

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0076c.8
(415 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
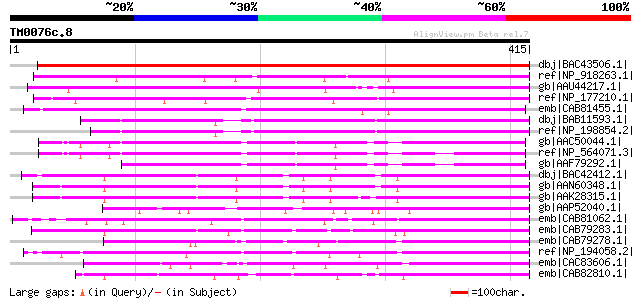
dbj|BAC43506.1| putative protein kinase [Arabidopsis thaliana] g... 564 e-159
ref|NP_918263.1| putative protein kinase [Oryza sativa (japonica... 301 2e-80
gb|AAU44217.1| hypothetical protein [Oryza sativa (japonica cult... 296 8e-79
ref|NP_177210.1| protein kinase family protein [Arabidopsis thal... 286 1e-75
emb|CAB81455.1| serine/threonine kinase-like protein [Arabidopsi... 267 5e-70
dbj|BAB11593.1| receptor-like serine/threonine kinase [Arabidops... 263 7e-69
ref|NP_198854.2| protein kinase family protein [Arabidopsis thal... 254 3e-66
gb|AAC50044.1| receptor-like serine/threonine kinase [Arabidopsi... 232 2e-59
ref|NP_564071.3| serine/threonine protein kinase (RKF2) [Arabido... 208 3e-52
gb|AAF79292.1| F14D16.24 [Arabidopsis thaliana] 205 2e-51
dbj|BAC42412.1| putative receptor-like protein kinase 4 RLK4 [Ar... 167 4e-40
gb|AAN60348.1| unknown [Arabidopsis thaliana] 167 6e-40
gb|AAK28315.1| receptor-like protein kinase 4 [Arabidopsis thali... 162 1e-38
gb|AAP52040.1| putative receptor-like protein kinase 4 [Oryza sa... 161 3e-38
emb|CAB81062.1| receptor protein kinase-like protein [Arabidopsi... 159 2e-37
emb|CAB79283.1| serine /threonine kinase-like protein [Arabidops... 158 3e-37
emb|CAB79278.1| putative protein [Arabidopsis thaliana] gi|30212... 155 2e-36
ref|NP_194058.2| protein kinase family protein [Arabidopsis thal... 155 2e-36
emb|CAC83606.1| putative receptor-like serine-threonine protein ... 154 6e-36
emb|CAB82810.1| protein kinase-like [Arabidopsis thaliana] gi|15... 152 2e-35
>dbj|BAC43506.1| putative protein kinase [Arabidopsis thaliana]
gi|15223169|ref|NP_177209.1| protein kinase family
protein [Arabidopsis thaliana]
gi|12325044|gb|AAG52471.1| putative protein kinase;
37247-34801 [Arabidopsis thaliana]
gi|25387056|pir||B96729 hypothetical protein F24J13.9
[imported] - Arabidopsis thaliana
Length = 649
Score = 564 bits (1453), Expect = e-159
Identities = 272/395 (68%), Positives = 325/395 (81%), Gaps = 2/395 (0%)
Query: 23 LLKFLLLSAT-VMAEPRAKTVQITCAKQLEHNTTIFVPNFVSTMEKISEQMRTDGFGTAV 81
LL FLL S + + RA+ V++TC+ LEHN T +VPNFV+TMEKIS Q++T GFG A+
Sbjct: 16 LLMFLLSSLRQITGDARARAVKVTCSPLLEHNETAYVPNFVATMEKISTQVQTSGFGVAL 75
Query: 82 AGTGPDTNYGLAQCYGDLSLLDCVLCYAEARTVLPQCFPYNGGRIYLDGCFMRAENYSFF 141
GTGPD NYGLAQCYGDL L DCVLCYAEART+LPQC+P NGGRI+LDGCFMRAENYSF+
Sbjct: 76 TGTGPDANYGLAQCYGDLPLNDCVLCYAEARTMLPQCYPQNGGRIFLDGCFMRAENYSFY 135
Query: 142 NEYTGPGDRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPNNKGYAKGVVAVSGTANESAYV 201
NEY GP D VCGNTTRKN TF A +Q + +AV +A GYA+ + +ESA+V
Sbjct: 136 NEYKGPEDSIVCGNTTRKNKTFGDAVRQGLRNAVTEASGTGGYARASAKAGESESESAFV 195
Query: 202 LADCWRTLDSSSCKACLENASSSIL-GCLPWSEGRALNTGCFMRYSDTDFLNKEAENGSS 260
LA+CWRTL SCK CLENAS+S++ GCLPWSEGRAL+TGCF+RYSD DFLNK NG S
Sbjct: 196 LANCWRTLSPDSCKQCLENASASVVKGCLPWSEGRALHTGCFLRYSDQDFLNKIPRNGRS 255
Query: 261 GGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKLAKTLQHNSLNFKY 320
G+V+VIVV+V+SSVVV ++GV + Y+ KRR I++KRRGS D +K+AKTL+ +SLNFKY
Sbjct: 256 RGSVVVIVVSVLSSVVVFMIGVAVSVYICKRRTIKRKRRGSKDVEKMAKTLKDSSLNFKY 315
Query: 321 STLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISS 380
STL++ATGSF +NKLGQGGFGTVYKGVL DGR+IA+KRL++NNRHRA DFYNEVN+IS+
Sbjct: 316 STLEKATGSFDNANKLGQGGFGTVYKGVLPDGRDIAVKRLFFNNRHRATDFYNEVNMIST 375
Query: 381 VEHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
VEHKNLVRLLGCSCSGPESLLVYE+L NKSLD FI
Sbjct: 376 VEHKNLVRLLGCSCSGPESLLVYEYLQNKSLDRFI 410
>ref|NP_918263.1| putative protein kinase [Oryza sativa (japonica cultivar-group)]
Length = 663
Score = 301 bits (772), Expect = 2e-80
Identities = 174/424 (41%), Positives = 241/424 (56%), Gaps = 32/424 (7%)
Query: 20 VFLLLKFLLLSATVMAEPRAKTVQITCAKQLEHNTTIFVPNFVSTMEKISEQMRTDGFGT 79
+ LLL L L +PR + CA + NFV M+ ++ + +GFGT
Sbjct: 6 LLLLLALLCLCGGAAGDPRTAVARQECAPGGAVSGPALADNFVPAMDDLNSNVSANGFGT 65
Query: 80 AVAGT----GPDTNYGLAQCYGDLSLLDCVLCYAEARTVLPQCFPYNGGRIYLDGCFMRA 135
+ GT P+ +GL QCY DLS +DC LC+AE R++LP+C+P GGR+YLDGCF R
Sbjct: 66 SAVGTTAGLNPNAVFGLGQCYRDLSPVDCKLCFAEVRSLLPKCYPSAGGRLYLDGCFGRY 125
Query: 136 ENYSFFNEYTGPGDRAVCG----------NTTRKNSTFQAAAKQAVLSAVQDAP------ 179
NYSFF+E GP D CG N T N A A +A L+ V
Sbjct: 126 ANYSFFSETLGPDDAVTCGVVGGGGAGGGNYTGANPRGFADAVRAALANVTGVAASAAVP 185
Query: 180 -NNKGYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWS-EGRAL 237
GYA G + G +A+ LA CW +L++++C CL A+++ C P + EGRAL
Sbjct: 186 GGGDGYAVGSASAGGA---TAFALAQCWGSLNATACGQCLRAAAAAAARCAPAAAEGRAL 242
Query: 238 NTGCFMRYSDTDF--LNKEAENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQ 295
TGC++RYS F LN A +GSS N +V ++ + SS+ V+ I+ WK++ +
Sbjct: 243 YTGCYLRYSTRLFWNLNSTAGSGSSRHNDVVWIL-LGSSLGAFVIVFIVVFLAWKKKIFR 301
Query: 296 KKRRGS----YDPDKLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLAD 351
K+R D + + +SLNFKY L +AT F +NKLGQG +G V+K +L D
Sbjct: 302 NKKRSKSFIDIYGDGVPVRIAQSSLNFKYEELRKATNYFDPANKLGQGSYGAVFKAILLD 361
Query: 352 GREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSL 411
G+++A+KRL+ N R F+NEV +IS V HKNLV+LLGCS +GPESLLVYE+ NKSL
Sbjct: 362 GKQVAVKRLFLNTREWVDQFFNEVELISQVRHKNLVKLLGCSVNGPESLLVYEYYFNKSL 421
Query: 412 DGFI 415
D F+
Sbjct: 422 DLFL 425
>gb|AAU44217.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 702
Score = 296 bits (758), Expect = 8e-79
Identities = 176/420 (41%), Positives = 242/420 (56%), Gaps = 26/420 (6%)
Query: 15 SSQCFVFLLLKFLLLSATVMAEPRAKTVQIT--CAKQLEHNTTIFVPNFVSTMEKISEQM 72
++ F LLL + +A AK V + C + F +FV+T+E I + +
Sbjct: 10 AAAAFRCLLLVVVAAAADDAGGAAAKPVILATVCGNTTAADQEGFDVSFVNTLELIYQNV 69
Query: 73 RTDGFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEARTVLPQCFPYNGGRIYLDGCF 132
GFG A +G G DT YGL QC G LS DC LCYA++R LP C P GGRIYLDGCF
Sbjct: 70 TRSGFGAAASGEGADTVYGLGQCMGYLSPTDCQLCYAQSRVKLPHCLPATGGRIYLDGCF 129
Query: 133 MRAENYSFFNEYTGPGDRAVCGNTTRKNSTFQAAAKQAVLSAVQDA-PNNKGYAKGVVAV 191
+R +F T D AVC N T + AA A+L V A P + Y +
Sbjct: 130 LRYGADNFTAAATDASDTAVCSNATVSSPASFAATSAALLRNVTAAAPGARDYYYYSASS 189
Query: 192 SGTANE------SAYVLADCWRTLDSSSCKACLENASSSILG-CLPWS-EGRALNTGCFM 243
S +A+ Y A CWR+L++++C AC+ A ++G CLP + EG LN GC +
Sbjct: 190 SSSASALPSVSPRVYAAAQCWRSLNATACAACVATARDRVVGRCLPRAAEGYGLNAGCVV 249
Query: 244 RYSDTDFL---NKEAENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRG 300
RYS F N A GSS +++++V+A V + V+G+ A VW + + RR
Sbjct: 250 RYSTQPFYLPANAAAAAGSSTRHIVIVVIASVFCALA-VIGI---ALVWAK---MRNRRN 302
Query: 301 SYDPD-----KLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREI 355
+ D ++ +T+ + L+FKY L +AT F++ NKLGQGG+G+VYKGVL DGREI
Sbjct: 303 DHHDDMDGSSEIIRTIAASQLSFKYEELCKATDDFNQINKLGQGGYGSVYKGVLLDGREI 362
Query: 356 AIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
A+KRL++N R A F+NEV ++S V+HKNLV+LLGCS GPESLLVYE+L N SLD ++
Sbjct: 363 AVKRLFFNTREWADQFFNEVRLVSQVQHKNLVKLLGCSIEGPESLLVYEYLCNTSLDHYL 422
>ref|NP_177210.1| protein kinase family protein [Arabidopsis thaliana]
gi|12325043|gb|AAG52470.1| putative protein kinase;
41292-38663 [Arabidopsis thaliana]
gi|25387052|pir||C96729 hypothetical protein F24J13.10
[imported] - Arabidopsis thaliana
Length = 646
Score = 286 bits (731), Expect = 1e-75
Identities = 159/408 (38%), Positives = 236/408 (56%), Gaps = 17/408 (4%)
Query: 20 VFLLLKFLLLSATVMAEPRAKTVQITCAKQLE--HNTTIFVPNFVSTMEKISEQMRTDGF 77
+ LL+ L +S +V ++ R TV C + ++FV NF++ M+ +S + G+
Sbjct: 6 LLLLILGLFISPSV-SDSRGDTVAQICNNRTTTPQQRSLFVTNFLAAMDAVSPLVEAKGY 64
Query: 78 GTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEARTVLPQCFPYN----GGRIYLDGCFM 133
G V GTG T Y +C DL DC LC+A+ + +P+C P+ GG+++ DGC++
Sbjct: 65 GQVVNGTGNLTVYAYGECIKDLDKKDCDLCFAQIKAKVPRCLPFQKGTRGGQVFSDGCYI 124
Query: 134 RAENYSFFNEYTGPGDRAVCGN---TTRKNSTFQAAAKQAVLSAVQDAPNNKGYAKGVVA 190
R ++Y+F+NE DR VC T + F+ A + V + +A N G+ G V
Sbjct: 125 RYDDYNFYNETLSLQDRTVCAPKEITGVNRTVFRDNAAELVKNMSVEAVRNGGFYAGFV- 183
Query: 191 VSGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSEGRALNTGCFMRYSDTDF 250
N + + LA CW TL+ S C CL AS I CL EGR L+ GC+MR+S F
Sbjct: 184 --DRHNVTVHGLAQCWETLNRSGCVECLSKASVRIGSCLVNEEGRVLSAGCYMRFSTQKF 241
Query: 251 LNKEAEN---GSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKL 307
N + G+ G N + +++AV SSVV V+ V ++ K+R+ KK+R L
Sbjct: 242 YNNSGNSTSDGNGGHNHLGVILAVTSSVVAFVLLVSAAGFLLKKRHA-KKQREKKQLGSL 300
Query: 308 AKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHR 367
++L F Y L+RAT F + NKLGQGG G+VYKGVL +G+ +A+KRL++N +
Sbjct: 301 FMLANKSNLCFSYENLERATDYFSDKNKLGQGGSGSVYKGVLTNGKTVAVKRLFFNTKQW 360
Query: 368 AADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
F+NEVN+IS V+HKNLV+LLGCS +GPESLLVYE++ N+SL ++
Sbjct: 361 VDHFFNEVNLISQVDHKNLVKLLGCSITGPESLLVYEYIANQSLHDYL 408
>emb|CAB81455.1| serine/threonine kinase-like protein [Arabidopsis thaliana]
gi|15235391|ref|NP_194596.1| protein kinase family
protein [Arabidopsis thaliana] gi|7488334|pir||T10661
serine/threonine-specific protein kinase homolog
T5F17.120 - Arabidopsis thaliana
Length = 625
Score = 267 bits (682), Expect = 5e-70
Identities = 155/415 (37%), Positives = 227/415 (54%), Gaps = 19/415 (4%)
Query: 12 SLISSQCFVFLLLKFLLLSATVMAEPRAKTVQITCAKQLEHNTTIFVPNFVSTMEKISEQ 71
+LISS V L LL + M R + C N + ++ ++ I
Sbjct: 4 TLISSLAVVLPLT--LLAPSMSMKISRIDVLGYICNNGTVSNEEAYRRSYQINLDAIRGD 61
Query: 72 MRTDGFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEARTVLPQCFPYNGGRIYLDGC 131
MR FGT G P+ Y L+QC DLS +C LC++ A +L QCFP GG +LDGC
Sbjct: 62 MRHVKFGTHEHGDPPERMYVLSQCVSDLSSDECSLCWSRATDLLSQCFPATGGWFHLDGC 121
Query: 132 FMRAENYSFFNEYTGPGDRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPNNKGYAKGVVAV 191
F+RA+NYSF+ E D +C + K++ F+ K+ S V+ AP ++G++ VA
Sbjct: 122 FVRADNYSFYQEPVSHQDTKICASDKEKSAEFKGLVKEVTKSIVEAAPYSRGFS---VAK 178
Query: 192 SGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSEGRALNTGCFMRYSDTDFL 251
G + + Y L CWRTL+ CK CL + + S+ CLP EG ALN GC++RYS+ F
Sbjct: 179 MGIRDLTVYGLGVCWRTLNDELCKLCLADGALSVTSCLPSKEGFALNAGCYLRYSNYTFY 238
Query: 252 NKEAENGSSGGNVMVIVVAVVSSVVVTVV--GVIIGAYVWKRRYIQKKRRGS-------- 301
N+ S + + V+S V V + G G + R +KK +G+
Sbjct: 239 NERGLLAMSFTKENLTYIFVISMVGVLAIAAGFWCGKCFYMRTSPKKKIKGTKTKKFHLF 298
Query: 302 ----YDPDKLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAI 357
+ + + + + ++F+YSTL +AT +F+ES KLG GG+G V+KG L+DGREIAI
Sbjct: 299 GHLRIEKESESICTESHLMSFEYSTLKKATNNFNESCKLGVGGYGEVFKGTLSDGREIAI 358
Query: 358 KRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLD 412
KRL+ + + + +NE+++IS +HKNLVRLLGC + S +VYEFL N SLD
Sbjct: 359 KRLHVSGKKPRDEIHNEIDVISRCQHKNLVRLLGCCFTNMNSFIVYEFLANTSLD 413
>dbj|BAB11593.1| receptor-like serine/threonine kinase [Arabidopsis thaliana]
Length = 651
Score = 263 bits (672), Expect = 7e-69
Identities = 142/365 (38%), Positives = 207/365 (55%), Gaps = 23/365 (6%)
Query: 57 FVPNFVSTMEKISEQMRTDGFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEARTVLP 116
++P FV M +S ++ T F T + Y L QC+ DLS DC LCYA ART +P
Sbjct: 53 YIPTFVEDMHSLSLKLTTRRFATESLNSTTSI-YALIQCHDDLSPSDCQLCYAIARTRIP 111
Query: 117 QCFPYNGGRIYLDGCFMRAENYSFFNE-YTGPGDRAVCGNTTRKNSTF-----QAAAKQA 170
+C P + RI+LDGCF+R E Y F++E + D C N T + F + AA+ A
Sbjct: 112 RCLPSSSARIFLDGCFLRYETYEFYDESVSDASDSFSCSNDTVLDPRFGFQVSETAARVA 171
Query: 171 VLSAVQDAPNNKGYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLP 230
V KG V+G + LA CW +L C+ CLE A + C+
Sbjct: 172 V-------------RKGGFGVAG--ENGVHALAQCWESLGKEDCRVCLEKAVKEVKRCVS 216
Query: 231 WSEGRALNTGCFMRYSDTDFLNKEAENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWK 290
EGRA+NTGC++RYSD F N + + ++VA+V + V+ +++ YV
Sbjct: 217 RREGRAMNTGCYLRYSDHKFYNGDGHHKFHVLFNKGVIVAIVLTTSAFVMLILLATYVIM 276
Query: 291 RRYIQKKRRGSYDPDKLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLA 350
+ + K ++ + +++ ++ FKY TL++AT F LGQGG GTV+ G+L
Sbjct: 277 TK-VSKTKQEKRNLGLVSRKFNNSKTKFKYETLEKATDYFSHKKMLGQGGNGTVFLGILP 335
Query: 351 DGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKS 410
+G+ +A+KRL +N R +F+NEVN+IS ++HKNLV+LLGCS GPESLLVYE++PNKS
Sbjct: 336 NGKNVAVKRLVFNTRDWVEEFFNEVNLISGIQHKNLVKLLGCSIEGPESLLVYEYVPNKS 395
Query: 411 LDGFI 415
LD F+
Sbjct: 396 LDQFL 400
>ref|NP_198854.2| protein kinase family protein [Arabidopsis thaliana]
Length = 591
Score = 254 bits (650), Expect = 3e-66
Identities = 139/357 (38%), Positives = 202/357 (55%), Gaps = 23/357 (6%)
Query: 65 MEKISEQMRTDGFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEARTVLPQCFPYNGG 124
M +S ++ T F T + Y L QC+ DLS DC LCYA ART +P+C P +
Sbjct: 1 MHSLSLKLTTRRFATESLNSTTSI-YALIQCHDDLSPSDCQLCYAIARTRIPRCLPSSSA 59
Query: 125 RIYLDGCFMRAENYSFFNE-YTGPGDRAVCGNTTRKNSTF-----QAAAKQAVLSAVQDA 178
RI+LDGCF+R E Y F++E + D C N T + F + AA+ AV
Sbjct: 60 RIFLDGCFLRYETYEFYDESVSDASDSFSCSNDTVLDPRFGFQVSETAARVAV------- 112
Query: 179 PNNKGYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSEGRALN 238
KG V+G + LA CW +L C+ CLE A + C+ EGRA+N
Sbjct: 113 ------RKGGFGVAG--ENGVHALAQCWESLGKEDCRVCLEKAVKEVKRCVSRREGRAMN 164
Query: 239 TGCFMRYSDTDFLNKEAENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKR 298
TGC++RYSD F N + + ++VA+V + V+ +++ YV + + K +
Sbjct: 165 TGCYLRYSDHKFYNGDGHHKFHVLFNKGVIVAIVLTTSAFVMLILLATYVIMTK-VSKTK 223
Query: 299 RGSYDPDKLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIK 358
+ + +++ ++ FKY TL++AT F LGQGG GTV+ G+L +G+ +A+K
Sbjct: 224 QEKRNLGLVSRKFNNSKTKFKYETLEKATDYFSHKKMLGQGGNGTVFLGILPNGKNVAVK 283
Query: 359 RLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
RL +N R +F+NEVN+IS ++HKNLV+LLGCS GPESLLVYE++PNKSLD F+
Sbjct: 284 RLVFNTRDWVEEFFNEVNLISGIQHKNLVKLLGCSIEGPESLLVYEYVPNKSLDQFL 340
>gb|AAC50044.1| receptor-like serine/threonine kinase [Arabidopsis thaliana]
Length = 617
Score = 232 bits (591), Expect = 2e-59
Identities = 146/411 (35%), Positives = 221/411 (53%), Gaps = 45/411 (10%)
Query: 24 LKFLLLSATVMAEPRAKTVQITCAKQLEHNTT--IFVPNFVSTMEKISEQMRTDGFG--T 79
+ FL+L ATV + ++++ + C + L+H+ + F+ M +++ + D +
Sbjct: 14 VSFLVLLATVGSSSSSESL-LNC-QPLDHHLVNPSRLLGFLRAMSSVNDFITNDKLWVVS 71
Query: 80 AVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEARTVLPQCFPYNGGRIYLDGCFMRAENYS 139
++ P Y QC DLS+ DC C+ E+R L + GRI+ D CF+R ++
Sbjct: 72 SITDVSPPI-YVFLQCREDLSVSDCRHCFNESRLELERNVRVPVGRIHSDRCFLRFDDRD 130
Query: 140 FFNEYTGPG-DRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPNNKGYAKGVVAVSGTANES 198
F E+ P D+A C T F +A+++ A N G+ A S E+
Sbjct: 131 FSEEFVDPTFDKANCEETGTGFGEFWRFLDEALVNVTLKAVKNGGFG----AASVIKTEA 186
Query: 199 AYVLADCWRTLDSSSCKACLENASSSILGCLPWSEGRALNTGCFMRYSDTDFLNKEAENG 258
Y LA CW+TLD ++C+ CL NA SS+ C E RA TGC+++YS F + +A
Sbjct: 187 VYALAQCWQTLDENTCRECLVNARSSLRAC-DGHEARAFFTGCYLKYSTHKFFDDDAAEH 245
Query: 259 S-----------------SGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGS 301
S +V + +A +S ++T +G I R + +KR+
Sbjct: 246 KPDADQRNFIRSSFFPHLSDRDVTRLAIAAISLSILTSLGAFISY-----RRVSRKRKAQ 300
Query: 302 YDPDKLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLY 361
+ +NFKY L++AT SFH+S KLGQGG G+VYKG+L DGR +A+K+L+
Sbjct: 301 VP----------SCVNFKYEMLEKATESFHDSMKLGQGGAGSVYKGILPDGRIVAVKKLF 350
Query: 362 YNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLD 412
+N R A F+NEVN+IS V+HKNLVRLLGCS GP+SLLVYE++ N+SLD
Sbjct: 351 FNTREWADQFFNEVNLISGVQHKNLVRLLGCSIEGPKSLLVYEYVHNRSLD 401
>ref|NP_564071.3| serine/threonine protein kinase (RKF2) [Arabidopsis thaliana]
Length = 600
Score = 208 bits (529), Expect = 3e-52
Identities = 139/410 (33%), Positives = 212/410 (50%), Gaps = 59/410 (14%)
Query: 24 LKFLLLSATVMAEPRAKTVQITCAKQLEHNTT--IFVPNFVSTMEKISEQMRTDGFG--T 79
+ FL+L ATV + ++++ + C + L+H+ + F+ M +++ + D +
Sbjct: 14 VSFLVLLATVGSSSSSESL-LNC-QPLDHHLVNPSRLLGFLRAMSSVNDFITNDKLWVVS 71
Query: 80 AVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEARTVLPQCFPYNGGRIYLDGCFMRAENYS 139
++ P Y QC DLS+ DC C+ E+R L + +GGRI+ D CF+R ++
Sbjct: 72 SITDVSPPI-YVFLQCREDLSVSDCRHCFNESRLELERKCSGSGGRIHSDRCFLRFDDRD 130
Query: 140 FFNEYTGPG-DRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPNNKGYAKGVVAVSGTANES 198
F E+ P D+A C T F +A+++ A N G+ A S E+
Sbjct: 131 FSEEFVDPTFDKANCEETGTGFGEFWRFLDEALVNVTLKAVKNGGFG----AASVIKTEA 186
Query: 199 AYVLADCWRTLDSSSCKACLENASSSILGCLPWSEGRALNTGCFMRYSDTDFLNKEAENG 258
Y LA CW+TLD ++C+ CL NA SS+ C E RA TGC+++YS F + AE+
Sbjct: 187 VYALAQCWQTLDENTCRECLVNARSSLRAC-DGHEARAFFTGCYLKYSTHKFFDDAAEHK 245
Query: 259 S----------------SGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSY 302
S +V + +A +S ++T +G I R + +KR+
Sbjct: 246 PDADQRNFIRSSFFPHLSDRDVTRLAIAAISLSILTSLGAFISY-----RRVSRKRKAQV 300
Query: 303 DPDKLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYY 362
+ +NFKY L++AT SFH+S KLGQGG A+K+L++
Sbjct: 301 P----------SCVNFKYEMLEKATESFHDSMKLGQGG---------------AVKKLFF 335
Query: 363 NNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLD 412
N R A F+NEVN+IS V+HKNLVRLLGCS GP+SLLVYE++ N+SLD
Sbjct: 336 NTREWADQFFNEVNLISGVQHKNLVRLLGCSIEGPKSLLVYEYVHNRSLD 385
>gb|AAF79292.1| F14D16.24 [Arabidopsis thaliana]
Length = 668
Score = 205 bits (521), Expect = 2e-51
Identities = 126/340 (37%), Positives = 179/340 (52%), Gaps = 52/340 (15%)
Query: 90 YGLAQCYGDLSLLDCVLCYAEARTVLPQCFPYNGGRIYLDGCFMRAENYSFFNEYTGPG- 148
Y QC DLS+ DC C+ E+R L + +GGRI+ D CF+R ++ F E+ P
Sbjct: 149 YVFLQCREDLSVSDCRHCFNESRLELERKCSGSGGRIHSDRCFLRFDDRDFSEEFVDPTF 208
Query: 149 DRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPNNKGYAKGVVAVSGTANESAYVLADCWRT 208
D+A C T F +A+++ A N G+ A S E+ Y LA CW+T
Sbjct: 209 DKANCEETGTGFGEFWRFLDEALVNVTLKAVKNGGFG----AASVIKTEAVYALAQCWQT 264
Query: 209 LDSSSCKACLENASSSILGCLPWSEGRALNTGCFMRYSDTDFLNKEAENGS--------- 259
LD ++C+ CL NA SS+ C E RA TGC+++YS F + AE+
Sbjct: 265 LDENTCRECLVNARSSLRAC-DGHEARAFFTGCYLKYSTHKFFDDAAEHKPDADQRNFIR 323
Query: 260 -------SGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKLAKTLQ 312
S +V + +A +S ++T +G I R + +KR+
Sbjct: 324 SSFFPHLSDRDVTRLAIAAISLSILTSLGAFISY-----RRVSRKRKAQVP--------- 369
Query: 313 HNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFY 372
+ +NFKY L++AT SFH+S KLGQGG A+K+L++N R A F+
Sbjct: 370 -SCVNFKYEMLEKATESFHDSMKLGQGG---------------AVKKLFFNTREWADQFF 413
Query: 373 NEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLD 412
NEVN+IS V+HKNLVRLLGCS GP+SLLVYE++ N+SLD
Sbjct: 414 NEVNLISGVQHKNLVRLLGCSIEGPKSLLVYEYVHNRSLD 453
>dbj|BAC42412.1| putative receptor-like protein kinase 4 RLK4 [Arabidopsis thaliana]
gi|29029020|gb|AAO64889.1| At4g23180 [Arabidopsis
thaliana] gi|30686087|ref|NP_567679.2| receptor-like
protein kinase 4, putative (RLK4) [Arabidopsis thaliana]
Length = 669
Score = 167 bits (424), Expect = 4e-40
Identities = 133/435 (30%), Positives = 190/435 (43%), Gaps = 41/435 (9%)
Query: 10 VVSLISSQCFVFLLLKFLLLSATVMAEPRAKTVQITCAKQLEHNTTIFVPNFVSTM-EKI 68
++S SS F+FL F L++ ++ V TC + + N + T+ +
Sbjct: 11 IMSYYSSFFFLFL---FSFLTSFRVSAQDPTYVYHTCQNTANYTSNSTYNNNLKTLLASL 67
Query: 69 SEQMRT--DGFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEA-RTVLPQCFPYNGGR 125
S + + GF A G PD GL C GD+S C C + A L +C
Sbjct: 68 SSRNASYSTGFQNATVGQAPDRVTGLFNCRGDVSTEVCRRCVSFAVNDTLTRCPNQKEAT 127
Query: 126 IYLDGCFMRAENYSFFNEYTGPGDRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPN---NK 182
+Y D C +R N + + G + NT S VL + A N
Sbjct: 128 LYYDECVLRYSNQNILSTLITTGG-VILVNTRNVTSNQLDLLSDLVLPTLNQAATVALNS 186
Query: 183 GYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSE--GRALNTG 240
G + TA +S Y L C L C CL+ ++ +P R +N
Sbjct: 187 SKKFGTRKNNFTALQSFYGLVQCTPDLTRQDCSRCLQ----LVINQIPTDRIGARIINPS 242
Query: 241 CFMRYSDTDFLNKEA------------------ENGSSGGNVMVIVVAVVSSVVVTVVGV 282
C RY F + A GN V+V+A+V ++V V+
Sbjct: 243 CTSRYEIYAFYTESAVPPPPPPPSISTPPVSAPPRSGKDGNSKVLVIAIVVPIIVAVLLF 302
Query: 283 IIGAYVWKRRYIQKKRRGSYDPDKLA--KTLQHNSLNFKYSTLDRATGSFHESNKLGQGG 340
I G RR R+ Y P A +SL Y T+ AT F ESNK+GQGG
Sbjct: 303 IAGYCFLTRR----ARKSYYTPSAFAGDDITTADSLQLDYRTIQTATDDFVESNKIGQGG 358
Query: 341 FGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESL 400
FG VYKG L+DG E+A+KRL ++ +F NEV +++ ++H+NLVRLLG G E +
Sbjct: 359 FGEVYKGTLSDGTEVAVKRLSKSSGQGEVEFKNEVVLVAKLQHRNLVRLLGFCLDGEERV 418
Query: 401 LVYEFLPNKSLDGFI 415
LVYE++PNKSLD F+
Sbjct: 419 LVYEYVPNKSLDYFL 433
>gb|AAN60348.1| unknown [Arabidopsis thaliana]
Length = 658
Score = 167 bits (423), Expect = 6e-40
Identities = 133/426 (31%), Positives = 184/426 (42%), Gaps = 39/426 (9%)
Query: 19 FVFLLLKFLLLSATVMAEPRAKTVQITCAKQLEHNTTIFVPNFVSTM-EKISEQMRT--D 75
F FL L L S V A+ V TC + + N + T+ +S + +
Sbjct: 7 FFFLFLFSFLTSFRVSAQDPTY-VYHTCQNTANYTSNSTYNNNLKTLLASLSSRNASYST 65
Query: 76 GFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEA-RTVLPQCFPYNGGRIYLDGCFMR 134
GF A G PD GL C GD+S C C + A L +C +Y D C +R
Sbjct: 66 GFQNATVGQAPDRVTGLFNCRGDVSTEVCRRCVSFAVNDTLTRCPNQKEATLYYDECVLR 125
Query: 135 AENYSFFNEYTGPGDRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPN---NKGYAKGVVAV 191
N + + G + NT S VL + A N G
Sbjct: 126 YSNQNILSTLITTGG-VILVNTRNVTSNQLDLLSDLVLPTLNQAATVALNSSKKFGTRKN 184
Query: 192 SGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSE--GRALNTGCFMRYSDTD 249
+ TA +S Y L C L C CL+ ++ +P R +N C RY
Sbjct: 185 NFTALQSFYGLVQCTPDLTRQDCSRCLQ----LVINQIPTDRIGARIINPSCTSRYEIYA 240
Query: 250 FLNKEA------------------ENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKR 291
F + A GN V+V+A+V ++V V+ I G R
Sbjct: 241 FYTESAVPPPPPPPSICTPPVSAPPRSGKDGNSKVLVIAIVVPIIVAVLLFIAGYCFLTR 300
Query: 292 RYIQKKRRGSYDPDKLA--KTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVL 349
R R+ Y P A +SL Y T+ AT F ESNK+GQGGFG VYKG L
Sbjct: 301 R----ARKSYYTPSAFAGDDITTADSLQLDYRTIQTATDDFVESNKIGQGGFGEVYKGTL 356
Query: 350 ADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNK 409
+DG E+A+KRL ++ +F NEV +++ ++H+NLVRLLG G E +LVYE++PNK
Sbjct: 357 SDGTEVAVKRLSKSSGQGEVEFKNEVVLVAKLQHRNLVRLLGFCLDGEERVLVYEYVPNK 416
Query: 410 SLDGFI 415
SLD F+
Sbjct: 417 SLDYFL 422
>gb|AAK28315.1| receptor-like protein kinase 4 [Arabidopsis thaliana]
Length = 658
Score = 162 bits (411), Expect = 1e-38
Identities = 132/427 (30%), Positives = 186/427 (42%), Gaps = 41/427 (9%)
Query: 19 FVFLLLKFLLLSATVMAEPRAKTVQITCAKQLEHNTTIFVPNFVSTM-EKISEQMRT--D 75
F FL L L S V A+ V TC + + N + T+ +S + +
Sbjct: 7 FFFLFLFSFLTSFRVSAQDPTY-VYHTCQNTANYTSNSTYNNNLKTLLASLSSRNASYST 65
Query: 76 GFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEA-RTVLPQCFPYNGGRIYLDGCFMR 134
GF A G PD GL C GD+S C C + A L +C +Y D C +R
Sbjct: 66 GFQNATVGQAPDRVTGLFNCRGDVSTEVCRRCVSFAVNDTLTRCPNQKEATLYYDECVLR 125
Query: 135 AENYSFFNEYTGPGDRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPN---NKGYAKGVVAV 191
N + + G + NT S VL + A N G
Sbjct: 126 YSNQNILSTLITTGG-VILVNTRNVTSNQLDLLSDLVLPTLNQAATVALNSSKKFGTRKN 184
Query: 192 SGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSE--GRALNTGCFMRYSDTD 249
+ TA +S Y L C L C CL+ ++ +P R +N C RY
Sbjct: 185 NFTALQSFYGLVQCTPDLTRQDCSRCLQ----LVINQIPTDRIGARIINPSCTSRYEIYA 240
Query: 250 FLNKEA------------------ENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKR 291
F + A GN V+V+A+V ++V V + I Y
Sbjct: 241 FYTESAVPPPPPPPSISTPPVSAPPRSEKEGNSKVLVIAIVVPIIVAV-RLFIAGYC--- 296
Query: 292 RYIQKKRRGSYD-PDKLA--KTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGV 348
++ ++ R SY P A +SL Y T+ AT F ESNK+GQGGFG VYKG
Sbjct: 297 -FLTRRARKSYSTPSAFAGDDITTADSLQLDYRTIQTATDDFVESNKIGQGGFGEVYKGT 355
Query: 349 LADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPN 408
L+DG E+A+KRL ++ +F NEV +++ ++H+NLVRLLG G E +LVYE++PN
Sbjct: 356 LSDGTEVAVKRLSKSSGQGEVEFKNEVVLVAKLQHRNLVRLLGFCLDGEERVLVYEYVPN 415
Query: 409 KSLDGFI 415
KSLD F+
Sbjct: 416 KSLDYFL 422
>gb|AAP52040.1| putative receptor-like protein kinase 4 [Oryza sativa (japonica
cultivar-group)] gi|37530902|ref|NP_919753.1| putative
receptor-like protein kinase 4 [Oryza sativa (japonica
cultivar-group)] gi|18642685|gb|AAK02023.2| Putative
receptor-like protein kinase 4 [Oryza sativa]
Length = 640
Score = 161 bits (408), Expect = 3e-38
Identities = 128/395 (32%), Positives = 186/395 (46%), Gaps = 72/395 (18%)
Query: 75 DGFGTAVAGTGPDTNYGLAQCYGDLSLL----DCVLC-YAEARTVLPQCFPYNGGRIYLD 129
D F TA AG PD Y LA C GD++ DCV + +AR P IY D
Sbjct: 55 DHFATATAGQAPDAAYALALCRGDVANATACGDCVAASFQDARRTCPSD---KSATIYYD 111
Query: 130 GCFMR------------AENYSFF---NEYTGPGDRAVCGNTTRKNSTFQAAAKQAVLSA 174
C +R EN + F N+ GD AV R+ T A A
Sbjct: 112 DCLLRFAGDDFLAAPNITENATLFQAWNQQNITGDAAVAAANVRELLTVTARTAAAAA-- 169
Query: 175 VQDAPNNKGYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLE------NASSSI-LG 227
+ +A G + S + ++ Y LA C L + C ACL+ N+++S+ LG
Sbjct: 170 -------RRFATGFMDGSSESKQTLYSLAQCTPDLAAGDCLACLQRLIAMVNSTTSVRLG 222
Query: 228 CLPWSEGRALNTGCFMRYSDTDFLNKEAEN-----GSSGGNVMVI-----------VVAV 271
GR L C +R+ F E GS+ + V+A
Sbjct: 223 ------GRVLLLRCNLRFEAFVFYAGEPTRRVSPPGSTPAPDSIAPTKNRKKSKSWVIAA 276
Query: 272 VSSVVVTVVGVIIGAYV--WKRRY------IQKKRRGSYDPDKLAKTLQHNSLNF---KY 320
+++ V VV +I Y W RR+ +++KR + D+L ++ F ++
Sbjct: 277 IAAPVAAVVLCLIVCYYCRWSRRFRKDRVRLREKRSRRFRGDELICEMEGEISEFSVFEF 336
Query: 321 STLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISS 380
+ +AT +F E NKLG+GGFG VYKG+ ++G EIA+KRL ++ +F NEV +I+
Sbjct: 337 REVIKATDNFSEENKLGEGGFGPVYKGLFSEGLEIAVKRLASHSGQGFLEFKNEVQLIAK 396
Query: 381 VEHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
++H+NLVRLLGC G E +LVYE+LPNKSLD +I
Sbjct: 397 LQHRNLVRLLGCCSQGEEKILVYEYLPNKSLDFYI 431
>emb|CAB81062.1| receptor protein kinase-like protein [Arabidopsis thaliana]
gi|15234659|ref|NP_192429.1| protein kinase family
protein [Arabidopsis thaliana] gi|25387047|pir||D85065
receptor protein kinase-like protein [imported] -
Arabidopsis thaliana
Length = 675
Score = 159 bits (402), Expect = 2e-37
Identities = 141/453 (31%), Positives = 210/453 (46%), Gaps = 61/453 (13%)
Query: 3 LSQKMKRVVSLISSQCFVFLLLKFLLLSATVMAEPRAKTVQITCAKQLEHNTTIFVPN-- 60
+S K VSL S F+F++LK TV ++P T + NTT + N
Sbjct: 1 MSSCFKSSVSLFS--VFLFMILK------TVTSDP-------TYLYHICPNTTTYSRNSS 45
Query: 61 FVSTMEKISEQMRTDG------FGTAVAGTGPDTN--YGLAQCYGDLSLLDCVLCYA-EA 111
+++ + + + + F A AG D+N YG+ C GD+S C C A A
Sbjct: 46 YLTNLRTVLSSLSSPNAAYASLFDNAAAGEENDSNRVYGVFLCRGDVSAEICRDCVAFAA 105
Query: 112 RTVLPQCFPYNGGRIYLDGCFMRAENYSFFNEYT-GPGDRAVCGNTTRKN--STFQAAAK 168
L +C I+ D C +R N S + PG +N S F +
Sbjct: 106 NETLQRCPREKVAVIWYDECMVRYSNQSIVGQMRIRPGVFLTNKQNITENQVSRFNESLP 165
Query: 169 QAVLS-AVQDAPNNKGYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLENASSSILG 227
++ AV+ A +++ +A + T ++ Y L C L + C++CL + +
Sbjct: 166 ALLIDVAVKAALSSRKFA--TEKANFTVFQTIYSLVQCTPDLTNQDCESCLRQVINYLPR 223
Query: 228 CLPWSEG-RALNTGCFMRYSDTDF----------------------LNKEAENGSSGGNV 264
C S G R + C RY F LN +E G G N+
Sbjct: 224 CCDRSVGGRVIAPSCSFRYELYPFYNETIAAAPMAPPPSSTVTAPPLNIPSEKGK-GKNL 282
Query: 265 MVIVVAVVSSVVVTVVGVIIGAYVW--KRRYIQKKRRGSYDPDKLAKTLQHNSLNFKYST 322
VIV A+ +V V+V +++GA W RR K + D D+ T +L F++S
Sbjct: 283 TVIVTAI--AVPVSVCVLLLGAMCWLLARRRNNKLSAETEDLDEDGIT-STETLQFQFSA 339
Query: 323 LDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSVE 382
++ AT F ESNKLG GGFG VYKG L G +AIKRL + A +F NEV++++ ++
Sbjct: 340 IEAATNKFSESNKLGHGGFGEVYKGQLITGETVAIKRLSQGSTQGAEEFKNEVDVVAKLQ 399
Query: 383 HKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
H+NL +LLG G E +LVYEF+PNKSLD F+
Sbjct: 400 HRNLAKLLGYCLDGEEKILVYEFVPNKSLDYFL 432
>emb|CAB79283.1| serine /threonine kinase-like protein [Arabidopsis thaliana]
gi|3021280|emb|CAA18475.1| serine /threonine kinase-like
protein [Arabidopsis thaliana]
gi|15236447|ref|NP_194059.1| protein kinase, putative
[Arabidopsis thaliana] gi|7488190|pir||T04845 probable
serine/threonine-specific protein kinase (EC 2.7.1.-)
F21P8.170 - Arabidopsis thaliana
Length = 656
Score = 158 bits (399), Expect = 3e-37
Identities = 129/423 (30%), Positives = 190/423 (44%), Gaps = 34/423 (8%)
Query: 18 CFVFLLLKFLLLSATVMAEPRAKTVQITCAKQLEHNTTIFVPNFVSTMEKISEQMRT--D 75
CF+FL L + S T A+ + + + N + + +S + +
Sbjct: 6 CFIFLFLFSFITSFTASAQNPFYLYHNCSITTTYSSNSTYSTNLKTLLSSLSSRNASYST 65
Query: 76 GFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYA-EARTVLPQCFPYNGGRIYLDGCFMR 134
GF A AG PD GL C G++S C C A L +C Y + C +R
Sbjct: 66 GFQNATAGQAPDMVTGLFLCRGNVSPEVCRSCIALSVNESLSRCPNEREAVFYYEQCMLR 125
Query: 135 AENYSFFNEYTGPGDRAVCGNTTRKNSTFQAAAKQAVLS-----AVQDAPNNKGYAKGVV 189
N + + G N S Q + VL+ A++ A + K +A V
Sbjct: 126 YSNRNILSTLNTDGG-VFMQNARNPISVKQDRFRDLVLNPMNLAAIEAARSIKRFA--VT 182
Query: 190 AVSGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSEGRALNTGCFMRYSDTD 249
A +S Y + C L C CL+ + + + GR C RY + +
Sbjct: 183 KFDLNALQSLYGMVQCTPDLTEQDCLDCLQQSINQVT--YDKIGGRTFLPSCTSRYDNYE 240
Query: 250 FLNKEAENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKL-- 307
F N+ N GGN VIV+AVV V +TV+ ++ A+ RR +KK G+ K+
Sbjct: 241 FYNEF--NVGKGGNSSVIVIAVV--VPITVLFLLFVAFFSVRRAKRKKTIGAIPLFKVKR 296
Query: 308 ---------AKTLQHN------SLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADG 352
A+T + SL F + + AT F NKLGQGGFG VYKG G
Sbjct: 297 KETEVTEPPAETTDGDDITTAGSLQFDFKAIVAATDIFLPINKLGQGGFGEVYKGTFPSG 356
Query: 353 REIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLD 412
++A+KRL N+ +F NEV +++ ++H+NLV+LLG G E +LVYEF+PNKSLD
Sbjct: 357 VQVAVKRLSKNSGQGEKEFENEVVVVAKLQHRNLVKLLGYCLEGEEKILVYEFVPNKSLD 416
Query: 413 GFI 415
F+
Sbjct: 417 YFL 419
>emb|CAB79278.1| putative protein [Arabidopsis thaliana] gi|3021275|emb|CAA18470.1|
putative protein [Arabidopsis thaliana]
gi|7485942|pir||T04840 hypothetical protein F21P8.120 -
Arabidopsis thaliana
Length = 485
Score = 155 bits (392), Expect = 2e-36
Identities = 120/370 (32%), Positives = 171/370 (45%), Gaps = 41/370 (11%)
Query: 76 GFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYA-EARTVLPQCFPYNGGRIYLDGCFMR 134
GF +A AG PD GL C GD+S C C A L QC Y D C +R
Sbjct: 49 GFQSARAGQAPDRVTGLFLCRGDVSPAVCRNCVAFSINDTLVQCPSERKSVFYYDECMLR 108
Query: 135 AENYSFFNE--YTGP-----GDRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPNNKGYAKG 187
+ + + Y G G+ ++ N + F ++ +P K Y
Sbjct: 109 YSDQNILSTLAYDGAWIRMNGNISIDQNQMNRFKDFVSSTMNQAAVKAASSPR-KFYT-- 165
Query: 188 VVAVSGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSEGRALNTGCFMRY-- 245
V + TA ++ Y L C L C +CLE SS L L + GR L + C RY
Sbjct: 166 -VKATWTALQTLYGLVQCTPDLTRQDCFSCLE--SSIKLMPLYKTGGRTLYSSCNSRYEL 222
Query: 246 --------------------SDTDFLNKEAENGSSGGNVMVIVVAVVSSVVVTVVGVIIG 285
S T + + G S N V+VVA+V +++V + +I G
Sbjct: 223 FAFYNETTVRTQQAPPPLPPSSTPLVTSPSLPGKSW-NSNVLVVAIVLTILVAALLLIAG 281
Query: 286 AYVWKRRYIQKKRRGSYDPDKLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVY 345
KR ++D D + SL Y + AT F E+NK+GQGGFG VY
Sbjct: 282 YCFAKRVKNSSDNAPAFDGDDITT----ESLQLDYRMIRAATNKFSENNKIGQGGFGEVY 337
Query: 346 KGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEF 405
KG ++G E+A+KRL ++ +F NEV +++ ++H+NLVRLLG S G E +LVYE+
Sbjct: 338 KGTFSNGTEVAVKRLSKSSGQGDTEFKNEVVVVAKLQHRNLVRLLGFSIGGGERILVYEY 397
Query: 406 LPNKSLDGFI 415
+PNKSLD F+
Sbjct: 398 MPNKSLDYFL 407
>ref|NP_194058.2| protein kinase family protein [Arabidopsis thaliana]
Length = 645
Score = 155 bits (392), Expect = 2e-36
Identities = 128/424 (30%), Positives = 196/424 (46%), Gaps = 35/424 (8%)
Query: 12 SLISSQCFVFLLLKFLLLSATVMAEPRAK-----TVQITCAKQLEHNTTIFVPNFVSTME 66
SLIS F+FL FL S T A+ +V T + ++T + +S++
Sbjct: 3 SLIS---FIFL---FLFSSITASAQNTFYLYHNCSVTTTFSSNSTYSTNL--KTLLSSLS 54
Query: 67 KISEQMRTDGFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEA-RTVLPQCFPYNGGR 125
++ + GF TA AG PD GL C D+S C C A L +C G
Sbjct: 55 SLNASSYSTGFQTATAGQAPDRVTGLFLCRVDVSSEVCRSCVTFAVNETLTRCPKDKEGV 114
Query: 126 IYLDGCFMRAENYSF---FNEYTGPGDRAVCGNTTRKNSTFQAAAKQAV-LSAVQDAPNN 181
Y + C +R N + N G ++ + K F+ + L+AV+ A +
Sbjct: 115 FYYEQCLLRYSNRNIVATLNTDGGMFMQSARNPLSVKQDQFRDLVLTPMNLAAVEAARSF 174
Query: 182 KGYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSEGRALNTGC 241
K +A V + A++S Y + C L C CL+ + + GR L C
Sbjct: 175 KKWA--VRKIDLNASQSLYGMVRCTPDLREQDCLDCLKIGINQVT--YDKIGGRILLPSC 230
Query: 242 FMRYSDTDFLNKE--------AENGSSGGNVMVIVVAVVSSVVVT--VVGVIIGAYVWKR 291
RY + F N+ + GGN VI++AVV + V ++ + +
Sbjct: 231 ASRYDNYAFYNESNVGTPQDSSPRPGKGGNSSVIIIAVVVPITVLFLLLVAVFSVRAKNK 290
Query: 292 RYIQKKRRGSYDPDKLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLAD 351
R + +K + D + + SL F + ++ AT F NKLGQGGFG VYKG L+
Sbjct: 291 RTLNEKEPVAEDGNDITTA---GSLQFDFKAIEAATNCFLPINKLGQGGFGEVYKGTLSS 347
Query: 352 GREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSL 411
G ++A+KRL + +F NEV +++ ++H+NLV+LLG G E +LVYEF+PNKSL
Sbjct: 348 GLQVAVKRLSKTSGQGEKEFENEVVVVAKLQHRNLVKLLGYCLEGEEKILVYEFVPNKSL 407
Query: 412 DGFI 415
D F+
Sbjct: 408 DHFL 411
>emb|CAC83606.1| putative receptor-like serine-threonine protein kinase [Solanum
tuberosum]
Length = 676
Score = 154 bits (388), Expect = 6e-36
Identities = 114/394 (28%), Positives = 177/394 (43%), Gaps = 50/394 (12%)
Query: 60 NFVSTMEKISEQMRTDGFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYAEARTVLPQCF 119
N + + +S ++ GF A G D + C GD+ L DC C + Q
Sbjct: 46 NLNTLLTSLSSKIDNYGFYNASIGQNSDRASVIVLCRGDVELADCRGCVDNVVQKIAQLC 105
Query: 120 PYNGGRIY--LDGCFMRAENYSFFNE--------YTGPGDRAVCGNTTRKNSTFQAAAKQ 169
P N ++ DGC ++ N S + P + G ++ +
Sbjct: 106 P-NQKEVFGGYDGCMLQYSNQSILETTSFSLKYYFWNPANATKPGEFNQELGRLLENLRD 164
Query: 170 AVLSAVQDAPNNKGYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLENASSSI--LG 227
AV D P K YA G +G ++ Y L C L SC +CL +A ++
Sbjct: 165 R---AVDDGPLQK-YASG--NATGPDFQAIYALVQCTPDLSRQSCFSCLSDAYGNMPRCP 218
Query: 228 CLPWSEGRALNTGCFMRYSDTDFL-----------------------NKEAENGSSGGNV 264
CL GR + C RY + F +K G
Sbjct: 219 CLGKRGGRIIGVRCNFRYESSRFFEDVPLEAPPPAGNDNTTVPTGTDDKTVPTGEDDKTT 278
Query: 265 MVIVVAVVSSVVVTVVGVIIGAYVWKRR---YIQKKRRGSYDPDKLAKTLQHNSLNFKYS 321
I++ VVS+V V ++ + I + +RR + + + S D +A++ Q++ +S
Sbjct: 279 RTIIIIVVSTVTVVILIICIAVILIRRRKRKLVNEIQSTSVDDTSIAESFQYD-----FS 333
Query: 322 TLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSV 381
+ AT F ++NKLG+GGFG VYKG L +G+E+A+KRL ++ +F NEV +++ +
Sbjct: 334 AIRAATDDFSDANKLGEGGFGPVYKGKLQNGQEVAVKRLSADSGQGDLEFKNEVLLVARL 393
Query: 382 EHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
+H+NLVRLLG G E LLVYEF+PN SLD F+
Sbjct: 394 QHRNLVRLLGFCLDGTERLLVYEFVPNASLDHFL 427
>emb|CAB82810.1| protein kinase-like [Arabidopsis thaliana]
gi|15231262|ref|NP_190172.1| receptor-like protein
kinase, putative [Arabidopsis thaliana]
gi|11346406|pir||T47526 protein kinase-like -
Arabidopsis thaliana
Length = 676
Score = 152 bits (383), Expect = 2e-35
Identities = 125/406 (30%), Positives = 181/406 (43%), Gaps = 58/406 (14%)
Query: 53 NTTIFVPNFVSTMEKISEQMRT--DGFGTAVAGTGPDTNYGLAQCYGDLSLLDCVLCYA- 109
N+T F N + + +S + + GF TA AG PD GL C GD+S C C A
Sbjct: 46 NSTYFT-NLKTLLSSLSSRNASYSTGFQTATAGQAPDRVTGLFLCRGDVSQEVCRNCVAF 104
Query: 110 EARTVLPQCFPYNGGRI-YLDGCFMRAENYSFFNEYTGPGDRAVCGNTTRKNST------ 162
+ L C PYN + Y D C +R + + + T G + +S
Sbjct: 105 SVKETLYWC-PYNKEVVLYYDECMLRYSHRNILSTVTYDGSAILLNGANISSSNQNQVDE 163
Query: 163 FQAAAKQAVLSAVQDAPNN--KGYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLEN 220
F+ + A +A N+ K Y + V+ + Y+L C L C CL+
Sbjct: 164 FRDLVSSTLNLAAVEAANSSKKFYTRKVITP-----QPLYLLVQCTPDLTRQDCLRCLQK 218
Query: 221 ASSSILGCLPWSEGRALNTGCFMRYSDTDFLNKEAENGSS-------------------- 260
+ + L GR C RY + F N+ A SS
Sbjct: 219 SIKGM--SLYRIGGRFFYPSCNSRYENYSFYNETATRSSSPPSLPPRSTPQQQLKLAPPP 276
Query: 261 -------GGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKLAKTLQH 313
G N VI+V VV + + ++ V A+ R KK R +Y+ + L +
Sbjct: 277 LISERGKGRNSSVIIVVVVPIIALLLLFV---AFFSLRA---KKTRTNYEREPLTEESDD 330
Query: 314 ----NSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAA 369
SL F + ++ AT F E+NKLGQGGFG VYKG+ G ++A+KRL +
Sbjct: 331 ITTAGSLQFDFKAIEAATNKFCETNKLGQGGFGEVYKGIFPSGVQVAVKRLSKTSGQGER 390
Query: 370 DFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
+F NEV +++ ++H+NLVRLLG E +LVYEF+PNKSLD FI
Sbjct: 391 EFANEVIVVAKLQHRNLVRLLGFCLERDERILVYEFVPNKSLDYFI 436
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 694,441,035
Number of Sequences: 2540612
Number of extensions: 28948073
Number of successful extensions: 86231
Number of sequences better than 10.0: 7325
Number of HSP's better than 10.0 without gapping: 2969
Number of HSP's successfully gapped in prelim test: 4356
Number of HSP's that attempted gapping in prelim test: 78278
Number of HSP's gapped (non-prelim): 7913
length of query: 415
length of database: 863,360,394
effective HSP length: 130
effective length of query: 285
effective length of database: 533,080,834
effective search space: 151928037690
effective search space used: 151928037690
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0076c.8


