

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0076a.6
(123 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
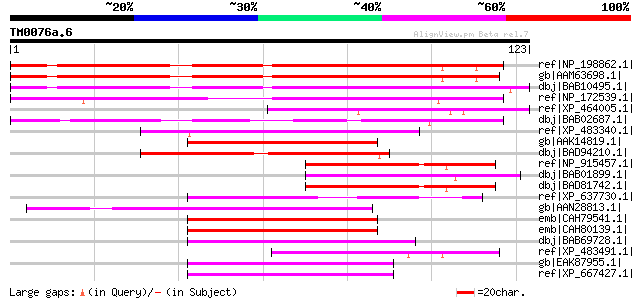
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_198862.1| expressed protein [Arabidopsis thaliana] 106 1e-22
gb|AAM63698.1| unknown [Arabidopsis thaliana] 104 4e-22
dbj|BAB10495.1| unnamed protein product [Arabidopsis thaliana] 93 1e-18
ref|NP_172539.1| expressed protein [Arabidopsis thaliana] gi|463... 78 6e-14
ref|XP_464005.1| unknown protein [Oryza sativa (japonica cultiva... 59 4e-08
dbj|BAB02687.1| unnamed protein product [Arabidopsis thaliana] g... 51 7e-06
ref|XP_483340.1| unknown protein [Oryza sativa (japonica cultiva... 49 2e-05
gb|AAK14819.1| hsp70-like protein [Plasmodium chabaudi] 49 3e-05
dbj|BAD94210.1| hypothetical protein [Arabidopsis thaliana] 49 3e-05
ref|NP_915457.1| P0406G08.25 [Oryza sativa (japonica cultivar-gr... 49 4e-05
dbj|BAB01899.1| unnamed protein product [Arabidopsis thaliana] 49 4e-05
dbj|BAD81742.1| hypothetical protein [Oryza sativa (japonica cul... 49 4e-05
ref|XP_637730.1| structural maintenance of chromosome protein [D... 49 4e-05
gb|AAN28813.1| At3g25910/MPE11_6 [Arabidopsis thaliana] gi|92795... 48 6e-05
emb|CAH79541.1| hypothetical protein PC000351.03.0 [Plasmodium c... 48 6e-05
emb|CAH80139.1| hypothetical protein PC000739.03.0 [Plasmodium c... 48 6e-05
dbj|BAB69728.1| hypothetical protein [Macaca fascicularis] 47 1e-04
ref|XP_483491.1| FtsJ cell division protein-like [Oryza sativa (... 46 2e-04
gb|EAK87955.1| membrane associated thioredoxin [Cryptosporidium ... 46 2e-04
ref|XP_667427.1| transmembrane protein 17 [Cryptosporidium homin... 46 2e-04
>ref|NP_198862.1| expressed protein [Arabidopsis thaliana]
Length = 112
Score = 106 bits (265), Expect = 1e-22
Identities = 62/119 (52%), Positives = 77/119 (64%), Gaps = 11/119 (9%)
Query: 1 MGFSEKSQQVEERGVESESRKWVITGIALRRPLKPIYTTTPVEKTEQHEEEEEEEEEETF 60
MGFS+KSQ E G+ES+ +KWVI GI++R LKP+ T K E E EE+ +
Sbjct: 1 MGFSKKSQF--EGGLESDGKKWVIAGISIRASLKPVKT-----KLRAPPEIVTEVEEDCY 53
Query: 61 MEEEEEECSTTPKGEEARIPATLKCPPPPRKPKPSLKCNFR-GGEFFTPP-DLEAVFIR 117
EEEECSTTP +E +IP L+CPP PRK +P+LKC EFFTPP DLE VF+R
Sbjct: 54 --NEEEECSTTPTAKETKIPELLECPPAPRKRRPALKCRSNVVREFFTPPSDLETVFLR 110
>gb|AAM63698.1| unknown [Arabidopsis thaliana]
Length = 112
Score = 104 bits (260), Expect = 4e-22
Identities = 61/118 (51%), Positives = 76/118 (63%), Gaps = 11/118 (9%)
Query: 1 MGFSEKSQQVEERGVESESRKWVITGIALRRPLKPIYTTTPVEKTEQHEEEEEEEEEETF 60
MGFS+KSQ E G+ES+ +KWVI GI++R LKP+ T K E E EE+ +
Sbjct: 1 MGFSKKSQF--EGGLESDGKKWVIAGISIRASLKPVKT-----KLRAPPEIVTEVEEDCY 53
Query: 61 MEEEEEECSTTPKGEEARIPATLKCPPPPRKPKPSLKCNFR-GGEFFTPP-DLEAVFI 116
EEEECSTTP +E +IP L+CPP PRK +P+LKC EFFTPP DLE VF+
Sbjct: 54 --NEEEECSTTPTAKETKIPELLECPPAPRKRRPALKCRSNVVREFFTPPSDLETVFL 109
>dbj|BAB10495.1| unnamed protein product [Arabidopsis thaliana]
Length = 118
Score = 93.2 bits (230), Expect = 1e-18
Identities = 55/124 (44%), Positives = 74/124 (59%), Gaps = 10/124 (8%)
Query: 1 MGFSEKSQQVEERGVESESRKWVITGIALRRPLKPIYTTTPVEKTEQHEEEEEEEEEETF 60
MGFS+KSQ E G+ES+ +KWVI GI++R LKP+ T K E E EE+ +
Sbjct: 1 MGFSKKSQF--EGGLESDGKKWVIAGISIRASLKPVKT-----KLRAPPEIVTEVEEDCY 53
Query: 61 MEEEEEECSTTPKGEEARIPATLKCPPPPRKPKPSLKCNFRGGEFFTPPDLEAVFIR-HV 119
EEEECSTTP +E +IP L+CPP PRK +P+LKC ++ P ++ + V
Sbjct: 54 --NEEEECSTTPTAKETKIPELLECPPAPRKRRPALKCRSNVKILYSEPWIKPTETKGEV 111
Query: 120 ERAN 123
RAN
Sbjct: 112 TRAN 115
>ref|NP_172539.1| expressed protein [Arabidopsis thaliana] gi|46359777|gb|AAS88752.1|
At1g10690 [Arabidopsis thaliana]
gi|45825137|gb|AAS77476.1| At1g10690 [Arabidopsis
thaliana] gi|4874279|gb|AAD31344.1| ESTs gb|T75618 and
gb|AA404816 come from this gene. [Arabidopsis thaliana]
gi|25372981|pir||C86240 hypothetical protein [imported]
- Arabidopsis thaliana
Length = 110
Score = 77.8 bits (190), Expect = 6e-14
Identities = 46/121 (38%), Positives = 63/121 (52%), Gaps = 19/121 (15%)
Query: 1 MGFSEKSQQVEERGVE--SESRKWVITGIALRRPLKPIYTTTPVEKTEQHEEEEEEEEEE 58
MG+S K + + ++ +KWVI GI R PLK I + V TE
Sbjct: 1 MGYSGKPHHQLDGEIRESTDGKKWVIAGIPSRSPLKQINLSPGVTVTET----------- 49
Query: 59 TFMEEEEEECSTTPKGEEARIPATLKCPPPPRKPKPSLKCNF--RGGEFFTPPDLEAVFI 116
EE+++C TTP RIP CP P+K KPSLKC++ ++F+PPDLE VFI
Sbjct: 50 ----EEQDQCPTTPTAVSVRIPRVPPCPAAPKKRKPSLKCSYVTVTRDYFSPPDLETVFI 105
Query: 117 R 117
+
Sbjct: 106 Q 106
>ref|XP_464005.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|41052563|dbj|BAD07745.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 140
Score = 58.5 bits (140), Expect = 4e-08
Identities = 36/79 (45%), Positives = 42/79 (52%), Gaps = 17/79 (21%)
Query: 62 EEEEEECSTTPKGEEARIPA-TLKCPPPPRKPKPSLKCNFRGG---------------EF 105
EEEEEE TTP+GEE R+PA CPP P+KP+ + GG EF
Sbjct: 62 EEEEEEEVTTPRGEECRLPAEAATCPPAPKKPRTAAVAIVAGGGRRCNCCDDDGGDSLEF 121
Query: 106 F-TPPDLEAVFIRHVERAN 123
F P DLEAVF V +AN
Sbjct: 122 FRVPADLEAVFANRVAKAN 140
>dbj|BAB02687.1| unnamed protein product [Arabidopsis thaliana]
gi|15232223|ref|NP_189400.1| hypothetical protein
[Arabidopsis thaliana]
Length = 99
Score = 50.8 bits (120), Expect = 7e-06
Identities = 39/119 (32%), Positives = 56/119 (46%), Gaps = 24/119 (20%)
Query: 1 MGFSEKSQQVEERGVESESRKWVITGIALRRPLKPIYTTTPVEKTEQHEEEEEEEEEETF 60
MG S+KSQ E ++++ +K++ ++R LKP+ KT+ + E E E+
Sbjct: 1 MGISKKSQVSRE--LDTDGKKFIFAKTSIRASLKPV-------KTKLIKPERESEDG--- 48
Query: 61 MEEEEEECSTTPKGEEARIPATLKCPPPPRKPKPSLKC--NFRGGEFFTPPDLEAVFIR 117
TP A+ P +CP PRK P LKC N R F P D E VFI+
Sbjct: 49 -------ICITPTARGAKTP---ECPAAPRKRPPVLKCQNNIRIEYFVPPSDFELVFIQ 97
>ref|XP_483340.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|42408757|dbj|BAD09993.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 126
Score = 49.3 bits (116), Expect = 2e-05
Identities = 27/70 (38%), Positives = 35/70 (49%), Gaps = 4/70 (5%)
Query: 32 PLKPIYTTTP----VEKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEARIPATLKCPP 87
PL PI TT P + +E +++EEEE TTP EE+R+ A CPP
Sbjct: 16 PLPPIKTTAPPPPPARTSSSTASPAPSDEARAPLKKEEEEEPTTPTSEESRLRAPTVCPP 75
Query: 88 PPRKPKPSLK 97
PRKP + K
Sbjct: 76 APRKPARTAK 85
>gb|AAK14819.1| hsp70-like protein [Plasmodium chabaudi]
Length = 407
Score = 48.9 bits (115), Expect = 3e-05
Identities = 26/45 (57%), Positives = 29/45 (63%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEARIPATLKCPP 87
EKTE+ EEEEEEEEEE EEEEEE P+ E T +CPP
Sbjct: 14 EKTEEEEEEEEEEEEEEEEEEEEEEEQDDPEELELIKEETEECPP 58
>dbj|BAD94210.1| hypothetical protein [Arabidopsis thaliana]
Length = 101
Score = 48.9 bits (115), Expect = 3e-05
Identities = 27/61 (44%), Positives = 37/61 (60%), Gaps = 5/61 (8%)
Query: 32 PLKPIYTTTPVEKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEARIPATLKCP--PPP 89
P K + PV+ + E+EE++E+ +EEEE+ TTPKGEE RIP +CP P P
Sbjct: 4 PEKSLKVRIPVKAPVSSKPEKEEQQEK---KEEEEDRFTTPKGEEFRIPPLFECPLAPEP 60
Query: 90 R 90
R
Sbjct: 61 R 61
>ref|NP_915457.1| P0406G08.25 [Oryza sativa (japonica cultivar-group)]
Length = 92
Score = 48.5 bits (114), Expect = 4e-05
Identities = 27/48 (56%), Positives = 29/48 (60%), Gaps = 4/48 (8%)
Query: 71 TPKGEEARIPATLKCPPPPRKPKPSLKCNFRG---GEFFTPPDLEAVF 115
TPK EE RIPATL CP PRK P RG +F PPDLEA+F
Sbjct: 35 TPKREECRIPATLPCPAAPRKAVPDFG-KRRGPPKNGYFQPPDLEALF 81
>dbj|BAB01899.1| unnamed protein product [Arabidopsis thaliana]
Length = 135
Score = 48.5 bits (114), Expect = 4e-05
Identities = 22/53 (41%), Positives = 32/53 (59%), Gaps = 2/53 (3%)
Query: 71 TPKGEEARIPATLKCPPPPRKPKPSLKCNFRGGE--FFTPPDLEAVFIRHVER 121
TPK +++RIP L CPP P+K + S C R + FF PP++E F+ +R
Sbjct: 83 TPKAKKSRIPEMLTCPPAPKKQRVSKNCVLRRRQIVFFAPPEIELFFVNAHDR 135
>dbj|BAD81742.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 175
Score = 48.5 bits (114), Expect = 4e-05
Identities = 27/48 (56%), Positives = 29/48 (60%), Gaps = 4/48 (8%)
Query: 71 TPKGEEARIPATLKCPPPPRKPKPSLKCNFRG---GEFFTPPDLEAVF 115
TPK EE RIPATL CP PRK P RG +F PPDLEA+F
Sbjct: 35 TPKREECRIPATLPCPAAPRKAVPDFG-KRRGPPKNGYFQPPDLEALF 81
>ref|XP_637730.1| structural maintenance of chromosome protein [Dictyostelium
discoideum] gi|60466163|gb|EAL64226.1| structural
maintenance of chromosome protein [Dictyostelium
discoideum]
Length = 1415
Score = 48.5 bits (114), Expect = 4e-05
Identities = 31/70 (44%), Positives = 36/70 (51%), Gaps = 19/70 (27%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEARIPATLKCPPPPRKPKPSLKCNFRG 102
E+ E+ EEEEEEEEEE EEEEEE TPK PPP K KPS +
Sbjct: 33 EEEEEEEEEEEEEEEEEKEEEEEEEEEETPK---------KPMPPPQSKAKPSSQ----- 78
Query: 103 GEFFTPPDLE 112
PPD++
Sbjct: 79 -----PPDIK 83
>gb|AAN28813.1| At3g25910/MPE11_6 [Arabidopsis thaliana] gi|9279596|dbj|BAB01054.1|
unnamed protein product [Arabidopsis thaliana]
gi|15983783|gb|AAL10488.1| AT3g25910/MPE11_6
[Arabidopsis thaliana] gi|18404727|ref|NP_566784.1|
expressed protein [Arabidopsis thaliana]
Length = 372
Score = 47.8 bits (112), Expect = 6e-05
Identities = 30/82 (36%), Positives = 40/82 (48%), Gaps = 5/82 (6%)
Query: 5 EKSQQVEERGVESESRKWVITGIALRRPLKPIYTTTPVEKTEQHEEEEEEEEEETFMEEE 64
E ++ E V+S+S T + L T + E+ EEE EEEEEE +EEE
Sbjct: 101 ESNEPTEMEDVDSDS-----TAVNLLGEAASEITVVDLSDGERGEEEVEEEEEEVVVEEE 155
Query: 65 EEECSTTPKGEEARIPATLKCP 86
EE TT + +E P L CP
Sbjct: 156 EEGIVTTEEDQEKNKPQKLTCP 177
>emb|CAH79541.1| hypothetical protein PC000351.03.0 [Plasmodium chabaudi]
Length = 285
Score = 47.8 bits (112), Expect = 6e-05
Identities = 25/45 (55%), Positives = 29/45 (63%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEARIPATLKCPP 87
EKTE+ EEEEEEEE+E EEEEEE P+ E T +CPP
Sbjct: 72 EKTEEEEEEEEEEEDEEEEEEEEEEEQDDPEELELIKEETEECPP 116
>emb|CAH80139.1| hypothetical protein PC000739.03.0 [Plasmodium chabaudi]
Length = 250
Score = 47.8 bits (112), Expect = 6e-05
Identities = 25/45 (55%), Positives = 29/45 (63%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEARIPATLKCPP 87
EKTE+ EEEEEEEE+E EEEEEE P+ E T +CPP
Sbjct: 72 EKTEEEEEEEEEEEDEEEEEEEEEEEQDDPEELELIKEETEECPP 116
>dbj|BAB69728.1| hypothetical protein [Macaca fascicularis]
Length = 641
Score = 46.6 bits (109), Expect = 1e-04
Identities = 25/54 (46%), Positives = 32/54 (58%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEARIPATLKCPPPPRKPKPSL 96
E+ E+ EEEEEEEEEE EEEEEE +GE++ P+ P +P P L
Sbjct: 427 EEAEEEEEEEEEEEEEEEEEEEEEEPQQGGQGEKSATPSQKILDPNTGEPAPVL 480
>ref|XP_483491.1| FtsJ cell division protein-like [Oryza sativa (japonica
cultivar-group)] gi|42761378|dbj|BAD11646.1| FtsJ cell
division protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 73
Score = 46.2 bits (108), Expect = 2e-04
Identities = 27/60 (45%), Positives = 33/60 (55%), Gaps = 6/60 (10%)
Query: 63 EEEEECSTTPKGEEARIPATLKCPPPPRKPK---PSLKCNFR---GGEFFTPPDLEAVFI 116
EEE + TP+ EE RIP +CP PPRK P L R G +F PPDLE +F+
Sbjct: 2 EEELQGWETPRREECRIPVMPQCPAPPRKRPVVLPELGKERREPPKGRYFQPPDLELLFV 61
>gb|EAK87955.1| membrane associated thioredoxin [Cryptosporidium parvum]
gi|66357966|ref|XP_626161.1| membrane associated
thioredoxin [Cryptosporidium parvum]
Length = 363
Score = 46.2 bits (108), Expect = 2e-04
Identities = 25/49 (51%), Positives = 29/49 (59%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEARIPATLKCPPPPRK 91
E+ E+ EEEEEEEEEE EEEEEE + EE P LK P R+
Sbjct: 286 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEAPKQLKTPQRGRR 334
Score = 39.7 bits (91), Expect = 0.017
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
++ E HEEEEEEEEEE EEEEEE + EE
Sbjct: 256 DECEDHEEEEEEEEEEEEEEEEEEEEEEEEEEEE 289
Score = 38.9 bits (89), Expect = 0.029
Identities = 21/40 (52%), Positives = 24/40 (59%)
Query: 37 YTTTPVEKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
Y+ E E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 253 YSDDECEDHEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 292
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 275 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 308
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 266 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 299
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 262 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 295
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 267 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 300
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 268 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 301
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 273 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 306
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 265 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 298
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 272 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 305
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 264 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 297
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 263 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 296
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 276 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 309
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 277 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 310
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 270 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 303
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 271 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 304
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 274 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 307
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 269 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 302
Score = 35.8 bits (81), Expect = 0.24
Identities = 17/35 (48%), Positives = 22/35 (62%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEA 77
E+ E+ EEEEEEEEEE EE ++ T +G A
Sbjct: 301 EEEEEEEEEEEEEEEEEEEEEAPKQLKTPQRGRRA 335
>ref|XP_667427.1| transmembrane protein 17 [Cryptosporidium hominis]
gi|54658565|gb|EAL37199.1| transmembrane protein 17
[Cryptosporidium hominis]
Length = 366
Score = 46.2 bits (108), Expect = 2e-04
Identities = 25/49 (51%), Positives = 29/49 (59%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEARIPATLKCPPPPRK 91
E+ E+ EEEEEEEEEE EEEEEE + EE P LK P R+
Sbjct: 289 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEAPKQLKTPQRGRR 337
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 274 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 307
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 275 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 308
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 271 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 304
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 263 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 296
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 276 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 309
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 262 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 295
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 278 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 311
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 266 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 299
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 269 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 302
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 267 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 300
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 279 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 312
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 270 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 303
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 268 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 301
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 277 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 310
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 273 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 306
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 265 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 298
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 264 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 297
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 272 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 305
Score = 38.5 bits (88), Expect = 0.038
Identities = 20/34 (58%), Positives = 23/34 (66%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 280 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 313
Score = 38.1 bits (87), Expect = 0.049
Identities = 20/35 (57%), Positives = 23/35 (65%)
Query: 42 VEKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
V+ E+ EEEEEEEEEE EEEEEE + EE
Sbjct: 259 VDHEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 293
Score = 35.8 bits (81), Expect = 0.24
Identities = 17/35 (48%), Positives = 22/35 (62%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEEA 77
E+ E+ EEEEEEEEEE EE ++ T +G A
Sbjct: 304 EEEEEEEEEEEEEEEEEEEEEAPKQLKTPQRGRRA 338
Score = 35.4 bits (80), Expect = 0.32
Identities = 19/34 (55%), Positives = 21/34 (60%)
Query: 43 EKTEQHEEEEEEEEEETFMEEEEEECSTTPKGEE 76
E + EEEEEEEEEE EEEEEE + EE
Sbjct: 257 ECVDHEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 290
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.309 0.130 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 259,101,421
Number of Sequences: 2540612
Number of extensions: 13228723
Number of successful extensions: 339205
Number of sequences better than 10.0: 4300
Number of HSP's better than 10.0 without gapping: 3040
Number of HSP's successfully gapped in prelim test: 1398
Number of HSP's that attempted gapping in prelim test: 234103
Number of HSP's gapped (non-prelim): 38044
length of query: 123
length of database: 863,360,394
effective HSP length: 99
effective length of query: 24
effective length of database: 611,839,806
effective search space: 14684155344
effective search space used: 14684155344
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0076a.6


