L L+P LF DY LRVFG AC+PHLRP
Sbjct: 633 YSSMPLKFWDEAFQAAVYLINRTPTKLLQFLTPLEHLFNQTPDYSSLRVFGCACWPHLRP 692
Query: 871 YNSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTLFSS 925
YN+ KL FRS++C FLGYS+ HKG+KCL S GR+YIS+DV F+E FP++ L ++
Sbjct: 693 YNTHKLQFRSKQCTFLGYSTLHKGFKCLDPSTGRVYISRDVIFDETNFPFAKLHTN 748
Score = 87.0 bits (214), Expect = 5e-15
Identities = 77/326 (23%), Positives = 134/326 (40%), Gaps = 59/326 (18%)
Query: 35 KLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDE-----DRANSVENPAFLVW 89
KL SN WK V VR RL+ L + P ++ ++ +V NP F W
Sbjct: 21 KLSKSNHALWKAQVMAAVRGARLEGHLTGATKTPNALITTTAGDKGEKEVTVRNPEFDDW 80
Query: 90 EQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSITK 149
D + +LLS+L+ VL V C + W+ + + + + + A+ R L + K
Sbjct: 81 VATDQQVLGFLLSTLARDVLAQVATCGTAAAAWQMLEEMYSSVTRARFINTRIALSNTKK 140
Query: 150 GSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTSREFPCSIS 209
G+ + ++Y+ ++K++ + + + G V + + I GL + Y +++ + ++ ++
Sbjct: 141 GTLSINEYVSKMKALADEMTAAGKIVDDDDLISYIIAGLDDTYEPVISTIVGKD-TMTLG 199
Query: 210 QAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQAPPP 269
+A + +++ E RL LR SS
Sbjct: 200 EAYSQLLSFE---QRLALRHGGDSS----------------------------------- 221
Query: 270 VAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGRGSYNNRGR-- 327
VN +R G + R + + R RGG NN GRG NN G
Sbjct: 222 ---------VNLANR-GRGGGGGQQRGG--NTGNGGRGRGGNNNGANRGRGRGNNGGARP 269
Query: 328 -GGDRSSVQCQICHKYGHDASVCYYR 352
GG + +CQ+C+K GH C+YR
Sbjct: 270 PGGVDNRPKCQLCYKRGHTVINCWYR 295
>gb|AAA57005.1| copia-like retrotransposon Hopscotch polyprotein [Zea mays]
gi|7444442|pir||T02087 gag/pol polyprotein - maize
retrotransposon Hopscotch
Length = 1439
Score = 371 bits (953), Expect = e-101
Identities = 197/456 (43%), Positives = 277/456 (60%), Gaps = 35/456 (7%)
Query: 469 WHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSASFSSPIHTQTTLLL 528
W+ D+GAT H+T D ++ TG DQ+ NG G+ I +IG+A + + +L L
Sbjct: 323 WYTDTGATDHITGDLDRLTMHDKYTGTDQIIAANGTGMTISNIGNAIVPT---SSRSLHL 379
Query: 529 KNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDG 588
+++L VP+ KNL+SV + DN V+ EFH H + + T LL G DGLY
Sbjct: 380 RSVLHVPSTHKNLISVHRLTNDNDVFIEFHSSHFLIKDRQTKAVLLHGKC-RDGLYPLPP 438
Query: 589 MHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETL 648
P + + FS S VP + WH RLGHP + +
Sbjct: 439 H---------PDLRLKHNFS---------------STRVP----LEHWHKRLGHPSRDIV 470
Query: 649 QSALTTCKIS-VPHKSKYTICHSCCVAKSHRLPSSASSTLYTAPLELIFADLWGPASIES 707
++ + + + S ++C +C AK+H+LP + S + +APL LIF+D++GPA I+S
Sbjct: 471 HRVISNNNLPCLSNNSTTSVCDACLQAKAHQLPYTISMSQSSAPLMLIFSDVFGPA-IDS 529
Query: 708 SAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLKIKSVQTDGGTEFK 767
Y Y+++ +D +S+FTWIYLL+ KS+ F +FQ +VE FG KI + Q+D G E++
Sbjct: 530 FGRYKYYVSFIDDYSKFTWIYLLRHKSDVYKSFCEFQHLVERMFGRKIIAFQSDWGGEYE 589
Query: 768 PLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLTAT 827
L HF +GI H+++CPHTH QNG+ ERKHRHIVE GL+LLA ++MP YWDHAFL A
Sbjct: 590 KLNAHFKTIGIHHQVSCPHTHQQNGAAERKHRHIVEVGLALLAQSSMPLKYWDHAFLAAV 649
Query: 828 FLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLG 887
+LINR P+ + + +P HKL DY LR+FG AC+P+LRPYN KL FRS CVFLG
Sbjct: 650 YLINRTPSKTIAHDTPLHKLTGATPDYSSLRIFGCACWPNLRPYNQHKLQFRSTRCVFLG 709
Query: 888 YSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTL 922
YS+ HKG+KCL S GRIYIS+DV F+E FP+++L
Sbjct: 710 YSNMHKGFKCLDISTGRIYISRDVVFDEHVFPFASL 745
Score = 124 bits (310), Expect = 3e-26
Identities = 94/353 (26%), Positives = 155/353 (43%), Gaps = 56/353 (15%)
Query: 1 MAEEASPTQIPTPTPISVAIPQLSSTSLVHKLLLKLDDSNFLSWKQHVEGIVRTHRLQSF 60
MA ++S + PT ++ + + KL N+L WK V +R +L
Sbjct: 1 MAMQSSLSTSAIPTSFAIPVSE------------KLTKGNYLLWKAQVLPAIRAAQLDDI 48
Query: 61 LLHQPEIPPRFLSD-EDRANSVENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSW 119
L PP+ +SD DR +V NPA+ W D A+ +LLSSLS VL +VVNC S
Sbjct: 49 LTGVEICPPKTISDASDRTVTVANPAYGRWIARDQAVLGYLLSSLSREVLSSVVNCSTSA 108
Query: 120 QIWEAVLDFFHAHSLAQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYRE 179
+W + + + +HS A+ R L + KG+ + ++Y +++ + L + G P+ E
Sbjct: 109 SVWTTLSEMYSSHSRARKVNTRIALATTKKGASSVAEYFAKMRGFADELGAAGKPLDDEE 168
Query: 180 HLEAIFDGLREDYSHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFL 239
+ + GL ED++ L+T V +R P + ++++E R+ H + S+ +
Sbjct: 169 FVSFLLTGLDEDFNPLVTAVVARSDPITPGDLYTQLLSYENRM-------HLQTGSSSLM 221
Query: 240 AQATAPPQSTPQAPPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDR 299
QS A ++P +S S R + R R
Sbjct: 222 -------------------QSSANARSPG-----------RGMSWGRSGGRGF-SRGRGR 250
Query: 300 SRSDDNRDRGGYNNRGRGGRGSYNNRGRGGDRSSVQCQICHKYGHDASVCYYR 352
R RGG+ + GRG +Y+ S +CQ+C + GH A C+YR
Sbjct: 251 GRGP---SRGGFQSFGRG--NNYSGATDADTSSRPRCQVCSRVGHTALNCWYR 298
>dbj|BAA78426.1| polyprotein [Arabidopsis thaliana]
Length = 1475
Score = 369 bits (947), Expect = e-100
Identities = 273/924 (29%), Positives = 418/924 (44%), Gaps = 165/924 (17%)
Query: 35 KLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRANSVENPAFLVWEQHDS 94
KL +N+L W + V + + L FL +PP + + A NP + W + D
Sbjct: 25 KLTSTNYLMWSRQVHALFDGYELAGFLDGSTPMPPATIGTD--AVPRVNPDYTRWRRQDK 82
Query: 95 ALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSITKGSKNT 154
+++ +L ++S SV P V + QIWE + + S TQLR++L+ TKG+K
Sbjct: 83 LIYSAILGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRTQLKQWTKGAKTI 142
Query: 155 SDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTSREFPCSISQAEAM 214
DY++ + + L +G P+ + E +E + + L +DY ++ + +++ P S+++
Sbjct: 143 DDYMQGFITRFDQLALLGKPMDHDEQVERVLENLPDDYKPVIDQIAAKDTPPSLTEIHER 202
Query: 215 VIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQAPPPVAIAP 274
+I E++L L + + P
Sbjct: 203 LINQESKLLALNSAE------------------------------------------VVP 220
Query: 275 PSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGRGSYNNRGRGGDRSSV 334
++ V R + +R +R D+R+ +++N + G R R
Sbjct: 221 ITANV-VTHRNTNTNRNQNNRGDNRNYNNNNNRSNSWQPSSSGSRSD----NRQPKPYLG 275
Query: 335 QCQICHKYGHDASVCYYRGNSTSPATNSQPNMTNQAPAPVAPTFGFGSSFGMMPQFGYSP 394
+CQIC GH A C P + + TNQ Q SP
Sbjct: 276 RCQICSVQGHSAKRC--------PQLHQFQSTTNQ-------------------QQSTSP 308
Query: 395 FRGFQPYGAPSPFGMSFPNSGFGSGSPYASGGMFTPPGYGFGSGYGFGLPPRAPARHIRA 454
F +Q P + SPY + G A H
Sbjct: 309 FTPWQ------------PRANLAVNSPYNANNWLLDSG----------------ATH--- 337
Query: 455 PQTMLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSA 514
+ S + L P +G D I+DG ++ PI GSA
Sbjct: 338 ---HITSDFNNLSFHQPYTGG------DDVMIADGSTI--------------PITHTGSA 374
Query: 515 SFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLL 574
S + + +L L +L VPNI KNL+SV + N V EF P V +T LL
Sbjct: 375 SLPT---SSRSLDLNKVLYVPNINKNLISVYRLCNTNRVSVEFFPASFQVKDLNTGVPLL 431
Query: 575 RGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYD 634
+G D LY + P++S+ ++S ASP S +
Sbjct: 432 QGKT-KDELYEW---------------PIASSQAVSMFASPC------------SKATHS 463
Query: 635 LWHSRLGHPHYETLQSALTTCKISVPHKS-KYTICHSCCVAKSHRLPSSASSTLYTAPLE 693
WHSRLGHP L S ++ + V + S K C C + KSH++P S S+ + PLE
Sbjct: 464 SWHSRLGHPSLAILNSVISNHSLPVLNPSHKLLSCSDCFINKSHKVPFSNSTITSSKPLE 523
Query: 694 LIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGL 753
I++D+W + I S Y Y++ VD F+R+TW+Y LK+KS+ FI F+++VE +F
Sbjct: 524 YIYSDVWS-SPILSIDNYRYYVIFVDHFTRYTWLYPLKQKSQVKDTFIIFKSLVENRFQT 582
Query: 754 KIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+I ++ +D G EF L + + GI+H + PHT NG ERKHRHIVE GL+LL+ A+
Sbjct: 583 RIGTLYSDNGGEFVVLRDYLSQHGISHFTSPPHTPEHNGLSERKHRHIVEMGLTLLSHAS 642
Query: 814 MPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNS 873
+P +YW +AF A +LINRLPTP+L SP+ KLF P +Y+ L+VFG ACYP LRPYN
Sbjct: 643 VPKTYWPYAFSVAVYLINRLPTPLLQLQSPFQKLFGQPPNYEKLKVFGCACYPWLRPYNR 702
Query: 874 SKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTL-FSSEFPASN 931
KL +S++C F+GYS + Y CL GR+Y S+ VQF+E FP+ST F
Sbjct: 703 HKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFDERCFPFSTTNFGVSTSQEQ 762
Query: 932 PSSVSSVIPVISYVPSLQQPIPSP 955
S + P + +P+ +P+P
Sbjct: 763 RSDSAPNWPSHTTLPTTPLVLPAP 786
>ref|NP_915223.1| putative rice retrotransposon retrofit gag/pol polyprotein [Oryza
sativa (japonica cultivar-group)]
gi|20161626|dbj|BAB90546.1| putative rice
retrotransposon retrofit gag/pol polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1448
Score = 369 bits (946), Expect = e-100
Identities = 212/535 (39%), Positives = 297/535 (54%), Gaps = 59/535 (11%)
Query: 415 GFGSGSPYASGGMFTPPGYGFGSGYGFGLPPRAPARHIR-------------------AP 455
G G G+P GG G G G G G R P + A
Sbjct: 252 GGGRGAPRGRGGGGQGRG---GHGRGTGGQDRRPTCQVCFKRGHTAADCWYRFDEDYVAD 308
Query: 456 QTMLASAPSTLG---NWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIG 512
+ ++A+A ++ G NW+ D+GAT H+T + ++ G +Q++ +G G+ I IG
Sbjct: 309 EKLVAAATNSYGIDTNWYIDTGATDHITGELEKLTTKEKYNGGEQIHTASGAGMDISHIG 368
Query: 513 SASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKT 572
+P + L N+L VP KNL+S S+ A DN + E H ++ Q T
Sbjct: 369 HTIVHTP---SRNIHLNNVLYVPQAKKNLISASQLAADNSAFLELHSKFFSIKDQVTRDV 425
Query: 573 LLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTA 632
LL G GLY +P S S +K A A +S
Sbjct: 426 LLEGKC-RHGLYP---------------IPKSFGRSTNKQALGAAKLSLSR--------- 460
Query: 633 YDLWHSRLGHPHYETLQSALTTCKISVPHKS-KYTICHSCCVAKSHRLPSSASSTLYTAP 691
WHSRLGHP ++ ++ + +S ++C++C AKSH+LP S+++ P
Sbjct: 461 ---WHSRLGHPSLPIVKQVISRNNLPCSVESVNQSVCNACQEAKSHQLPYIRSTSVSQFP 517
Query: 692 LELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKF 751
LEL+F+D+WGPA ES Y+++ +D FS+FTWIYLLK KSE F +FQA+VE F
Sbjct: 518 LELVFSDVWGPAP-ESVGRNKYYVSFIDDFSKFTWIYLLKYKSEVFEKFKEFQALVERMF 576
Query: 752 GLKIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLAC 811
KI ++QTD G E++ L F ++GI H ++CPHTH QNGS ERKH+HI+E GLSLL+
Sbjct: 577 DRKIIAMQTDWGGEYQKLNSFFAQIGIDHHVSCPHTHQQNGSAERKHKHIIEVGLSLLSY 636
Query: 812 ANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPY 871
A+MP +WD AF+ AT+LINR+P+ + N +P KLF DY LRVF C+PHL PY
Sbjct: 637 ASMPLKFWDEAFVAATYLINRIPSKTIQNSTPLEKLFNQKPDYSSLRVFSCTCWPHLHPY 696
Query: 872 NSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTLFSS 925
N+ KL FRS++CVFLG+S+ HKG+K L S GR+YIS+DV F+E FP+STL S+
Sbjct: 697 NTHKLQFRSKQCVFLGFSTHHKGFKFLDVSSGRVYISRDVVFDENVFPFSTLHSN 751
Score = 99.8 bits (247), Expect = 7e-19
Identities = 79/328 (24%), Positives = 135/328 (41%), Gaps = 51/328 (15%)
Query: 30 HKLLLKLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSD--EDRANSVENPAFL 87
H + KL +N WK V V RL +L + P +S + + + NPAF
Sbjct: 19 HSVSEKLGKANHALWKAQVSAAVHGARLLGYLNGDIKAPNAEISVTIDGKTTTKPNPAFE 78
Query: 88 VWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSI 147
WE +D + +LLSSLS VL V C+ + + W + + + A++ R L +
Sbjct: 79 DWEANDQLVLGYLLSSLSRDVLIQVATCKTAAEAWRNIEALYSTGTRARAVNTRLALTNT 138
Query: 148 TKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTSREFPCS 207
KG+ ++Y+ +++++ + + + G P+ ++ I GL ED+S +++ + ++ P +
Sbjct: 139 KKGTMKIAEYVAKMRALCDEMAAGGRPLDEEGLVQYIIAGLNEDFSPIVSNLCNKSDPIT 198
Query: 208 ISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQAP 267
+ + + ++ E LD L + +AF+A
Sbjct: 199 VGELYSQLVNFETLLD---LYRSTGQGGAAFVANRGR----------------------- 232
Query: 268 PPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDN---RDRGGYNNRGRGGRGSYNN 324
G R + D R RGG +GRGG G
Sbjct: 233 ---------------GGGGGGGRGNNNNSDGGGGGGGRGAPRGRGG-GGQGRGGHG---- 272
Query: 325 RGRGGDRSSVQCQICHKYGHDASVCYYR 352
RG GG CQ+C K GH A+ C+YR
Sbjct: 273 RGTGGQDRRPTCQVCFKRGHTAADCWYR 300
>ref|XP_476197.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|46981313|gb|AAT07631.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
gi|46981245|gb|AAT07563.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1431
Score = 365 bits (938), Expect = 5e-99
Identities = 217/537 (40%), Positives = 293/537 (54%), Gaps = 58/537 (10%)
Query: 408 GMSFPNSGFGSGSPYASG-GMFTPPGYGFGSGYGFGLPPRAPARHIRA------------ 454
G S N G G G G G P G G G G P H R
Sbjct: 232 GSSTGNRGGGRGGFGRGGHGRGAPSGASQGRGRGNDTRPVCQVCHKRGHVASDCWHRYDD 291
Query: 455 ---PQTMLASAPSTL----GNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLP 507
P L A + NW+ D+GAT H+T ++ G DQ++ +G+G
Sbjct: 292 SYVPDEKLGGAATYAYGVDTNWYVDTGATDHITGQLDKLTTKERYKGTDQIHTASGEGTS 351
Query: 508 IQSIGSASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQ 567
I+ +G A +P H L LKN+L VP KNLVSV K DN + E H + + +
Sbjct: 352 IKHVGHAIVPTPSHP---LHLKNVLHVPEAAKNLVSVHKLVADNYAFLEIHGKYFLIKDK 408
Query: 568 DTCKTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNV 627
T +T+L G GLY +P S+ + A+P+
Sbjct: 409 ATRRTILEGPCRR-GLYP---------------LPARSSLRQAFVATPS----------- 441
Query: 628 PSPTAYDLWHSRLGHPHYETLQSALTTCKISVPHKS-KYTICHSCCVAKSHRLPSSASST 686
+ WH RLGHP + L+ K+ S ++C +C AK H+LP S++
Sbjct: 442 -----FVRWHGRLGHPSKPIVLRILSQNKLPCLSNSVNESVCDACQQAKCHQLPFPRSTS 496
Query: 687 LYTAPLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAM 746
+ PLELI +D+WGPAS ES Y+++ +D +S+F WIY LK KSE F +FQ +
Sbjct: 497 VSNNPLELIHSDVWGPAS-ESVGAKKYYVSFIDDYSKFVWIYFLKHKSEVFQKFHEFQKL 555
Query: 747 VELKFGLKIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGL 806
VE +F KI S+QTD G E+ L F ++GI+H ++CPHTH QNG+VERKHRHIVE GL
Sbjct: 556 VERQFDRKILSMQTDWGGEYLKLHTFFNEIGISHHVSCPHTHQQNGAVERKHRHIVEVGL 615
Query: 807 SLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYP 866
SLLA A+MP +WD AFL AT+LINRLP+ V+ +P +LFK DYK LR FG AC+P
Sbjct: 616 SLLAHASMPLKFWDEAFLAATYLINRLPSKVIKFDTPLERLFKQTPDYKSLRTFGCACWP 675
Query: 867 HLRPYNSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTL 922
+LRPYN+ KL FRS++CVFLG+S+ HKG+KCL S GR+Y+S+DV F+E FP++ L
Sbjct: 676 NLRPYNTHKLQFRSKQCVFLGFSNIHKGFKCLDVSTGRVYVSRDVTFDEQVFPFANL 732
Score = 87.4 bits (215), Expect = 3e-15
Identities = 81/332 (24%), Positives = 133/332 (39%), Gaps = 79/332 (23%)
Query: 35 KLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRANS------VENPAFLV 88
KL N W + +R +L+ ++ P + ED + NP +
Sbjct: 22 KLTKQNHPLWAAQILTTLRGAQLEEHIVSTTAAPAAEIEKEDGDKDKKTKIVIPNPEYKT 81
Query: 89 WEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSIT 148
W D + ++ SSLS VL V + + Q W + D F +S+ +I
Sbjct: 82 WFVQDQQVLGFIFSSLSREVLQQVAGARTAAQAWNMIDDMF---------SCKSKAGTIN 132
Query: 149 KGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLM--TIVTSREFPC 206
KG + S+Y+ +++S+ + + + G P+ E + I +GL ++ + + T+R P
Sbjct: 133 KGPMSMSEYIAKMRSLADKMAATGKPLDEEELVAYIINGLDSEFDAAVEGLMATARIAPV 192
Query: 207 SISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQA 266
SIS + ++++E +R+R+RQ A+L
Sbjct: 193 SISHVYSQLLSYE---NRIRIRQ-------AYL--------------------------- 215
Query: 267 PPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGR------G 320
++ N +R G R S NR GG GRGG G
Sbjct: 216 ---------TTSANAANRGGG--------RGGRGSSTGNRG-GGRGGFGRGGHGRGAPSG 257
Query: 321 SYNNRGRGGDRSSVQCQICHKYGHDASVCYYR 352
+ RGRG D V CQ+CHK GH AS C++R
Sbjct: 258 ASQGRGRGNDTRPV-CQVCHKRGHVASDCWHR 288
>ref|NP_915573.1| putative gag/pol polyprotein [Oryza sativa (japonica
cultivar-group)]
Length = 1449
Score = 365 bits (937), Expect = 7e-99
Identities = 198/462 (42%), Positives = 275/462 (58%), Gaps = 43/462 (9%)
Query: 468 NWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSASFSSPIHTQTTLL 527
NW+ D+GAT HVT + + G+DQV+ +G G+ I IG + +P +
Sbjct: 319 NWYVDTGATDHVTGELEKLIVRDRYKGHDQVHTASGAGMEISHIGHSIVKTP---SRDIH 375
Query: 528 LKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFD 587
L+N+L VP KNLVS + DN + E + + + T K LLRG LY+
Sbjct: 376 LRNILHVPKANKNLVSAQRLVSDNSAFMELYSKYFNLKDLATKKLLLRGPCRGR-LYA-- 432
Query: 588 GMHSSNFHKDLPQVP-----VSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWHSRLGH 642
LP P + +SKP+ ++ WHSRLGH
Sbjct: 433 ----------LPSTPPHAHSTKEVYGVSKPS-------------------FERWHSRLGH 463
Query: 643 PHYETLQSALTTCKIS-VPHKSKYTICHSCCVAKSHRLPSSASSTLYTAPLELIFADLWG 701
P ++ ++ + + +K ++C +C AKSH+LP S SS++ + PLELI++D+WG
Sbjct: 464 PSSPIVEKVISKNNLPCLVESNKQSVCDACQKAKSHQLPYSISSSISSHPLELIYSDVWG 523
Query: 702 PASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLKIKSVQTD 761
PA S G Y+++ +D FS+FTWIYL+K KSE F +FQA+VE F KI ++Q+D
Sbjct: 524 PAPT-SVGGKQYYVSFIDDFSKFTWIYLIKFKSEVIQKFHEFQALVERLFNRKILAMQSD 582
Query: 762 GGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDH 821
G E++ L F K+GITH ++CPHTH QNGS ERKHRHIVE GL+LLA ++MP +WD
Sbjct: 583 WGGEYEKLNSFFTKIGITHHVSCPHTHQQNGSAERKHRHIVEMGLALLAYSSMPLKFWDE 642
Query: 822 AFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSSKLSFRSQ 881
AF +A +LINR P+ +L +P +LFK DY IL VFG AC+PHLRPYN+ KL FRS+
Sbjct: 643 AFTSAVYLINRTPSRLLKYATPLERLFKQTPDYNILCVFGCACWPHLRPYNARKLQFRSK 702
Query: 882 ECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTL 922
+C FLGYS HKG+KCL S GR+YIS+DV F E FP+++L
Sbjct: 703 QCTFLGYSPLHKGFKCLDPSTGRVYISRDVIFYENVFPFASL 744
Score = 88.6 bits (218), Expect = 2e-15
Identities = 83/328 (25%), Positives = 137/328 (41%), Gaps = 57/328 (17%)
Query: 35 KLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLS-----DEDRANSVENPAFLVW 89
KL +N+ WK V VR RL+ L P +S + D+A NPA+ W
Sbjct: 15 KLAKNNYALWKAQVLASVRGARLEGHLTGTTAAPAITISVPGEKEGDKATRAANPAYDEW 74
Query: 90 EQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSITK 149
D + LLS+LS VL V C + W + + + + + A+ R L + K
Sbjct: 75 VATDQQILGLLLSTLSKDVLAQVATCGTAAAAWSMLEEMYTSMTRARFINTRIALSNTKK 134
Query: 150 GSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTSREFPCSIS 209
G + ++Y+ +++++ + + + G V + + I GL + Y +++ + + P S
Sbjct: 135 GDLSITEYVAKMRALGDDMTAAGKVVDDEDLISYIIAGLDDTYEPVISSIVGKSEPMSFG 194
Query: 210 QAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQAPPP 269
+A + +++ E R + LR H S SA LA
Sbjct: 195 EAFSQLLSFEQR-NNLR---HGGES-SANLANRGR------------------------- 224
Query: 270 VAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYN--NRGRGGRGSYN-NRG 326
TG + ++R R + GG N NRG+GGRG+ N G
Sbjct: 225 -------------GTTGGNGGQRGRGGNNRGRGGN----GGNNSANRGKGGRGNGGFNGG 267
Query: 327 R--GGDRSSVQCQICHKYGHDASVCYYR 352
R GG + +CQ+C+K GH C+YR
Sbjct: 268 RQGGGVDTRPKCQLCYKRGHTVINCWYR 295
>gb|AAC02669.1| polyprotein [Arabidopsis thaliana]
Length = 1451
Score = 364 bits (935), Expect = 1e-98
Identities = 209/508 (41%), Positives = 289/508 (56%), Gaps = 36/508 (7%)
Query: 458 MLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSASFS 517
M+++ P GNW DSGATHH+T D + ++ G+++V + +G GLPI GSA
Sbjct: 322 MVSATPYNSGNWLLDSGATHHLTSDLNNLALHQPYNGDEEVTIADGSGLPISHSGSALLP 381
Query: 518 SPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGS 577
+P + L LK++L VP+I KNL+SV + NGV EF P H V T LL+G
Sbjct: 382 TPTRS---LALKDVLYVPDIQKNLISVYRMCNTNGVSVEFFPAHFQVKDLSTGARLLQGK 438
Query: 578 LGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWH 637
++ LY + PV+S+ + S ASP T ++PS WH
Sbjct: 439 TKNE-LYEW---------------PVNSSIATSMFASPTPKT------DLPS------WH 470
Query: 638 SRLGHPHYETLQSALTTCKISVPHK-SKYTICHSCCVAKSHRLPSSASSTLYTAPLELIF 696
+RLGHP L++ ++ + + H +C C + KSH+LP +++ + PLE ++
Sbjct: 471 ARLGHPSLPILKALISKFSLPISHSLQNQLLCSDCSINKSHKLPFYSNTIASSHPLEYLY 530
Query: 697 ADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLKIK 756
D+W + I S Y Y+L VD ++R+TW+Y L++KS+ +FI F A+VE KF KI
Sbjct: 531 TDVW-TSPITSIDNYKYYLVIVDHYTRYTWLYPLRKKSQVREMFITFTALVENKFKFKIG 589
Query: 757 SVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPT 816
++ +D G EF + GI+H T PHT NG ERKHRHIVETGL+LL+ A+MP
Sbjct: 590 TLYSDNGGEFIAMRSFLASHGISHMTTPPHTPELNGISERKHRHIVETGLTLLSTASMPK 649
Query: 817 SYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSSKL 876
YW +AF TA +LINR+ TPVLGN SPY KLF P +Y LR+FG C+P LRPY + KL
Sbjct: 650 EYWSYAFATAVYLINRMLTPVLGNESPYVKLFGQPPNYLKLRIFGCLCFPWLRPYTAHKL 709
Query: 877 SFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTLFSSEFPASNPSSV 935
RS CV LGYS S Y CL + GR+Y S+ VQF E FP+ST S P S+P
Sbjct: 710 DNRSVPCVLLGYSLSQSAYLCLDRATGRVYTSRHVQFAESSFPFSTTSPSVTPPSDPPLS 769
Query: 936 SSVIPVISYVPSLQQPIPSPHHSHPTAS 963
PV VP L +P+ + S P+ S
Sbjct: 770 QDTRPV--SVPLLARPLTTAPPSSPSCS 795
Score = 100 bits (250), Expect = 3e-19
Identities = 88/371 (23%), Positives = 156/371 (41%), Gaps = 56/371 (15%)
Query: 1 MAEEASPTQIPTPTPISVAIPQLSSTSLVHKLLLKLDDSNFLSWKQHVEGIVRTHRLQSF 60
M++ SP + I+V+ L + ++ + + +L DSNF+ W + V ++ + L +
Sbjct: 1 MSDSTSPVSVSET--IAVSTSTLLNVNMTN--VTRLTDSNFVMWSRQVHALLDGYDLAGY 56
Query: 61 LLHQPEIPPRFLSDEDRANSVENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQ 120
+ IP + D + N + +W++ D +++ LL ++S SV P + S +
Sbjct: 57 VDGSIPIPTPTRTTADGVVTTNND-YTLWKRQDKLIYSALLGAISLSVQPLLSKANTSAE 115
Query: 121 IWEAVLDFFHAHSLAQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREH 180
IWE + F S A QLR +L+ TKG+K+ Y + + + L +G E
Sbjct: 116 IWETLSSTFANPSWAHVQQLRQQLKQWTKGTKSIVTYFQGFTTRFDHLALLGKAPEREEQ 175
Query: 181 LEAIFDGLREDYSHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLA 240
+E I GL EDY ++ + RE P ++++ +I HE +L A A
Sbjct: 176 IELILGGLPEDYKTVVDQIEGRENPPALTEVLEKLINHEVKL--------------AAKA 221
Query: 241 QATAPPQSTPQAPPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRS 300
+AT+ P + ++ VNY G+++ R + R+
Sbjct: 222 EATSVPVT---------------------------ANAVNY---RGNNNNNNNSRSNGRN 251
Query: 301 RSDDNRDRGGYNNRGRGGRGSYNNRGRGGDRSSVQCQICHKYGHDASVCYYRGNSTSPAT 360
S N N++ R Y R G +CQIC +GH A C
Sbjct: 252 NSRGN--TSWQNSQSTSNRQQYTPRPYQG-----KCQICSVHGHSARRCPQLQQHAGSYA 304
Query: 361 NSQPNMTNQAP 371
++Q + + AP
Sbjct: 305 SNQSSSASYAP 315
>gb|AAC02666.1| polyprotein [Arabidopsis thaliana]
Length = 1451
Score = 364 bits (935), Expect = 1e-98
Identities = 209/508 (41%), Positives = 289/508 (56%), Gaps = 36/508 (7%)
Query: 458 MLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSASFS 517
M+++ P GNW DSGATHH+T D + ++ G+++V + +G GLPI GSA
Sbjct: 322 MVSATPYNSGNWLLDSGATHHLTSDLNNLALHQPYNGDEEVTIADGSGLPISHSGSALLP 381
Query: 518 SPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGS 577
+P + L LK++L VP+I KNL+SV + NGV EF P H V T LL+G
Sbjct: 382 TPTRS---LALKDVLYVPDIQKNLISVYRMCNTNGVSVEFFPAHFQVKDLSTGARLLQGK 438
Query: 578 LGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWH 637
++ LY + PV+S+ + S ASP T ++PS WH
Sbjct: 439 TKNE-LYEW---------------PVNSSIATSMFASPTPKT------DLPS------WH 470
Query: 638 SRLGHPHYETLQSALTTCKISVPHK-SKYTICHSCCVAKSHRLPSSASSTLYTAPLELIF 696
+RLGHP L++ ++ + + H +C C + KSH+LP +++ + PLE ++
Sbjct: 471 ARLGHPSLPILKALISKFSLPISHSLQNQLLCSDCSINKSHKLPFYSNTIASSHPLEYLY 530
Query: 697 ADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLKIK 756
D+W + I S Y Y+L VD ++R+TW+Y L++KS+ +FI F A+VE KF KI
Sbjct: 531 TDVW-TSPITSIDNYKYYLVIVDHYTRYTWLYPLRKKSQVREMFITFTALVENKFKFKIG 589
Query: 757 SVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPT 816
++ +D G EF + GI+H T PHT NG ERKHRHIVETGL+LL+ A+MP
Sbjct: 590 TLYSDNGGEFIAMRSFLASHGISHMTTPPHTPELNGISERKHRHIVETGLTLLSTASMPK 649
Query: 817 SYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSSKL 876
YW +AF TA +LINR+ TPVLGN SPY KLF P +Y LR+FG C+P LRPY + KL
Sbjct: 650 EYWSYAFATAVYLINRMLTPVLGNESPYVKLFGQPPNYLKLRIFGCLCFPWLRPYTAHKL 709
Query: 877 SFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTLFSSEFPASNPSSV 935
RS CV LGYS S Y CL + GR+Y S+ VQF E FP+ST S P S+P
Sbjct: 710 DNRSVPCVLLGYSLSQSAYLCLDRATGRVYTSRHVQFAESSFPFSTTSPSVTPPSDPPLS 769
Query: 936 SSVIPVISYVPSLQQPIPSPHHSHPTAS 963
PV VP L +P+ + S P+ S
Sbjct: 770 QDTRPV--SVPLLARPLTTAPPSSPSCS 795
Score = 101 bits (251), Expect = 2e-19
Identities = 88/371 (23%), Positives = 156/371 (41%), Gaps = 56/371 (15%)
Query: 1 MAEEASPTQIPTPTPISVAIPQLSSTSLVHKLLLKLDDSNFLSWKQHVEGIVRTHRLQSF 60
M++ SP + I+V+ L + ++ + + +L DSNF+ W + V ++ + L +
Sbjct: 1 MSDSTSPVSVSET--IAVSTSTLLNVNMTN--VTRLTDSNFVMWSRQVHALLDGYDLAGY 56
Query: 61 LLHQPEIPPRFLSDEDRANSVENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQ 120
+ IP + D + N + +W++ D +++ LL ++S SV P + S +
Sbjct: 57 IDGSIPIPTPTRTTADGVVTTNND-YTLWKRQDKLIYSALLGAISLSVQPLLSKANTSAE 115
Query: 121 IWEAVLDFFHAHSLAQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREH 180
IWE + F S A QLR +L+ TKG+K+ Y + + + L +G E
Sbjct: 116 IWETLSSTFANPSWAHVQQLRQQLKQWTKGTKSIVTYFQGFTTRFDHLALLGKAPEREEQ 175
Query: 181 LEAIFDGLREDYSHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLA 240
+E I GL EDY ++ + RE P ++++ +I HE +L A A
Sbjct: 176 IELILGGLPEDYKTVVDQIEGRENPPALTEVLEKLINHEVKL--------------AAKA 221
Query: 241 QATAPPQSTPQAPPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRS 300
+AT+ P + ++ VNY G+++ R + R+
Sbjct: 222 EATSVPVT---------------------------ANAVNY---RGNNNNNNNSRSNGRN 251
Query: 301 RSDDNRDRGGYNNRGRGGRGSYNNRGRGGDRSSVQCQICHKYGHDASVCYYRGNSTSPAT 360
S N N++ R Y R G +CQIC +GH A C
Sbjct: 252 NSRGN--TSWQNSQSTSNRQQYTPRPYQG-----KCQICSVHGHSARRCPQLQQHAGSYA 304
Query: 361 NSQPNMTNQAP 371
++Q + + AP
Sbjct: 305 SNQSSSASYAP 315
>gb|AAT85031.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1437
Score = 364 bits (934), Expect = 1e-98
Identities = 225/582 (38%), Positives = 313/582 (53%), Gaps = 68/582 (11%)
Query: 394 PFRGFQPYGAPSPFGMSFPNSGFGSGSPYASGGMFTPPGYGFGSGYGFGLPPRAPARHIR 453
P RG G S G P++G G G P G G G P+ + R
Sbjct: 231 PQRGGSRSGGDSNRGRGAPSNGANRGR-----GRGNPSGGRANVGGGTDNRPKCQLCYKR 285
Query: 454 A---------------PQTMLASAPSTLG---NWHPDSGATHHVTHDASAISDGMSVTGN 495
P A + G NW+ D+GAT HVT + ++ GN
Sbjct: 286 GHTVCDCWYRYDENFVPDERFAGTAVSYGVDTNWYLDTGATDHVTGELDKLTVRDKYHGN 345
Query: 496 DQVYMGNGQGLPIQSIGSASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYF 555
DQV+ +G G+ I IG++ +P L LK++L VP KNLVS K DN +
Sbjct: 346 DQVHTASGAGMEISHIGNSVVKTP---SRNLHLKDVLYVPKANKNLVSAYKLTSDNLAFI 402
Query: 556 EFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASP 615
E + + +TLLRG GLY+ SS+ H+ V + ++KP+
Sbjct: 403 ELYRKFFFIKDLAMRRTLLRGRCHK-GLYALPSP-SSHHHQ------VKQVYGVTKPS-- 452
Query: 616 AVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETLQSALTTCKISVPHKSKY-TICHSCCVA 674
++ WHSRLGHP Y ++ + + + S+ ++C +C A
Sbjct: 453 -----------------FERWHSRLGHPSYTVVEKVIKSQNLPCLDVSEQVSVCDACQKA 495
Query: 675 KSHRL--PSSASSTLYTAPLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKR 732
KSH+L P S S + Y PLEL+F+D+WGPA +S Y+++ +D +S+FTWIYLLK
Sbjct: 496 KSHQLSFPKSTSESKY--PLELVFSDVWGPAP-QSVGNNKYYVSFIDDYSKFTWIYLLKY 552
Query: 733 KSETSTVFIQFQAMVELKFGLKIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNG 792
KSE F +FQ++VE F KI ++QTD G E++ L F K+GITH ++CPHTH QNG
Sbjct: 553 KSEVFDKFHEFQSLVERLFNRKIVAMQTDWGGEYQKLHSFFNKVGITHHVSCPHTHQQNG 612
Query: 793 SVERKHRHIVETGLSLLACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPA 852
S ERKHRHIVE GL+LLA ++MP +W AFL+A +LINR P+ VL ++SP +L
Sbjct: 613 SAERKHRHIVEVGLALLAYSSMPLKFWGEAFLSAVYLINRTPSRVLHDVSPLERLLGHKP 672
Query: 853 DYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQ 911
DY LRVFG AC+P+LRPYN KL FRS C FLGYS+ HKG+KCL S GR+YIS+DV
Sbjct: 673 DYNALRVFGCACWPNLRPYNKHKLQFRSTTCTFLGYSTLHKGFKCLDPSTGRVYISRDVV 732
Query: 912 FNEFKFPYSTLFSSEFPASNPSSVSSVIPVISYVPSLQQPIP 953
F+E +FP++ L +P+ + + I+ VP L +P
Sbjct: 733 FDETQFPFTKL--------HPNVGAKLRAEIALVPELAASLP 766
Score = 114 bits (285), Expect = 3e-23
Identities = 83/325 (25%), Positives = 135/325 (41%), Gaps = 57/325 (17%)
Query: 35 KLDDSNFLSWKQHVEGIVRTHRLQSFLL--HQPEIPPRFLSDEDRANSVENPAFLVWEQH 92
KL SN WK + +R RL+ L QP P + ++ V NP + W
Sbjct: 21 KLGKSNHAVWKAQILATIRGARLEGHLTGDDQPPAPILRRKEGEKEVVVSNPEYEEWVAT 80
Query: 93 DSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSITKGSK 152
D + +LLSS++ +L V C+ + W + F + + A++ R L ++ KG
Sbjct: 81 DQQVLAYLLSSMTKDLLVQVATCRTAASAWSMIQGMFGSMTRARTINTRLSLSTLQKGDM 140
Query: 153 NTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTSREFPCSISQAE 212
N + Y+ +++++ + L+++G PV E + IF GL +++ +++ + R P +I +
Sbjct: 141 NITTYVGKMRALADDLMAVGKPVDDDELIGYIFAGLDDEFEPVISTIVGRPDPVTIGETY 200
Query: 213 AMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQAPPPVAI 272
A +I+ E RL R
Sbjct: 201 AQLISFEQRLAHRR---------------------------------------------- 214
Query: 273 APPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGRGSYNNRG-----R 327
+ S VN SR+ R R RS D NR RG +N GRG N G
Sbjct: 215 SGDQSSVNSASRS----RGQPQRGGSRSGGDSNRGRGAPSNGANRGRGRGNPSGGRANVG 270
Query: 328 GGDRSSVQCQICHKYGHDASVCYYR 352
GG + +CQ+C+K GH C+YR
Sbjct: 271 GGTDNRPKCQLCYKRGHTVCDCWYR 295
>gb|AAC02664.1| polyprotein [Arabidopsis thaliana]
Length = 1451
Score = 362 bits (930), Expect = 4e-98
Identities = 208/508 (40%), Positives = 289/508 (55%), Gaps = 36/508 (7%)
Query: 458 MLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSASFS 517
M+++ P GNW DSGATHH+T + + ++ G+++V + +G GLPI GSA
Sbjct: 322 MVSATPYNSGNWLLDSGATHHLTSNLNNLALHQPYNGDEEVTIADGSGLPISHSGSALLP 381
Query: 518 SPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGS 577
+P + L LK++L VP+I KNL+SV + NGV EF P H V T LL+G
Sbjct: 382 TPTRS---LALKDVLYVPDIQKNLISVYRMCNTNGVSVEFFPAHFQVKDLSTGARLLQGK 438
Query: 578 LGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWH 637
++ LY + PV+S+ + S ASP T ++PS WH
Sbjct: 439 TKNE-LYEW---------------PVNSSIATSMFASPTPKT------DLPS------WH 470
Query: 638 SRLGHPHYETLQSALTTCKISVPHK-SKYTICHSCCVAKSHRLPSSASSTLYTAPLELIF 696
+RLGHP L++ ++ + + H +C C + KSH+LP +++ + PLE ++
Sbjct: 471 ARLGHPSLPILKALISKFSLPISHSLQNQLLCSDCSINKSHKLPFYSNTIASSHPLEYLY 530
Query: 697 ADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLKIK 756
D+W + I S Y Y+L VD ++R+TW+Y L++KS+ +FI F A+VE KF KI
Sbjct: 531 TDVW-TSPITSIDNYKYYLVIVDHYTRYTWLYPLRKKSQVREMFITFTALVENKFKFKIG 589
Query: 757 SVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPT 816
++ +D G EF + GI+H T PHT NG ERKHRHIVETGL+LL+ A+MP
Sbjct: 590 TLYSDNGGEFIAMRSFLASHGISHMTTPPHTPELNGISERKHRHIVETGLTLLSTASMPK 649
Query: 817 SYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSSKL 876
YW +AF TA +LINR+ TPVLGN SPY KLF P +Y LR+FG C+P LRPY + KL
Sbjct: 650 EYWSYAFATAVYLINRMLTPVLGNESPYVKLFGQPPNYLKLRIFGCLCFPWLRPYTAHKL 709
Query: 877 SFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTLFSSEFPASNPSSV 935
RS CV LGYS S Y CL + GR+Y S+ VQF E FP+ST S P S+P
Sbjct: 710 DNRSVPCVLLGYSLSQSAYLCLDRATGRVYTSRHVQFAESSFPFSTTSPSVTPPSDPPLS 769
Query: 936 SSVIPVISYVPSLQQPIPSPHHSHPTAS 963
PV VP L +P+ + S P+ S
Sbjct: 770 QDTRPV--SVPLLARPLTTAPPSSPSCS 795
Score = 100 bits (250), Expect = 3e-19
Identities = 88/371 (23%), Positives = 156/371 (41%), Gaps = 56/371 (15%)
Query: 1 MAEEASPTQIPTPTPISVAIPQLSSTSLVHKLLLKLDDSNFLSWKQHVEGIVRTHRLQSF 60
M++ SP + I+V+ L + ++ + + +L DSNF+ W + V ++ + L +
Sbjct: 1 MSDSTSPVSVSET--IAVSTSTLLNVNMTN--VTRLTDSNFVMWSRQVHALLDGYDLAGY 56
Query: 61 LLHQPEIPPRFLSDEDRANSVENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQ 120
+ IP + D + N + +W++ D +++ LL ++S SV P + S +
Sbjct: 57 VDGSIPIPTPTRTTADGVVTTNND-YTLWKRQDKLIYSALLGAISLSVQPLLSKANTSAE 115
Query: 121 IWEAVLDFFHAHSLAQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREH 180
IWE + F S A QLR +L+ TKG+K+ Y + + + L +G E
Sbjct: 116 IWETLSSTFANPSWAHVQQLRQQLKQWTKGTKSIVTYFQGFTTRFDHLALLGKAPEREEQ 175
Query: 181 LEAIFDGLREDYSHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLA 240
+E I GL EDY ++ + RE P ++++ +I HE +L A A
Sbjct: 176 IELILGGLPEDYKTVVDQIEGRENPPALTEVLEKLINHEVKL--------------AAKA 221
Query: 241 QATAPPQSTPQAPPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRS 300
+AT+ P + ++ VNY G+++ R + R+
Sbjct: 222 EATSVPVT---------------------------ANAVNY---RGNNNNNNNSRSNGRN 251
Query: 301 RSDDNRDRGGYNNRGRGGRGSYNNRGRGGDRSSVQCQICHKYGHDASVCYYRGNSTSPAT 360
S N N++ R Y R G +CQIC +GH A C
Sbjct: 252 NSRGN--TSWQNSQSTSNRQQYTPRPYQG-----KCQICSVHGHSARRCPQLQQHAGSYA 304
Query: 361 NSQPNMTNQAP 371
++Q + + AP
Sbjct: 305 SNQSSSASYAP 315
>gb|AAC02672.1| polyprotein [Arabidopsis arenosa] gi|7522104|pir||T31353
polyprotein - Arabidopsis arenosa Evelknievel
retrotransposon (fragment)
Length = 1390
Score = 359 bits (921), Expect = 5e-97
Identities = 209/508 (41%), Positives = 286/508 (56%), Gaps = 36/508 (7%)
Query: 458 MLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSASFS 517
M+++ P GNW DSGATHH+T D + ++ G ++V + +G GLPI GSA
Sbjct: 322 MVSATPYNSGNWLLDSGATHHLTSDLNNLALHQPYNGGEEVTIADGSGLPISHSGSALLP 381
Query: 518 SPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGS 577
+P + L LK++L VP+I KNL+SV + NGV EF P H V T LL+G
Sbjct: 382 TPTRS---LDLKDVLYVPDIQKNLISVYRMCNTNGVSVEFFPAHFQVKDLSTGARLLQGK 438
Query: 578 LGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWH 637
++ LY + PV+S+ + S ASP T ++PS WH
Sbjct: 439 TKNE-LYEW---------------PVNSSIATSMFASPTPKT------DLPS------WH 470
Query: 638 SRLGHPHYETLQSALTTCKISVPHK-SKYTICHSCCVAKSHRLPSSASSTLYTAPLELIF 696
+RLGHP L++ ++ + + H +C C + KSH+LP +++ + PLE ++
Sbjct: 471 ARLGHPSLPILKTLISKFSLPISHSLQNQLLCSDCSINKSHKLPFYSNTIASSHPLEYLY 530
Query: 697 ADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLKIK 756
D+W + I S Y Y+L VD ++R+TW+Y L++KS+ FI F A+VE KF KI
Sbjct: 531 TDVW-TSPITSIDNYKYYLVIVDHYTRYTWLYPLRQKSQVRETFITFTALVENKFKSKIG 589
Query: 757 SVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPT 816
++ +D G EF L GI+H T PHT NG ERKHRHIVETGL+LL+ A+M
Sbjct: 590 TLYSDNGGEFIALRSFLASHGISHMTTPPHTPELNGISERKHRHIVETGLTLLSTASMSK 649
Query: 817 SYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSSKL 876
YW +AF TA +LINR+ TPVLGN SPY KLF P +Y LRVFG C+P LRPY + KL
Sbjct: 650 EYWSYAFTTAVYLINRMLTPVLGNESPYMKLFGQPPNYLKLRVFGCLCFPWLRPYTAHKL 709
Query: 877 SFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTLFSSEFPASNPSSV 935
RS CV LGYS S Y CL + GR+Y S+ VQF E FP+ST S P S+P
Sbjct: 710 DNRSMPCVLLGYSLSQSAYLCLDRATGRVYTSRHVQFAESIFPFSTTSPSVTPPSDPPLS 769
Query: 936 SSVIPVISYVPSLQQPIPSPHHSHPTAS 963
P+ VP L +P+ + S P+ S
Sbjct: 770 QDTRPI--SVPILARPLTTAPPSSPSCS 795
Score = 104 bits (259), Expect = 3e-20
Identities = 88/362 (24%), Positives = 150/362 (41%), Gaps = 54/362 (14%)
Query: 12 TPTPISVAIPQLSSTSLVHKL--LLKLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPP 69
+P S I +ST L + + +L DSNF+ W + V ++ + L ++ IPP
Sbjct: 6 SPASASETIAVSTSTLLNINMTNVTRLTDSNFVMWSRQVHALLDGYDLAGYVDGSVPIPP 65
Query: 70 RFLSDEDRANSVENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFF 129
+ +D + N + +W++ D +++ LL ++S SV P + S ++WE + F
Sbjct: 66 PTRTTDDGVVTTNND-YTLWKRQDKLVYSALLGAISLSVQPLLSKANTSAEVWETLSSTF 124
Query: 130 HAHSLAQSTQLRSELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLR 189
S A QLR +L+ TKG+K+ Y + + + L +G E +E I GL
Sbjct: 125 ANPSWAHVQQLRQQLKQWTKGTKSVVTYFQGFTTRFDHLALLGKAPEREEQIELILGGLP 184
Query: 190 EDYSHLMTIVTSREFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQST 249
EDY ++ + SRE P ++++ +I HE +L A A+AT+ T
Sbjct: 185 EDYKTVVDQIESRENPPALTEVLEKLINHEVKL--------------AAKAEATSSVPIT 230
Query: 250 PQAPPLVPSQSPAPPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRG 309
A VNY +++ R++ R +
Sbjct: 231 ANA--------------------------VNYRGNNNNNNSRSNGRNNSRGNT------S 258
Query: 310 GYNNRGRGGRGSYNNRGRGGDRSSVQCQICHKYGHDASVCYYRGNSTSPATNSQPNMTNQ 369
NN+ R Y R G +CQIC +GH A C ++Q + ++
Sbjct: 259 WQNNQSTTNRQQYTPRPYQG-----KCQICSVHGHSARRCPQLQQHAGSYASNQSSSSSY 313
Query: 370 AP 371
AP
Sbjct: 314 AP 315
>gb|AAF99727.1| F17L21.7 [Arabidopsis thaliana]
Length = 1534
Score = 348 bits (894), Expect = 6e-94
Identities = 209/509 (41%), Positives = 284/509 (55%), Gaps = 47/509 (9%)
Query: 469 WHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSASFSSPIHTQTTLLL 528
W DSGATHH+T D + ++ G ++V + +G LPI GS++ S+ +L L
Sbjct: 424 WLLDSGATHHLTTDLNNLALHQPYNGGEEVTIADGSTLPITHTGSSTLSTQ---SRSLAL 480
Query: 529 KNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLLRGSLGSDGLYSFDG 588
N+L VPN+ KNL+SV K N V EF P H V T LL+G D LY +
Sbjct: 481 NNILYVPNLHKNLISVYKLCNANKVSVEFFPAHFQVKDLSTGARLLQGRT-KDELYEW-- 537
Query: 589 MHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYDLWHSRLGHPHYETL 648
PV S +S ASP T +PS WHSRLGHP L
Sbjct: 538 -------------PVPSNTPISLFASPTPKT------TLPS------WHSRLGHPSPPVL 572
Query: 649 QSALTTCKISVPHKS-KYTICHSCCVAKSHRLPSSASSTLYTAPLELIFADLWGPASIES 707
+S ++ + V + S K+ C C + KSH+LP +++ + PLE +++D+W + + S
Sbjct: 573 KSLVSQFSLPVSNSSQKHFPCSHCLINKSHKLPFYSNTIISYTPLEYVYSDVW-TSPVTS 631
Query: 708 SAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGLKIKSVQTDGGTEFK 767
+ Y+L VD ++R+TW+Y LK+KS+ F+ F+A+VE +F KI+++ +D G EF
Sbjct: 632 VDNFKYYLILVDHYTRYTWLYPLKQKSQVRETFVAFKALVENRFQTKIRTLYSDNGGEFI 691
Query: 768 PLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACANMPTSYWDHAFLTAT 827
L L GI+H + PHT NG ERKHRHI+ETGL+LL A++PTSYW +AF TA
Sbjct: 692 ALRQFLLTHGISHLTSLPHTPEHNGIAERKHRHILETGLTLLTQASIPTSYWTYAFGTAV 751
Query: 828 FLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNSSKLSFRSQECVFLG 887
+LINRLP+ VL N SPY KLFK +Y LRVFG +C+P LRPY + KL RSQ CVFLG
Sbjct: 752 YLINRLPSSVLNNESPYSKLFKTSPNYLKLRVFGCSCFPWLRPYTNHKLERRSQPCVFLG 811
Query: 888 YSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTLFSSEFPASNPSSVS-------SVI 939
YS + Y CL S GR+Y S+ VQF E +FP+S + S+P S S I
Sbjct: 812 YSLTQSAYLCLDRSSGRVYTSRHVQFVEDQFPFSISDTHSVSNSSPEEASPSCHQPPSRI 871
Query: 940 PVISYVPSLQQ------PIPSPHHSHPTA 962
P+ S P L Q P+ S H P A
Sbjct: 872 PIQSSSPPLVQAPSSLPPLSSDSHRRPNA 900
Score = 108 bits (269), Expect = 2e-21
Identities = 84/349 (24%), Positives = 156/349 (44%), Gaps = 60/349 (17%)
Query: 25 STSLVHKLLL---KLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRANSV 81
STSL++ + KL SNFL W++ V+ ++ + L ++ +PP ++
Sbjct: 113 STSLLNVNMTNVTKLTSSNFLMWRRQVQALLNGYDLTGYIDGSIVVPPATITANGAVTV- 171
Query: 82 ENPAFLVWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLR 141
NPAF W++ D +++ LL ++S SV P + S +IW ++D + S + QLR
Sbjct: 172 -NPAFKHWQRQDQLIYSALLGAISISVQPILSRTTTSAEIWTKLMDTYAKPSWSHIQQLR 230
Query: 142 SELRSITKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTS 201
+++ K +K+ ++ + + + L +G P+ E +E I +GL +DY ++ +
Sbjct: 231 QQIKQWKKDTKSIDEFFQGLVMRFDQLALLGKPMESEEQMEVIVEGLSDDYKQVIDQIQG 290
Query: 202 REFPCSISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSP 261
RE P S+++ ++ HE +L Q AASS +P +
Sbjct: 291 REVPPSLTEIHEKLLNHEVKL------QAAASS---------------------LPISAN 323
Query: 262 APPQAPPPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGRGS 321
A PP + + + ++ R ++ +N +RG N+ +
Sbjct: 324 AASYRPPA-------------------NNKHNNSNNYRGQNRNNNNRGA-NSYQQPRNDQ 363
Query: 322 YNNRGRGGDRSSVQCQICHKYGHDASVC---YYRGNSTSPATNSQPNMT 367
++RG G +CQIC +GH A C G ++P+ + PN T
Sbjct: 364 PSSRGYQG-----KCQICGVFGHSARRCSQLQMSGAYSTPSPSQYPNAT 407
>gb|AAB82754.1| retrofit [Oryza longistaminata] gi|7444451|pir||T10728 probable
gag/pol polyprotein - long-staminate rice
retrotransposon retrofit
Length = 1445
Score = 346 bits (888), Expect = 3e-93
Identities = 209/538 (38%), Positives = 292/538 (53%), Gaps = 58/538 (10%)
Query: 413 NSGFGSGSPYASGGMFTPPGYGFGSGYGFGLPPRAPARHIR------------------- 453
NSG G G G G G G G G G R P +
Sbjct: 244 NSGGGGGRSAPGGRGSGSQGRG-GRGRGTGGQDRRPTCQVCFKRGHTAADCWYRFDEDYV 302
Query: 454 APQTMLASAPSTLG---NWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQS 510
A + ++A+A ++ G NW+ D+GAT H+T + ++ G +Q++ +G G+ I
Sbjct: 303 ADEKLVAAATNSYGIDTNWYIDTGATDHITGELEKLTTKEKYNGGEQIHTASGAGMDISH 362
Query: 511 IGSASFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTC 570
IG +P + L N+L VP KNL+S S+ A DN + E H ++ Q T
Sbjct: 363 IGHTIVHTP---SRNIHLNNVLYVPQAKKNLISASQLAADNSAFLELHSKFFSIKDQVTR 419
Query: 571 KTLLRGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSP 630
LL G GLY + +K Q ++ SLS+
Sbjct: 420 DVLLEGKC-RHGLYPIPKFFGRSTNK---QALGAAKLSLSR------------------- 456
Query: 631 TAYDLWHSRLGHPHYETLQSALTTCKISVPHKS-KYTICHSCCVAKSHRLPSSASSTLYT 689
WHSRLGHP ++ ++ + +S ++C++C AKSH+LP S+++
Sbjct: 457 -----WHSRLGHPSLPIVKQVISRNNLPCSVESVNQSVCNACQEAKSHQLPYIRSTSVSQ 511
Query: 690 APLELIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVEL 749
PLEL+F+D+WGPA ES Y+++ +D FS+FTWIYLLK KSE F +FQA+VE
Sbjct: 512 FPLELVFSDVWGPAP-ESVGRNKYYVSFIDDFSKFTWIYLLKYKSEVFEKFKEFQALVER 570
Query: 750 KFGLKIKSVQTDG-GTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSL 808
F KI ++QTD G ++ L F ++G+ QNGS ERKHRHIVE GLSL
Sbjct: 571 MFDRKIIAMQTDWRGGRYQKLNSFFAQIGLIIMCHVLTLIRQNGSAERKHRHIVEVGLSL 630
Query: 809 LACANMPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHL 868
L+ A+MP +WD AF+ AT+LINR+P+ + N +P KLF DY LRVFG AC+PHL
Sbjct: 631 LSYASMPLKFWDEAFVAATYLINRIPSKTIQNSTPLEKLFNQKPDYSSLRVFGCACWPHL 690
Query: 869 RPYNSSKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTLFSS 925
RPYN+ KL FRS++CVFLG+S+ HKG+KCL S GR+YIS+DV F+E FP+STL S+
Sbjct: 691 RPYNTHKLQFRSKQCVFLGFSTHHKGFKCLDVSSGRVYISRDVVFDENVFPFSTLHSN 748
Score = 104 bits (260), Expect = 2e-20
Identities = 80/325 (24%), Positives = 138/325 (41%), Gaps = 49/325 (15%)
Query: 30 HKLLLKLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSD--EDRANSVENPAFL 87
H + KL +N WK V VR RL +L + P LS + + + NPAF
Sbjct: 19 HSVSEKLGKANHALWKAQVSAAVRGARLLGYLNGDIKAPDAELSVTIDGKTTTKPNPAFE 78
Query: 88 VWEQHDSALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSI 147
WE +D + +LLSSLS VL V C+ + + W ++ + + A++ R L +
Sbjct: 79 DWEANDQLVLGYLLSSLSRDVLIQVATCKTAAEAWRSIEALYSTGTRARAVNTRLALTNT 138
Query: 148 TKGSKNTSDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTSREFPCS 207
KG+ ++Y+ +++++ + + + G P+ + ++ I GL ED+S +++ + ++ P +
Sbjct: 139 KKGTMKIAEYVAKMRALGDEMAAGGHPLDEEDLVQYIIAGLNEDFSPIVSNLCNKSDPIT 198
Query: 208 ISQAEAMVIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQAP 267
+ + + ++ E LD L + +AF+A
Sbjct: 199 VGELYSQLVNFETLLD---LYRSTGQGGAAFVA--------------------------- 228
Query: 268 PPVAIAPPSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGRGSYNNRGR 327
N G R + RG ++GRGGRG RG
Sbjct: 229 ------------NRGRGGGGGGRGNNNNSGGGGGRSAPGGRGS-GSQGRGGRG----RGT 271
Query: 328 GGDRSSVQCQICHKYGHDASVCYYR 352
GG CQ+C K GH A+ C+YR
Sbjct: 272 GGQDRRPTCQVCFKRGHTAADCWYR 296
>emb|CAB77781.1| putative polyprotein of LTR transposon [Arabidopsis thaliana]
gi|3924609|gb|AAC79110.1| putative polyprotein of LTR
transposon [Arabidopsis thaliana] gi|7444420|pir||T01397
LTR gag/pol polyprotein homolog T4I9.16 - Arabidopsis
thaliana
Length = 1456
Score = 342 bits (878), Expect = 5e-92
Identities = 267/924 (28%), Positives = 405/924 (42%), Gaps = 184/924 (19%)
Query: 35 KLDDSNFLSWKQHVEGIVRTHRLQSFLLHQPEIPPRFLSDEDRANSVENPAFLVWEQHDS 94
KL +N+L W + V + + L FL +PP + + A NP + W + D
Sbjct: 25 KLTSTNYLMWSRQVHALFDGYELAGFLDGSTPMPPATIGTD--AVPRVNPDYTRWRRQDK 82
Query: 95 ALFTWLLSSLSPSVLPTVVNCQHSWQIWEAVLDFFHAHSLAQSTQLRSELRSITKGSKNT 154
+++ +L ++S SV P V + QIWE + + S TQLR R
Sbjct: 83 LIYSAILGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRFITR--------- 133
Query: 155 SDYLKRIKSIVNALISIGDPVTYREHLEAIFDGLREDYSHLMTIVTSREFPCSISQAEAM 214
+ L +G P+ + E +E + + L +DY ++ + +++ P S+++
Sbjct: 134 ----------FDQLALLGKPMDHDEQVERVLENLPDDYKPVIDQIAAKDTPPSLTEIHER 183
Query: 215 VIAHEARLDRLRLRQHAASSPSAFLAQATAPPQSTPQAPPLVPSQSPAPPQAPPPVAIAP 274
+I E++L L + + P
Sbjct: 184 LINRESKLLALNSAE------------------------------------------VVP 201
Query: 275 PSSEVNYISRTGSDDRAYEDRDDDRSRSDDNRDRGGYNNRGRGGRGSYNNRGRGGDRSSV 334
++ V R + +R +R D+R+ +++N + G R R
Sbjct: 202 ITANV-VTHRNTNTNRNQNNRGDNRNYNNNNNRSNSWQPSSSGSRSD----NRQPKPYLG 256
Query: 335 QCQICHKYGHDASVCYYRGNSTSPATNSQPNMTNQAPAPVAPTFGFGSSFGMMPQFGYSP 394
+CQIC GH A C P + + TNQ Q SP
Sbjct: 257 RCQICSVQGHSAKRC--------PQLHQFQSTTNQ-------------------QQSTSP 289
Query: 395 FRGFQPYGAPSPFGMSFPNSGFGSGSPYASGGMFTPPGYGFGSGYGFGLPPRAPARHIRA 454
F +Q P + SPY + G A H
Sbjct: 290 FTPWQ------------PRANLAVNSPYNANNWLLDSG----------------ATH--- 318
Query: 455 PQTMLASAPSTLGNWHPDSGATHHVTHDASAISDGMSVTGNDQVYMGNGQGLPIQSIGSA 514
+ S + L P +G D I+DG ++ PI GSA
Sbjct: 319 ---HITSDFNNLSFHQPYTGG------DDVMIADGSTI--------------PITHTGSA 355
Query: 515 SFSSPIHTQTTLLLKNLLLVPNITKNLVSVSKFAQDNGVYFEFHPFHCTVNSQDTCKTLL 574
S + + +L L +L VPNI KNL+SV + N V EF P V +T LL
Sbjct: 356 SLPT---SSRSLDLNKVLYVPNIHKNLISVYRLCNTNRVSVEFFPASFQVKDLNTGVPLL 412
Query: 575 RGSLGSDGLYSFDGMHSSNFHKDLPQVPVSSAFSLSKPASPAVNTIVSTSNNVPSPTAYD 634
+G D LY + P++S+ ++S ASP S +
Sbjct: 413 QGKT-KDELYEW---------------PIASSQAVSMFASPC------------SKATHS 444
Query: 635 LWHSRLGHPHYETLQSALTTCKISVPHKS-KYTICHSCCVAKSHRLPSSASSTLYTAPLE 693
WHSRLGHP L S ++ + V + S K C C + KSH++P S S+ + PLE
Sbjct: 445 SWHSRLGHPSLAILNSVISNHSLPVLNPSHKLLSCSDCFINKSHKVPFSNSTITSSKPLE 504
Query: 694 LIFADLWGPASIESSAGYSYFLTCVDAFSRFTWIYLLKRKSETSTVFIQFQAMVELKFGL 753
I++D+W + I S Y Y++ VD F+R+TW+Y LK+KS+ FI F+++VE +F
Sbjct: 505 YIYSDVWS-SPILSIDNYRYYVIFVDHFTRYTWLYPLKQKSQVKDTFIIFKSLVENRFQT 563
Query: 754 KIKSVQTDGGTEFKPLLPHFLKLGITHRLTCPHTHHQNGSVERKHRHIVETGLSLLACAN 813
+I ++ +D G EF L + + GI+H + PHT NG ERKHRHIVE GL+LL+ A+
Sbjct: 564 RIGTLYSDNGGEFVVLRDYLSQHGISHFTSPPHTPEHNGLSERKHRHIVEMGLTLLSHAS 623
Query: 814 MPTSYWDHAFLTATFLINRLPTPVLGNLSPYHKLFKVPADYKILRVFGSACYPHLRPYNS 873
+P +YW +AF A +LINRLPTP+L SP+ KLF P +Y+ L+VFG ACYP LRPYN
Sbjct: 624 VPKTYWPYAFSVAVYLINRLPTPLLQLQSPFQKLFGQPPNYEKLKVFGCACYPWLRPYNR 683
Query: 874 SKLSFRSQECVFLGYSSSHKGYKCL-ASDGRIYISKDVQFNEFKFPYSTL-FSSEFPASN 931
KL +S++C F+GYS + Y CL GR+Y S+ VQF+E FP+ST F
Sbjct: 684 HKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFDERCFPFSTTNFGVSTSQEQ 743
Query: 932 PSSVSSVIPVISYVPSLQQPIPSP 955
S + P + +P+ +P+P
Sbjct: 744 RSDSAPNWPSHTTLPTTPLVLPAP 767
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.331 0.142 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,701,459,639
Number of Sequences: 2540612
Number of extensions: 122655893
Number of successful extensions: 770424
Number of sequences better than 10.0: 3103
Number of HSP's better than 10.0 without gapping: 986
Number of HSP's successfully gapped in prelim test: 2261
Number of HSP's that attempted gapping in prelim test: 729300
Number of HSP's gapped (non-prelim): 22765
length of query: 1571
length of database: 863,360,394
effective HSP length: 142
effective length of query: 1429
effective length of database: 502,593,490
effective search space: 718206097210
effective search space used: 718206097210
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 82 (36.2 bits)
Lotus: description of TM0079b.1


